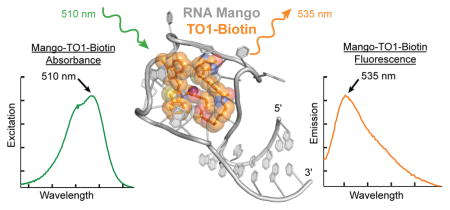
Abstract
Genetically encoded fluorescent protein tags revolutionized proteome studies, while the lack of intrinsically fluorescent RNAs has hindered transcriptome exploration. Among several RNA-fluorophore complexes that potentially address this problem, RNA Mango has an exceptionally high affinity for its thiazole orange (TO)-derived fluorophore, TO1-Biotin (Kd ~3 nM), and in complex with related ligands, is one of the most red-shifted fluorescent macromolecular tags known. To elucidate how this small aptamer exhibits such properties, which make it well suited for studying low-copy cellular RNAs, we determined its 1.7 Å resolution co-crystal structure. Unexpectedly, the entire ligand, including TO, biotin, and the linker connecting them, abuts one of the near-planar faces of the three-tiered G-quadruplex. The two heterocycles of TO are held in place by two loop adenines and make a 45° angle with respect to each other. Minimizing this angle would increase quantum yield and further improve this tool for in vivo RNA visualization.
Graphical Abstract
INTRODUCTION
RNA Mango1 is an aptamer2 selected in vitro to bind to the thiazole orange (TO) derivative TO1-Biotin (Fig. 1a). It is one of several recently discovered RNAs that bind small molecule fluorophores and markedly enhance their fluorescence. The malachite green (MG) aptamer3 increases the fluorescence of its bound fluorophore by 2400-fold4, while Spinach5 enhances the fluorescence of small-molecule analogs of the intrinsic fluorophore of green fluorescent protein (GFP) ~2000 times6. Like GFP and its homologs, which as genetically encoded tags have revolutionized the study of proteins7, these aptamer-fluorophore complexes have found use in imaging as fusions to cellular RNAs. However, the properties of existing fluorogenic RNAs could still be improved for live cell applications. For instance, the MG aptamer-dye complex8 is cytotoxic when illuminated (indeed, it was originally selected as an agent for laser RNA ablation), and Spinach is limited by its modest fluorophore affinity (dissociation constant, Kd, ~ 300–500 nM), its rapid loss of fluorescence, and its tendency to misfold5,9–11.
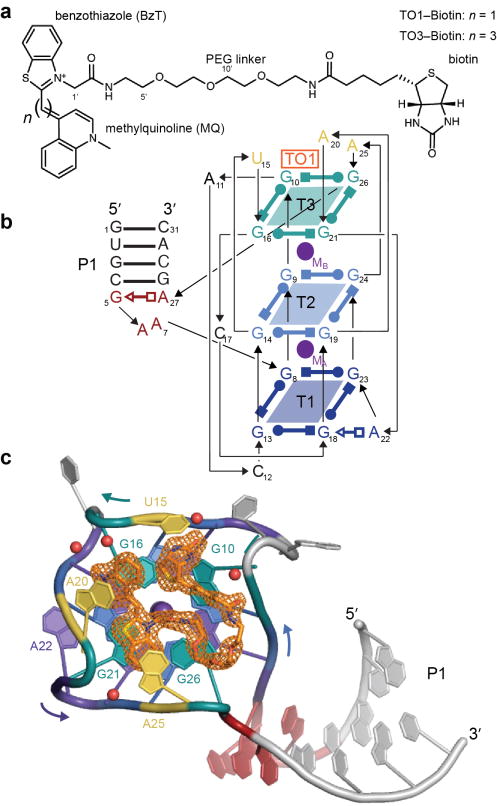
Figure 1.
Overall structure of RNA Mango in complex with TO1-Biotin. (a) Chemical structures of TO1-Biotin and TO3-Biotin (ref. 1). The two compounds differ in having one or three benzylidene (methine) carbons, respectively, connecting the two heterocycles of their TO moieties. PEG, polyethyleneglycol linker. (b) Secondary structure of the RNA Mango-TO1-Biotin complex. Thin lines with arrowheads denote connectivity. Base pairs are represented using Leontis-Westhof symbols39. The location of the fluorophore and two K+ ions (TO1, MA and MB, respectively) are indicated. Except where noted, this color scheme is used throughout. (c) Cartoon representation of the RNA Mango-TO1-complex. Curved arrows indicate direction of chain, 5′ to 3′. Orange mesh depicts a simulated-annealing omit |Fo|-|Fc| map (TO1-biotin was omitted from the calculation using the final refined atomic coordinates) contoured at 1.5 σ. Purple and red spheres represent K+ ions and water molecules, respectively.
RNA Mango enhances the fluorescence of TO1-Biotin by 1100-fold. As a tag for RNA visualization it overcomes several of the shortcomings of previously described fluorogenic RNAs. Because the efficiency of a fluorescent aptamer is directly proportional to the affinity for its fluorophore1, affinity was an explicit consideration in the selection for RNA Mango. As a result, its Kd for its cognate ligand (~ 3 nM) is considerably smaller than the corresponding Kd’s of Spinach and its derivatives. This high affinity facilitates imaging of low copy RNAs while allowing cells to be subjected to more dilute fluorophore. Under such conditions, TO1-Biotin exhibits low background from non-specific binding to cellular nucleic acids, and no detectable toxicity1.
Fluorescent tags that can be excited by long-wavelength visible light and fluoresce in the near IR are attractive because of greater tissue depth penetration and contrast12. The excitation and emission maxima of the TO1-Biotin complex of RNA Mango (510 and 535 nm, respectively) are only modestly red-shifted compared to those of Spinach (469 and 501 nm). Although with lower affinity, RNA Mango will bind and stimulate fluorescence of other TO-related fluorophores with more extended conjugation. Thus, when bound to TO3-Biotin (Fig. 1a; KD ~8 nM), RNA Mango exhibits excitation and emission maxima at 637 and 658 nm, respectively. The latter is 10 nm beyond that of the far-red fluorescent protein mPlum (ref. 12), and makes this RNA-dye complex one of the most red-shifted macromolecular tags described to date1,4,5.
Reselection experiments following partial randomization of RNA Mango revealed an invariant 23-nucleotide (nt) core that, when flanked by nucleotides presumed to form a short duplex, exhibited maximal fluorescence1. The conserved minimal core of RNA Mango contains a pattern of four consecutive guanine dinucleotides separated by short linkers that suggested the presence of a G-quadruplex13,14 structure. This, and the results of biochemical, mutagenesis and spectroscopic analyses led to the suggestion that the functional core of RNA Mango is an all-parallel, two-tiered G-quadruplex, one face of which binds TO1-Biotin by sequestering it with nucleotides from the “propeller” loops connecting adjacent guanine stacks1 (Supplementary Results, Supplementary Fig. 1). Although the fluorogenic aptamer Spinach also folds around a G-quadruplex15,16 (which provides one of the binding faces for its cognate fluorophore, DFHBI), that RNA is larger (~100 nt), and because its quadruplex is flanked on both sides by A-form duplexes, its connectivity is considerably more complicated than the simple parallel G-quadruplex proposed for RNA Mango. The complex connectivity and relatively large size of Spinach may contribute to its propensity to misfold9. RNA Mango, which is among the smallest fluorogenic RNA-dye complexes reported to date, may therefore be a better tag for biological RNAs6.
