Summary
The relevance of topographic cues for commitment of induced pluripotent stem cells (iPSCs) is largely unknown. In this study, we demonstrate that groove-ridge structures with a periodicity in the submicrometer range induce elongation of iPSC colonies, guide the orientation of apical actin fibers, and direct the polarity of cell division. Elongation of iPSC colonies impacts also on their intrinsic molecular patterning, which seems to be orchestrated from the rim of the colonies. BMP4-induced differentiation is enhanced in elongated colonies, and the submicron grooves impact on the spatial modulation of YAP activity upon induction with this morphogen. Interestingly, TAZ, a YAP paralog, shows distinct cytoskeletal localization in iPSCs. These findings demonstrate that topography can guide orientation and organization of iPSC colonies, which may affect the interaction between mechanosensors and mechanotransducers in iPSCs.
Keywords: iPSC, topography, biomaterials, microtopography, laser beam interference, pattern, mechanobiology, cytoskeleton, YAP, TAZ
Graphical Abstract
Highlights
-
•
Morphology of iPSC colonies can be controlled by nanotopography
-
•
Micropatterns direct cell division plane and orientation of apical actin fibers
-
•
The shape of colonies impacts on their self-organized patterning
-
•
TAZ is associated with the actin cytoskeleton at the rim of iPSC colonies
Topography can impact on growth and differentiation of iPSCs. Abagnale et al. used laser beam interference to generate submicron grooves that guide the shape of iPSC colonies, thereby modulating the spatial distribution of pluripotency markers, induction with BMP4, in vitro differentiation, and subcellular localization of the mechanotransducers YAP and TAZ.
Introduction
Biophysical properties of the microenvironment, such as stiffness and topography, modulate differentiation and self-renewal ability of stem cells (Dalby et al., 2007, Engler et al., 2006). Structural features of the extracellular landscape can be mimicked in vitro by grooves and pores to control cell shape (Clark et al., 1991, Curtis and Varde, 1964, Dunn and Heath, 1976, Oakley and Brunette, 1993). Interaction of cells with surface topography results in forces that trigger intracellular responses such as proliferation and directed differentiation (Abagnale et al., 2015, Unadkat et al., 2011). Topographical cues are also relevant for the modulation of pluripotent stem cells (PSCs), which possess the ability to self-renew infinitely and have the potential to develop into every cell type of the human body (Takahashi et al., 2007). For example, it has been suggested that mechanical stimulation of embryonic stem cells (ESCs) can mimic the embryonic microenvironment and thereby influence the expression of pluripotency markers and cell fate decisions (Sun et al., 2012, Sun et al., 2014). Other reports showed that surface roughness affects the spreading of human ESCs (Chen et al., 2012) and that defined structures can drive lineage-specific differentiation (Lee et al., 2010, McFarlin et al., 2009, Pan et al., 2013). Nonetheless, a comparative study of how groove-ridge structures of different sizes impact on induced PSCs (iPSCs) has so far not been reported. Moreover, it remains to be elucidated whether microtopographic stimuli affect the self-organization within iPSC colonies.
It has been recently described that expression of pluripotency factors is heterogeneous within iPSC colonies and that this heterogeneity may relate to the inherent diversity of human embryonic cells prior to gastrulation (Nazareth et al., 2013, Warmflash et al., 2014). A better understanding of how topography modulates the spatial organization of pluripotent cells is particularly relevant in early embryonic development, since it can drive the establishment of body axis and the formation of early anatomical structures (Keller et al., 2003). Changes in the geometry of pluripotent cells might alter such cell-cell interactions and the distribution of morphogenetic factors.
In mesenchymal stem cells the transcriptional coactivators YAP and TAZ function as mechanotransducers that translate physical stimuli, such as substrate elasticities and cellular density, into control of cellular growth and differentiation (Dupont et al., 2011, Halder et al., 2012). There is accumulating evidence that YAP and TAZ are generally involved in the regulation of cellular polarity and tissue homeostasis (Pan, 2007, Yu et al., 2015) and that they are crucial for the maintenance of pluripotency (Lian et al., 2010, Varelas et al., 2008). Nevertheless, it remains largely unclear whether YAP and TAZ are directly involved in interaction with the cytoskeleton and recognition of topographic stimuli (Raghunathan et al., 2014). Moreover, YAP and TAZ might also be involved in regulation of cell-cell interaction and spatial conformation within iPSC colonies.
In this study we have used groove-ridge structures in the submicrometer range that were generated with laser interference technology to investigate the impact of surface topography on the morphology of individual iPSCs. Moreover, we have analyzed how surface topography impacts on the organization of iPSC colonies with regard to expression patterns of pluripotency markers, activity of YAP and TAZ, and differentiation in response to morphogen stimulation.
Results
Laser-Ablated Submicron Grooves Induce Cellular and Nuclear Elongation of Individual iPSCs
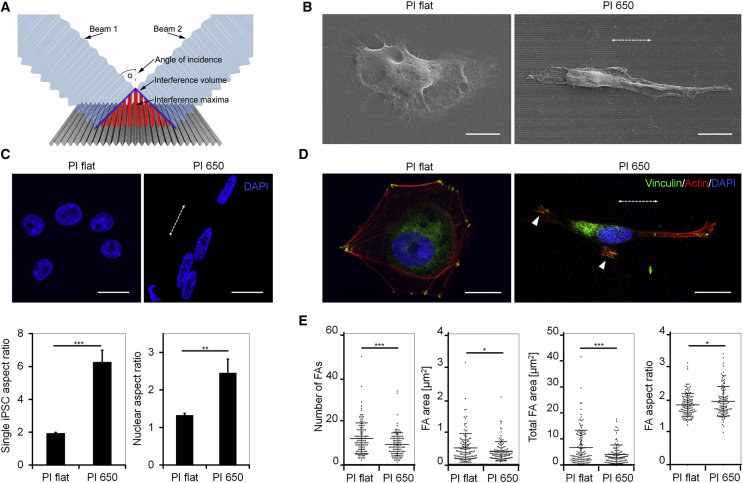
In this work we employed multibeam laser interference to structure a polyimide foil with grooves with a periodicity of 650 nm and height of 200 nm (PI 650; Figure 1A). Polyimide is a biocompatible plastic polymer with good mechanical properties (Julien et al., 2011) and it is relatively resistant to higher temperatures, which makes it ideal for applications involving laser ablation. Atomic force microscopy (AFM) measurements of PI 650 validated the homogeneity of surface structures (Figures S1A and S1B). These substrates were highly anisotropic compared with the flat polyimide (PI flat), as indicated by contact angle measurements (Figure S1C) (Good, 1992). When single iPSCs were seeded on PI 650 they aligned parallel to the submicron structures and showed both cellular (Figure 1B) and nuclear elongation (Figure 1C). Fluorescence staining of individual iPSCs revealed an organized cytoskeleton with actin fibers extending along the grooves (Figure 1D). Vinculin staining indicated that focal adhesions are reduced in size and number on the structures, and possess increased aspect ratio compared with the flat material (Figure 1E). Thus, topography in the submicrometer range affects cellular and nuclear morphology of single iPSCs and modulates the focal adhesion compartment.
Figure 1.
Submicron Grooves Affect Morphology of Single iPSCs
(A) Schematic representation of the two-beam interference structuring of grooves in polyimide with a periodicity of 650 nm (PI 650).
(B) Scanning electron microscopy images of individual iPSCs on flat polyimide and PI 650. Scale bars, 10 μm.
(C) Analysis of cellular and nuclear aspect ratios of individual iPSCs (n = 3 independent biological replica; ∗∗p < 0.01, ∗∗∗p < 0.001, unpaired Student's t test). Scale bars, 20 μm.
(D) Confocal microscopy of actin (red) and vinculin (green) in single iPSCs. Arrowhead indicates elongated focal adhesions. Data are presented as mean ± SD. Scale bars, 20 μm.
