Summary
Growing evidence implicates the importance of glia, particularly astrocytes, in neurological and psychiatric diseases. Here, we describe a rapid and robust method for the differentiation of highly pure populations of replicative astrocytes from human induced pluripotent stem cells (hiPSCs), via a neural progenitor cell (NPC) intermediate. We evaluated this protocol across 42 NPC lines (derived from 30 individuals). Transcriptomic analysis demonstrated that hiPSC-astrocytes from four individuals are highly similar to primary human fetal astrocytes and characteristic of a non-reactive state. hiPSC-astrocytes respond to inflammatory stimulants, display phagocytic capacity, and enhance microglial phagocytosis. hiPSC-astrocytes also possess spontaneous calcium transient activity. Our protocol is a reproducible, straightforward (single medium), and rapid (<30 days) method to generate populations of hiPSC-astrocytes that can be used for neuron-astrocyte and microglia-astrocyte co-cultures for the study of neuropsychiatric disorders.
Keywords: human induced pluripotent stem cell, iPSC, astrocyte
Highlights
-
•
hiPSC-derived astrocyte populations generated from 42 NPC lines
-
•
Transcriptomic analysis shows hiPSC-astrocytes resemble primary human astrocytes
-
•
hiPSC-astrocyte transcription is consistent with a non-reactive state
-
•
hiPSC-astrocytes undergo inflammatory response and enhance microglial phagocytosis
Brennand, Goate, and colleagues report a rapid and robust method for the differentiation of highly pure populations of replicative astrocytes from human induced pluripotent stem cells (hiPSCs) via a neural progenitor cell (NPC) intermediate. hiPSC-astrocytes resemble primary human fetal astrocytes, have a transcriptional signature consistent with a non-reactive state, respond to inflammatory stimulants, and enhance microglial phagocytosis.
Introduction
Astrocytes are the most abundant cell type in the CNS, rivaling the diversity of neurons in cellular morphologies, gene expression profiles, developmental origins, physiological properties, functions, and responses to injury and disease (Zhang and Barres, 2010). Within the human brain, astrocytes have a variety of essential functions including glutamate biology, axonal guidance, trophic support, inflammatory response and wound healing, formation of the blood-brain barrier, and neuronal synapse formation, and plasticity (Barres, 2008, Eroglu and Barres, 2010, Freeman and Rowitch, 2013). Although the full contribution of astrocytes to neurological disease remains unresolved, astrocyte cell-autonomous deficits have been implicated in a variety of neurological disorders (Seifert et al., 2006, Tong et al., 2014). The most significant genetic risk factor for Alzheimer's disease (AD), apolipoprotein E4 (APOE4), is predominantly synthesized and secreted by astrocytes (Xu et al., 2006). Furthermore, astrocytes derived from human induced pluripotent stem cell (hiPSC)- or mouse-based models of amyotrophic lateral sclerosis (Di Giorgio et al., 2008, Marchetto et al., 2008, Papadeas et al., 2011), Rett syndrome (Ballas et al., 2009), and Huntington disease (Bradford et al., 2009) damage neurons in co-culture or after transplantation.
Evolutionarily, the astrocyte-to-neuron ratio increases from low vertebrates to rodents and to primates (Sherwood et al., 2006). Human cortical astrocytes are larger, structurally more complex and diverse, and propagate calcium waves 4-fold faster than their rodent counterparts (Oberheim et al., 2009). Transplantation of human glia into mice enhances activity-dependent plasticity and learning (Bi et al., 2013). Given the unique biology of human astrocytes, it is critical that improved human-specific cell-based systems be established to enable the study of human astrocyte function in health and disease.
Because of their ability to model all of the (known and unknown) genetic risk factors underlying neuropsychiatric disease, hiPSCs are routinely used as a source of various types of neurons and astrocytes for study (Mertens et al., 2016). Current hiPSC-based methods for the differentiation of astrocytes typically rely on either a neural progenitor cell (NPC) (Haidet-Phillips et al., 2014, Krencik et al., 2011, McGivern et al., 2013, Serio et al., 2013, Shaltouki et al., 2013) or oligodendrocyte progenitor cell (Jiang et al., 2013) intermediate. While it has been widely demonstrated that hiPSCs can be differentiated to functional astrocytes for cell-based models of neuropsychiatric disorders in vitro (Haidet-Phillips et al., 2014, McGivern et al., 2013, Serio et al., 2013, Shaltouki et al., 2013) or engraftment in vivo (Chen et al., 2015, Haidet-Phillips et al., 2014, Jiang et al., 2013, Krencik et al., 2011), existing methods are slow (up to 6 months) (Jiang et al., 2013, Krencik et al., 2011, Shaltouki et al., 2013) and/or require sorting to reduce heterogeneity (Chaboub and Deneen, 2013, Yuan et al., 2011). Here, we screened a number of published protocols, along with commercially available media for primary human astrocyte culture, identifying a robust and straightforward differentiation protocol for generating astrocytes from hiPSCs. By co-culture with microglia, we compared the function of primary human fetal astrocytes and hiPSC-astrocytes in assays for neuroinflammatory response, phagocytosis, and spontaneous calcium activity, concluding that hiPSC-astrocytes are highly similar to their primary counterparts. Altogether, our rapid differentiation protocol, co-culture strategy, and scalable phenotypic assays will serve as a robust platform for queries of healthy and diseased human astrocytes.
Results
30-Day Exposure of hiPSC-Derived NPCs to Commercial Astrocyte Media Is Sufficient to Robustly Generate hiPSC-Astrocytes
We first screened 11 different media conditions on forebrain-patterned NPCs (Brennand et al., 2015, Brennand and Gage, 2011) derived from hiPSCs (Table 1). The screening conditions, based on recently published hiPSC-astrocyte differentiation protocols (Chen et al., 2015, Haidet-Phillips et al., 2014, Jiang et al., 2013, Krencik et al., 2011, McGivern et al., 2013, Serio et al., 2013, Shaltouki et al., 2013), included different combinations of fibroblast growth factor 2 (FGF2) (Haidet-Phillips et al., 2014), ciliary neurotrophic factor (CNTF), (Krencik et al., 2011, Shaltouki et al., 2013), bone morphogenetic protein 4 (BMP4) (Han et al., 2013, Jiang et al., 2013, Shaltouki et al., 2013), fibroblast bovine serum (FBS) (Han et al., 2013, Shaltouki et al., 2013), neuregulin (Pinkas-Kramarski et al., 1994, Shaltouki et al., 2013), insulin (Heni et al., 2011), and ascorbic acid (AA) (Palm et al., 2015), as well as three commercial astrocyte media (ScienCell, Gibco, and Lonza) for the culture of primary human fetal astrocytes (Table 1). Screening criteria included immunoreactivity for two classical markers of astrocyte identity, S100β and glial fibrillary acidic protein (GFAP) (Ludwin et al., 1976), astrocyte morphology, survival, replicative ability, and cell line variability (Table S1; Figure S1A). When tested on NPCs, most conditions resulted in limited cell proliferation and expression of neuronal markers (Table S1); however, two commercial media, ScienCell and Lonza, yielded S100β- and GFAP-positive astrocyte-like cells (Figures S1B–S1D). These results were confirmed across four representative NPC lines by both flow cytometry and immunocytochemistry by 30 days (Figures 1A and S1E–S1G). Culture of NPCs in both media, when combined with low initial seeding density (nearly single cells: 15,000 cells/cm2) and minimal serum exposure (1%–2%), resulted in astrocyte morphology within 10 days (Figure S1H); star-shaped astrocyte morphologies were evident within 30 days (Figure S1I). Although ScienCell and Lonza astrocyte media showed equivalent efficiencies (Figures S1B–S1D), ScienCell medium was selected owing to its lower cost and relative simplicity.
Table 1.
Screening Conditions for Astrocyte Differentiation
| ID | Reference | Basal Medium | Supplement 1 | Supplement 2 | Growth Factors | |||
|---|---|---|---|---|---|---|---|---|
| 1 | Brennand and Gage (2011) | DMEM/F12 | N2, B27 | GlutaMAX + sodium bicarbonate + sodium pyruvate | BDNF | GDNF | cAMP | AA |
| 2 | ScienCell 1801 | ND (AM) | AGS | 2% FBS | ||||
| 3 | Thermo Fisher A1261301 | DMEM | N2 | GlutaMAX + D-glucose + sodium pyruvate | 10% FBS | |||
| 4 | Lonza CC-3186 | ND | – | L-glutamine | EGF | 3% FBS | insulin + AA | |
| 5 | C.M.K., unpublished data | neural basal glutamine | 2× N2 | FGF2 + CNTF | ||||
| 6 | modified from C.M.K., unpublished data | neural basal glutamine | N2, B27 | GlutaMAX, NEAA, bME | FGF2 + CNTF | |||
| 7 | Krencik et al. (2011) | DMEM/F12 | N2, B27 | GlutaMAX, NEAA, bME | FGF2 + CNTF | |||
| 8 | Jiang et al. (2013) | DMEM/F12 | N2, B27 | GlutaMAX, NEAA, bME | FGF2 + CNTF | BMP4 | ||
| 9 | reduction of % FBS from no. 10 | DMEM/F12 | N2, B27 | GlutaMAX, NEAA, bME | FGF2 + CNTF | BMP4 | 3% FBS | |
| 10 | Roybon et al. (2013) | DMEM/F12 | N2, B27 | GlutaMAX, NEAA, bME | FGF2 + CNTF | BMP4 | 10% FBS | |
| 11 | Shaltouki et al. (2013) | DMEM/F12 | N2, B27 | GlutaMAX, NEAA, bME | FGF2 + CNTF | BMP4 | 3% FBS | neuregulin |
| 12 | adding insulin + AA to no. 9 | DMEM/F12 | N2, B27 | GlutaMAX, NEAA, bME | FGF2 + CNTF | BMP4 | 3% FBS | insulin + AA |
The 11 screened conditions of astrocyte differentiation from hiPSCs, including 3 commercially available astrocyte media (nos. 2–4) and 8 conditions modified from published protocols (nos. 5–12), with a forebrain neuron differentiation protocol (no. 1). NEAA, non-essential amino acid; bME, 2-mercaptoethanol; AA, ascorbic acid; ND, non-disclosure; AM, astrocyte medium; AGS, astrocyte growth supplement.
