Abstract
Deubiquitylating enzymes (DUBs) remove ubiquitin (Ub) from various cellular proteins and render eukaryotic ubiquitylation a dynamic process. The misregulation of protein ubiquitylation is associated with many human diseases, and there is an urgent need to identify specific DUBs associated with therapeutically relevant targets of Ub. We report the development of two facile selenocysteine-based strategies to generate the DUB probe dehydroalanine (Dha). Optimized oxidative or alkylative elimination of Se yielded Dha at the C-terminus of Ub. The high utility of alkylative elimination, which is compatible with multiple thiols in Ub targets, was demonstrated by generating a probe derived from the Ub ligase tripartite motif protein 25 (TRIM-25). Successful capture of the TRIM-25-associated DUB, ubiquitin-specific protease 15, demonstrated the versatility of our chemical strategy for identifying target-specific DUBs.
There are ~100 known deubiquitylating enzymes (DUBs) in humans that are involved in a wide range of cellular processes, including the processing of ubiquitin (Ub) itself prior to its conjugation with target proteins.1 About 80 DUBs, constituting four of the five known DUB classes, employ an active-site cysteine thiol to hydrolyze the isopeptide linkage between Ub and its various targets.2 In order to elucidate the specific functions and cellular targets of this large majority of human DUBs, we sought to develop a generalizable strategy for the installation of covalent-capture probes in various ubiquitylated substrates.3,4
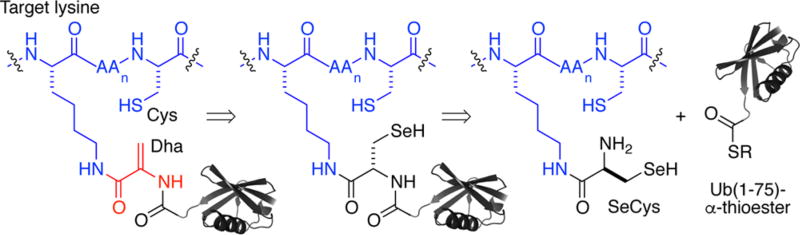
Several methods exist for the incorporation of thiol-reactive electrophilic probes at the C-terminus of Ub or within dimeric forms of Ub.5–7 However, the challenges associated with their chemoselective installation limits their application to proteins that lack cysteine. For example, the Michael acceptor dehydroalanine (Dha) was recently reported as a probe for DUB activity.6,7 Dha is readily generated by the bisalkylation and subsequent elimination of cysteine side chains.8 Although the chemical orthogonality of cysteine with other amino acids is appealing, a DUB probe that incorporates additional cysteines from the target of Ub is challenging to produce by current technologies. Pioneering efforts by Ploegh and co-workers, who employed the direct amidation of truncated Ub C-terminal azides with glycine-derived electrophiles,9 are in principle compatible with cysteine-containing target peptides. However, the presence of additional lysine residues in such targets is an inherent challenge toward selective amidation of the Ub azide. We addressed this key limitation for DUB probes by developing a chemical strategy to generate Dha that is compatible with multiple cysteine and lysine residues.
Toward our goal, we identified the 21st naturally occurring amino acid, selenocysteine (SeCys), as a potential handle for the site-specific introduction of Dha in ubiquitylated substrates.10 Strategies to generate Dha from SeCys have employed oxidative elimination of its arylated11 or alkylated derivatives.12 However, we noted that oxidative elimination of a single unmodified SeCys in the presence of five Cys residues was seen in the mammalian housekeeping enzyme glutathione peroxidase 1 (GPX1).13,14 Quaderer and Hilvert also reported hydrogen peroxide-mediated oxidative elimination of the diselenide form of a 16-residue integrin-binding cyclic RGD peptide.15 This led us to consider replacing Gly76 in Ub with SeCys as a latent precursor to Dha. We envisioned using SeCys as a handle for site-specific substrate ubiquitylation,16 followed by its chemoselective elimination to generate Dha (Scheme 1).