(E) Analysis of focal adhesions on PI flat and PI 650 (n = 3 independent biological replica; ∗p < 0.05, ∗∗∗p < 0.001, unpaired Student's t test).
The dotted white arrow always indicates the direction of the grooves; nuclei are stained with DAPI (blue).
Surface Topography Impacts on the Morphology of iPSC Colonies by Directing the Cell Division Plane
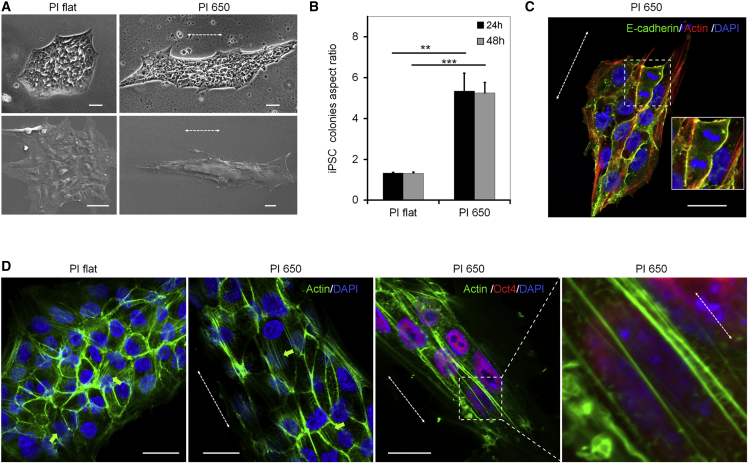
iPSCs typically grow as tightly packed colonies, similar to epiblast cells within the developing embryo (Zou and Niswander, 1996). When seeded on flat polyimide substrates, iPSC colonies displayed the classical round morphology (Figure 2A) and were positive for OCT4 (POU5F1), NANOG, and E-cadherin (Figure S2A). Notably, on grooves with a periodicity of 650 nm these colonies acquired an elongated shape (Figures 2A and 2B), which did not interfere with the expression of these three pluripotency factors (Figure S2B). We also observed that the aspect ratio of elongated colonies remained constant, at about 6:1, as they grew over time, which was in a range similar to that previously observed for single iPSCs on PI 650 (Figure 2B). On the other hand, elongation parallel to grooves was scarcely observed for individual cells localized within iPSC colonies. Therefore, the mechanism of how an iPSC colony consisting of thousands of cells elongates cannot simply be attributed to elongation of individual cells. We hypothesized that the polarity of mitosis might be influenced by surface topography. In fact the plane of cell division was found to be perpendicular to the direction of the grooves already at a single-cell level (Movie S1) and the same was observed within colonies (Figure 2C). Furthermore we observed apical actin fibers, which span above the nuclei and are orientated parallel to the submicron grooves (Figure 2D). On the other hand, the complex network of basal actin stress fibers and of actin filaments at cell-cell contacts did not seem to be modified by the structures (Figures S2C and S2D). These results indicate that topographic surface patterns can control the morphology of iPSC colonies by directing cell division planes and orientation of apical actin fibers.
Figure 2.
The Shape of iPSC Colonies Is Controlled by Submicron Topography
(A) Phase-contrast (top) and scanning electron microscopy images (bottom) of round and elongated iPSC colonies on PI flat and PI 650, respectively. Scale bars, 40 μm.
(B) Aspect ratio of the colonies either 24 or 48 hr after seeding (n = 3 independent biological replica; ∗∗p < 0.01, ∗∗∗p < 0.001, unpaired Student's t test). Data are presented as mean ± SD.
(C) Representative images showing how topography directs the cell division plane perpendicularly to the submicron grooves; inlay highlights a mitosis. Scale bar, 40 μm.
(D) iPSCs have been stained for the actin cytoskeleton (green) and the pluripotency marker OCT4 (red) with DAPI (blue) as nuclear counterstaining. Green arrows highlight apical actin fibers. The dotted white arrows indicate the direction of the grooves. Scale bars, 20 μm.
Elongation of iPSC Colonies on Different Substrates and Structures
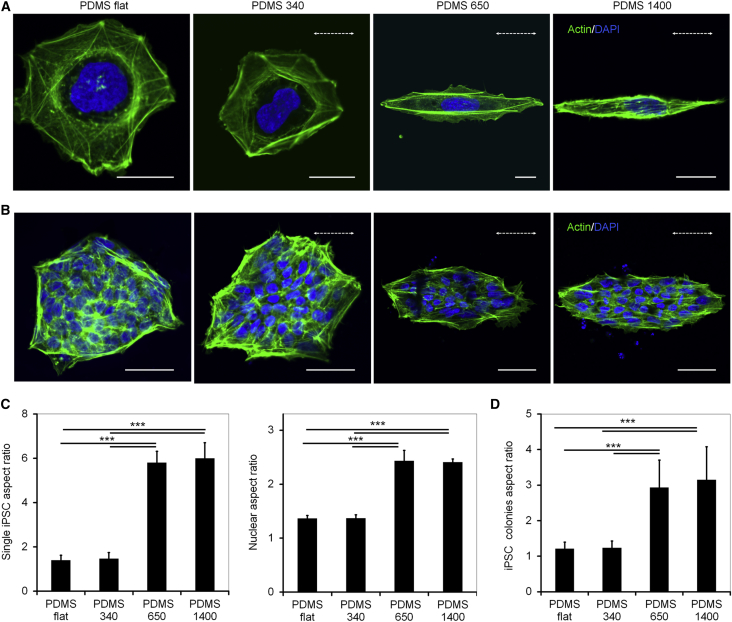
Subsequently, we analyzed whether similar effects on iPSC colonies were also observed on grooved structures of other dimensions or on different biomaterials. To this end, we generated submicron grooves featuring periodicities of 340 nm, 650 nm, and 1,400 nm on a polydimethylsiloxane (PDMS) by a strain-oxidation-release procedure with different air plasma oxidation times (Zhou et al., 2015). These materials are subsequently referred to as PDMS 340, PDMS 650, and PDMS 1400, respectively (Figures S3A–S3C). AFM measurements demonstrated that the periodicity of the structures was directly related to groove depth (Figure S3D). Single iPSCs seeded on PDMS 650 and PDMS 1400 clearly aligned along the patterns and displayed similar cellular and nuclear aspect ratios as previously observed on PI 650 (Figures 3A and 3C). Furthermore, the geometry of iPSC colonies was significantly altered by PDMS 650 and PDMS 1400, albeit to a lesser degree than on PI 650 (Figure 3B). Notably, on PDMS 340 neither individual iPSCs nor iPSC colonies were elongated and instead displayed a round shape as observed on flat PDMS. This similar threshold may indicate that the molecular processes for elongation of individualized cells and colonies are related.
Figure 3.
Structures with Diverse Size Induced Distinct Morphological Changes in iPSCs
(A and B) Confocal microscopy images of single iPSCs (A) and iPSC colonies (B) seeded on PDMS substrates that were structured with grooves of different periodicities. Cells were stained for actin (green) and nuclei (blue). The dotted white arrows indicate the direction of the submicron grooves. Scale bars, 20 μm in (A) and 50 μm in (B).
(C and D) Analysis of nuclear and cellular aspect ratio for single iPSCs (C) and whole iPSC colonies (D) when seeded on the PDMS submicron-grooved substrates. Data are presented as mean ± SD (n = 3 independent biological replicates, ∗∗∗p < 0.001).