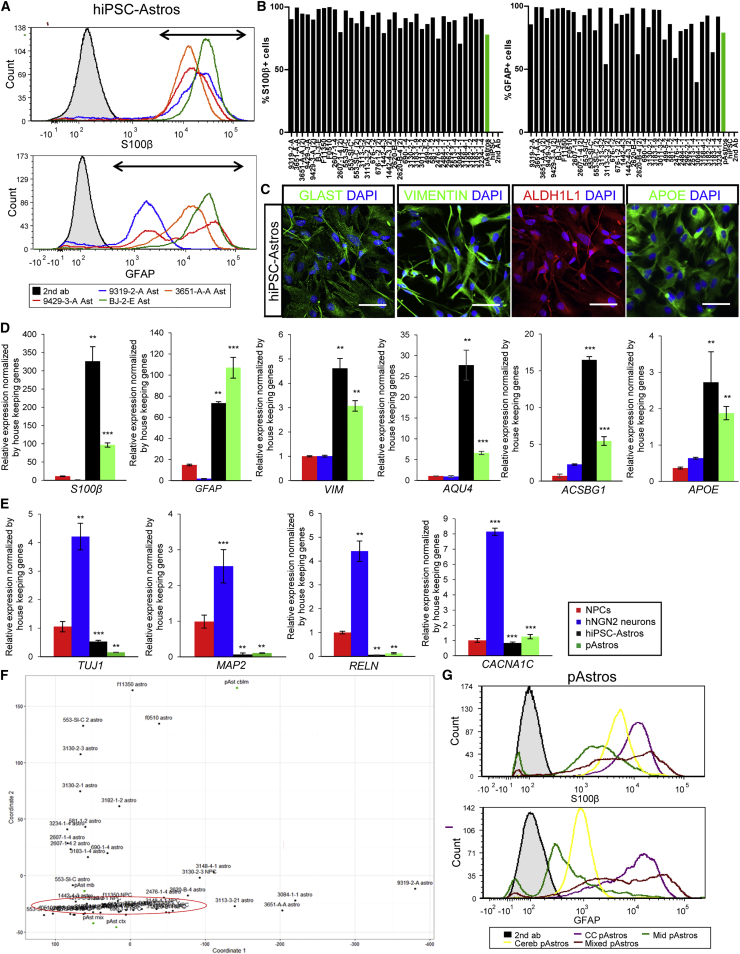
Figure 1.
Rapid Differentiation of hiPSC-Derived NPCs to Astrocyte-like Identity
(A) Representative flow-cytometry analysis of S100β (top) and GFAP (bottom) for four 30-day hiPSC-astrocyte differentiations. Arrows indicate the cells positive for each marker protein. Appropriate secondary-only control is shown in black.
(B) Graphs of flow-cytometry analysis across 35 hiPSC-astrocyte differentiations from 26 NPC lines from three independent hiPSC cohorts. S100β (left) and GFAP (right) immunostaining is shown, with primary human fetal astrocytes (positive control) and hiPSCs (negative control).
(C) Representative immunofluorescence images of hiPSC-astrocytes stained for astrocyte markers, glutamate transporters GLAST (green), VIMENTIN (green), ALDH1L1 (red), and APOE (green). Scale bars, 100 μm.
(D) mRNA levels of astrocyte markers: S100β, GFAP, VIM, AQU4, ACSBG1, and APOE in hiPSC-astrocytes (n = 3 from four different lines) and primary human fetal astrocytes (pAstrocytes; n = 3 from cerebral cortex astrocytes). Primer sequences are listed in Table S3. n, the number of independent experiments.
(E) mRNA levels of neuronal markers: TUJ1, MAP2AB, RELN, and CACNA1C in hiPSC-astrocytes (n = 3 from four different lines) and pAstrocytes (n = 3 from cerebral cortex astrocytes), relative to hNGN2-induced neurons (n = 3 from two lines). Primer sequences are listed in Table S3.
(F) Principal component analysis of lineage-specific marker expression in 23 pairs of hiPSC-astrocytes and isogenic source NPCs, together with four pAstrocyte lines isolated from fetal cerebral cortex, midbrain, cerebellum, and whole brain.
(G) Flow-cytometry analysis of S100β (top) and GFAP (bottom) for pAstrocytes from the cerebral cortex, midbrain, cerebellum, and whole brain. Appropriate secondary-only control is shown in black. CC, cerebral cortex; Mid, midbrain; Cereb, cerebellum; Mixed, whole brain; hiPSC-Astros, hiPSC-astrocytes; pAstros, primary human fetal astrocytes.
Data are presented as mean ± SD using two-tailed homoscedastic Student's t test. ∗∗p < 0.01, ∗∗∗p < 0.001.
We next tested the efficacy of this protocol across 42 NPC lines from 30 individuals (including healthy controls as well as cases with schizophrenia and frontotemporal dementia; 16 male and 14 female) generated as part of three unique hiPSC cohorts (reprogrammed and differentiated through different protocols in independent laboratories) (Table S2). All 42 NPC lines ultimately yielded replicative cells with an astrocyte-like morphology (although not necessarily on the first attempt), with an average composition of 90% S100β+ and 82% GFAP+ cells by flow cytometry, relative to the appropriate secondary antibody control (Figures 1A and 1B; hiPSCs served as a negative control, lacking S100β- and GFAP-positive cells), and confirmed by immunocytochemistry (Figures 1A, 1C, S1J, and S1K). As with primary astrocytes (Figures 1G and S1O), there is substantial variability in mean fluorescent intensity between S100β- and GFAP-positive hiPSC-astrocyte populations (Figure 1A).
Within 30 days, hiPSC-astrocytes were immunopositive for the astrocyte markers ALDH1L1 (Cahoy et al., 2008) and Vimentin (VIM) (Schnitzer et al., 1981), as well as the glutamate transporters GLAST (EAAT1 and GLUT1) (Rothstein et al., 1994) (Figures 1C, S1L, and S1M). Compared with NPCs or excitatory neurons (hNGN2-induced neurons) (Brennand et al., 2015), hiPSC-astrocytes expressed GFAP (Ludwin et al., 1976), S100β (Ludwin et al., 1976), VIM (Schnitzer et al., 1981), AQU4 (Hubbard et al., 2015), ACSBG1 (Chaboub and Deneen, 2013), and APOE (Boyles et al., 1985) by qPCR (Figure 1D), although expression levels between individual hiPSC-astrocytes and primary astrocyte lines varied substantially. In addition, hiPSC-astrocytes expressed low levels of the neuronal markers TUJ1, MAP2AB, RELN, and CACNA1C (Figure 1E).
Across a larger panel of neural lineage markers (Table S4) in 23 hiPSC-astrocyte lines and their isogenic NPC lines, principal component analysis (PCA) revealed that the NPCs grouped together, while the astrocytes (both hiPSC-derived and primary human fetal astrocytes) were more dispersed (Figures 1F and S1N). Because it was apparent in our hiPSC-astrocytes as well as the primary human fetal astrocytes (obtained from different donors and brain regions), this variability in lineage marker expression may reflect inter-individual variability and/or differences in regional patterning (Figures 1G and S1O).
hiPSC-Astrocytes and Primary Human Fetal Astrocytes Share Similar Transcriptional Profiles
To query the extent to which global gene expression in hiPSC-astrocytes resembles primary human fetal brain astrocytes, we performed RNA sequencing (RNA-seq) transcriptomic analyses on four control hiPSC-astrocytes and primary human fetal astrocytes from two brain regions (cerebral cortex and midbrain), together with isogenic hiPSC-derived NPCs and neurons (Figures 2 and S2), comparing them with in vivo human and rodent astrocyte transcriptomic datasets (Figures 2, 3, S2, and S3). hiPSC-astrocytes showed transcriptional profiles most similar to those of fetal brain astrocytes. Using PCA and hierarchical clustering, all four hiPSC-astrocytes clustered together with the primary human fetal astrocytes and distinct from the NPC and neuron clusters (Figures 2A, 2B, and S2A).