Scheme 1.
Retrosynthetic Analysis of a Site-Specifically Ubiquitylated Target Containing Cysteine and Dehydroalanine (Dha) (AA = Amino Acid)
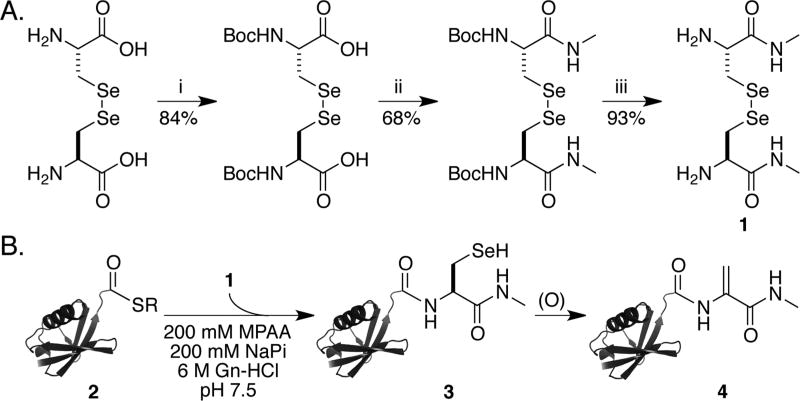
In order to test our ability to generate Dha by the oxidative elimination of SeCys in sufficient quantities for biochemical applications, we first synthesized the compound l-selenocystine-N-methylamide (1) (Supporting Information and Scheme 2A). This was ligated with the Ub(1–75)-α-thioester 2, which was generated by thiolysis of a Ub(1–75)–intein fusion protein with the sodium salt of 2-mercaptoethanesulfonic acid (Figure S1 and Scheme 2B).17 The ligation product, Ub-SeCys-N-methylamide (3), was oxidized with 1 molar equiv of NaIO4 at pH 5.5 to yield multiply oxidized intermediates (Figure S2A). After 2 h of oxidation, the addition of 6 M Gn-HCl facilitated in situ elimination to yield Ub-Dha-N-methylamide (4) over 48 h in 37% purified yield (Figure 1A,B). Surprisingly, analysis of the in situ elimination reaction consistently revealed a population of doubly (M + 32 Da) and triply oxidized species (M + 48 Da) that failed to eliminate Se (Figure S2B).
Scheme 2.
(A) Synthesis of l-Selenocystine-N-methylamide (1) and (B) Semisynthesis of Ub-Dha-N-methylamide (4)a
a(A) (i) Boc2O/TEA, H2O. (ii) CH3NH2–HCl, EDCI, HOBt, TEA, DMF. (iii) TFA, DCM, 25 °C. (B) Expressed protein ligation of Ub(1–75)-α-thioester with in situ-generated l-SeCys-N-methylamide followed by oxidative elimination to generate Ub-Dha-N-methylamide.
Figure 1.