Spatial Distribution of Pluripotency Markers Is Affected by Colony Geometry
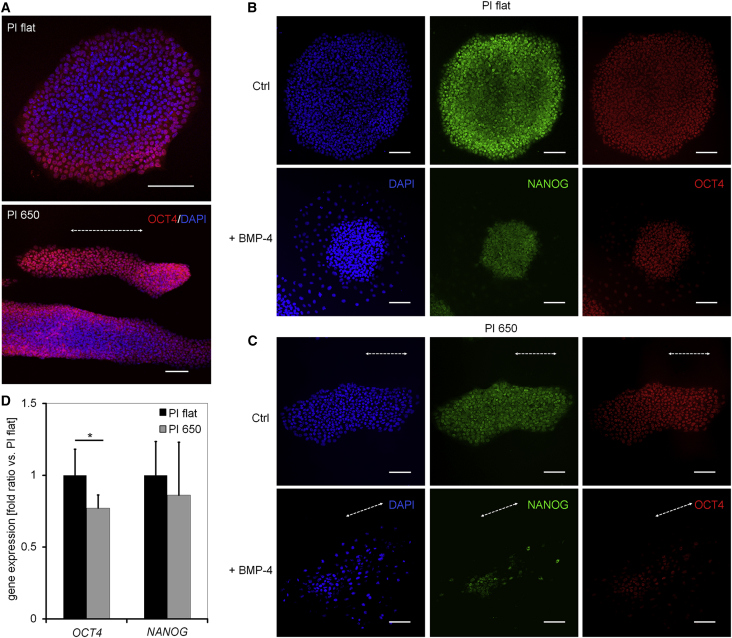
Next we analyzed whether the geometry of iPSCs impacts on the distribution of pluripotency-related proteins within the colonies. As described before, iPSCs seeded on non-structured polystyrene coverslips or flat polyimide display higher expression of pluripotency markers at the outer region of colonies compared with the center (Figure S4A). In iPSC colonies seeded on the PI 650, the expression pattern of OCT4 was elongated similarly to the morphology of the entire colony (Figure 4A). The same was true for NANOG, which is strongly expressed in cells at the rim of the colonies (Figures 4B and 4C). Furthermore, we observed a similar patterning in iPSC colonies seeded on flat and structured PDMS substrates (Figure S4B). These results indicate that the geometry of the colony is relevant for the self-organized heterogeneous expression of pluripotency factors. To rule out the possibility that these effects are only observed under specific culture conditions we performed additional experiments in TeSR-E8, a chemically defined culture medium specifically developed for culture of human PSCs (Chen et al., 2011). iPSCs cultured in TeSR-E8 displayed the same heterogeneous patterns of pluripotency markers as previously observed in iPS-Brew medium (Figure S4C).
Figure 4.
Submicron Structures Affect Patterning of iPSC Colonies
(A) Spatial heterogeneity of OCT4 (red) expression in round versus elongated iPSC colonies as shown by immunofluorescence staining. Scale bars, 100 μm.
(B and C) Confocal microscopy images showing of the distribution the pluripotency markers NANOG (green) OCT4 (red) in pluripotent colonies and upon induction with BMP4 for 48 hr on flat (B) and grooved polyimide (C). The dotted white arrows indicate the direction of the grooves; nuclei are stained with DAPI (blue). Scale bars, 100 μm.
(D) qRT-PCR analysis of expression levels of OCT4 and NANOG in iPSCs after exposure to 50 ng/μL BMP4 for 17 hr. Data are presented as mean ± SD. GAPDH was used as housekeeping gene (n = 3 independent biological replicates, ∗p < 0.05).
The elongated morphology of iPSC colonies might also alter the response to biochemical signals. We have exemplarily analyzed the effect of bone morphogenetic proteins (BMPs) on the differentiation of iPSCs, since these factors play a crucial role in early embryonic differentiation (Richter et al., 2014). Exposure to BMP4 led to decreased expression of NANOG and OCT4, which started at the rim of colonies and successively proceeded toward the cells in the center. A similar differentiation pattern was also observed in TeSR-E8 medium (Figure S4D). Importantly, in elongated colonies the loss of these pluripotency markers was more pronounced compared with round ones (Figures 4B and 4C). Moreover, qRT-PCR analysis confirmed that OCT4 and NANOG are downregulated faster on PI 650 compared with PI flat after stimulation with BMP4 (Figure 4D). Our data indicate that changes in colony morphology, elicited by submicron surface topography, impact on differentiation patterns in response to morphogens.
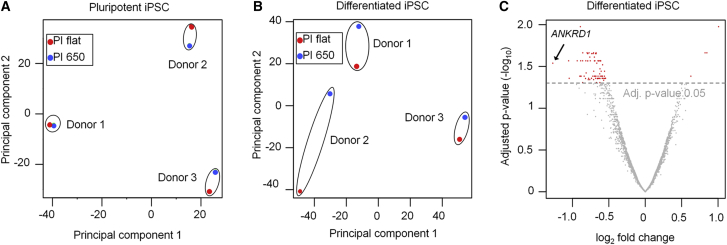
We followed the hypothesis that submicron topography directly modulates the transcriptional program of iPSCs. Single iPSCs were seeded on either flat or structured polyimide, and gene expression profiles were compared after 3 days. None of the genes revealed significant differential gene expression when cells were kept under culture conditions that maintained pluripotency (Figure 5A). Additionally we analyzed gene expression changes during 2 weeks of multilineage differentiation. In the course of differentiation many genes were significantly up- and downregulated, and this was more pronounced on PI flat than on PI 650 (Figures S5A–S5C). Furthermore, pairwise comparison of iPSCs that were differentiated on structured versus flat biomaterials revealed significant downregulation of 80 genes and upregulation of 4 genes (adjusted p < 0.05; Figures 5B and 5C; Table S1). Gene Ontology analysis demonstrated that differentially expressed genes were particularly enriched in the categories of extracellular matrix remodeling and tissue morphogenesis (Figure S5D). Of note, the biggest fold change on PI 650 versus PI flat after differentiation was observed in ANKRD1, which is a well-characterized readout of YAP/TAZ activity (Aragona et al., 2013).
Figure 5.
Genome-wide Analysis of the iPSC Transcriptome on Flat and Structured Polyimide
(A) Principal component analysis (PCA) based on gene expression data of iPSCs in pluripotent state. The iPSC lines cluster together, whereas surface topography has little impact (n = 3 independent biological replicates).
(B) PCA based on gene expression data of iPSCs after 2 weeks of differentiation on either PI flat or PI 650: cells cultured on the submicron structures have higher PC2 scores than cells of the same donor cultured on flat surfaces (n = 3 independent biological replicates).
(C) Differentiation iPSCs on the two substrates resulted in significant differences of gene expression: 80 are downregulated and 4 are upregulated on PI 650 versus PI flat (n = 3 independent biological replicates; adjusted p < 0.05, paired limma t test).