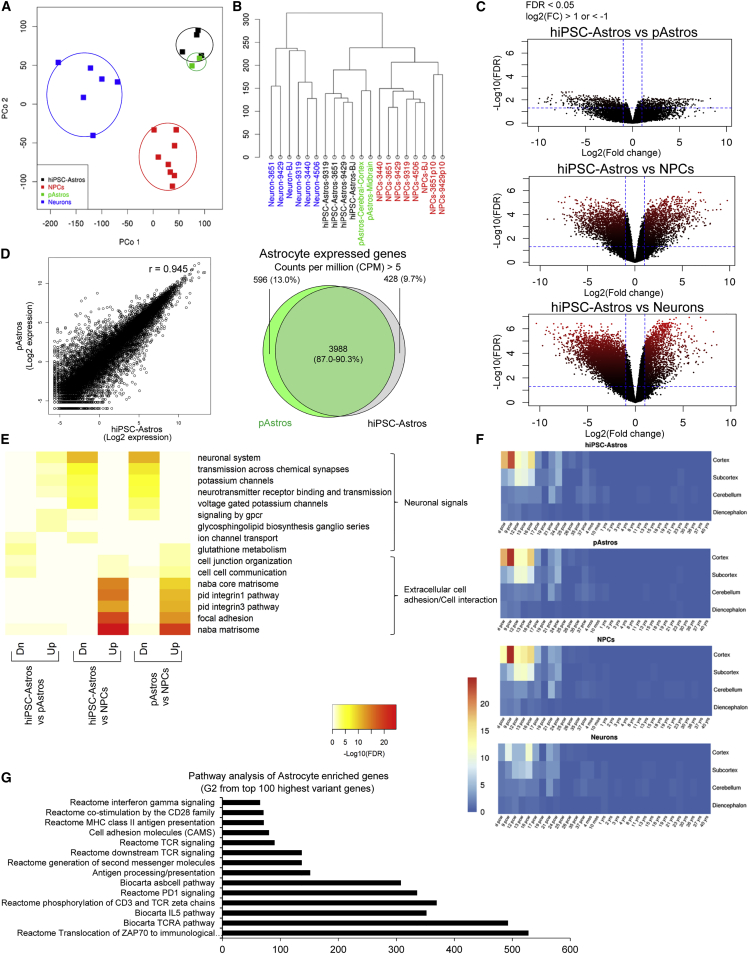
Figure 2.
Transcriptional Profile of hiPSC-Astrocytes and Primary Human Fetal Astrocytes
RNA-seq analysis of hiPSC-derived NPCs (n = 8), neurons (n = 6), and astrocytes (n = 4) together with pAstrocytes from human fetal cerebral cortex and midbrain.
(A and B) Principal component (P.Co) analysis (A) and clustering diagram (B) of hiPSC-derived NPCs, neurons, and astrocytes, together with pAstrocytes.
(C) Volcano plot comparison of hiPSC-astrocytes to pAstrocytes (top), as well as to hiPSC-derived NPCs (middle) and neurons (bottom). Average log2(fold change) versus −log10(FDR) is shown for all genes. Genes upregulated and downregulated by 2-fold change and FDR < 0.05 are labeled by red dots. The number of genes differentially expressed between different cell types is indicated by the red color density and quantified in Figure S2B.
(D) Scatterplot (left) comparing gene expression in hiPSC-astrocytes and pAstrocytes. r represents the Spearman correlation coefficient. Venn diagram (right) of overlapping gene expression (CPM > 5) between hiPSC-astrocytes and pAstrocytes.
(E) Functional pathway enrichment analysis of differentially expressed genes between hiPSC-astrocytes and pAstrocytes (left), hiPSC-astrocytes and NPCs (middle), and pAstrocytes and NPCs (right); hiPSC-astrocytes and pAstrocytes express increased extracellular cell communication signals, but decreased neuronal signals relative to NPCs.
(F) Heatmap produced by Wilcoxon's rank-sum comparisons of hiPSC-derived NPCs, neurons, and astrocytes, as well as pAstrocytes, relative to the Allen BrainSpan Atlas.
(G) Fold enrichment from functional pathway analysis of astrocyte-enriched genes, group G2 sorted from top 100 most variable genes from Figure S3A and Table 2.
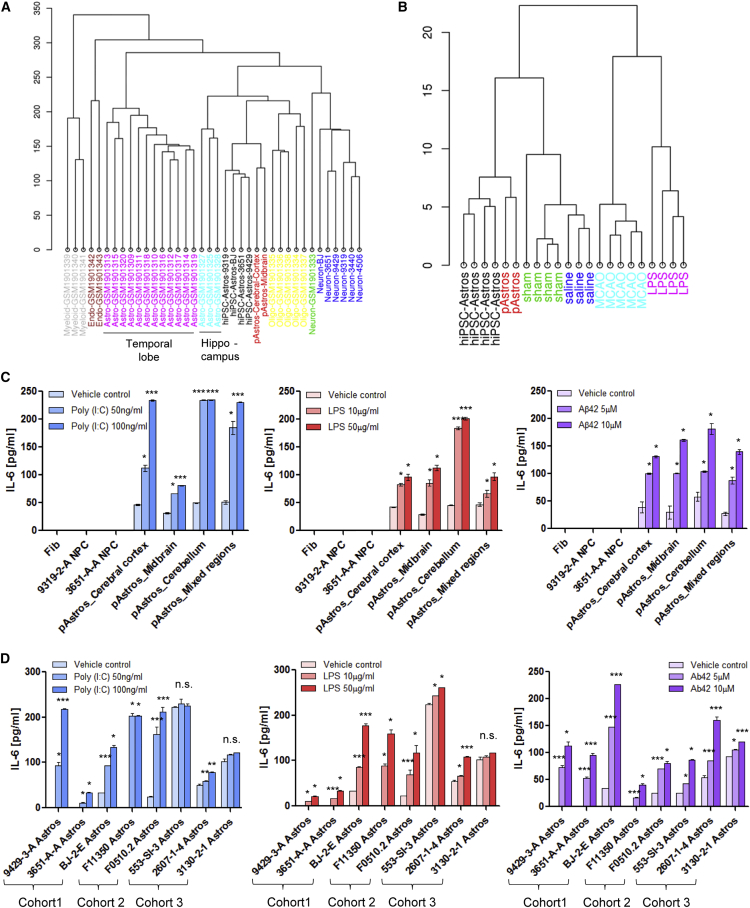
Figure 3.
Characterization of the Neuroinflammatory Status and Reactivity of hiPSC-Astrocytes
(A) Cluster analysis of hiPSC-derived NPCs, neurons, and astrocytes, as well as pAstrocytes, combined with a human adult brain dataset (Zhang et al., 2016). hiPSC-Astros, hiPSC-astrocytes; pAstros, primary human fetal astrocytes; FDR, false discovery rate; CPM, counts per million.
(B) Cluster diagram of hiPSC-astrocytes and pAstrocytes compared with the astrocyte reactivity dataset (Zamanian et al., 2012), which was sorted by reactivity genes enriched in the A1 (LPS-treatment), A2 (MCAO ischemia), and pan-reactive phenotypes, or related controls (saline and sham, respectively).
(C) IL-6 secretion from pAstrocytes (cerebral cortex, midbrain, cerebellum and whole brain [mixed regions]) and negative controls (fibroblasts and NPCs), after 24-hr treatment with 50 ng/mL or 100 ng/mL poly(I:C), 10 μg/mL or 50 μg/mL LPS, or 5 μM or 10 μM Aβ42 and its relative vehicle controls (saline for poly(I:C) and LPS, and Tris-HCl [pH 8] for Aβ(1–42)), as measured by ELISA.
(D) IL-6 secretion following 24-hr treatment with poly(I:C), LPS, and Aβ42 across hiPSC-astrocyte differentiations from nine NPC lines from three independent hiPSC cohorts. hiPSC-Astros, hiPSC-astrocytes; pAstros, primary human fetal astrocytes.
Data are presented as mean ± SD using one-way ANOVA with Tukey multiple comparison test. n.s., not significant; ∗p < 0.05, ∗∗∗p < 0.001.
There were nearly 6-fold fewer differentially expressed genes (DEGs) between hiPSC-astrocytes and primary human fetal astrocytes (900 genes) than between hiPSC-astrocytes and isogenic hiPSC-derived neurons (10,000 genes) or NPCs (5,500 genes) (Figures 2C and S2B). hiPSC-astrocytes are highly similar to primary human fetal astrocytes (r = 0.945); the majority of both expressed genes (counts per million [CPM] > 1) and enriched genes (CPM > 5) were shared between hiPSC-astrocytes and primary human fetal astrocytes (87.0% and 90.3%, respectively) (Figure 2D). Functional enrichment analyses (using MSigDB) demonstrated that signals regulating neuronal maturation, such as synapse or ion channel formation, were downregulated in hiPSC-astrocytes and primary human fetal astrocytes, whereas signals promoting extracellular cell adhesion and interaction were upregulated (Figure 2E). When we specifically considered just the top 100 most varying genes distinguishing hiPSC-astrocytes from hiPSC-derived NPCs and neurons, functional enrichment analysis identified a group of 19 genes related to reactivity, cytokine, interferon, T cell receptor (TCR), and antigen-processing signaling that were enriched in astrocytes (G2) (Figures 3A and S2C; Table 2).