To elucidate how a small aptamer RNA can bind a TO-derived fluorophore with a 3 nM Kd and enhance its fluorescence over 1000-fold, we have now determined the crystal structure of the 1:1 complex between RNA Mango and TO1-Biotin at 1.73 Å resolution. Our structure reveals that the core of this aptamer is a three-tiered G-quadruplex of mixed parallel and antiparallel connectivity. Unexpectedly, the entire fluorophore, including the PEG linker and the biotin, rather than just TO, associate intimately with the RNA. Our co-crystal structure provides a framework for understanding the photophysics of this fluorogenic RNA, and is also the starting point for devising fluorophore and RNA variants with improved properties.
RESULTS
Overall structure of RNA Mango bound to TO1-Biotin
A 31-nt construct, comprised of the 23-nt RNA Mango core1 flanked by eight ribonucleotides predicted to extend it proximally with a four base-pair duplex, was co-crystallized with TO1-Biotin (Supplementary Fig. 2). The co-crystal structure was solved by the single-wavelength anomalous dispersion (SAD) method using data from an iridium derivative (Online Methods and Supplementary Table 1). The unbiased 1.6 Å-resolution experimental electron density map (Supplementary Fig. 2d) was of high quality, allowing unambiguous tracing of the RNA, and immediately revealing the location and conformation of the bound TO1-Biotin. The structure of RNA Mango loosely resembles a kite, with an A-form duplex “tail” projecting away from a G-quadruplex “wing” (Fig. 1b,c). TO1-Biotin binds on one of the faces of the quadruplex.
The asymmetric unit (ASU) of our co-crystals contains two copies of the RNA Mango-TO1-Biotin complex. In one of them, the duplex portion lacks electron density. Since gel-electrophoretic analysis shows that the RNA in the crystals is intact (not shown), the second duplex moiety is presumed disordered, and was not modeled (Supplementary Fig. 2d). The quadruplex moieties of the two RNA Mango molecules in the ASU are very similar, superimposing with a root mean square difference (rmsd) of 0.12 Å (for 19 C1′ atom and two K+ ion pairs). The two RNA Mango quadruplexes in the ASU stack on each other using their faces opposite from where TO1-Biotin binds (Supplementary Fig. 3). This inter-protomer interface contains an octacoordinate K+ ion equidistant from the two quadruplexes, and is relatively large (538 Å2 total buried solvent-accessible surface area) suggesting that RNA Mango may dimerize in solution. We therefore examined the RNA-ligand complex by dynamic light scattering and analytical ultracentrifugation (Online Methods and Supplementary Fig. 4). The sedimentation and translational diffusion coefficients of our RNA Mango crystallization construct (calculated monomer molecular mass = 10.4 kDa) in the presence of TO1-Biotin are 2.55 S and 1.10 × 10−6 cm2s−1, respectively. By the Svedberg equation, and assuming a partial specific volume of 0.53 cm3g−1 (ref. 17), the apparent molecular mass of the complex in solution is 10.6 kDa. Our experiments indicate that the complex is monomeric in solution at concentrations as high as 0.5 mM.
The RNA Mango G-quadruplex is three-tiered
The RNA Mango G-quadruplex is comprised of three G-quartet tiers (T1, T2, T3; Fig. 1,2). T1 and T2 are connected in parallel, and all guanine residues in these tiers are in the anti conformation. In T1, the four nucleotides have alternating 2′-endo and 3′-endo puckers, while in T2, all four guanines adopt the 2′-endo pucker. T3, the third quartet, is unusual in that three of its four guanines (G16, G21, G26) are antiparallel to the adjacent guanines in T2 (these three residues all have anti-glycosidic bond angles and adopt the 3′-endo pucker), while the remaining guanine (G10) is parallel relative to T2 (and adopts a syn-glycosidic angle and a 3′-endo pucker). While the nucleobases of T1, T2 and the three antiparallel guanines of T3 are nearly coplanar within their corresponding tiers, the base of G10 is buckled, its long axis ~30° from the mean plane defined by the other guanines of T3. This unusual orientation of G10 allows its exocyclic amine to hydrogen bond to its own pro-SP non-bridging phosphate oxygen (Fig. 2d), and improves its packing against the biotin of the fluorophore (Fig. 1c).
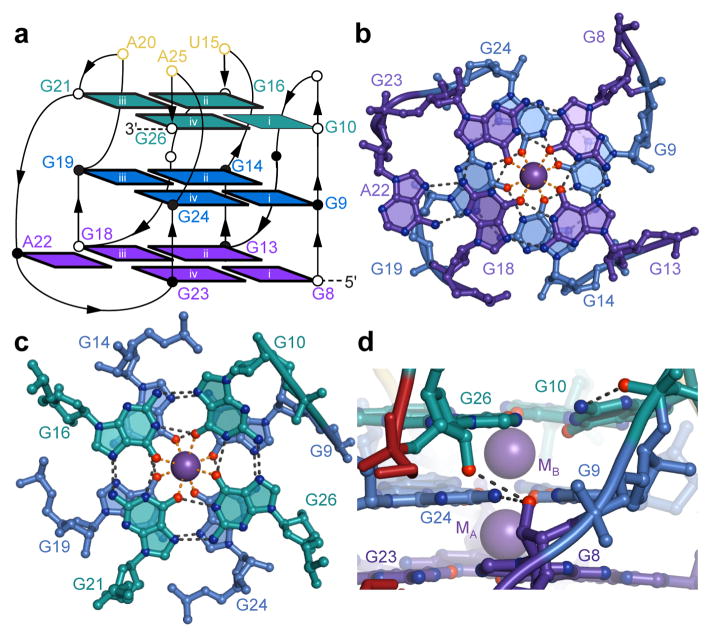
Figure 2.
Structure of the G-quadruplex core of RNA Mango. (a) Connectivity and stereochemistry of the RNA Mango G-quadruplex. Except for G10, which adopts the syn- conformation, all nucleotides are anti- (dark nucleobase outlines). Circles denote the pucker of successive backbone riboses (open and black circles, 3′-endo and 2′-endo, respectively). The four guanine stacks are denoted by white lower-case Roman numerals. (b) Ball-and-stick representation of the T1 and T2 tiers and the K+ ion MA. Black and orange dashed lines represent hydrogen bonding and inner-sphere cation coordination, respectively. (c) Tiers T2 and T3 and the K+ ion MB. (d) Detail of a side-view of the quadruplex showing a ribose-zipper-like interaction between G8, G24 and G26 (in tiers T1, T2, and T3, respectively). Buckling of G10 allows its nucleobase to hydrogen bond to its backbone phosphate.