YAP and TAZ Show Distinct Localizations within iPSC Colonies
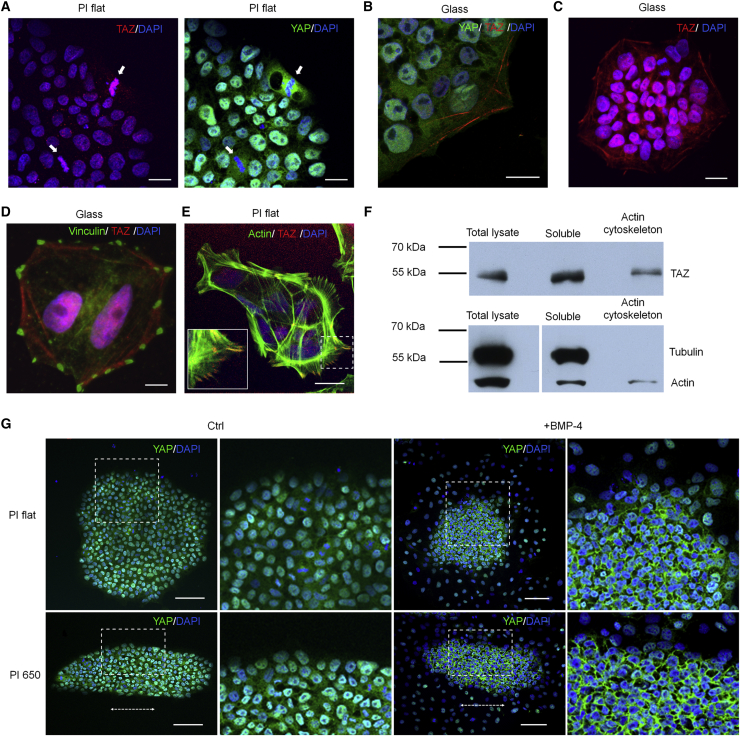
The transcriptional coactivators YAP and TAZ play important roles for mechanotransduction (Dupont et al., 2011, Kanai et al., 2000). Nonetheless, little is known about their function in pluripotent cells. Within iPSC colonies YAP and TAZ are predominantly localized in the nucleus, which may reflect their activity as transcription factors. Notably, during mitosis TAZ was tightly associated with condensed chromatin, whereas YAP was predominantly confined to the cytoplasm, and this was consistently observed in more than 250 cell divisions analyzed (Figures 6A, S6A, and S6B). Furthermore, we report that TAZ, but not YAP, was associated with filamentous structures that might resemble actin fibers (Figures 6B and S6C). Such TAZ localization was particularly evident in cells at the outer rim of iPSC colonies (Figure 6C). In contrast, YAP did not co-stain with cytoskeletal structures. These results indicate that the two paralogs TAZ and YAP might exert different functions.
Figure 6.
Distribution of YAP and TAZ in iPSCs
(A) Immunofluorescence images showing distinct distribution of YAP (green) and TAZ (red) in dividing iPSCs (white arrows). Scale bars, 20 μm.
(B) Immunostainings showing the presence of the mechanotransducer TAZ (red) but not of YAP (green) in filamentous structures similar to stress fibers. Scale bar, 20 μm.
(C) Confocal image displaying stronger cytoskeletal localization for TAZ (red) at the rim of an iPSC colony. Scale bar, 20 μm.
(D) Immunofluorescence image showing TAZ filaments (red) spanning between focal adhesion sites (green). Scale bar, 10 μm.
(E) Confocal microscopy image of iPSCs showing TAZ (red) localization at the tip of the actin fibers (green). Scale bar, 20 μm.
(F) Western blot shows that TAZ is present in the protein fraction of iPSCs associated to the actin cytoskeleton.
(G) Immunostaining showing YAP (green) localization in iPSCs before and after induction with BMP4 on both flat and grooved polyimide. Nuclei are always counterstained with DAPI (blue), and the dotted arrows indicate the direction of the grooves. Scale bars, 100 μm.
We subsequently wanted to further investigate the filamentous localization of TAZ. Previous studies have described such localization at cell-cell junctions (Zhao et al., 2011), whereas we also observed prominent TAZ expression close to focal adhesions of individual iPSCs (Figure 6D) and at the tip of actin filaments (Figure 6E; Movie S2). Western blot analysis validated that TAZ was present in the protein fraction that is bound to actin (Figure 6F). To gain additional insight into the subcellular localization of TAZ, we overexpressed a TAZ-GFP fusion protein in iPSCs and analyzed it by total internal reflection fluorescence (TIRF) microscopy. This analysis confirmed that TAZ is localized in filaments within 70 nm of the basal plasma membrane (Movie S3) at a region where recognition of topographic stimuli takes place (Kanchanawong et al., 2010). Furthermore, the GFP-tagged TAZ revealed transient association with condensed chromatin during mitosis, as previously observed in immunofluorescence microscopy (Movie S4). These results indicate that TAZ can interact with the actin cytoskeleton and components of focal adhesions. Interestingly, we observed that vinculin was partly detected in the nuclei of iPSCs. This was observed by immunostaining and validated by western blot analysis of subcellular fractions of iPSCs (Figures S6D and S6E). It is therefore conceivable that nuclear transfer of vinculin is due to its association with TAZ.
YAP, on the other hand, seemed to modulate BMP4-induced differentiation in relation to the spatial organization of the colonies. Upon BMP4 exposure, YAP was predominantly confined to the cytoplasm in cells of the inner portion of the colonies (Figure 6G) while in cells of the outer rim it was exclusively nuclear. This differential localization of YAP in the middle versus outside of the colonies was also validated in TeSR-E8 medium (Figure S6F). Morphological changes of iPSC colonies that were triggered by surface topography were also reflected in spatial expression of YAP: on polyimide with submicron structures the stimulation with BMP4 resulted in an elongated inner portion of the colony where YAP was translocated to the cytoplasm. Hence, surface topography may, either directly or indirectly, influence localization of YAP in response to morphogens, depending on the position of the target cells within the colony.
Discussion
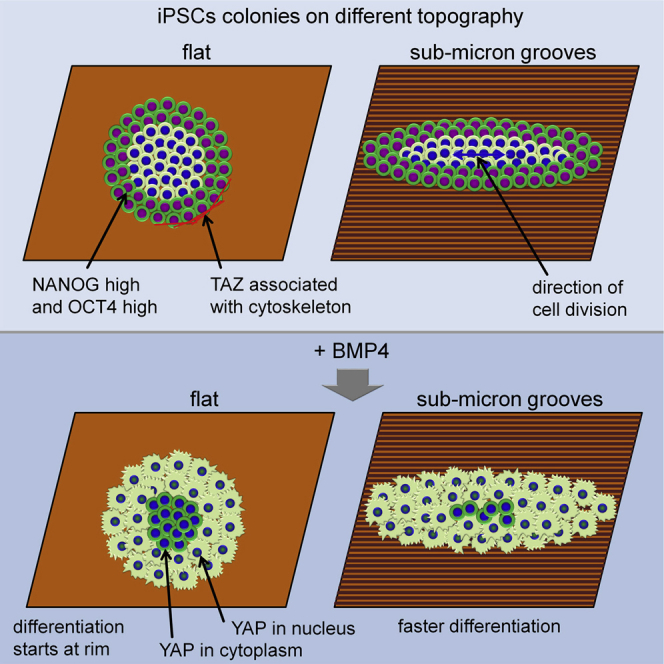
Surface topography guides not only morphology and migration of individual iPSCs but also impacts on the architecture and self-organization of iPSC colonies. Elongated colonies have distinct expression patterns of pluripotency markers and are more susceptible to morphogen-induced differentiation. In principle, the grooves might directly influence cellular differentiation or might exert indirect effects via reorganization of cell-cell interactions.
Different microfabrication processes can be employed to generate highly reproducible substrates with precise topographies in the submicrometer range. Laser technology, and in particular multibeam interference, is used to ablate the surface of a substrate and allows the direct structuring of most types of materials. In contrast to common laser techniques, laser interference is not limited by light diffraction and hence enables generation of subwavelength structures (Burrow and Gaylord, 2011). Production of submicron grooves in pre-strained PDMS substrates by air plasma treatment followed by their release is relatively easy to implement and is cost effective (Zhou et al., 2015). Furthermore, homogeneous patterning of the entire surface is facilitated and gradients can be obtained. The technique is initially limited to PDMS substrates, but it is possible to translate the structures into other materials by imprinting lithography (Zhou et al., 2016). On the other hand, the wrinkle or grating structures are less controlled and are more susceptible to batch-to-batch variation than surface structures generated by multibeam interference.