Table 2.
Annotation of the Top 100 Most Variable Genes
| MSigDB | Group | Overlap Size | Group Size | MSigDB Size | FE | p Value | FDR | Overlap Genes |
|---|---|---|---|---|---|---|---|---|
| Antigen processing and presentation | G2 | 7 | 19 | 57 | 151.3 | 1.71 × 10−14 | 8.49 × 10−11 | CD74; HLA-DOA; HLA-DQA1; HLA-DQB1; HLA-DRA; HLA-DRB1; HLA-DRB5 |
| Cell adhesion molecules (CAMS) | G2 | 6 | 19 | 115 | 64.3 | 3.17 × 10−10 | 3.61 × 10−7 | HLA-DOA; HLA-DQA1; HLA-DQB1; HLA-DRA; HLA-DRB1; HLA-DRB5 |
| Reactome MHC class II antigen presentation | G2 | 5 | 19 | 86 | 71.6 | 6.64 × 10−9 | 6.94 × 10−6 | CD74; HLA-DOA; HLA-DQA1; HLA-DRB1; HLA-DRB5 |
| Reactome translocation of ZAP70 to immunological synapse | G2 | 3 | 19 | 7 | 528 | 1.58 × 10−8 | 1.53 × 10−5 | HLA-DQA1; HLA-DRB1; HLA-DRB5 |
| Reactome phosphorylation of CD3 and TCR ζ chains | G2 | 3 | 19 | 10 | 369.6 | 5.42 × 10−8 | 4.85 × 10−5 | HLA-DQA1; HLA-DRB1; HLA-DRB5 |
| Reactome PD1 signaling | G2 | 3 | 19 | 11 | 336 | 7.45 × 10−8 | 6.22 × 10−5 | HLA-DQA1; HLA-DRB1; HLA-DRB5 |
| Reactome generation of second messenger molecules | G2 | 3 | 19 | 18 | 205.3 | 3.67 × 10−7 | 0.000287 | HLA-DQA1; HLA-DRB1; HLA-DRB5 |
| Reactome downstream TCR signaling | G2 | 3 | 19 | 27 | 136.9 | 1.31 × 10−6 | 0.000965 | HLA-DQA1; HLA-DRB1; HLA-DRB5 |
| Neuron differentiation | G1 | 4 | 28 | 73 | 45.8 | 1.68 × 10−6 | 0.001205 | CNTN4; LMX1B; OTX2; SLIT1 |
| Generation of neurons | G1 | 4 | 28 | 79 | 42.3 | 2.31 × 10−6 | 0.001574 | CNTN4; LMX1B; OTX2; SLIT1 |
| Axon guidance | G1 | 3 | 28 | 22 | 114 | 2.32 × 10−6 | 0.001574 | CNTN4; OTX2; SLIT1 |
| Neurogenesis | G1 | 4 | 28 | 89 | 37.6 | 3.73 × 10−6 | 0.002458 | CNTN4; LMX1B; OTX2; SLIT1 |
| Nervous system development | G1 | 6 | 28 | 366 | 13.7 | 3.95 × 10−6 | 0.002538 | CNTN4; LMX1B; LY6H; OTX2; SHOX2; SLIT1 |
| Reactome TCR signaling | G2 | 3 | 19 | 41 | 90.1 | 4.74 × 10−6 | 0.002876 | HLA-DQA1; HLA-DRB1; HLA-DRB5 |
| Nervous system development | S1 | 5 | 17 | 366 | 18.8 | 4.82 × 10−6 | 0.002876 | CNTN4; LMX1B; LY6H; SHOX2; SLIT1 |
| Biocarta TCRA pathway | G2 | 2 | 19 | 5 | 492.8 | 6.23 × 10−6 | 0.003549 | HLA-DRA; HLA-DRB1 |
| Reactome co-stimulation by the CD28 family | G2 | 3 | 19 | 52 | 71.1 | 9.77 × 10−6 | 0.005322 | HLA-DQA1; HLA-DRB1; HLA-DRB5 |
| Reactome interferon-γ signaling | G2 | 3 | 19 | 57 | 64.8 | 1.29 × 10−5 | 0.006553 | HLA-DQA1; HLA-DRB1; HLA-DRB5 |
| Biocarta IL-5 pathway | G2 | 2 | 19 | 7 | 352 | 1.31 × 10−5 | 0.006553 | HLA-DRA; HLA-DRB1 |
| Axonogenesis | G1 | 3 | 28 | 42 | 59.7 | 1.71 × 10−5 | 0.007939 | CNTN4; OTX2; SLIT1 |
| Biocarta asbcell pathway | G2 | 2 | 19 | 8 | 308 | 1.74 × 10−5 | 0.007939 | HLA-DRA; HLA-DRB1 |
| Neuron differentiation | S1 | 3 | 17 | 73 | 56.6 | 1.92 × 10−5 | 0.00858 | CNTN4; LMX1B; SLIT1 |
MSigDB (http://software.broadinstitute.org/gsea/msigdb) is a collection of gene annotations, including gene ontology, and functional pathways. Overlap size: the number of genes shared between a query gene set (e.g., genes in group 1, G1) and a gene set in MSigDB (e.g., antigen processing and presentation). Group size: the number of genes in a query gene set. MSigDB size: the number of genes in MSigDB gene set. FE, fold enrichment; FDR, false discovery rate.
To assess the extent to which hiPSC-astrocytes are related to human brain astrocytes in vivo, we compared expression profiles of hiPSC-derived astrocytes, NPCs, and neurons from four healthy controls, as well as primary human fetal astrocytes, with the Allen BrainSpan Atlas of the Developing Human Brain (http://www.brainspan.org) (Miller et al., 2014) using a Spearman rank correlation analysis. Heatmaps generated through a Wilcoxon's rank-sum test revealed that gene expression in hiPSC-astrocytes and primary human fetal astrocytes was highly correlated with gene expression in human fetal cortical brain tissue (8–16 weeks post conception) (Figure 2F). Unsupervised hierarchical cell-type-specific cluster analysis demonstrated that hiPSC-astrocytes closely cluster with astrocytes purified by immunopanning from human brain tissue, particularly hippocampal astrocytes, rather than oligodendrocytes, endothelial cells, myeloid cells, or neurons (Zhang et al., 2016) (Figure 3A); pathway analysis further revealed that many astrocyte-enriched genes were related to immune signaling (Figure 2G).
Expression Profile of hiPSC-Astrocytes Resembles Quiescent Astrocytes
To determine whether the gene expression pattern of our control hiPSC-astrocytes more resembles quiescent or reactive astrocytes, we compared them with an RNA-seq dataset of “proinflammatory” A1-type and “immunoregulatory” A2-type murine astrocytes, as well as their saline- and sham-treated controls (Zamanian et al., 2012). (Because these are in vivo cell types, whereby A1-type astrocytes are induced by a bacterial lipopolysaccharide [LPS] infection and A2-type astrocytes by middle cerebral artery occlusion [MCAO], no comparable dataset exists for human astrocytes.) hiPSC-astrocyte gene expression best clusters with the control conditions (Figures 3B and S3A), suggesting that hiPSC-astrocytes may be closer to the quiescent state than to reactive astrocytes.
hiPSC-Astrocytes Secrete Cytokines in Response to Inflammatory Stimuli
A key effector of the astrocyte neuroinflammatory response in neurodegenerative diseases is interleukin 6 (IL-6) (Zhao and Schwartz, 1998). Consistent with effects observed in primary human fetal astrocytes (Figure 3C), 24-hr treatment with polyinosinic-polycytidylic acid (poly(I:C)) (50 ng/mL or 100 ng/mL), LPS (10 μg/mL or 50 μg/mL), and β-amyloid 42 (Aβ42) (5 μM or 10 μM) led to dose-dependent increases in IL-6 secretion in control hiPSC-astrocytes (Figure 3D), but not the source hiPSC-derived NPCs (Figure 3C).