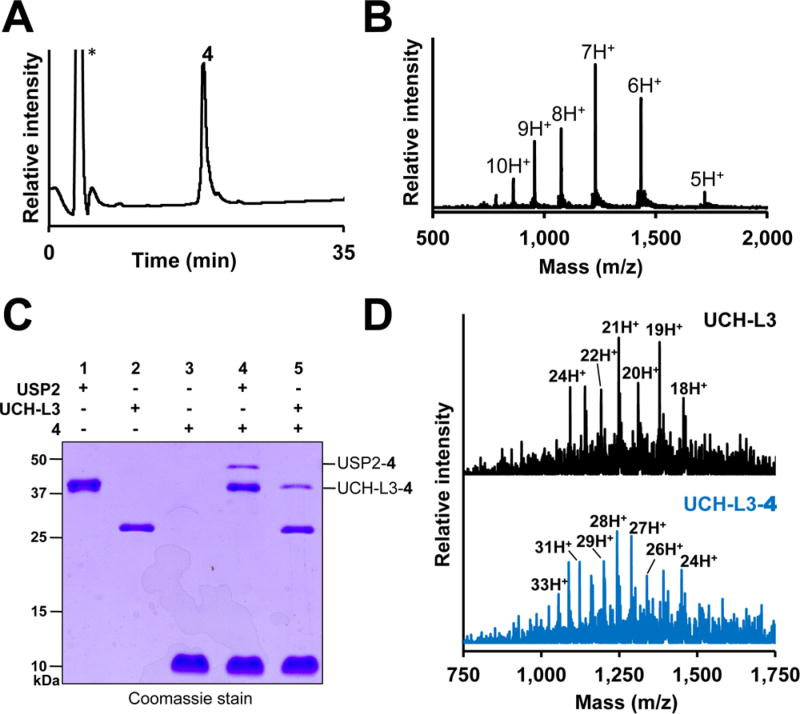
Covalent capture of DUBs with Ub-Dha-N-methylamide (4). (A) C18 RP-HPLC chromatogram of 4. The asterisk indicates the injection peak. (B) ESI-MS of 4. Calculated [M + H]+ 8590.8 Da, observed 8593.1 ± 2.2 Da. (C) 15% SDS-PAGE of 4 incubated with USP2 (lane 4) and UCH-L3 (lane 5). (D) ESI-MS of UCH-L3 (top) and the UCH-L3–4 adduct (bottom). Calculated for UCH-L3 [M + H]+ 26 183 Da, observed 26 194.1 ± 4.4 Da. Calculated for UCH-L3–4 [M + H]+ 34 774 Da, observed 34 774.0 ± 4.9 Da.
Incomplete Dha formation from the oxidized sample may be due to double oxidation occurring predominantly at the N-terminal Met1 of Ub and only single oxidation of SeCys. In order to interrogate the oxidation states of Met1 and SeCys76, we employed a top-down tandem mass spectrometry approach.18 Fragmentation of the isolated (M + 48 Da) species revealed singly oxidized b ions and doubly oxidized y ions, indicating that double oxidation was favored at SeCys76 over Met1 (Figure S2C). Consistent with this observation, the HPLC-purified (M + 48 Da) species underwent slow elimination over several days to yield the singly oxidized form of 4 (Figure S2D). Hence, we conclude that β-elimination of the doubly oxidized Se is a slow step in Dha formation from SeCys in proteins. Importantly, circular dichroism (CD) spectroscopy confirmed that purified probe 4 was structurally similar to unmodified Ub (Figure S3).
Next we tested the utility of 4 for capturing the active-site thiol in DUBs from two distinct families, namely, ubiquitin C-terminal hydrolase-L3 (UCH-L3) and ubiquitin-specific protease 2 (USP2). UCH-L3 removes Ub from small-molecule substrates,19 and USP2 disassembles polyubiquitin chains from several protein targets, including the p53 ubiquitin ligase Mdm2.20 Incubation of 4 with UCH-L3 and USP2 at 37 °C for 12 h led to the appearance of higher-molecular-weight species corresponding to the respective DUB–4 adducts (Figure 1C). The UCH-L3–4 adduct was further confirmed by LC-ESI-MS (Figure 1D). Altogether, our results demonstrated that the oxidative elimination methodology yields functional probes of active DUBs.