We demonstrate that submicron topography induces an increase in the aspect ratio of individual iPSCs and of iPSC colonies, and this was observed on polyimide and PDMS. Notably, elongation of individual iPSCs was similar in PI 650 and PDMS 650. However, iPSCs do not normally grow as dissociated cells; they persist in this state only for few hours before they undergo mitosis and grow eventually into a colony (Movie S1). The higher aspect ratio of individualized iPSCs might impact on their actin-myosin contractility, which was shown to be responsible for reduced viability of dissociated pluripotent cells (Chen et al., 2010). In contrast, elongation of colonies may rather be attributed to defined cell division planes and apical actin fibers, although the molecular mechanisms for mechanosensing might be the same. On PDMS 340 we did not observe elongation of individual iPSCs or colonies. It needs to be taken into account that for cell-substrate interactions not only the periodicity of the structures, but also their depth, is relevant (Lamers et al., 2010, Wojciak-Stothard et al., 1996) as well as the elastic modulus of the material and its chemical composition (Discher et al., 2005). Perhaps the periodicity of 340 nm is below the threshold that can be sensed by the cells, or the grooves are too shallow to be detected (about 13 nm depth). Either way, neither individualized iPSCs nor iPSC colonies seem to be affected by these very small structures. The height of PDMS 650 patterns is about 60–70 nm, which is in a range similar to the diameters of collagen fibers (Hulmes, 2002, Schmitt et al., 1942); therefore, the extracellular matrix components may also feature similar topography. Furthermore, as mechanical stress is generated not only at the cell-material interface but also at cell-cell contacts (Chu et al., 2004), the effects of surface topography are also influenced by the interplay of cells on structured materials. Traction forces between neighboring cells might mitigate the morphological changes elicited by the grooves. In fact, cells within the colonies display a more round shape, whereas individual cells revealed extensive elongation along the submicron grooves.
The higher expression of pluripotency factors at the outer region of iPSC colonies was similarly observed in two different culture media and on three different materials: polystyrene, polyimide, and PDMS. Thus, this patterning appears to be orchestrated by cell-cell interaction, rather than by material properties such as surface chemistry or elasticity. Hence, it would be interesting to compare gene expression at different regions of the same colony by laser microdissection to better understand the molecular differences. Surface topography did not interfere with pluripotency of iPSC colonies, and we did not observe significant gene expression changes when individual iPSCs were seeded on either PI flat or PI 650. These results indicate that surface structures have relatively little direct impact on the molecular makeup of iPSCs while they are in a pluripotent state.
Differentiation of iPSC colonies seems to be initiated at the rim, where the cells are more likely to undergo reversible epithelial-to-mesenchymal transition compared with those at the center (Chen et al., 2014). It has to be taken into account that culture media have a significant impact on cellular behavior. In fact, other authors reported that BMP4-induced differentiation of human ESCs varies under different defined culture conditions for pluripotent cells (Yu et al., 2011). However, when comparing two different commercially available media for the maintenance of iPSCs, we did not find differences in the intrinsic patterning of the colonies nor in the loss of pluripotency markers initiated by BMP4 treatment. This finding seems to be consistent with those of other studies (Warmflash et al., 2014). In elongated colonies the rim region is larger as compared with round colonies of comparable size, and this might be the reason why morphogen-driven differentiation was faster in elongated colonies. Alternatively cell-cell interactions, e.g., via paracrine effects, might affect differentiation kinetics. This finding is in line with a recent study that described self-organization of cellular architecture at different developmental stages of human blastocysts (Deglincerti et al., 2016). Taken together, it appears likely that our submicron grooves do not directly influence differentiation but rather control the spatial conformation of iPSC colonies, which then impacts on cell fate decisions.
Several studies indicated that YAP plays an important role during lineage specification (Pan, 2010). It is intriguing that upon BMP4 stimulation YAP reveals nuclear localization at the rim, whereas it is rather cytoplasmic at the center of differentiating iPSC colonies. Thus, it is conceivable that the different regions are more prone to differentiate into one or the other lineage. YAP might translate positional information in human pluripotent cells, similarly to what was previously observed in murine models (Nishioka et al., 2009). Furthermore, YAP was reported to be localized in the nucleus within the inner cell mass of human blastocytes and functions as important regulator of naive state (Qin et al., 2016). On the other hand TAZ, a YAP paralog, displayed unique spatial expression, which might indicate a different function: we describe that TAZ strongly co-localizes with actin filaments and cell-material adhesion sites in iPSCs. It is therefore conceivable that TAZ shuttles between the molecular machinery that recognizes topographical cues and the nucleus, where these stimuli are translated into the expression of specific genes.
In conclusion, surface topography in the submicrometer range affects morphology and migration of individual iPSCs. Furthermore, such structures guide the organization of iPSC colonies by directing apical actin fibers and cell division planes. Our work indicates that this indirect impact on cell-cell interaction and self-organization within iPSC colonies is relevant for differentiation processes and seems to involve the YAP/TAZ pathways. Tailored biomaterials may therefore mimic topographic cues of the extracellular matrix to modulate cell-cell interaction and thereby support directed differentiation.
Experimental Procedures
Two-Beam Laser Interference
Polyimide substrates (PI-2611, HD Microsystems) with a sample size of 20 × 20 mm were structured using a diode pumped Q301-HD Nd:YAG laser (JDS Uniphase) operating at a wavelength of 355 nm with a pulse duration of 38 ns and a coherence length of 5.2 mm, as described previously (Abagnale et al., 2015). The laser output is measured with a QE12LP-S-MP energy detector (Gentec Electro Optics) and a beam profiler (Ophir-Spiricon), while the ablation area is measured by a VKX 5000 digital microscope (Keyence). The effective interference contrast was determined to be 0.72 (Steger et al., 2014). The incidence angle of beams was set to 15.87° relative to surface normal, which results in periodicity of the interference pattern of 650 nm (Steger et al., 2013). The topography of generated structures was assessed by a Rados N8 atomic force microscope (Bruker Corporation).
Generation and Characterization of the PDMS Substrates
PDMS substrates were generated as described previously (Zhou et al., 2015, Zhou et al., 2016). In brief, PDMS pre-polymer and crosslinker (Sylgard 184; Dow Corning) were mixed in 10:1 ratio (w/w). PDMS substrates (9.5 × 9.5 cm, ∼1.5 mm thickness) were then stretched uniaxially to 130% of their original length. The samples were oxidized between 60 and 500 s in air plasma under pressure of 14 torr (0.2 mbar; Plasma Activate Flecto 10 USB). Removal of strain evoked wrinkle formation of different periodicities and height depending on the oxidation time. All samples were post-treated with air plasma for 10 min to ensure equal oxidation and Young's modulus values (62 MPa). AFM profiles were obtained using a Nanoscope V Dimension 3100 microscope (Veeco), whereas the stiffness analysis was performed on a Catalyst Nanoscoop V instrument (Bruker). Bruker SCANASYST-AIR (0.4 N m−1) and NP (0.017 N m−1) cantilevers made from silicon nitride with silicon tips were used before each measurement.
Induced Pluripotent Stem Cells
In this work we used two iPSC lines derived from bone marrow-derived human mesenchymal stromal cells, and one iPSC line obtained from human dermal fibroblasts (Willmann et al., 2013). Pluripotency was assessed by immunophenotypic analysis, multilineage differentiation, and Epi-Pluri-Score (Lenz et al., 2015). iPSCs were cultured in 6-well plates coated with 0.5 μg/cm2 of vitronectin (STEMCELL Technologies) and maintained in iPS-Brew medium (Miltenyi Biotec) supplemented with 1% penicillin/streptomycin (Life Technologies). Furthermore, we performed additional experiments with TeSR E8 (STEMCELL) to exclude that the observations were only related to specific culture conditions. iPSC differentiation was induced by addition of 50 ng/mL BMP4 (Miltenyi Biotec) for 48 hr.