To more fully characterize the release of inflammatory mediators from control hiPSC-astrocytes following 24-hr treatment with 5 μM Aβ42, a main component of the amyloid plaques in AD, we measured proinflammatory (IL-1β, IL-4, IL-6, IL-8, IL-10, and tumor necrosis factor α) and anti-inflammatory cytokines (IL-1α, IL-2, IL-12, IL-17α, interferon-γ, and granulocyte macrophage colony-stimulating factor) by Multi-Analyte ELISArray, confirming that Aβ42 treatment primarily increased IL-6 secretion (Garwood et al., 2011), as well as a second proinflammatory cytokine, IL-8 (Figures S3B and S3C). We also measured 36 cytokines, chemokines, and acute-phase proteins using the Proteome Profiler Human Cytokine Array (Table S5) in baseline conditions and following 24-hr treatment with 5 μM Aβ42 (Figures 3D and S3C–S3E); Aβ42 increased IL-6 release in both hiPSC-astrocytes and primary human fetal astrocytes (Figure S3D). Together, our findings indicate that hiPSC-astrocytes are capable of secreting proinflammatory cytokines in response to neuroinflammatory cues.
hiPSC-Astrocytes Display Phagocytic Capacity and Promote Phagocytic Function of Microglia
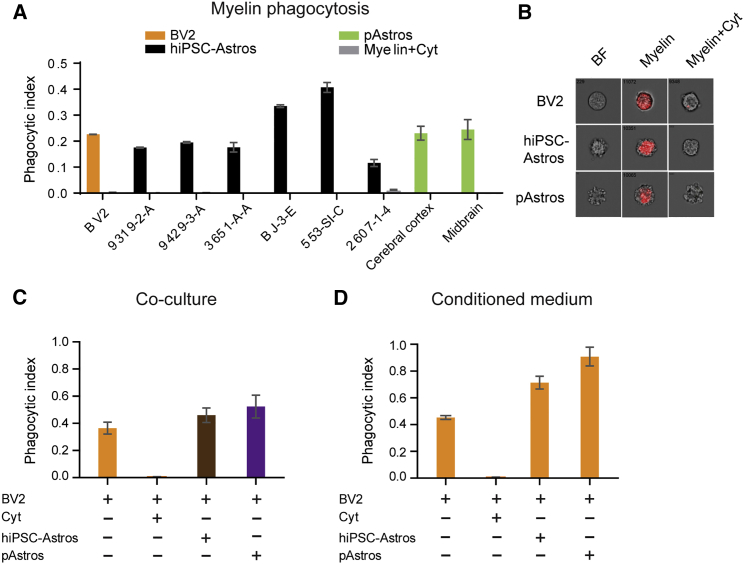
Astrocytes can phagocytose and degrade β-amyloid in AD (Chung et al., 2013, Wyss-Coray et al., 2003). We used flow cytometry to examine the ability of control hiPSC-astrocytes to phagocytose pHrodo red conjugated myelin or zymosan bioparticles (pHrodo red dye fluoresces red in the acidic environment of phagosomes; zymosan is an agonist of Toll-like receptor 2 [TLR2] [Marinelli et al., 2015, Richard et al., 2008]). hiPSC-astrocytes, primary human fetal astrocytes, and BV2 microglia showed a similar capacity to internalize myelin purified from brain homogenates, while zymosan bioparticle uptake was much greater in microglia (consistent with higher levels of TLR2 reported in microglia) (Jana et al., 2008) (Figures 4A, 4B, and S4A–S4C; Movie S1. Phagocytosis of Myelin in BV2 Microglia, Movie S2. Phagocytosis of Myelin in hiPSC-Astrocytes, Movie S3. Phagocytosis of Myelin in Primary Astrocytes). The specificity of bioparticle uptake was confirmed by treating cells with cytochalasin D, an inhibitor of β-actin polymerization that reduces phagocytosis (Figures 4A and 4B).
Figure 4.
Impact of hiPSC-Astrocytes on the Phagocytic Capacity of BV2 Microglial Cells
(A) Phagocytic indices of BV2 cells, hiPSC-astrocytes, and pAstrocytes incubated with 20 μg of pHrodo-labeled myelin for 3 hr and analyzed by flow cytometry.
(B) Amnis images representative of pHrodo red myelin after engulfment in hiPSC-astrocytes, pAstrocytes, and BV2 cells.
(C) Phagocytic indices of BV2 microglia co-cultured with hiPSC-astrocytes and pAstrocytes that were treated with 30 μg of pHrodo-labeled zymosan for 3 hr and analyzed by flow cytometry, F(3, 33) = 79.33.
(D) Phagocytic indices of BV2 microglia treated with astrocyte conditioned medium for 20 hr and then incubated with 30 μg of pHrodo-labeled zymosan for 3 hr for analysis by flow cytometry, F(3, 13) = 30.80. Data are representative of three independent experiments from 6–8 different control hiPSC-astrocytes and 2–4 different pAstrocytes and are shown as mean ± SEM. Similar significance was obtained in two other independent experiments. hiPSC-Astros, hiPSC-astrocytes; pAstros, primary human fetal astrocytes. Treatment with 2 μM cytochalasin D (Cyt) was used as a negative control for phagocytosis inhibition.
Although microglia are the major phagocytic cells in the brain, astrocytes mediate microglial response to inflammatory stimuli (Krencik et al., 2011, Lee and Landreth, 2010, Skripuletz et al., 2013). We repeated the pHrodo red zymosan assay using microglia-astrocyte co-cultures; both hiPSC-astrocytes and primary human fetal astrocytes enhanced phagocytic capacity of BV2 mouse microglial cells (Figures 4C and S4D). We discriminated microglia and astrocyte phagocytic activity by labeling CD11b- and GFAP-positive cells, respectively (Figure S4E). After 3 hr of labeling, only the CD11b+ BV2 microglial cells phagocytosed the pHrodo red zymosan bioparticles (Figure S4E) Finally, we treated BV2 microglial cells with astrocyte conditioned medium (ACM) for 24 hr prior to analyzing their phagocytic capacity; pretreatment of BV2 microglial cells with ACM from either hiPSC-astrocytes or primary human fetal astrocytes significantly increased microglial phagocytic capacity (Figures 4D and S4F). Taken together, these findings show that both hiPSC-astrocytes and primary human fetal astrocytes secrete factors that increase the capacity of microglia to phagocytose zymosan bioparticles.
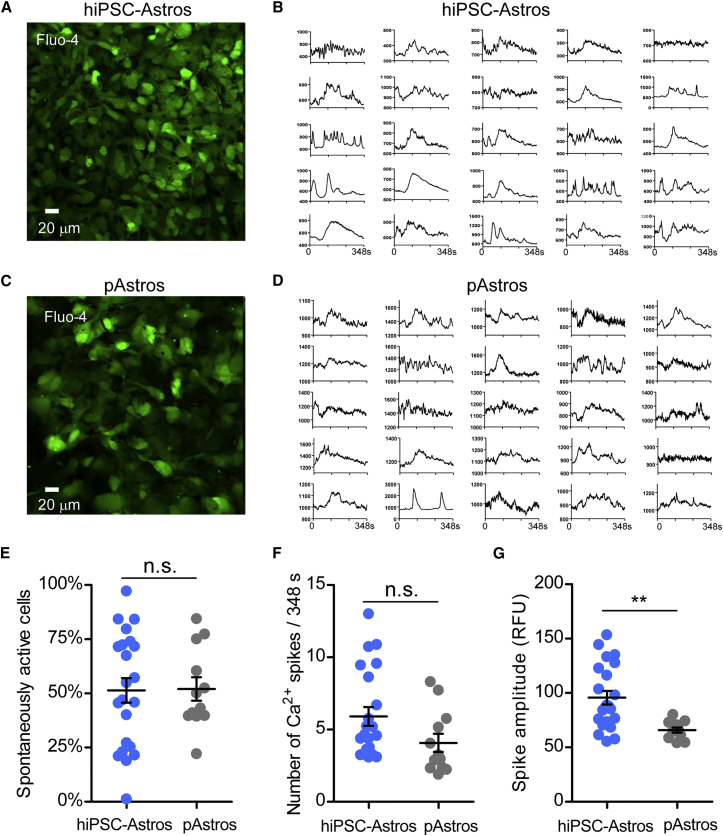
hiPSC-Astrocytes Display Spontaneous Calcium Transient Activity
We next evaluated whether control hiPSC-astrocytes exhibit spontaneous calcium transients, as has been described for astrocytes (Volterra et al., 2014). We used the calcium indicator Fluo-4AM to monitor calcium signaling under basal conditions and in response to a pulse of extracellular glutamate (3 μM) (Zhang et al., 2016). A single pulse of glutamate produced a slow calcium response in both hiPSC-astrocytes and primary human fetal astrocytes (Figure 5). In addition, some cells exhibited spontaneous calcium spikes, suggesting the presence of a network of connected astrocytes (Scemes and Giaume, 2006) (Figures 5A–5D). To quantify these responses, we measured the frequency of spontaneous activity, the number of spontaneous spikes per time, and the amplitude of the calcium spike (Figures 5E–5G). There was no statistical difference in the frequency and number of spikes between hiPSC-astrocytes and primary astrocytes. Interestingly, the amplitude of the spontaneous calcium spike was significantly higher in hiPSC-astrocytes compared with primary astrocytes. Taken together, the excitability of hiPSC-astrocytes was largely indistinguishable from that of primary human astrocytes.
Figure 5.
Spontaneous and Glutamate-Responsive Calcium Transients in hiPSC-Astrocytes and Primary Astrocytes
(A and C) Representative Fluo-4-stained hiPSC-astrocytes and pAstrocytes. Note similarity in shape of cells.
(B and D) Plots of fluorescence (RFUs) versus time for 25 regions of interest from hiPSC-astrocytes and pAstrocytes.
(E) Average number of spontaneously active cells per field (318 μm2) for hiPSC-astrocytes (n = 22 fields from four different lines) and pAstrocytes (n = 12 fields from three different preparations of cerebral cortex astrocytes). n, the number of fields.