RNA Mango folds into its mixed parallel and antiparallel G-quadruplex using six loops to connect the twelve guanine residues of its three G-quartets. Nucleotides in four of these loops make noteworthy interactions in addition to providing connectivity. A22, which is part of a propeller loop connecting T3 with T1, augments T1 into a pentad by binding the sugar edge of G18 with its Hoogsteen face. U15, A20 and A25, each of which forms one of three loops that allow the RNA chain to reverse direction above T3, participate in forming the TO1-Biotin binding site (Fig. 1b,c).
Typical for a G-quadruplex, RNA Mango coordinates K+ ions between successive G-quartets (Fig. 2b,c). Two octacoordinate K+ ions lie in the central channel of the quadruplex and are 2.8 ± 0.1 Å (mean ± s.d.) from the eight carbonyl oxygen atoms of the guanines from their respective flanking tiers. Two ribose-zipper-like interactions appear to stitch together the three G-quartets. The 2′-OH of T1 residues G8 and G18 are 3.0 Å from the exocyclic amines of T2 residues G14 and G24, respectively, and also 3.0 Å from the 2′-OH of T3 residues G16 and G26, respectively (Fig. 2d). These contacts are made possible by the 3′-endo puckers of T1 guanosines G8 and G18. A third potential stabilizing feature of the G-quadruplex is that the 5′ and 3′ ends of the quadruplex, in tiers T1 and T3, respectively, are kept in proximity by connecting with the duplex portion of RNA Mango.
A tetraloop-like junction connects two domains
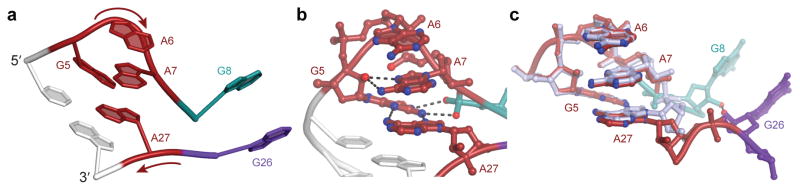
No tertiary interactions join the A-form duplex and the quadruplex of RNA Mango; they connect solely through a junction that resembles a GAAA tetraloop (Fig. 3). Canonically, such tetraloops feature a sugar-edge-to-Hoogsteen closing base pair between first and fourth residues, a chain direction reversal between the guanine and the first adenine, and stacking of the three adenines on the 3′ side18. In the case of RNA Mango, G5 and A27 make the closing base pair, and the chain reverses between G5 and A6. A6 then stacks on A7 on the 3′ side of the loop. However, rather than stacking underneath A7, the next residue of RNA Mango (G8) points ~120° away and forms part of the G-quadruplex. The fourth position in the tetraloop is instead occupied by A27, which lies immediately 3′ to the G-quadruplex (Fig. 3a) rather than being adjacent in sequence to the second adenine of the GAAA motif. Thus, the G-quadruplex interrupts the GAAA tetraloop-like element of RNA Mango between the second and third adenines. As in a canonical GAAA tetraloop, the 2′-OH of the guanosine (G5) hydrogen bonds with the N7 of the second adenine (A7), and the Watson-Crick face of this guanine hydrogen bonds to the phosphate that follows the second adenine. In the junction, this is the phosphate of A27, rather than that of the next residue in the RNA chain (G8). Despite the interruption in the backbone, the GAAA motif of RNA Mango superimposes closely (rmsd = 0.29 Å for all non-hydrogen atoms, excluding the phosphate of A27) on a conventional tetraloop19 (Fig. 3c). Unlike many GAAA tetraloops, which are involved in A-minor and stacking tertiary interactions20–23, the junction of RNA Mango makes none (except crystal contacts, Supplementary Fig. 3a,c).
Figure 3.
The duplex-quadruplex junction of RNA Mango resembles a GAAA tetraloop. (a) Cartoon representation of the junction with one flanking Watson-Crick base pair from the duplex (gray), and adjacent residues from the G-quadruplex (G8 and G26). b) Hydrogen bonding pattern within the junction. (c) Junction of RNA Mango superimposed on a canonical GAAA tetraloop (gray; PDB 4FNJ; ref. 20).
Structure of the TO1-Biotin binding site
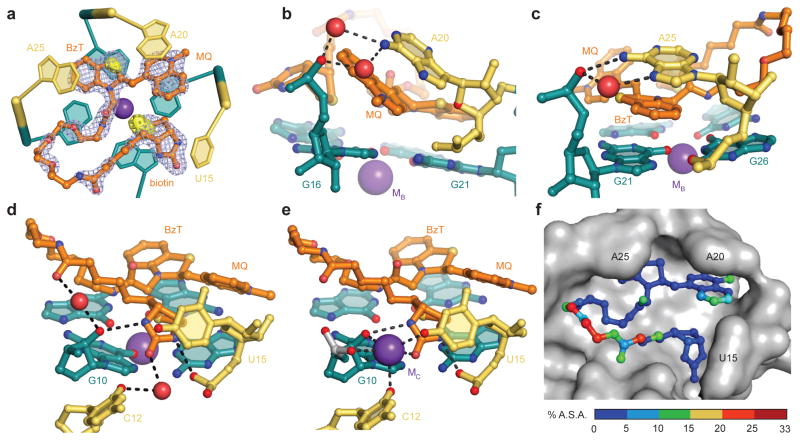
When bound to RNA Mango, TO1-Biotin adopts a ring-like conformation in which the methylquinoline of TO1 is in van der Waals contact with the tetrahydrothiophene of biotin. Except for four atoms in the PEG linker (7′, 13′, 14′, and 17′), electron density for the entire RNA-bound TO1-Biotin was well defined in unbiased residual Fourier syntheses. The conformation of the TO1-Biotin was confirmed by strong anomalous difference Fourier features corresponding to the sulfur atoms in the benzothiazole and tetrahydrothiophene (Fig. 4a). The two TO1-Biotin molecules in the ASU adopt nearly identical conformations (rmsd 0.12 Å for all non-hydrogen atoms). The fluorophore lies on top of the T3 tier of the G-quadruplex of RNA Mango, and each of its three heterocycles makes a stacking interaction with a nucleobase from an RNA loop: the methylquinoline with A20, the benzothiazole with A25 and the biotin with U15 (Fig. 4).
Figure 4.