Immunofluorescence Staining
Samples were fixed with 4% paraformaldehyde (Carl Roth) for 20 min, blocked with 1% BSA in Tris-buffered saline for 40 min, washed three times in 1× PBS (Life Technologies), and stained with primary antibody (OCT4, sc9081, Santa Cruz Biotechnology; Nanog, ab62734, Abcam; E-cadherin, AF648, R&D Systems; YAP, sc101199, Santa Cruz; TAZ, ab84927, Abcam) overnight at 4°C. All antibodies were diluted 1:50 in PBS containing 0.1% Triton X-100. Subsequently the samples were washed three times with PBS and incubated with secondary antibodies (Alexa Fluor, Life Technologies; diluted 1:200 in PBS containing 0.1% Triton X-100 and DAPI [Molecular Probes, 1:1,000 dilution]) for 1 hr.
Microscopy and Image Analysis
Acquisition of immunofluorescence images was performed with an LSM 710 confocal microscope (Carl Zeiss). Aspect ratio analysis was performed with ImageJ (https://imageJ.nih.gov./ij/) by dividing the major axis by the minor axis of cells and nuclei. For focal adhesion analysis, images of vinculin staining were converted to binary images in ImageJ and then analyzed with the SOFAST plugin. Phase-contrast images were obtained using an EVOS fl microscope (Life Technologies). Scanning electron microscopy images of iPSCs were acquired with an LEO 1455 EP microscope (Carl Zeiss) on fixed cells (4% paraformaldehyde) that were coated with a gold layer using a Sputter Coater 108auto (Cressington Scientific) as described previously (Abagnale et al., 2015). Time-lapse movies were taken with an Axio Observer Z1 inverted microscope (Carl Zeiss) equipped with heating stage and CO2 controller. For TIRF (Movie S3), GFP-TAZ was imaged using a 100×/1.46 numerical aperture (NA) objective. GFP was excited using 488-nm laser line at 10% of its nominal output power. The depth of the evanescent field was about 70 nm. Images were acquired every 15 s using a CCD camera (Evolve; Photometrics) driven by ZEN software (Carl Zeiss). For time-lapse movies (Movie S4), GFP-TAZ was imaged using a 20×/0.5 NA objective. GFP was excited using an HXP 120 illumination source in combination with filter set 52 (Carl Zeiss). Images were acquired every minute using a CCD camera (Evolve; Photometrics) driven by ZEN software (Carl Zeiss).
Gene Expression Profiling
iPSC colonies were dissociated into single cells with Accutase (Innovative Cell Technologies) and then either cultured for 3 days on vitronectin coated materials in iPSCs culture medium or for 14 days in medium that supports their spontaneous differentiation (KO-DMEM 80%, serum replacement 20%, 2 mM L-glutamine, 100 U/mL penicillin-streptomycin, non-essential amino acids 1%; all reagents from Life Technologies). Subsequently the cells were harvested and total RNA was isolated using a Nucleospin RNA plus kit (Macheray Nagel). Quality of RNA was assessed with a NanoDrop 2000 spectrophotometer (Thermo Scientific). Hybridization on Gene Chip Human Transcriptome Arrays 2.0 (Affymetrix) was performed at Life&Brain (Bonn, Germany) according to the manufacturer’s guidelines. We performed normalization by Robust Multichip Average algorithm (RMA; oligo package in R) and differential expression was analyzed with paired limma t test (Ritchie et al., 2015). To this end, we focused on gene with higher variance than 0.05 across different arrays, and did multiple-test correction by Benjamini-Hochberg method with cutoff of adjusted p value of <0.05. Gene ontology analysis was performed with the gProfile tool (Reimand et al., 2016). The microarray data are accessible in the NCBI GEO (http://www.ncbi.nlm.nih.gov/geo/) through accession number GEO: GSE84848.
qRT-PCR Analysis for the Expression of Pluripotency Markers
RNA was isolated with TRIzol reagent (Sigma-Aldrich) and its quality assessed with a NanoDrop ND2000 (Thermo Scientific). cDNA was generated with a High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Darmstadt, Germany). Semi-quantitative RT-PCR was carried out with power SYBR Green master mix (Applied Biosystems) for the following genes: OCT4 (forward 5′-GGG GGT TCT ATT TGG GAA GGT A-3′, reverse 5′-ACC CAC TTC TGC AGC AAG GG-3′), NANOG (forward 5′-CAG AAG GCC TCA GCA CCT AC-3′, reverse 5′-ATT GTT CCA GGT CTG GTT GC-3′), and GAPDH (forward 5′-GAA GGT GAA GGT CGG AGT C-3′, reverse 5′-GAA GAT GGT GAT GGG ATT TC-3′).
Western Blotting
Cytoplasmic and nuclear fractions were prepared as previously described in detail (Suzuki et al., 2010). In brief, cells were washed with PBS (37°C) and lysed for 10 min on ice with cytoskeleton lysis buffer (CLB; 0.5% Triton X-100, 10 mM 1,4-piperazinediethanesulfonic acid [pH 7.4], 150 mM NaCl, 5 mM EGTA, 5 mM glucose, 5 mM magnesium chloride, supplemented with 0.5 mM sodium fluoride, 1 mM sodium orthovanadate, and protease inhibitors cocktail). Afterward the Triton soluble fraction, corresponding to the cytosolic fraction, was collected and clarified at 10,000 × g for 10 min at 4°C. The cell monolayer was then washed twice with ice-cold cell lysis buffer and incubated with RIPA buffer for 15 min on ice. After the cells were scraped off, the RIPA buffer (Triton-insoluble or cytoskeletal fraction) was clarified at 12,000 × g for 10 min at 4°C. Both fractions were resolved by SDS-PAGE and probed by western blotting with antibodies against TAZ (1:1,000; V386, Cell Signaling Technology), actin (1:10,000; clone AC-74, Sigma-Aldrich), tubulin (clone YL1/2 [Gamper et al., 2016]), and vinculin (1:1,000; clone hVin-1, Sigma-Aldrich).
Generation of TAZ-GFP Fusion Protein
The open reading frame (ORF) of human WWTR1 (TAZ) was amplified by PCR from the 3xFLAG pCMV5-TOPO TAZ WT plasmid (Addgene; plasmid #24809) using the following primers: forward 5′-AAT TAG GAA TTC CAT GAA TCC GGC CTC GGC GCC C-3′ (to introduce an EcoRI site) and reverse 5′-GAT GGA TCC CTT ACA GCC AGG TTA GAA AGG GCT C-3′ (to introduce a BamHI site). The amplified product was cloned into the pWPXL-GFP plasmid (kindly provided by Trono Lab, EPFL, Lausanne, Switzerland) to generate WWTR1-GFP. The integritiy of the ORF was verified by DNA sequencing. The pWPXL WWTR1-GFP plasmid was then transfected into HEK293T cells together with pMD2G and psPAX vectors (kindly provided by Trono Lab) to produce lentiviral particles that were used to infect iPSCs. GFP-positive clones were then manually picked and expanded.
Author Contributions
G. Abagnale performed most of the experiments and, together with W.W., designed the study and wrote the paper. A.S. provided support with the live-cell imaging, focal adhesion analysis, and image processing. M.S. and A.G. generated and characterized the polyimide substrates. Q.Z. and P.v.R. generated and characterized the PDMS substrates. G. Aydin performed the western blots. C.S. generated the TAZ-GFP plasmid. G.M.-N. assisted in the acquisition of confocal microscopy images. C.-C.K., I.G.C., and M.Z. carried out the bioinformatics analysis. All authors contributed to the writing of the manuscript.
Acknowledgments
This work was supported by the German Research Foundation (DFG; WA 1706/3-2), within the Boost-Fund Project “MechCell” of the excellence initiative of RWTH Aachen University (T11-2), and by the Interdisciplinary Center for Clinical Research (IZKF), within the faculty of Medicine at the RWTH Aachen University. We are particularly grateful to the Immunohistochemistry and Confocal Microscopy Unit, a core facility of IZKF. We would like to thank Herbert Horn-Solle (ILT) for performing the scanning electron microscopy images. Q.Z. is very grateful for financial support from the China Scholarship Council (201406630003).