(F) Average number of calcium spikes per 348-s trace for hiPSC-astrocytes (n = 22) and pAstrocytes (n = 12).
(G) Average amplitude of calcium spike in each 348-s trace for hiPSC-astrocytes (n = 22) and pAstrocytes (n = 12). Peak excludes the amplitude of the glutamate-induced spike. Each point represents the average multiple regions of interest per 318 μm2. Line shows mean ± SEM. hiPSC-Astros, hiPSC-astrocytes, pAstros, primary human fetal astrocytes. Using a two-tailed Student's t test, n.s., not significant; ∗∗p < 0.05.
Discussion
We screened 11 methods to differentiate hiPSC-derived NPCs into astrocytes, identifying a medium with low FBS (1%–2%) that produces populations of hiPSC-astrocytes within 30 days. Although this is a commercial medium sold for the culture of primary astrocytes, it has not previously been demonstrated to support differentiation of hiPSC to astrocytes. Our method is fast and robust: unlike previous reports, it does not require prolonged culture (∼6 months) or a serial sorting process (Krencik et al., 2011, Shaltouki et al., 2013, Yuan et al., 2011). This protocol was evaluated across 42 NPC lines from 30 individuals (16 males and 14 females) generated from three independent hiPSC cohorts (Table S2). We caution that the quality of the starting NPC population is a critical predictor of success and note that for a few particularly intransient NPC lines, starting from very low-passage stocks proved critical to the ultimate successful differentiation of hiPSC-astrocytes. By both fluorescence-activated cell sorting and qPCR, GFAP seems to be a more variable marker of astrocyte fate (Figures 1A and 1B) and we recommend instead using S100β when evaluating hiPSC-astrocyte populations; moreover, more than any single gene, using panels of markers and/or global transcriptomic analysis more fully reveals the extent of astrocyte patterning of each line. We also suggest that hiPSC-astrocyte response to inflammatory stimuli might represent a simple and easy platform by which to confirm the functionality of hiPSC-astrocytes; astrocytes undergo stimulation-dependent cytokine secretion, while NPCs do not secrete cytokines.
To our knowledge, our method is distinct from others recently reported (Chen et al., 2015, Haidet-Phillips et al., 2014, Jiang et al., 2013, Krencik et al., 2011, McGivern et al., 2013, Serio et al., 2013, Shaltouki et al., 2013), its major advantage being the ease by which hiPSC-astrocytes can be concurrently differentiated from many NPCs from multiple individuals, owing to the simplicity of the protocol, and furthermore that astrocytes can be expanded for months and cryopreserved, serving as a source of cells to support an array of experiments. Therefore, because our protocols for NPC and astrocyte culture are robust and easily scaled, this methodology is both preferable for large case-control studies from dozens of idiopathic patients and highly amenable to automated culture and future high-throughput drug screens using patient-derived astrocytes (Xu et al., 2016).
We have established a platform for querying astrocyte-specific contributions to disease predisposition in hiPSC-based models. hiPSC-astrocytes closely resemble primary human fetal astrocytes, purified adult brain astrocytes (Zhang et al., 2016), and brain tissue homogenate (Miller et al., 2014) (Figures 2 and 3A). Although the transcriptional profile of our hiPSC-astrocytes most closely resembles a quiescent state (Figure 3B), hiPSC-astrocytes secrete various cytokines and chemokines in response to neuroinflammatory stimuli (Figures 3C and 3D). hiPSC-astrocytes displayed phagocytic capacity (Figure 4A) and enhanced the phagocytic function of microglia in a co-culture assay (Figure 4C). They also showed spontaneous calcium transient activity and responses to glutamate stimulus (Figure 5). Moreover, because nearly pure populations of excitatory neurons and astrocytes from the same individual, hiPSC clone, and even NPC batch can now be compared, our protocol may reduce experimental variation in studies of the cell-autonomous and non-cell-autonomous factors underlying disease predisposition.
While many studies of neurodegeneration and synaptic dysfunction use either primary mouse or human fetal glia for human neuron co-culture (Johnson et al., 2007, Kiskinis et al., 2014, Verheyen et al., 2015), these methods are limited by species-specific differences in astrocyte functions and limited access to human fetal samples. Our hiPSC-astrocytes overcome these drawbacks, making possible hiPSC-based astrocyte-neuron (Jiang et al., 2013) and astrocyte-microglia (Muffat et al., 2016, Pandya et al., 2017) co-culture platforms for uncovering disease-related mechanisms in vitro, which should help to reveal how the crosstalk between these three neural cell types contributes to neurological and psychiatric disease.
Experimental Procedures
Human subject work on these de-identified control hiPSCs was approved by the Institutional Review Board at Icahn School of Medicine at Mount Sinai. Control mouse brain tissue was obtained from the Center for Comparative Medicine and Surgery at Icahn School of Medicine at Mount Sinai.
NPCs and Astrocyte Cell Culture
Forebrain NPCs were maintained at high density, grown on Matrigel (BD Bioscience) in NPC medium (DMEM/F12, 1× N2, 1× B27-RA [Invitrogen], 1 mg/mL laminin [Invitrogen], and 20 ng/mL FGF2 [Invitrogen]), and split approximately 1:3 to 1:4 every week with Accutase (Millipore). NPCs could be expanded up to 14 passages. Forebrain NPCs were differentiated to astrocytes by seeding dissociated single cells at 15,000 cells/cm2 density on Matrigel-coated plates in astrocyte medium (ScienCell: 1801, astrocyte medium [1801-b], 2% fetal bovine serum [0010], astrocyte growth supplement [1852] and 10 U/mL penicillin/streptomycin solution [0503]). Initial NPC quality, seeding density, and single-cell dissociation are critical, particularly during the first 30 days of differentiation, in order to efficiently generate hiPSC-astrocytes. See detailed protocol for astrocyte differentiation from hiPSC-derived NPCs in the Supplemental Information.
Immunocytochemistry
Cultures were washed with PBS and fixed using 4% paraformaldehyde (no. 15714, Electron Microscopy Sciences) for 20 min, then washed three times with PBS 1× plus 0.01% Triton X-100 prior to preincubation with blocking solution made of PBS 1× plus 0.01% Triton X-100 and 10% donkey serum (Sigma-Aldrich), for at least 1 hr. Cultures were then incubated in primary antibody solution overnight at 4°C, followed by an incubation with secondary antibodies for 1 hr at room temperature. See Supplemental Experimental Procedures for antibody details and confocal microscopy.
Real-Time qPCR
The total RNA was extracted from cultured cells with an RNeasy mini kit (no. 74106, Qiagen) and reverse transcribed into cDNA with an iScript cDNA Synthesis kit (no. 1708890, Bio-Rad). cDNA was used as template for the qPCR using a 7900 Real-Time PCR system (Applied Biosystems) with Power SYBR Green PCR Master Mix. The primers used are listed in Table S3. Gene expression was analyzed using the ΔΔCt method. All results were normalized to GAPDH, β-ACTIN, and TBP (TATA-box binding protein) expression, and the values of uninduced fibroblasts were set to 1. Three replicates were used to determine the error bars. See the Supplemental Information for RNA expression studies for qRT-PCR and primer sequences.
Analysis of RNA-Seq Data
Pair-ended RNA-seq data was generated using the Illumina HiSeq 2500 platform for hiPSC-astrocytes (four lines), neurons (six lines), and NPCs (eight lines) (Table S2), as well as for two primary astrocyte samples. The pair-ended sequencing reads were aligned to human hg19 genome using Star Aligner (version 2.5.0b). Following read alignment, featureCounts (Liao et al., 2014) was used to quantify the gene expression at the gene level based on Ensembl gene model GRCh37.70. Gene expression data preprocessing and downstream analyses, including differential gene expression and functional enrichment, are detailed in Supplemental Experimental Procedures.
IL-6 ELISA, Multi-Analyte ELISArray, and Protein Array
hiPSC-astrocytes and primary astrocytes were seeded in astrocyte medium 1 day before the experiment. Cells were treated for 24 hr with 50 ng/mL or 100 ng/mL of poly(I:C) (InvivoGen, no. tlrl-pic), 10 μg/mL or 50 μg/mL of LPS (Sigma-Aldrich, no. L5886), and 5 μM or 10 μM human β-amyloid (Aβ42) (CaliforniaPeptide, no. 641-15) and vehicle control solutions (saline for poly(I:C) and LPS or Tris-HCl for Aβ42). Samples were analyzed with IL-6 ELISA assay (Affymetrix eBioscience, no. 88-7066), Human Inflammatory Cytokines Multi-Analyte ELISArray (Qiagen, no. MEH-004A), RT2 Profiler PCR Arrays (Qiagen), and Proteome Profiler Human Cytokine Array (R&D Systems, no. ARY005B). For a detailed protocol see Supplemental Experimental Procedures.