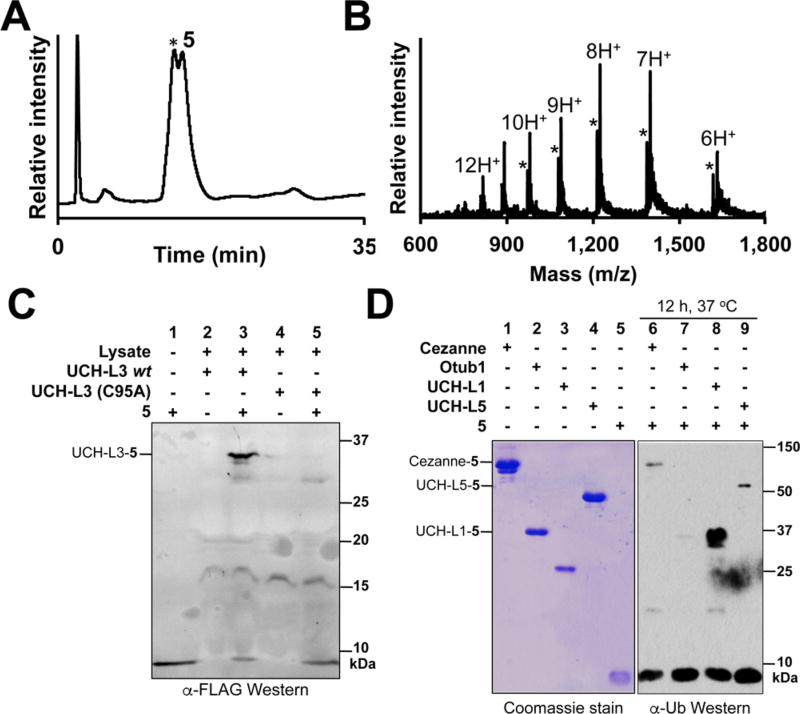
In order to facilitate the detection of DUB–probe conjugates in complex proteomes, we also generated 5, a FLAG epitopetagged form of Ub-Dha-N-methylamide (Figures S4 and S5). Although 5 was inseparable from unligated FLAG-Ub(1–75)-CO2H by HPLC (Figure 2A,B), we did not anticipate this to be a limitation, as eukaryotic proteomes also contain unconjugated Ub.21 However, one concern when using Dha as a thiol-reactive probe is the potential for nonspecific reactivity and quenching by cellular thiols. Indeed, van der Donk and co-workers reported the nonenzymatic addition of cysteine thiols to Dha at physiological pH.11 We therefore tested the ability of 5 to specifically label overexpressed UCH-L3 in an Escherchia coli lysate. We were pleased to observe robust labeling of UCH-L3 that was strictly dependent on the presence of the catalytic Cys95 (Figures 2C and S6).17
Figure 2.
Mechanism-based DUB labeling by FLAG-Ub-Dha-N-methylamide (5). (A) C18 RP-HPLC chromatogram of 5. The asterisk marks coeluting FLAG-Ub(1–75). (B) ESI-MS of 5. Calculated [M + H]+ 9788.1 Da, observed 9790.0 ± 1.8 Da. Asterisks indicate ions from FLAG-Ub(1–75). Calculated [M + H]+ 9705.0 Da, observed 9708.0 ± 2.8 Da. (C) Western Blot of E. coli lysates containing overexpressed wild-type UCH-L3 or UCH-L3(C95A) incubated with 5. The covalent DUB-5 adduct was detected with an anti-FLAG antibody (F3165, Sigma-Aldrich). (D) Western blot of Cezanne (lane 1), OTUB1 (lane 2), UCH-L1 (lane 3), and UCH-L5 (lane 4) incubated with 5. The DUB–5 adducts were detected with an anti-Ub antibody (P4D1, Santa Cruz Biotech).
We also compared the efficiency of UCH-L3 labeling by 5 with that by the commercially available DUB probe Ub C-terminal vinyl methyl ester (Ub-VME).4 Surprisingly, although we observed UCH-L3 labeling by excess 5 within 15 min (Figure S7A), the reaction products remained largely unchanged after 24 h of incubation (Figure S7B). In comparison, excess Ub-VME reacted with a majority of the UCH-L3 within 15 min (Figure S7A). However, increasing amounts of UCH-L3 did lead to increased labeling by 5, indicating that a fraction of UCH-L3 is reactive toward the probe (Figure S8).