Published: July 27, 2017
Footnotes
Supplemental Information includes six figures, one table, and four movies and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.06.016.
Supplemental Information
A single iPSC on PI 650 undergoing mitosis and displaying a division plane parallel to the topography.
A z-stack of an iPSC colony taken with a confocal microscope with atypical TAZ localization in filaments.
TIRF microscopy of iPSCs expressing a GFP tagged version of TAZ. The movie highlights how TAZ is localized in fibrils in living iPSCs.
In iPSCs expressing GFP-TAZ, this protein is specifically translocated to the nucleus during cell division.
References
- Abagnale G., Steger M., Nguyen V.H., Hersch N., Sechi A., Joussen S., Denecke B., Merkel R., Hoffmann B., Dreser A. Surface topography enhances differentiation of mesenchymal stem cells towards osteogenic and adipogenic lineages. Biomaterials. 2015;61:316–326. doi: 10.1016/j.biomaterials.2015.05.030. [DOI] [PubMed] [Google Scholar]
- Aragona M., Panciera T., Manfrin A., Giulitti S., Michielin F., Elvassore N., Dupont S., Piccolo S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell. 2013;154:1047–1059. doi: 10.1016/j.cell.2013.07.042. [DOI] [PubMed] [Google Scholar]
- Burrow G.M., Gaylord T.K. Multi-beam interference advances and applications: nano-electronics, photonic crystals, metamaterials, subwavelength structures, optical trapping, and biomedical structures. Micromachines. 2011;2:221. [Google Scholar]
- Chen G., Hou Z., Gulbranson D.R., Thomson J.A. Actin-myosin contractility is responsible for the reduced viability of dissociated human embryonic stem cells. Cell Stem Cell. 2010;7:240–248. doi: 10.1016/j.stem.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Gulbranson D.R., Hou Z., Bolin J.M., Ruotti V., Probasco M.D., Smuga-Otto K., Howden S.E., Diol N.R., Propson N.E. Chemically defined conditions for human iPSC derivation and culture. Nat. Methods. 2011;8:424–429. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Villa-Diaz L.G., Sun Y., Weng S., Kim J.K., Lam R.H., Han L., Fan R., Krebsbach P.H., Fu J. Nanotopography influences adhesion, spreading, and self-renewal of human embryonic stem cells. ACS Nano. 2012;6:4094–4103. doi: 10.1021/nn3004923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.G., Mallon B.S., Johnson K.R., Hamilton R.S., McKay R.D., Robey P.G. Developmental insights from early mammalian embryos and core signaling pathways that influence human pluripotent cell growth and differentiation. Stem Cell Res. 2014;12:610–621. doi: 10.1016/j.scr.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu Y.S., Thomas W.A., Eder O., Pincet F., Perez E., Thiery J.P., Dufour S. Force measurements in E-cadherin-mediated cell doublets reveal rapid adhesion strengthened by actin cytoskeleton remodeling through Rac and Cdc42. J. Cell Biol. 2004;167:1183–1194. doi: 10.1083/jcb.200403043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark P., Connolly P., Curtis A.S., Dow J.A., Wilkinson C.D. Cell guidance by ultrafine topography in vitro. J. Cell Sci. 1991;99(Pt 1):73–77. doi: 10.1242/jcs.99.1.73. [DOI] [PubMed] [Google Scholar]
- Curtis A.S., Varde M. Control of cell behavior: topological factors. J. Natl. Cancer Inst. 1964;33:15–26. [PubMed] [Google Scholar]
- Dalby M.J., Gadegaard N., Tare R., Andar A., Riehle M.O., Herzyk P., Wilkinson C.D.W., Oreffo R.O.C. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- Deglincerti A., Croft G.F., Pietila L.N., Zernicka-Goetz M., Siggia E.D., Brivanlou A.H. Self-organization of the in vitro attached human embryo. Nature. 2016;533:251–254. doi: 10.1038/nature17948. [DOI] [PubMed] [Google Scholar]
- Discher D.E., Janmey P., Wang Y.L. Tissue cells feel and respond to the stiffness of their substrate. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Dunn G.A., Heath J.P. A new hypothesis of contact guidance in tissue cells. Exp. Cell Res. 1976;101:1–14. doi: 10.1016/0014-4827(76)90405-5. [DOI] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- Gamper I., Fleck D., Barlin M., Spehr M., El Sayad S., Kleine H., Maxeiner S., Schalla C., Aydin G., Hoss M. GAR22beta regulates cell migration, sperm motility, and axoneme structure. Mol. Biol. Cell. 2016;27:277–294. doi: 10.1091/mbc.E15-06-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good R.J. Contact angle, wetting, and adhesion: a critical review. J. Adhes. Sci. Technol. 1992;6:1269–1302. [Google Scholar]
- Halder G., Dupont S., Piccolo S. Transduction of mechanical and cytoskeletal cues by YAP and TAZ. Nat. Rev. Mol. Cell Biol. 2012;13:591–600. doi: 10.1038/nrm3416. [DOI] [PubMed] [Google Scholar]
- Hulmes D.J.S. Building collagen molecules, fibrils, and suprafibrillar structures. J. Struct. Biol. 2002;137:2–10. doi: 10.1006/jsbi.2002.4450. [DOI] [PubMed] [Google Scholar]
- Julien S., Peters T., Ziemssen F., Arango-Gonzalez B., Beck S., Thielecke H., Buth H., Van Vlierberghe S., Sirova M., Rossmann P. Implantation of ultrathin, biofunctionalized polyimide membranes into the subretinal space of rats. Biomaterials. 2011;32:3890–3898. doi: 10.1016/j.biomaterials.2011.02.016. [DOI] [PubMed] [Google Scholar]
- Kanai F., Marignani P.A., Sarbassova D., Yagi R., Hall R.A., Donowitz M., Hisaminato A., Fujiwara T., Ito Y., Cantley L.C. TAZ: a novel transcriptional co-activator regulated by interactions with 14-3-3 and PDZ domain proteins. EMBO J. 2000;19:6778–6791. doi: 10.1093/emboj/19.24.6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanchanawong P., Shtengel G., Pasapera A.M., Ramko E.B., Davidson M.W., Hess H.F., Waterman C.M. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R., Davidson L.A., Shook D.R. How we are shaped: the biomechanics of gastrulation. Differentiation. 2003;71:171–205. doi: 10.1046/j.1432-0436.2003.710301.x. [DOI] [PubMed] [Google Scholar]
- Lamers E., van Horssen R., te Riet J., van Delft F.C., Luttge R., Walboomers X.F., Jansen J.A. The influence of nanoscale topographical cues on initial osteoblast morphology and migration. Eur. Cells Mater. 2010;20:329–343. doi: 10.22203/ecm.v020a27. [DOI] [PubMed] [Google Scholar]
- Lee M.R., Kwon K.W., Jung H., Kim H.N., Suh K.Y., Kim K., Kim K.S. Direct differentiation of human embryonic stem cells into selective neurons on nanoscale ridge/groove pattern arrays. Biomaterials. 2010;31:4360–4366. doi: 10.1016/j.biomaterials.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Lenz M., Goetzke R., Schenk A., Schubert C., Veeck J., Hemeda H., Koschmieder S., Zenke M., Schuppert A., Wagner W. Epigenetic biomarker to support classification into pluripotent and non-pluripotent cells. Sci. Rep. 2015;5:8973. doi: 10.1038/srep08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian I., Kim J., Okazawa H., Zhao J., Zhao B., Yu J., Chinnaiyan A., Israel M.A., Goldstein L.S., Abujarour R. The role of YAP transcription coactivator in regulating stem cell self-renewal and differentiation. Genes Dev. 2010;24:1106–1118. doi: 10.1101/gad.1903310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlin D.F., Finn K.J., Nealey P.F., Murphy C.J. Nanoscale through substratum topographic cues modulate human embryonic stem cell self-renewal. J. Biomim. Biomater. Tissue Eng. 2009;2:15–26. [Google Scholar]