Structural basis of TO1-Biotin recognition by RNA Mango. (a) Cartoon representation of the ligand binding site, superimposed on the |Fo| – |Fc| electron density map calculated prior to addition of the fluorophore to the crystallographic model (cyan mesh, 2 σ contour). The native anomalous difference Fourier synthesis is shown as a solid yellow surface (3 σ). (b) Detail of a ball-and-stick representation of the complex around the methylquinoline (MQ) of TO1-Biotin. (c) View around the benzothiazole (BzT) of TO1-Biotin. (d) View around the biotin of TO1-Biotin in chain A (Supplementary Fig. 3) of the ASU. (e) View around the biotin of TO1-Biotin in chain B (Supplementary Fig. 3) of the ASU. For clarity, the K+ ion MB is omitted. The K+ ion MC is only present in chain B. (f) Molecular surface of the ligand binding pocket of RNA Mango (grey) with a ball-and-stick representation of TO1-Biotin. The latter is colored according to the percentage of the surface area of each non-hydrogen atom that remains solvent-accessible upon complex formation.
Although sandwiched between the bases of G16 and A20, the methylquinoline of TO1 stacks extensively only with the latter, because it lies at an angle of ~45° with the guanine base (Fig. 4b). In contrast, the benzothiazole is nearly parallel to the bases of both G21 and A25 (Fig. 4c). G16 and G21 form an essentially flat surface. Therefore, the two TO heterocycles are at an angle of ~45° relative to each other (Fig. 4b,c,d,e). Neither the methylquinoline nor the benzothiazole have any hydrogen-bonding capability; they associate with the RNA solely through van der Waals contacts. The conformation of the polar PEG linker appears to be a consequence of van der Waals interactions with the RNA, and of anchoring of its ends by the benzothiazole and biotin. In addition to the stacking of its tetrahydrothiophene and the ureido rings on the TO methylquinoline and U15 nucleobase, respectively, the biotin makes several water-mediated hydrogen bonds. These are between its fatty acid carbonyl and the phosphates of G10 and A11, and the head group carbonyl and the phosphate of U15 as well as the O2 carbonyl of C12. Further, there are two direct hydrogen bonds between the RNA (2′-OH of G10 and the pro-RP non-bridging phosphate oxygen of U15) and the ureido nitrogens of biotin (Fig. 4d,e).
Intramolecular hydrogen bonding appears to stabilize the conformations of A20 and A25 of the ligand binding site of RNA Mango. Thus, the phosphate of A25 hydrogen bonds to the exocyclic amine of G26, the Hoogsteen face of A25 makes direct and water-mediated hydrogen bonds with the phosphate of G21, the phosphate of A20 hydrogen bonds with the exocyclic amine of G21, and its Hoogsteen face makes water-mediated hydrogen bonds with the phosphate of G16. Overall, in associating with RNA Mango, TO1-Biotin buries 645 Å2 (71%) of its solvent-accessible surface area with the benzothiazole, the biotin and the proximal part of the PEG linker being most deeply recessed into the RNA (Fig. 4f).
Functional importance of RNA Mango structural features
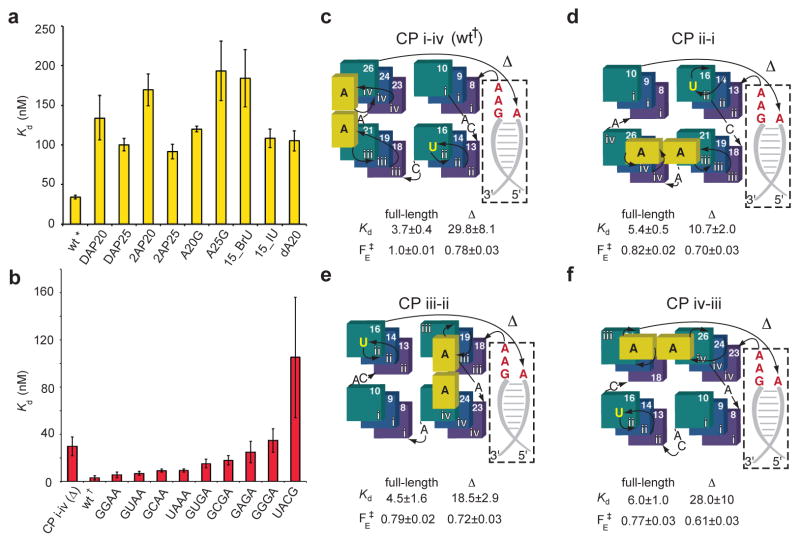
We performed nucleotide and single-atom substitutions to evaluate the importance of three salient molecular features of RNA Mango: the G-quadruplex, the TO1-Biotin binding site, and the GAAA tetraloop-like junction (Fig. 5). Consistent with loss of RNA Mango activity in the presence of Li+ (which destabilizes G-quadruplexes24–26), substitution of single guanines in each tetrad with 1-methylguanine resulted in 25–50-fold loss of affinity for TO1-Biotin. The deleterious effect decreased from T1 to T2 to T3 (G13, G14, G16), suggesting the quadruplex is stabilized by interactions with TO1-Biotin. Consistent with destabilization not affecting the structure of the bound fluorophore, at saturating ligand concentration, the fluorescence enhancement of the three mutants was comparable to that of wild-type (Supplementary Table 2). Substitutions of the three flap residues that stack on the fluorophore are also deleterious (Fig. 5a).
Figure 5.
Structure-based analysis of the RNA Mango-TO1-Biotin complex. (a) Effect on fluorophore-binding affinity of mutations in the ligand binding pocket (* Kds in panel (b) were measured using 31-nt RNA constructs with a 4 base pair duplex). (b) Effect on fluorophore-binding affinity of mutations in the tetraloop-like junction motif († measurements in this panel were with 39–40 nt RNA constructs each containing an 8 base pair duplex). (c) through (f), effect on Kd and relative fluorescence enhancement (FE‡) of circular permutation (CP) of the connectivity of RNA Mango by either moving the attachment point of the tetraloop-like junction in a full-length (FL) 39–40 nt construct, or by deleting the duplex and junction (Δ). Guanines of the three tiers are represented as squares, colored as in Fig. 1. The four guanine stacks are numbered as in Fig. 2a. The two adenosines that cover the two heterocycles of the TO moiety of the fluorophore are represented as yellow rectangles. All data are the mean of three independent trials ± s.d.
G-quadruplexes are intrinsically, if weakly, fluorescent27–29. To examine whether the intrinsic fluorescence of the G-quadruplex of RNA Mango could directly excite the bound TO1-Biotin fluorophore, we measured the fluorescence of RNA Mango in the absence of fluorophore (Supplementary Fig. 5). In its ligand-free state, the RNA fluoresces much more weakly than the complex, and its emission maximum at 410 nm is too blue-shifted to excite TO1-Biotin. Thus, while their close physical proximity implies that there is necessarily some degree of electronic coupling between the G-quadruplex and the TO1-Biotin, it is unlikely that the intrinsic fluorescence of the G-quadruplex of RNA Mango donates energy through FRET to TO1-Biotin.