Given the incomplete labeling of UCH-L3 by 5, we asked whether 5 may be useful for labeling DUBs and other cellular proteins at endogenous concentrations in DUB-labeling assays. We generated whole-cell lysates from human HeLa cells and incubated these with varying concentrations of 5. Gratifyingly, in Western blots with an anti-FLAG antibody, we observed labeling of endogenous proteins that increased with increasing amounts of 5 (Figure S9). Moreover, labeling by 5 was effectively competed with small-molecule DUB inhibitors and labeling by Ub-VME (Figure S10). We also identified the labeling of UCH-L3 in these lysates with an anti-UCH-L3 antibody (Figure S9). Thus, we conclude that despite a lower labeling efficiency than Ub-VME, the Ub–Dha probe captures human DUBs at endogenous concentrations and has the advantage of facile incorporation into ubiquitylated targets (Scheme 1). Indeed, it was recently shown that a similar Ub–Dha probe labels E1, E2, and E3 Ub ligases in living cells.7
Furthermore, we tested whether 5 would indiscriminately react with all DUBs or could select a subset of DUBs on the basis of their substrate specificity. Hence, we tested the DUBs cellular zinc finger anti-NF-κB (Cezanne), otubain 1 (OTUB1), and the ubiquitin C-terminal hydrolases UCH-L1 and UCH-L5. Incubation with 5 for 12 h led to labeling of UCH-L1 and UCH-L5 along with Cezanne. However, no significant labeling was observed with OTUB1 under our assay conditions (Figure 2D). OTUB1 cleaves Ub chains in a K48-linkage-specific manner, and binding to both proximal and distal ubiquitin monomers is required for its efficient activity.22 This requirement likely underlies its poor reactivity with the monomeric probe 5. The selective reactivity of 5 suggests that efficient binding in the DUB active site is a prerequisite for labeling and holds promise for future attempts to encode ubiquitin-substrate specificity in these probes.
Having established the mechanism-based reactivity of monomeric 5, we proceeded to test our ability to generate Dha in the presence of multiple Cys and Met residues. As a model substrate, we chose a 13-mer peptide sequence, Ac-KEAAVKTCLVCMA-CONH2, centered on Lys117 in the Ub E3 ligase tripartite motif protein 25 (TRIM-25). TRIM-25 ubiquitylates the RNA-sensing retinoic acid-inducible gene-1 (RIG-1)-like receptor and triggers the production of type 1 interferons in response to viral infection.23 Lys48-linked polyubiquitylation of TRIM-25 itself, however, inhibits its activity and thereby suppresses the immune response to viral pathogens. The DUB ubiquitin-specific peptidase 15 (Usp15) was implicated in deubiquitylating TRIM-25 and suppressing viral replication in cells by rescuing the RIG-1-mediated pathway.24 TRIM-25 is ubiquitylated at several lysines, including Lys117, and we wondered whether the ubiquitylated 13-mer TRIM-25 peptide bearing Dha would capture Usp15 in action.