- Nazareth E.J.P., Ostblom J.E.E., Lucker P.B., Shukla S., Alvarez M.M., Oh S.K.W., Yin T., Zandstra P.W. High-throughput fingerprinting of human pluripotent stem cell fate responses and lineage bias. Nat. Methods. 2013;10:1225–1231. doi: 10.1038/nmeth.2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishioka N., Inoue K., Adachi K., Kiyonari H., Ota M., Ralston A., Yabuta N., Hirahara S., Stephenson R.O., Ogonuki N. The Hippo signaling pathway components Lats and Yap pattern Tead4 activity to distinguish mouse trophectoderm from inner cell mass. Dev. Cell. 2009;16:398–410. doi: 10.1016/j.devcel.2009.02.003. [DOI] [PubMed] [Google Scholar]
- Oakley C., Brunette D.M. The sequence of alignment of microtubules, focal contacts and actin filaments in fibroblasts spreading on smooth and grooved titanium substrata. J. Cell Sci. 1993;106(Pt 1):343–354. doi: 10.1242/jcs.106.1.343. [DOI] [PubMed] [Google Scholar]
- Pan D. Hippo signaling in organ size control. Genes Dev. 2007;21:886–897. doi: 10.1101/gad.1536007. [DOI] [PubMed] [Google Scholar]
- Pan D. The hippo signaling pathway in development and cancer. Dev. Cell. 2010;19:491–505. doi: 10.1016/j.devcel.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F., Zhang M., Wu G., Lai Y., Greber B., Scholer H.R., Chi L. Topographic effect on human induced pluripotent stem cells differentiation towards neuronal lineage. Biomaterials. 2013;34:8131–8139. doi: 10.1016/j.biomaterials.2013.07.025. [DOI] [PubMed] [Google Scholar]
- Qin H., Hejna M., Liu Y., Percharde M., Wossidlo M., Blouin L., Durruthy-Durruthy J., Wong P., Qi Z., Yu J. YAP induces human naive pluripotency. Cell Rep. 2016;14:2301–2312. doi: 10.1016/j.celrep.2016.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan V.K., Dreier B., Morgan J.T., Tuyen B.C., Rose B.W., Reilly C.M., Russell P., Murphy C.J. Involvement of YAP, TAZ and HSP90 in contact guidance and intercellular junction formation in corneal epithelial cells. PLoS One. 2014;9:e109811. doi: 10.1371/journal.pone.0109811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimand J., Arak T., Adler P., Kolberg L., Reisberg S., Peterson H., Vilo J. g:Profiler—a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A., Valdimarsdottir L., Hrafnkelsdottir H.E., Runarsson J.F., Omarsdottir A.R., Ward-van Oostwaard D., Mummery C., Valdimarsdottir G. BMP4 promotes EMT and mesodermal commitment in human embryonic stem cells via SLUG and MSX2. Stem Cells. 2014;32:636–648. doi: 10.1002/stem.1592. [DOI] [PubMed] [Google Scholar]
- Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt F.O., Hall C.E., Jakus M.A. Electron microscope investigations of the structure of collagen. J. Cell. Physiol. 1942;20:11–33. [Google Scholar]
- Steger M., Boes S., Thilker S., Gillner A. Measuring method for the interference contrast of multi-beam-interference. J. Laser Micro Nanoen. 2014;9:225–229. [Google Scholar]
- Steger M., Hartmann C., Beckemper S., Holtkamp J., Gillner A. Fabrication of hierarchical structures by direct laser writing and multi-beam-interference. J. Laser Micro Nanoen. 2013;8:210–215. [Google Scholar]
- Sun Y., Villa-Diaz L.G., Lam R.H., Chen W., Krebsbach P.H., Fu J. Mechanics regulates fate decisions of human embryonic stem cells. PLoS One. 2012;7:e37178. doi: 10.1371/journal.pone.0037178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Yong K.M.A., Villa-Diaz L.G., Zhang X., Chen W., Philson R., Weng S., Xu H., Krebsbach P.H., Fu J. Hippo/YAP-mediated rigidity-dependent motor neuron differentiation of human pluripotent stem cells. Nat. Mater. 2014;13:599–604. doi: 10.1038/nmat3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Bose P., Leong-Quong R.Y., Fujita D.J., Riabowol K. REAP: a two minute cell fractionation method. BMC Res. Notes. 2010;3:294. doi: 10.1186/1756-0500-3-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Unadkat H.V., Hulsman M., Cornelissen K., Papenburg B.J., Truckenmuller R.K., Carpenter A.E., Wessling M., Post G.F., Uetz M., Reinders M.J. An algorithm-based topographical biomaterials library to instruct cell fate. Proc. Natl. Acad. Sci. USA. 2011;108:16565–16570. doi: 10.1073/pnas.1109861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varelas X., Sakuma R., Samavarchi-Tehrani P., Peerani R., Rao B.M., Dembowy J., Yaffe M.B., Zandstra P.W., Wrana J.L. TAZ controls Smad nucleocytoplasmic shuttling and regulates human embryonic stem-cell self-renewal. Nat. Cell Biol. 2008;10:837–848. doi: 10.1038/ncb1748. [DOI] [PubMed] [Google Scholar]
- Warmflash A., Sorre B., Etoc F., Siggia E.D., Brivanlou A.H. A method to recapitulate early embryonic spatial patterning in human embryonic stem cells. Nat. Methods. 2014;11:847–854. doi: 10.1038/nmeth.3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann C.A., Hemeda H., Pieper L.A., Lenz M., Qin J., Joussen S., Sontag S., Wanek P., Denecke B., Schuler H.M. To clone or not to clone? Induced pluripotent stem cells can be generated in bulk culture. PLoS One. 2013;8:e65324. doi: 10.1371/journal.pone.0065324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojciak-Stothard B., Curtis A., Monaghan W., MacDonald K., Wilkinson C. Guidance and activation of murine macrophages by nanometric scale topography. Exp. Cell Res. 1996;223:426–435. doi: 10.1006/excr.1996.0098. [DOI] [PubMed] [Google Scholar]
- Yu F.-X., Zhao B., Guan K.-L. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Pan G., Yu J., Thomson J.A. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8:326–334. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Li L., Lu Q., Wang L.H., Liu C.Y., Lei Q., Guan K.L. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev. 2011;25:51–63. doi: 10.1101/gad.2000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Kuhn P.T., Huisman T., Nieboer E., van Zwol C., van Kooten T.G., van Rijn P. Directional nanotopographic gradients: a high-throughput screening platform for cell contact guidance. Sci. Rep. 2015;5:16240. doi: 10.1038/srep16240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Wünnemann P., Kühn P.T., de Vries J., Helmin M., Böker A., van Kooten T.G., van Rijn P. Mechanical properties of aligned nanotopologies for directing cellular behavior. Adv. Mater. Inter. 2016;3:1600275. [Google Scholar]
- Zou H., Niswander L. Requirement for BMP signaling in interdigital apoptosis and scale formation. Science. 1996;272:738–741. doi: 10.1126/science.272.5262.738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A single iPSC on PI 650 undergoing mitosis and displaying a division plane parallel to the topography.
A z-stack of an iPSC colony taken with a confocal microscope with atypical TAZ localization in filaments.
TIRF microscopy of iPSCs expressing a GFP tagged version of TAZ. The movie highlights how TAZ is localized in fibrils in living iPSCs.
In iPSCs expressing GFP-TAZ, this protein is specifically translocated to the nucleus during cell division.