Although devoid of tertiary interactions, the GAAA tetraloop-like junction appears to be functionally important, as variant sequences isolated from randomization-reselection experiments all contained the sequence (G/U)AAA at this location1. Indeed, removing the GAAA stem from RNA Mango substantially disrupted function (Fig. 5b, Supplementary Table 2, Supplementary Table 3). We produced variant RNAs containing each of the eight GNRA tetraloops as well as the UAAA and UACG tetraloops. Dissociation constants for TO1-Biotin increased modestly from 3 nM to 10 nM through the GNRA tetraloop series N= A, G, U and C with R=A, while the series N= U, C, A, G with R=G had Kd values increasing nearly linearly from ~15 nM to ~35 nM. The binding affinity of the UAAA construct was intermediate between the two GNRA series, while the UACG (a member of the thermodynamically stable30 UNCG tetraloops) construct showed a significant decrease in binding affinity (Fig. 5b).
To further explore the importance of the GAAA junction, we circularly permuted its site of attachment to the G-quadruplex from its wild-type locus (G8 and G26 in stacks i and iv, respectively; stack numbering defined in Fig. 2a) to each of the other three faces of the G-quadruplex (Fig. 5c,d,e,f). Remarkably, all three circular permutations decreased binding affinity by less than 2-fold and had only marginal effects on fluorescence enhancement. Moreover, removing the junction sequence and stem from these circular permutations resulted in a further 2- to 8-fold decrease in binding affinity. Since these circular permutations systematically disrupted each of the three propeller loops connecting the adjacent guanine stacks of the G-quadruplex, neither the site of the GAAA junction insertion nor the detailed sequence of the propeller loops responsible for linking adjacent stacks of the G-quadruplex are essential for binding. Circular permutation and alteration of the loops did significantly impact fluorescence enhancement (Fig. 5c,d,e,f, Supplementary Table 2), reminiscent of the altered intrinsic fluorescence observed for loop connectivity in model G-quadruplexes29,31.
To examine the functional importance of the circularized conformation of TO1-Biotin in complex with RNA Mango, we determined the fluorescence enhancements and lifetimes of TO1-Desthiobiotin and TO1-Acetate and compared them to those of TO1-Biotin (Supplementary Fig. 6,7). The former results from removal of the sulfur from the biotin. On the basis of our structure, this would be expected to relax steric constraints on the methylquinoline, and indirectly add flexibility to the PEG linker. We find that the enhancement for TO1-Desthiobiotin is 50% larger than that of TO1-Biotin with a modest increase in lifetime of 0.25 ns. Removing the biotin and the PEG linker to yield TO1-Acetate has a large negative impact on fluorescence, decreasing the enhancement by an order of magnitude and the lifetime by ~0.5 ns. Thus, modest relief of steric constraints from the methylquinoline enhances fluorescence, but loss of the entire PEG linker and terminal moiety is detrimental. Furthermore, the PEG linker and biotin appear to have no effect on the binding of TO3-Biotin and its derivatives (Supplementary Fig. 6). TO3-Biotin cannot be modeled into the ligand binding pocket without steric clashes (Supplementary Fig. 8) suggesting that TO3 derivatives do not bind in a ring-like conformation.
DISCUSSION
Biochemical and sequence analyses suggested1 that RNA Mango would comprise a duplex and a two-tiered, all-parallel G-quadruplex, and that TO1-Biotin would bind on one of the faces of the latter interacting with conserved nucleotides found within the trinucleotide propeller loops1 (Supplementary Fig. 1). The G-quadruplex revealed by our co-crystal structure contains a third G-quartet of mixed parallel and antiparallel connectivity. The additional G-quartet effectively shortens three of the propeller loops to a single nucleotide each, and these lone nucleotides serve as flaps, each of which folds over one of the three heterocycles of TO1-Biotin. The majority of RNA G-quadruplexes described to date have simple, all-parallel connectivity14, but some in vitro selected aptamers have been found to be more complex. An RNA that binds to a peptide from the fragile-X mental retardation (FMR) protein is comprised of three G-quartets, two of which are parallel and one which is entirely antiparallel32,33. Spinach assembles around a two-tier G-quadruplex in which two of the guanine stacks are parallel, and the other two antiparallel15,16. In both of those RNAs, the G-quadruplex is juxtaposed to base quadruples or triples of mixed composition that enable the four strands of the quadruplex to connect to a canonical antiparallel duplex while maintaining base stacking throughout. The G-quadruplex of RNA Mango differs from those of the FMR protein-binding RNA and Spinach in that while having a mixed connectivity, all three of its tiers are comprised exclusively of guanines, and that connection to a duplex is achieved by a GAAA tetraloop-like junctional element, none of whose nucleotides stack or otherwise make tertiary interactions with the quadruplex.
Because TO1-Biotin was immobilized by binding it to streptavidin during in vitro selection, the close association of its biotin moiety with the G-quadruplex of RNA Mango was unexpected. Nonetheless, structure-activity studies1 support the importance of biotin, as well as the PEG linker, for fluorescence activation by this aptamer RNA. When the biotin in TO1-Biotin is replaced with an acetate (leaving the PEG linker intact), RNA Mango binds slightly more weakly but loses nearly a factor of two in fluorescence enhancement1. TO1-Acetate (which lacks both the PEG linker and the biotin) binds 10-fold more weakly than TO1-Biotin; removing the acetate to leave only TO1 reduces binding affinity 2-fold and lowers brightness by 6- to 7-fold (relative to TO1-Biotin)6. The PEG linker is near-identical between the two independent copies in the ASU of our crystals; this appears to be primarily a result of anchoring of its termini. RNA Mango binds to the biotin moiety using a combination of stacking between the flap U15 and the surface of the quadruplex, direct hydrogen bonding, and water- and metal ion-mediated interactions (the latter differ between the two copies in the ASU; Fig. 4d,e). Comparison with structures of a previously reported biotin aptamer34 and with streptavidin35,36 shows that RNA Mango exploits strategies used by both. Whereas the ureido carbonyl oxygen of biotin is recognized through metal ion-mediated contacts in both RNA Mango and the biotin aptamer, extensive direct hydrogen bonds with the biotin heterocycle and linker as well as the close shape complementarity are more reminiscent of streptavidin (Supplementary Fig. 9). Thus, the structure of RNA Mango-TO1-Biotin highlights the diversity of RNA-small molecule interactions that can result from in vitro selection.
