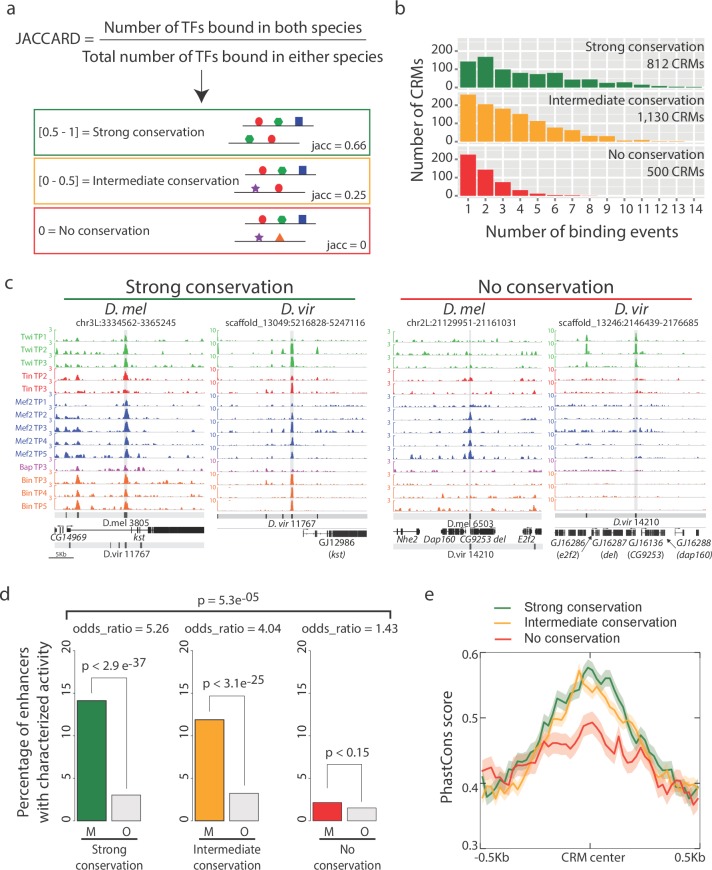
Figure 2. Enhancers with conserved binding are associated with functional elements.
(a) Jaccard similarity index was used to categorize CRMs into three groups based on their TF binding conservation. The schematic represents CRM pairs for each of the three groups with hypothetical combinatorial binding and the associated Jaccard Index. (b) Barplot representing the complexity of D. melanogaster CRMs, in term of number of binding events (x-axis), for each CRM binding conservation category. (c) Genome browser visualization of strongly conserved (left) and non-conserved (right) CRM pairs. D. virilis (D. vir) tracks display RPM normalized, input subtracted ChIP-seq signal. D. melanogaster (D. mel) tracks show log2(IP/mock) ChIP-chip signal. In D. mel, both D. mel and translated D. vir CRMs are shown along with D. mel gene models. In D. vir, D. vir CRMs and genes are shown with D. mel orthologous gene name depicted between parenthesis. The same genomic window size was used for both species, indicated in the lower left corner. (d) Fraction of mesodermal (‘M’, coloured) and non-mesodermal (‘O’ for ‘Other tissues’, gray) enhancers overlapping CRMs from the three categories defined in (a). Overall p-value corresponds to a two-sided proportions test, p-values within categories correspond to Fisher exact test. (e) CRMs with strong or intermediate conservation in their TF binding events have stronger sequence constrains than those with no conserved binding, based on their average PhastCons conservation scores. Solid lines represent the mean and shading delineates standard error.