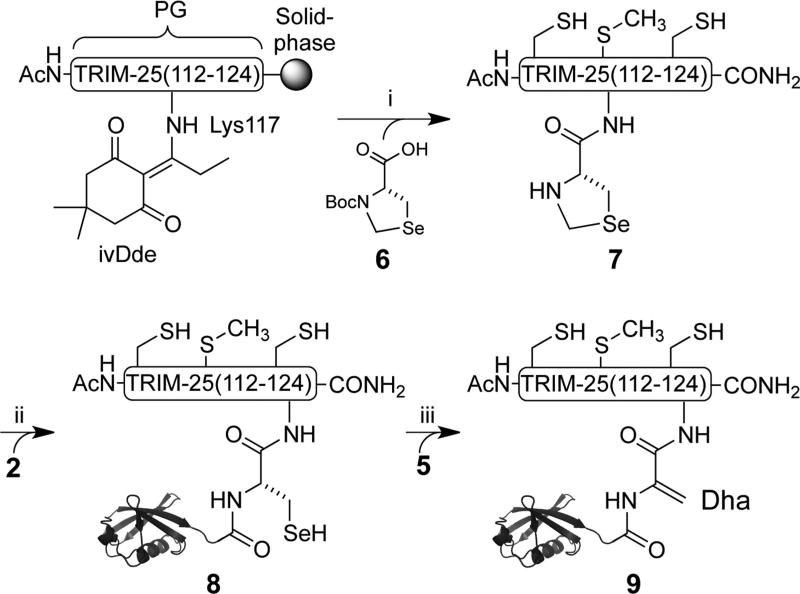
In order to facilitate the introduction of SeCys at Lys117 in the TRIM-25(112–124) peptide, we synthesized Boc-protected l-selenazolidine 6 (Supporting Information and Scheme S1).25 The orthogonally protected ε-amine of Lys117 was deprotected after peptide assembly on the solid phase and conjugated with 6 (Scheme 3). Subsequent acidolytic cleavage from the solid phase afforded the TRIM-25(112–124) peptide 7 linked to l-selenazolidine at Lys117 (Supporting Information and Figure S11). Deprotection of the selenazolidine with 1.6 M methoxylamine followed by expressed protein ligation (EPL) with 2 yielded the site-specifically ubiquitylated TRIM-25 peptide 8 bearing Ub Gly76SeCys (Figure S12). Subsequent attempts to selectively oxidize SeCys in good yields were unsuccessful and yielded a mixture of overoxidized species with as many as six extra oxygens per product (Figure S13). Therefore, we decided to explore selective alkylation of SeCys over Cys as an alternate approach for Se elimination.8
Scheme 3.
Synthesis of Ub-Dha-TRIM-25 (9)a
a(i) (1) 5% H2N–NH2 in DMF; (2) 6, DIC/Oxyma, 13%. (ii) (1) 5 M Gn-HCl, 1.6 M MeONH2, pH 5, 80%; (2) 2, 5% DMF, 6 M Gn-HCl, 100 mM MPAA, 100 mM NaPi, 1 mM EDTA, pH 7.5, 43%. (iii) 28 mM α,α′-dibromoadipoylbis(amide), pH 3.4, 0 °C, 12 h, 27%.
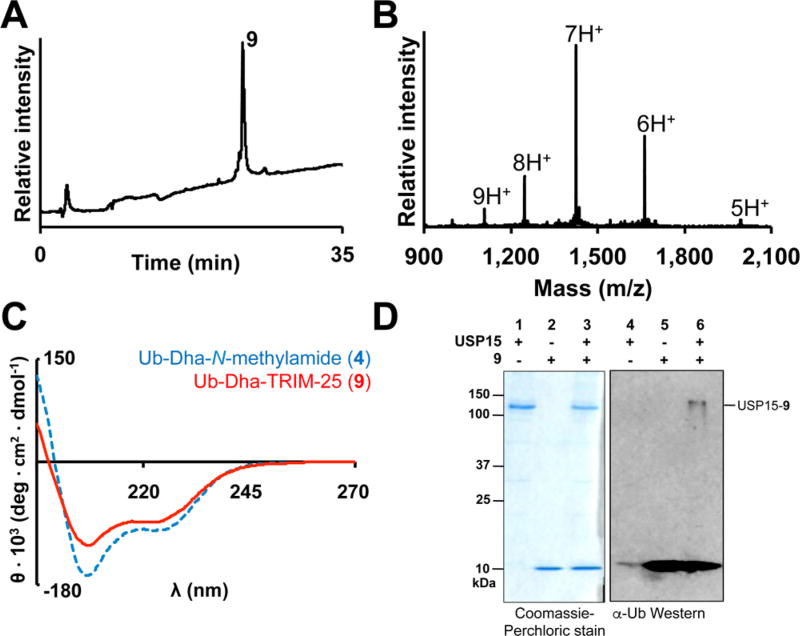
Peptide 8 was alkylated with 100 equiv of α,α′-dibromoadipoylbis(amide) at carefully controlled pH 3.4 on ice. At this lower pH, Cys was significantly less nucleophilic than SeCys (pKa ~ 8.0 vs ~4.0, respectively), leading to the formation of Ub-Dha-TRIM-25 probe 9 over 12 h in 27% purified yield (Figure 3A,B). Importantly, under these conditions we observed minimal thiol elimination. We further confirmed by CD spectroscopy that the presence of the TRIM-25 peptide did not inhibit Ub folding (Figure 3C). Finally, we tested the ability of probe 9 at 70 µM to label 1.2 µM Usp15 over the course of 12 h at 37 °C. Gratifyingly, we observed a new band corresponding to the Usp15–9 adduct by Western blot, which persisted in the presence of 10 mM DTT (Figure 3D). Moreover, labeling primarily occurred at the active-site Cys269 of Usp15 (Figure S14). Thus, our result suggests that Usp15 can deubiquitylate Lys117 along with other known lysine sites of deubiquitylation in TRIM-25.