Non-specific, low-affinity intercalation of TO into double-stranded DNA and RNA results in enhancement of its fluorescence37. Intercalation is thought to facilitate a conformation where the two heterocycles and the unsaturated linker (a single benzylidiene carbon in the case of TO1, Fig. 1a) are more coplanar than in solution, and restricting the mobility of the conjugated connection between the methylquinole and benzothiazole rings. This reduces nonradiative decay, thereby increasing quantum yield38. Acylation of the benzothiazole of TO1 to yield TO1-Acetate substantially decreases nonspecific DNA and RNA binding by TO1, presumably by disfavoring intercalation38. Because the TO moiety of TO1-Biotin has minimal hydrogen bonding potential, RNA Mango evolved a high affinity-binding strategy that combines shape complementarity to the fluorescent headgroup and polar interactions with the biotin. Our results indicate that fluorescence activation by RNA Mango takes advantage of the biotin sterically restricting the benzothiazole and methylquinoline heterocycles. The ring-like conformation of the RNA Mango-bound TO1-Biotin may function analogously to a spring lock washer, pushing the benzothiazole and methylquinoline against their respective binding pockets, thereby restricting their mobility. That the conformation of the TO in this complex is not optimal or a unique solution is consistent with our observation (Supplementary Fig. 6,7) that TO1-Desthiobiotin (which has a longer PEG linker than TO1-Biotin, but loses the sulfur atom from the biotin) produces higher fluorescence enhancement and longer fluorescence lifetime than TO1-Biotin. Likewise, substitution of the PEG-biotin of the fluorophore by a pyrazine can also result in efficiently fluorescent ligands1. Likely, the pyrazine interacts with an RNA nucleobase, thus anchoring the distal end of the fluorophore.
The Spinach fluorescent aptamer5 enforces a near-coplanar arrangement on its fluorophore (DFHBI), which also consists of two heterocyles linked by a benzylidine or methine carbon, by sandwiching it between a G-quadruplex and a base triple15,16, thereby achieving5 a high quantum yield (~0.74). RNA Mango-TO1-Biotin has a more modest quantum yield (~0.14, ref. 1), consistent with the ~45° angle between the heterocycles of its bound fluorophore (Fig. 4d,e). Substitution of the biotin of its ligand with desthiobiotin results in a 50% increase in fluorescence enhancement (Supplementary Fig. 6). Our structure suggests that this may reflect binding to the RNA that results in a higher degree of coplanarity of the two thiazole orange heterocycles. While TO1-Biotin binds weakly to Spinach, that aptamer can induce surprisingly bright fluorescence at saturation6. Together, these results suggest that further engineering of RNA Mango and optimization of its ligand may result in RNA tags with improved photophysical properties.
METHODS
RNA preparation
RNA constructs used in this study are listed in Supplementary Table 3. RNAs 1–14 were chemically synthesized (Dharmacon or IDT), deprotected as per manufacturers’ instructions, exchanged into 50 mM HEPES-KOH (pH 7.5), 150 mM KCl, and 10 μM EDTA through centrifugal ultrafiltration (3000 Da cutoff, Millipore), filtered (0.1 μm cutoff, Amicon Ultrafree-MC, Millipore) and stored at 4°C. RNAs 15–32 were transcribed in vitro from PCR templates essentially as described40. RNAs were purified by electrophoresis on 14% polyacrylamide (19:1 acrylamide/bisacrylamide), 1× TBE, 8 M urea gels, elecroeluted from gel slices, washed once with 1 M KCl, desalted by ultrafiltration (10,000 Da cutoff, Millipore), filtered, and stored in water at 4° C.
Crystallization and diffraction data collection
RNA 1 (in 50 mM HEPES-KOH (pH 7.5), 150 mM KCl, and 10 μM EDTA) was heated at 95°C for 3 min, and then kept at 21°C for 1 h. Equimolar TO1-Biotin (ref. 1) in the same buffer was added and the mixture (250 μM final RNA concentration) incubated at 21°C for 30 min. For crystallization, sitting drops prepared by mixing 0.5 μL each of the RNA-dye solution and the reservoir were equilibrated at 21°C against 125–150 μL of 50 mM HEPES-KOH (pH 7.5), 0.25 M KCl, 1.4 M ammonium acetate, and 2.5 – 5.0 mM BaCl2. Strongly fluorescent (500 nm illumination, Supplementary Fig. 2a,b) tetragonal bipyramidal crystals grew in 1–3 days to maximum dimensions of 220 × 80 × 80 μm3. Prior to harvesting, the reservoir solution was exchanged to 50 mM HEPES-KOH (pH 7.5), 250 mM KCl, 2.2 M ammonium formate and 10% (v/v) glycerol, and the drops allowed to equilibrate for 1 – 3 days. Crystal I was mounted in a nylon loop immediately after addition of 1 μL of reservoir to the drop and vitrified by plunging into liquid nitrogen. Crystal II was transferred to 2 μL of reservoir solution supplemented with 40 mM iridium hexammine and incubated for 1.5 hours. The crystal was vitrified after a 1 min back-soak in 2 μL of reservoir solution supplemented with 100 μM iridium hexammine. Diffraction data were collected at 100 K with 0.9792 Å X-radiation at beamline 24-ID-C of the Advanced Photon Source (APS) and reduced with the HKL package (ref. 41). Data collection statistics are summarized in Supplementary Table 1. Fluorescence of crystals was not visibly altered by X-ray exposure (Supplementary Fig. 2c).
Structure determination and refinement
Heavy atom substructure determination, SAD phasing and initial density modification using Crystal II data were performed using SHELXD (ref. 42) and the AutoSol module in Phenix (ref. 43). The highest quality map was obtained using a substructure with 5 Ir and a solvent content of 0.55. Phase extension44 to 1.6 Å resolution of the unmodified phase-probability distributions (mean overall figure of merit = 0.28 for all reflections between 46.7 and 2.5 Å resolution) against Crystal I amplitudes produced an electron density map (Supplementary Fig. 2d) into which 55 RNA residues could be built manually45. Simulated annealing, energy minimization and restrained individual atomic B-factor refinement43 produced a model with Rfree = 28.3 %. Addition of TO1-Biotin followed by further refinement and model building yielded the current crystallographic model (Supplementary Table 1) which contains 55 RNA residues (all residues of chain A and 24 residues of chain B), 7 K+ ions, and two TO1-Biotin and 170 water molecules), and has a mean coordinate precision (maximum likelihood estimate43) of 0.16 Å. Ions were identified as K+ based on coordination, location relative to quadruplex plane, and B-factors. Except where noted, all structure figures are based on RNA chain A of the ASU, and were prepared using PyMol (ref. 46).
Analytical Ultracentrifugation
RNA 1 was heated to 93 °C for 3 min in 20 mM MOPS-KOH (pH 7.0), 150 mM KCl, 10 μM EDTA and then kept at 21°C for 1 hour. RNA Mango at 4 μM was added to each cell with an equal concentration of TO1-Biotin being added to one of the cells. The reference cell for each sample was 20 mM MOPS pH 7.0, 150 mM KCl and 10 μM EDTA; TO1-Biotin was not added to either reference cell. 500 scans were collected and averaged on a Beckman XLI analytical ultracentrifuge. Absorbance was measured at 295 nM, under constant velocity, with a run speed of 60,000 RPM at 20 °C. The viscosity and density of the buffer were calculated to be 0.01015 P and 1.0068 g ml−1 respectively, by the Sednterp server (http://sednterp.unh.edu).