Phagocytosis Assay
Phagocytic capacity of hiPSC-astrocytes, primary astrocytes, and BV2 cells was analyzed by incubating cells with pHrodo-labeled zymosan (Thermo Fisher, P35364) or myelin purified from mouse brains, and then measured by flow cytometry. Detailed protocols for co-culture experiments and astrocyte conditioned media treatment are provided in the Supplemental Experimental Procedures.
Calcium Imaging
Cells were incubated with 2 μM Fluo-4AM (Molecular Probes) and 0.02% Pluronic F 127 detergent in Krebs HEPES buffer (KHB). Time-lapse image sequences (×40 magnification) were acquired at 0.9 Hz and analyzed by FluoroSNNAP software operated by MATLAB. For a detailed protocol see Supplemental Experimental Procedures.
Statistical Analysis
For all experiments, data are represented as mean ± SD or SEM of three to five biological replicates. Statistical significance was determined using a two-tailed homoscedastic Student's t test or one-way ANOVA and Tukey-Kramer post hoc test for differences of means between each group of data with parametric distribution. Significant comparisons are labeled in the figures as ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
Extended experimental procedures and any associated references are available in the online version of this paper.
Author Contributions
J.TCW, K.J.B., and A.M.G. designed the experiments and wrote the manuscript. J.TCW developed the astrocyte differentiation protocol. J.TCW, K.R.B., B.J.H., and R.A. (with H.P.) differentiated the hiPSC-astrocyte lines. M.W. and B.Z. conducted the RNA-seq analysis. J.TCW performed the neuroinflammatory assays. A.A.P., J.TCW, and S.I.M. performed the phagocytosis assays. J.TCW, A.A.P., and B.J.H. executed flow-cytometry analysis on astrocytes and NPCs. J.TCW and K.R.B. performed real-time qPCR and immunofluorescence staining. E.L. and P.A.S. conducted and analyzed the calcium imaging analysis. C.M.K. shared prepublication information about astrocyte differentiation conditions.
Acknowledgments
BV2 cell line was kindly provided by Marc Diamond (UT Southwestern Medical Center). We are grateful for the continuous support of the Flow Cytometry Core at the Icahn School of Medicine at Mount Sinai Hospital. This work was supported in part by the JPB Foundation (A.M.G.), the Rainwater Foundation (A.M.G.), NIMH R01MH101454 (K.J.B.), NIA U01P50AG005138-30-1 (Alzheimer's Disease Research Center: Pilot 30-1) (K.J.B.), NIA/NIH U01AG046170 (K.J.B., M.W., and B.Z.), a component of the AMP-AD Target Discovery and Preclinical Validation Project, NIMH R01MH11499 (P.A.S.), NIAAA R01AA018734 (P.A.S.), NIDA R01DA037170 (P.A.S.), K01AG046374 (C.M.K.), and the New York Stem Cell Foundation (K.J.B.) and Project ALS (H.P.). A.M.G. serves on the Scientific Advisory Board for Denali Therapeutics.
Published: July 27, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.06.018.
Contributor Information
Alison M. Goate, Email: alison.goate@mssm.edu.
Kristen J. Brennand, Email: kristen.brennand@mssm.edu.
Accession Numbers
The accession number for the RNA-seq data reported in this paper is GEO: GSE97904.
Supplemental Information
References
- Ballas N., Lioy D.T., Grunseich C., Mandel G. Non-cell autonomous influence of MeCP2-deficient glia on neuronal dendritic morphology. Nat. Neurosci. 2009;12:311–317. doi: 10.1038/nn.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barres B.A. The mystery and magic of glia: a perspective on their roles in health and disease. Neuron. 2008;60:430–440. doi: 10.1016/j.neuron.2008.10.013. [DOI] [PubMed] [Google Scholar]
- Bi F., Huang C., Tong J., Qiu G., Huang B., Wu Q., Li F., Xu Z., Bowser R., Xia X.G. Reactive astrocytes secrete lcn2 to promote neuron death. Proc. Natl. Acad. Sci. USA. 2013;110:4069–4074. doi: 10.1073/pnas.1218497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyles J.K., Pitas R.E., Wilson E., Mahley R.W., Taylor J.M. Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. J. Clin. Invest. 1985;76:1501–1513. doi: 10.1172/JCI112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford J., Shin J.Y., Roberts M., Wang C.E., Li X.J., Li S. Expression of mutant huntingtin in mouse brain astrocytes causes age-dependent neurological symptoms. Proc. Natl. Acad. Sci. USA. 2009;106:22480–22485. doi: 10.1073/pnas.0911503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand K.J., Gage F.H. Concise review: the promise of human induced pluripotent stem cell-based studies of schizophrenia. Stem Cells. 2011;29:1915–1922. doi: 10.1002/stem.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennand K., Savas J.N., Kim Y., Tran N., Simone A., Hashimoto-Torii K., Beaumont K.G., Kim H.J., Topol A., Ladran I. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol. Psychiatry. 2015;20:361–368. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboub L.S., Deneen B. Astrocyte form and function in the developing central nervous system. Semin. Pediatr. Neurol. 2013;20:230–235. doi: 10.1016/j.spen.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Qian K., Chen W., Hu B., Blackbourn L.W., Du Z., Ma L., Liu H., Knobel K.M., Ayala M. Human-derived neural progenitors functionally replace astrocytes in adult mice. J. Clin. Invest. 2015;125:1033–1042. doi: 10.1172/JCI69097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W.S., Clarke L.E., Wang G.X., Stafford B.K., Sher A., Chakraborty C., Joung J., Foo L.C., Thompson A., Chen C. Astrocytes mediate synapse elimination through MEGF10 and MERTK pathways. Nature. 2013;504:394–400. doi: 10.1038/nature12776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio F.P., Boulting G.L., Bobrowicz S., Eggan K.C. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Eroglu C., Barres B.A. Regulation of synaptic connectivity by glia. Nature. 2010;468:223–231. doi: 10.1038/nature09612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M.R., Rowitch D.H. Evolving concepts of gliogenesis: a look way back and ahead to the next 25 years. Neuron. 2013;80:613–623. doi: 10.1016/j.neuron.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garwood C.J., Pooler A.M., Atherton J., Hanger D.P., Noble W. Astrocytes are important mediators of Abeta-induced neurotoxicity and tau phosphorylation in primary culture. Cell Death Dis. 2011;2:e167. doi: 10.1038/cddis.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haidet-Phillips A.M., Roybon L., Gross S.K., Tuteja A., Donnelly C.J., Richard J.P., Ko M., Sherman A., Eggan K., Henderson C.E. Gene profiling of human induced pluripotent stem cell-derived astrocyte progenitors following spinal cord engraftment. Stem Cells Transl. Med. 2014;3:575–585. doi: 10.5966/sctm.2013-0153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Chen M., Wang F., Windrem M., Wang S., Shanz S., Xu Q., Oberheim N.A., Bekar L., Betstadt S. Forebrain engraftment by human glial progenitor cells enhances synaptic plasticity and learning in adult mice. Cell Stem Cell. 2013;12:342–353. doi: 10.1016/j.stem.2012.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heni M., Hennige A.M., Peter A., Siegel-Axel D., Ordelheide A.M., Krebs N., Machicao F., Fritsche A., Haring H.U., Staiger H. Insulin promotes glycogen storage and cell proliferation in primary human astrocytes. PLoS One. 2011;6:e21594. doi: 10.1371/journal.pone.0021594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard J.A., Hsu M.S., Seldin M.M., Binder D.K. Expression of the astrocyte water channel aquaporin-4 in the mouse brain. ASN Neuro. 2015;7 doi: 10.1177/1759091415605486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana M., Palencia C.A., Pahan K. Fibrillar amyloid-beta peptides activate microglia via TLR2: implications for Alzheimer's disease. J. Immunol. 2008;181:7254–7262. doi: 10.4049/jimmunol.181.10.7254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P., Chen C., Wang R., Chechneva O.V., Chung S.H., Rao M.S., Pleasure D.E., Liu Y., Zhang Q., Deng W. hESC-derived Olig2+ progenitors generate a subtype of astroglia with protective effects against ischaemic brain injury. Nat. Commun. 2013;4:2196. doi: 10.1038/ncomms3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M.A., Weick J.P., Pearce R.A., Zhang S.C. Functional neural development from human embryonic stem cells: accelerated synaptic activity via astrocyte coculture. J. Neurosci. 2007;27:3069–3077. doi: 10.1523/JNEUROSCI.4562-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiskinis E., Sandoe J., Williams L.A., Boulting G.L., Moccia R., Wainger B.J., Han S., Peng T., Thams S., Mikkilineni S. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell. 2014;14:781–795. doi: 10.1016/j.stem.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krencik R., Weick J.P., Liu Y., Zhang Z.J., Zhang S.C. Specification of transplantable astroglial subtypes from human pluripotent stem cells. Nat. Biotechnol. 2011;29:528–534. doi: 10.1038/nbt.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.Y., Landreth G.E. The role of microglia in amyloid clearance from the AD brain. J. Neural Transm. (Vienna) 2010;117:949–960. doi: 10.1007/s00702-010-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Ludwin S.K., Kosek J.C., Eng L.F. The topographical distribution of S-100 and GFA proteins in the adult rat brain: an immunohistochemical study using horseradish peroxidase-labelled antibodies. J. Comp. Neurol. 1976;165:197–207. doi: 10.1002/cne.901650206. [DOI] [PubMed] [Google Scholar]