Figure 3.
Labeling of Usp15 with Ub-Dha-TRIM25 (9). (A) RP-HPLC chromatograph of 9. (B) ESI-MS of 9. Calculated 9966.5 Da, observed 9968.0 ± 1.4 Da. (C) Comparison of the CD spectra of 9 and Ub-Dha-N-methylamide. (D) 10% SDS-PAGE (left) and Western blot (right) of 9 incubated with full-length USP15. The USP15–9 adduct was detected with an anti-Ub antibody (P4D1, Santa Cruz Biotech).
In conclusion, we have developed oxidative and alkylative β-elimination of SeCys as chemical strategies for the introduction of DUB-reactive Dha at the C-terminus of Ub. The latent reactivity of SeCys enables its application as a handle for the site-specific attachment of Ub to peptide targets and subsequently toward the chemoselective generation of Dha. The alkylative elimination strategy is particularly useful when multiple cysteines are present in the target of Ub and in principle may be extended to any ubiquitylated protein amenable to semisynthesis. Finally, we demonstrated the utility of our semisynthetic strategy by covalently labeling Usp15 with a ubiquitylated peptide derived from its proposed target, the E3 ligase TRIM-25. In the future, our chemoselective strategy will enable experiments aimed at identifying substrate-specific DUBs for therapeutically relevant ubiquitylated proteins.
Supplementary Material
Acknowledgments
We thank the Departments of Chemistry at the University of Washington and the University of Wisconsin for their generous support. We thank the NIH/NIGMS for grants to C.C. (R01GM110430) and E.R.S. (R01GM110543). C.E.W. is grateful for an NSF GRFP (DGH-1256082) and an ARCS Foundation Fellowship.
Footnotes
ASSOCIATED CONTENT
- Supporting figures and detailed experimental procedures (PDF)
The authors declare no competing financial interest.