Dynamic Light Scattering
7.8 g/l (~750 μM) of RNA Mango was heated to 93°C for 3 min in 20 mM MOPS pH, pH 7.0, 150 mM K+, 10 μM EDTA and cooled on bench for 30 min. TO1-Biotin was added to the RNA solution making final concentrations of 490 μM TO1-Biotin and 480 μM RNA Mango. The sample was filtered through a 0.1 μm spin filter (Millipore) prior to analysis. Light scattering was measured on a DYNA Pro NanoStar.
Fluorescence spectroscopy
RNA concentrations were determined by UV-spectrophotometry in buffer P (10 mM Na-phosphate, pH 7.2, 140 mM KCl, 1 mM MgCl2, 0.05% Tween 20; ref. 1) using nearest neighbor-corrected47 extinction coefficients (wild-type RNA Mango ε260 = 426.4 mM−1 cm−1). Buffer P was also used for dilutions of RNAs for fluorescent titrations. Prior to fluorescence measurements, RNAs were incubated at 95°C for 3 min and then kept at room temperature for 20–25 min. Fluorescence was measured on a Photon Technologies International/820 Photomultiplier Detection System with excitation and emission centered at 510 nm and 535 nm, respectively. The fluorescent signal was acquired over 125 seconds after addition and mixing of RNA. The fluorescence reading was taken as the average of the counts over the last 35 seconds. Dissociation constants for RNA Mango-TO1-Biotin complexes were determined by titrations where the concentration of TO1-Biotin was kept constant (40 nM) and RNA concentration was varied. The resulting curves were fit using the Hill equation, allowing both the Kd and Hill coefficient to float. Values reported are means ± standard deviation of three independent measurements. All measurements reported in Supplementary Table 2 were performed using Buffer P.
Fluorescence Lifetime
Fluorescence lifetime measurements were performed on a PTI EasyLife LS instrument in 20 mM MOPS pH 7.0, 150 mM KCl, 10 μM EDTA with 1 μM ligand and 10 μM of RNA 1. Excitation was performed with a 510 nm diode for all TO1 ligands with emission being detected at 535 nm using a 535 ± 6 nm filter. TO3 ligands were excited with a 635 nm diode and detected at 658 nm using a 660 ± 13 nm filter. Ludox colloidal silica suspension (Sigma-Aldrich) was used to determine the diode intensity and duration for each combination of diodes and filters.
Modeling of TO3 in Ligand Binding Pocket
TO3-Biotin coordinates were generated using phenix.elbow (ref. 43) and modeled into the electron density of TO1-Biotin using the final refined structure of Mango-TO1-Biotin using coot (ref. 45). Attempts to place the entire ligand into the binding pocket failed due to steric clashes. Given the fluorescence enhancement and fluorescence lifetimes of TO3-Biotin and its derivatives (Supplementary Fig. 6) the PEG-Biotin linker of TO3-Biotin likely does not interact with the binding pocket and was therefore omited from the binding model. TO3-methyl was generated using phenix.elbow and placed into the binding pocket using the TO1-Biotin electron density as a guide. The curent model has the benzothiazole, methylquinolin and the conjugated linker in plane, without any steric clashes with the RNA (Supplementary Fig. 8).
Data Availability
All data generated or analysed during this study are included in this published article (and its supplementary information files). Atomic coordinates and structure factor amplitudes for the RNA Mango-TO1-Biotin co-crystal structure have been deposited into the protein data bank with accession code 5V3F.
Supplementary Material
Acknowledgments
We thank the staff at beamlines 5.0.1 and 5.0.2 of the Advanced Light Source (ALS), Lawrence Berkeley National Laboratory, and beamline ID-24E of the Advanced Photon Source (APS), Argonne National Laboratory for crystallographic data collection; G. Piszczek (Biophysics Core, US National Heart, Lung and Blood Institute, NHLBI, National Institutes of Health (NIH)) for analytical ultracentrifugation and dynamic light scattering; D. Lee and R. Levine (NHLBI) for mass spectrometry; and S. Bachas, M. Chen, C. Fagan, C. Jones, T. Numata, D. Sen, L. Sjekloca, L. Truong, K. Warner, and J. Zhang for discussions, and an anonymous referee for suggesting the inclusion of Supplementary Fig. 8. This work was partly conducted at the ALS, on the on the Berkeley Center for Structural Biology Beamlines, and at the APS on the NE-CAT beamlines, which are supported by the NIH. Use of ALS and APS was supported by the US Department of Energy. P.J.U. was supported by an NSERC (Canada) operating grant. This work was supported in part by the intramural program of the NHLBI, NIH.
Footnotes
Accession codes
Protein Data Bank: Atomic coordinates and structure factor amplitudes for the RNA Mango-TO1-Biotin co-crystal structure have been deposited with accession code 5V3F.
AUTHOR CONTRIBUTIONS
P.J.U. and A.R.F. conceived the project; M.W.L. performed initial crystallization screens; S.C.Y.J. synthesized ligands; R.J.T., N.A.D. and M.W.L. carried out preparative biochemistry; R.J.T. performed crystallization, diffraction data collection, structure determination and refinement; R.J.T., N.A.D., S.S.S.P. and S.C.Y.J. performed structure-guided analyses; R.J.T. and A.R.F. prepared the manuscript with input from all authors.
COMPETING FINANCIAL INTERSTS
The authors declare no competing financial interests.