- Marchetto M.C., Muotri A.R., Mu Y., Smith A.M., Cezar G.G., Gage F.H. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Marinelli C., Di Liddo R., Facci L., Bertalot T., Conconi M.T., Zusso M., Skaper S.D., Giusti P. Ligand engagement of Toll-like receptors regulates their expression in cortical microglia and astrocytes. J. Neuroinflammation. 2015;12:244. doi: 10.1186/s12974-015-0458-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGivern J.V., Patitucci T.N., Nord J.A., Barabas M.E., Stucky C.L., Ebert A.D. Spinal muscular atrophy astrocytes exhibit abnormal calcium regulation and reduced growth factor production. Glia. 2013;61:1418–1428. doi: 10.1002/glia.22522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J., Marchetto M.C., Bardy C., Gage F.H. Evaluating cell reprogramming, differentiation and conversion technologies in neuroscience. Nat. Rev. Neurosci. 2016;17:424–437. doi: 10.1038/nrn.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.A., Ding S.L., Sunkin S.M., Smith K.A., Ng L., Szafer A., Ebbert A., Riley Z.L., Royall J.J., Aiona K. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206. doi: 10.1038/nature13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muffat J., Li Y., Yuan B., Mitalipova M., Omer A., Corcoran S., Bakiasi G., Tsai L.H., Aubourg P., Ransohoff R.M. Efficient derivation of microglia-like cells from human pluripotent stem cells. Nat. Med. 2016;22:1358–1367. doi: 10.1038/nm.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberheim N.A., Takano T., Han X., He W., Lin J.H., Wang F., Xu Q., Wyatt J.D., Pilcher W., Ojemann J.G. Uniquely hominid features of adult human astrocytes. J. Neurosci. 2009;29:3276–3287. doi: 10.1523/JNEUROSCI.4707-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm T., Bolognin S., Meiser J., Nickels S., Trager C., Meilenbrock R.L., Brockhaus J., Schreitmuller M., Missler M., Schwamborn J.C. Rapid and robust generation of long-term self-renewing human neural stem cells with the ability to generate mature astroglia. Sci. Rep. 2015;5:16321. doi: 10.1038/srep16321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandya H., Shen M.J., Ichikawa D.M., Sedlock A.B., Choi Y., Johnson K.R., Kim G., Brown M.A., Elkahloun A.G., Maric D. Differentiation of human and murine induced pluripotent stem cells to microglia-like cells. Nat. Neurosci. 2017;20:753–759. doi: 10.1038/nn.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadeas S.T., Kraig S.E., O'Banion C., Lepore A.C., Maragakis N.J. Astrocytes carrying the superoxide dismutase 1 (SOD1G93A) mutation induce wild-type motor neuron degeneration in vivo. Proc. Natl. Acad. Sci. USA. 2011;108:17803–17808. doi: 10.1073/pnas.1103141108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkas-Kramarski R., Eilam R., Spiegler O., Lavi S., Liu N., Chang D., Wen D., Schwartz M., Yarden Y. Brain neurons and glial cells express Neu differentiation factor/heregulin: a survival factor for astrocytes. Proc. Natl. Acad. Sci. USA. 1994;91:9387–9391. doi: 10.1073/pnas.91.20.9387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard K.L., Filali M., Prefontaine P., Rivest S. Toll-like receptor 2 acts as a natural innate immune receptor to clear amyloid beta 1-42 and delay the cognitive decline in a mouse model of Alzheimer's disease. J. Neurosci. 2008;28:5784–5793. doi: 10.1523/JNEUROSCI.1146-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein J.D., Martin L., Levey A.I., Dykes-Hoberg M., Jin L., Wu D., Nash N., Kuncl R.W. Localization of neuronal and glial glutamate transporters. Neuron. 1994;13:713–725. doi: 10.1016/0896-6273(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Roybon L., Lamas N.J., Garcia-Diaz A., Yang E.J., Sattler R., Jackson-Lewis V., Kim Y.A., Kachel C.A., Rothstein J.D., Przedborski S. Human stem cell-derived spinal cord astrocytes with defined mature or reactive phenotypes. Cell Rep. 2013;4:1035–1048. doi: 10.1016/j.celrep.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scemes E., Giaume C. Astrocyte calcium waves: what they are and what they do. Glia. 2006;54:716–725. doi: 10.1002/glia.20374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzer J., Franke W.W., Schachner M. Immunocytochemical demonstration of vimentin in astrocytes and ependymal cells of developing and adult mouse nervous system. J. Cell Biol. 1981;90:435–447. doi: 10.1083/jcb.90.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert G., Schilling K., Steinhauser C. Astrocyte dysfunction in neurological disorders: a molecular perspective. Nat. Rev. Neurosci. 2006;7:194–206. doi: 10.1038/nrn1870. [DOI] [PubMed] [Google Scholar]
- Serio A., Bilican B., Barmada S.J., Ando D.M., Zhao C., Siller R., Burr K., Haghi G., Story D., Nishimura A.L. Astrocyte pathology and the absence of non-cell autonomy in an induced pluripotent stem cell model of TDP-43 proteinopathy. Proc. Natl. Acad. Sci. USA. 2013;110:4697–4702. doi: 10.1073/pnas.1300398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaltouki A., Peng J., Liu Q., Rao M.S., Zeng X. Efficient generation of astrocytes from human pluripotent stem cells in defined conditions. Stem Cells. 2013;31:941–952. doi: 10.1002/stem.1334. [DOI] [PubMed] [Google Scholar]
- Sherwood C.C., Stimpson C.D., Raghanti M.A., Wildman D.E., Uddin M., Grossman L.I., Goodman M., Redmond J.C., Bonar C.J., Erwin J.M. Evolution of increased glia-neuron ratios in the human frontal cortex. Proc. Natl. Acad. Sci. USA. 2006;103:13606–13611. doi: 10.1073/pnas.0605843103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skripuletz T., Hackstette D., Bauer K., Gudi V., Pul R., Voss E., Berger K., Kipp M., Baumgartner W., Stangel M. Astrocytes regulate myelin clearance through recruitment of microglia during cuprizone-induced demyelination. Brain. 2013;136:147–167. doi: 10.1093/brain/aws262. [DOI] [PubMed] [Google Scholar]
- Tong X., Ao Y., Faas G.C., Nwaobi S.E., Xu J., Haustein M.D., Anderson M.A., Mody I., Olsen M.L., Sofroniew M.V. Astrocyte Kir4.1 ion channel deficits contribute to neuronal dysfunction in Huntington's disease model mice. Nat. Neurosci. 2014;17:694–703. doi: 10.1038/nn.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheyen A., Diels A., Dijkmans J., Oyelami T., Meneghello G., Mertens L., Versweyveld S., Borgers M., Buist A., Peeters P. Using human iPSC-derived neurons to model TAU aggregation. PLoS One. 2015;10:e0146127. doi: 10.1371/journal.pone.0146127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volterra A., Liaudet N., Savtchouk I. Astrocyte Ca(2)(+) signalling: an unexpected complexity. Nat. Rev. Neurosci. 2014;15:327–335. doi: 10.1038/nrn3725. [DOI] [PubMed] [Google Scholar]
- Wyss-Coray T., Loike J.D., Brionne T.C., Lu E., Anankov R., Yan F., Silverstein S.C., Husemann J. Adult mouse astrocytes degrade amyloid-beta in vitro and in situ. Nat. Med. 2003;9:453–457. doi: 10.1038/nm838. [DOI] [PubMed] [Google Scholar]
- Xu Q., Bernardo A., Walker D., Kanegawa T., Mahley R.W., Huang Y. Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J. Neurosci. 2006;26:4985–4994. doi: 10.1523/JNEUROSCI.5476-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Lee E.M., Wen Z., Cheng Y., Huang W.K., Qian X., Tcw J., Kouznetsova J., Ogden S.C., Hammack C. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan S.H., Martin J., Elia J., Flippin J., Paramban R.I., Hefferan M.P., Vidal J.G., Mu Y., Killian R.L., Israel M.A. Cell-surface marker signatures for the isolation of neural stem cells, glia and neurons derived from human pluripotent stem cells. PLoS One. 2011;6:e17540. doi: 10.1371/journal.pone.0017540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamanian J.L., Xu L., Foo L.C., Nouri N., Zhou L., Giffard R.G., Barres B.A. Genomic analysis of reactive astrogliosis. J. Neurosci. 2012;32:6391–6410. doi: 10.1523/JNEUROSCI.6221-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Barres B.A. Astrocyte heterogeneity: an underappreciated topic in neurobiology. Curr. Opin. Neurobiol. 2010;20:588–594. doi: 10.1016/j.conb.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S., Li G. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Schwartz J.P. Involvement of cytokines in normal CNS development and neurological diseases: recent progress and perspectives. J. Neurosci. Res. 1998;52:7–16. doi: 10.1002/(SICI)1097-4547(19980401)52:1<7::AID-JNR2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.