References
- 1.Komander D, Clague MJ, Urbé S. Nat. Rev. Mol. Cell Biol. 2009;10:550. doi: 10.1038/nrm2731. [DOI] [PubMed] [Google Scholar]
- 2.Nijman SMB, Luna-Vargas MPa, Velds A, Brummelkamp TR, Dirac AMG, Sixma TK, Bernards R. Cell. 2005;123:773. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 3.Ovaa H. Nat. Rev. Cancer. 2007;7:613–20. doi: 10.1038/nrc2128. [DOI] [PubMed] [Google Scholar]
- 4.Love KR, Pandya RK, Spooner E, Ploegh HL. ACS Chem. Biol. 2009;4:275. doi: 10.1021/cb9000348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li G, Liang Q, Gong P, Tencer AH, Zhuang Z. Chem. Commun. 2014;50:216. doi: 10.1039/c3cc47382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haj-Yahya N, Hemantha HP, Meledin R, Bondalapati S, Seenaiah M, Brik A. Org. Lett. 2014;16:540. doi: 10.1021/ol403416w. [DOI] [PubMed] [Google Scholar]
- 7.Mulder MP, Witting K, Berlin I, Pruneda JN, Wu KP, Chang JG, Merkx R, Bialas J, Groettrup M, Vertegaal AC, Schulman BA, Komander D, Neefjes J, El Oualid F, Ovaa H. Nat. Chem. Biol. 2016;12:523. doi: 10.1038/nchembio.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chalker JM, Gunnoo SB, Boutureira O, Gerstberger SC, Fernández-González M, Bernardes GJL, Griffin L, Hailu H, Schofield CJ, Davis BG. Chem. Sci. 2011;2:1666. [Google Scholar]
- 9.Borodovsky A, Kessler BM, Casagrande R, Overkleeft HS, Wilkinson KD, Ploegh HL. EMBO J. 2001;20:5187. doi: 10.1093/emboj/20.18.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu Y, van der Donk WA. Org. Lett. 2001;3:1189. doi: 10.1021/ol015648a. [DOI] [PubMed] [Google Scholar]
- 11.Okeley NM, Zhu Y, van Der Donk WA. Org. Lett. 2000;2:3603. doi: 10.1021/ol006485d. [DOI] [PubMed] [Google Scholar]
- 12.Seebeck FP, Szostak JW. J. Am. Chem. Soc. 2006;128:7150. doi: 10.1021/ja060966w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma S, Caprioli RM, Hill KE, Burk RF. J. Am. Soc. Mass Spectrom. 2003;14:593. doi: 10.1016/S1044-0305(03)00141-7. [DOI] [PubMed] [Google Scholar]
- 14.Rhee SG, Cho CS. Methods Enzymol. 2010;474:23. doi: 10.1016/S0076-6879(10)74002-7. [DOI] [PubMed] [Google Scholar]
- 15.Quaderer R, Hilvert D. Chem. Commun. 2002:2620. doi: 10.1039/b208288h. [DOI] [PubMed] [Google Scholar]
- 16.Weller CE, Pilkerton ME, Chatterjee C. Biopolymers. 2014;101:144. doi: 10.1002/bip.22253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee C, McGinty RK, Pellois JP, Muir TW. Angew. Chem. Int. Ed. 2007;46:2814. doi: 10.1002/anie.200605155. [DOI] [PubMed] [Google Scholar]
- 18.Scheffler K. Methods Mol. Biol. 2014;1156:465. doi: 10.1007/978-1-4939-0685-7_31. [DOI] [PubMed] [Google Scholar]
- 19.Popp MW, Artavanis-Tsakonas K, Ploegh HL. J. Biol. Chem. 2009;284:3593. doi: 10.1074/jbc.M807172200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stevenson LF, Sparks A, Allende-Vega N, Xirodimas DP, Lane DP, Saville MK. EMBO J. 2007;26:976. doi: 10.1038/sj.emboj.7601567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaiser SE, Riley BE, Shaler TA, Trevino RS, Becker CH, Schulman H, Kopito RR. Nat. Methods. 2011;8:691. doi: 10.1038/nmeth.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ritorto MS, Ewan R, Perez-Oliva AB, Knebel A, Buhrlage SJ, Wightman M, Kelly SM, Wood NT, Virdee S, Gray NS, Morrice NA, Alessi DR, Trost M. Nat. Commun. 2014;5:4763. doi: 10.1038/ncomms5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. Nature. 2007;446:916. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 24.Pauli E-K, Chan YK, Davis ME, Gableske S, Wang MK, Feister KF, Gack MU. Sci. Signaling. 2014;7:ra3. doi: 10.1126/scisignal.2004577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy PS, Dery S, Metanis N. Angew. Chem. Int. Ed. 2016;55:992. doi: 10.1002/anie.201509378. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.