References
- 1.Dolgosheina EV, et al. RNA mango aptamer-fluorophore: a bright, high-affinity complex for RNA labeling and tracking. ACS Chem Biol. 2014;9:2412–2420. doi: 10.1021/cb500499x. [DOI] [PubMed] [Google Scholar]
- 2.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 3.Grate D, Wilson C. Laser-mediated, site-specific inactivation of RNA transcripts. Proc Natl Acad Sci USA. 1999;96:6131–6136. doi: 10.1073/pnas.96.11.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babendure JR, Adams SR, Tsien RY. Aptamers switch on fluorescence of triphenylmethane dyes. J Am Chem Soc. 2003;125:14716–14717. doi: 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- 5.Paige JS, Wu KY, Jaffrey SR. RNA mimics of green fluorescent protein. Science. 2011;333:642–646. doi: 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jeng SCY, Chan HHY, Booy EP, McKenna SA, Unrau PJ. Fluorophore ligand binding and complex stabilization of the RNA Mango and RNA Spinach aptamers. RNA. 2016;22:1884–1892. doi: 10.1261/rna.056226.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Day RN, Davidson MW. The Fluorescent Protein Revolution. CRC Press; Boca Raton: 2014. [Google Scholar]
- 8.Baugh C, Grate D, Wilson C. 2.8 Å crystal structure of the malachite green aptamer. J Mol Biol. 2000;301:117–128. doi: 10.1006/jmbi.2000.3951. [DOI] [PubMed] [Google Scholar]
- 9.Strack RL, Disney MD, Jaffrey SR. A superfolding Spinach2 reveals the dynamic nature of trinucleotide repeat-containing RNA. Nature Methods. 2013;10:1219–1224. doi: 10.1038/nmeth.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang PC, et al. Photochemical properties of Spinach and its use in selective imaging. Chemical Science. 2013;4:2865–2873. [Google Scholar]
- 11.Han KY, Leslie BJ, Fei JY, Zhang JC, Ha T. Understanding the photophysics of the Spinach-DFHBI RNA aptamer-fluorogen complex to improve live-cell RNA Imaging. J Am Chem Soc. 2013;135:19033–19038. doi: 10.1021/ja411060p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escobedo JO, Rusin O, Lim S, Strongin RM. NIR dyes for bioimaging applications. Curr Op Chem Biol. 2010;14:64–70. doi: 10.1016/j.cbpa.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc Natl Acad Sci U S A. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Quadruplex DNA: sequence, topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warner KD, et al. Structural basis for activity of highly efficient RNA mimics of green fluorescent protein. Nature Struct Mol Biol. 2014;21:658–663. doi: 10.1038/nsmb.2865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang H, et al. A G-quadruplex–containing RNA activates fluorescence in a GFP-like fluorophore. Nature Chem Biol. 2014;10:686–691. doi: 10.1038/nchembio.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramesh A, Wakeman CA, Winkler WC. Insights into metalloregulation by M-box riboswitch RNAs via structural analysis of manganese-bound complexes. J Mol Biol. 2011;407:556–570. doi: 10.1016/j.jmb.2011.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fiore JL, Nesbitt DJ. An RNA folding motif: GNRA tetraloop–receptor interactions. Quarterly Rev Biophys. 2013;46:223–264. doi: 10.1017/S0033583513000048. [DOI] [PubMed] [Google Scholar]
- 19.Coonrod LA, Lohman JR, Berglund JA. Utilizing the GAAA tetraloop/receptor to facilitate crystal packing and determination of the structure of a CUG RNA helix. Biochemistry. 2012;51:8330–8337. doi: 10.1021/bi300829w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 21.Jaeger L, Michel F, Westhof E. Involvement of a GNRA tetraloop in long-range RNA tertiary interactions. J Mol Biol. 1994;236:1271–6. doi: 10.1016/0022-2836(94)90055-8. [DOI] [PubMed] [Google Scholar]
- 22.Costa M, Michel F. Frequent use of the same tertiary motif by self-folding RNAs. EMBO J. 1995;14:1276–1285. doi: 10.1002/j.1460-2075.1995.tb07111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci USA. 2001;98:4899–4903. doi: 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howard FB, Miles HT. Poly(inosinic acid) helices: essential chelation of alkali metal ions in the axial channel. Biochemistry. 1982;21:6736–6745. doi: 10.1021/bi00269a019. [DOI] [PubMed] [Google Scholar]
- 25.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 26.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 27.Mendez MA, Szalai VA. Fluorescence of unmodified oligonucleotides: A tool to probe G-quadruplex DNA structure. Biopolymers. 2009;91:841–850. doi: 10.1002/bip.21268. [DOI] [PubMed] [Google Scholar]
- 28.Dao NT, Haselsberger R, Michel-Beyerle ME, Phan AT. Following G-quadruplex formation by its intrinsic fluorescence. FEBS letters. 2011;585:3969–3977. doi: 10.1016/j.febslet.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 29.Kwok CK, Sherlock ME, Bevilacqua PC. Effect of loop sequence and loop length on the intrinsic fluorescence of G-quadruplexes. Biochemistry. 2013;52:3019–3021. doi: 10.1021/bi400139e. [DOI] [PubMed] [Google Scholar]
- 30.Molinaro M, Tinoco I. Use of ultra stable UNCG tetraloop hairpins to fold RNA structures: thermodynamic and spectroscopic applications. Nucleic Acids Res. 1995;23:3056–3063. doi: 10.1093/nar/23.15.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sherlock ME, et al. Steady-state and time-resolved studies into the origin of the intrinsic fluorescence of G-quadruplexes. J Phys Chem B. 2016;120:5146–5158. doi: 10.1021/acs.jpcb.6b03790. [DOI] [PubMed] [Google Scholar]
- 32.Phan AT, et al. Strucuture-function studies of FMRP RGG peptide recognition of an RNA duplex-quadruplex junction. Nature Struct Mol Biol. 2011;18:796–804. doi: 10.1038/nsmb.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vasilyev N, et al. Crystal structure reveals specific recognition of a G-quadruplex RNA by a β-turn in the RGG motif of FMRP. Proc Natl Acad Sci U A. 2015;112:E5391–E5400. doi: 10.1073/pnas.1515737112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nix J, Sussman D, Wilson C. The 1.3 Å crystal structure of a biotin-binding pseudoknot and the basis for RNA molecular recognition. J Mol Biol. 2000;296:1235–1244. doi: 10.1006/jmbi.2000.3539. [DOI] [PubMed] [Google Scholar]
- 35.Hendrickson WA, et al. Crystal structure of core streptavidin determined from multiwavelength anomalous diffraction of synchrotron radiation. Proc Natl Acad Sci USA. 1989;86:2190–2194. doi: 10.1073/pnas.86.7.2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weber PC, Ohlendorf DH, Wendoloski JJ. Structural origins of high-affinity biotin binding to streptavidin. Science. 1989;243:85–88. doi: 10.1126/science.2911722. [DOI] [PubMed] [Google Scholar]
- 37.Nygren J, Svanvik N, Kubista M. The interactions between the fluorescent dye thiazole orange and DNA. Biopolymers. 1998;46:39–51. doi: 10.1002/(SICI)1097-0282(199807)46:1<39::AID-BIP4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 38.Carreon JR, Mahon KP, Kelley SO. Thiazole orange–peptide conjugates: sensitivity of DNA binding to chemical structure. Organic Lett. 2004;6:517–519. doi: 10.1021/ol0362818. [DOI] [PubMed] [Google Scholar]
- 39.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao H, Edwards TE, Ferré-D’Amaré AR. Structural basis for specific, high-affinity tetracycline binding by an in vitro evolved aptamer and artificial riboswitch. Chem Biol. 2008;15:1125–1137. doi: 10.1016/j.chembiol.2008.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Otwinowski Z, Minor W. Processing of diffraction data collected in oscillation mode. Meth Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 42.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 43.Adams P, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abrahams JP, Leslie AGW. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. 1996;D52:30–42. doi: 10.1107/S0907444995008754. [DOI] [PubMed] [Google Scholar]
- 45.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos: 2002. [Google Scholar]
- 47.Puglisi JD, Tinoco I., Jr Absorbance melting curves of RNA. Methods in Enzymology. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analysed during this study are included in this published article (and its supplementary information files). Atomic coordinates and structure factor amplitudes for the RNA Mango-TO1-Biotin co-crystal structure have been deposited into the protein data bank with accession code 5V3F.