Abstract
Transient kinetic studies have shown that the uptake of the pheromone (bombykol) of the silkworm moth (Bombyx mori), by its pheromone-binding protein (PBP) BmorPBP, proceeds with an “on” rate of 0.068 ± 0.01 μM–1·s–1. With the high concentration of PBP in the sensillar lymph (10 mM), the half-life for the uptake of pheromone in vivo is ≈1 ms. A pH-dependent conformational change (BmorPBPB → BmorPBPA), associated with the release of pheromone, is a first-order reaction (k = 74.1 ± 0.32 s–1; t1/2, 9.3 ms). Under physiological conditions, both reactions proceed with half-life times on the order of milliseconds, as is required for odorant-oriented navigation in insects. Molecular interactions of bombykol with both native and mutated PBPs were analyzed by a novel binding assay. A recombinant protein with the native conformation (BmorPBP) showed high binding affinity (KD = 105 nM) at pH 7 but low affinity (KD = 1,600 nM) at pH 5, when tested at both low and high KCl concentrations. A protein with a C-terminal segment deleted (BmorPBPΔP129-V142) was found to bind bombykol at pH 7 and at pH 5 with the same affinity as the native protein at pH 7, indicating that the C-terminal segment is essential for preventing binding at low pH. Binding studies with three mutated proteins (BmorPBPW37F, BmorPBPW127F, and BmorPBPW37A) showed that replacing Trp-37 (with Phe or Ala) or Trp-127 (with Phe) did not affect the binding affinity to bombykol. Fluorescence studies shed light on the contributions of Trp-37 and Trp-127 emissions to the overall fluorescence.
Keywords: Bombyx mori pheromone-binding protein, bombykol, effect of C terminus on pheromone release, fast uptake of pheromone and delivery, mutated pheromone-binding proteins
For many insects, small-molecule signals communicate the availability of food, the presence of friends and foes, and the readiness to mate. In general, mate-finding is an essential prerequisite for exploring insects' enormous reproductive potential and, consequently, leads to their domination of the terrestrial world. To advertise their readiness to mate, female moths, for example, produce and release sex pheromones. Only minute amounts are released, so as to avoid chemical conspicuousness. Once released, the chemical signals are diluted in the environment and mixed with a myriad of physiologically irrelevant compounds, including pheromones from other species. Even though each species communicates with a specific pheromonic language, males can detect the low-level signals from conspecific females because of a highly developed olfactory system. To find females and successfully reproduce, males may have to make long-distance, odorant-oriented flights. Navigation requires a highly selective, sensitive, and dynamic sensory system for the detection of pheromones. Given the structure of pheromone plumes (1), insects have only a few milliseconds to reset their detectors while flying in the clean air spaces between pockets of chemical signals.
Pheromones are largely hydrophobic compounds, whereas pheromone receptors are surrounded by an aqueous solution (the sensillar lymph) and are isolated from the external environment. The crossing of this aqueous barrier to reach the odorant receptors is assisted by pheromone-binding proteins (PBPs). In a proposed model for the mode of action of PBPs (2–5), pheromones enter the sensillar lymph through small pores in the cuticle of the antennae (the sensillar wall), are solubilized upon being encapsulated in the binding cavity of PBPs, and are transported to the olfactory receptors. Bound pheromone molecules are protected from pheromone-degrading enzymes (6), which are proteins present in the sensillar lymph (2, 7). Upon interaction with negatively charged sites on or near the receptors, the PBP–pheromone complex undergoes a conformational change that leads to the ejection of the pheromone (3–5). In the PBP from the silkworm moth, Bombyx mori (BmorPBP), this pH-dependent conformational change (4, 5, 8) leads to the formation of a C-terminal α-helix in the acidic form of the protein (BmorPBPA) (5). The newly formed α-helix in Bmor-PBPA occupies the cavity that serves as a binding pocket in the basic form of the protein (BmorPBPB). In this model, the pheromone per se activates the receptor, thus initiating a cascade of events leading to neuronal activity and information being conveyed to the brain. In fact, a recently discovered pheromone receptor of the silkmoth responded to bombykol when expressed in Xenopus oocytes (9). Because this heterologous system is very likely devoid of odorant-binding proteins, these experiments indicate that the receptor is activated by pheromone per se. However, the onset of response and the overall kinetics of receptor activation and deactivation processes were much slower (on the scale of seconds) than those required to follow a pheromone plume (on the scale of milliseconds). Here, we have investigated the kinetics of pheromone binding and release by the PBP of the silkmoth as well as binding of bombykol to native and mutated BmorPBPs. Our data strongly suggest that PBPs are essential for the fast detection of pheromones during odorant-oriented navigation by insects.
Materials and Methods
Protein and Peptide Samples. The four pET (plasmid for expression) vectors for expression of recombinant BmorPBPΔP129-V142 (C terminus deleted), BmorPBPW127F (Trp-127 replaced by Phe), BmorPBPW37F (Trp-37 replaced by Phe), and BmorPBPW37A (Trp-37 replaced by Ala) were constructed with the QuikChange site-directed mutagenesis kit (Stratagene) by using the pETBmorPBP vector (8) as template DNA. Expression was performed in LB medium with transformed BL21(DE3) cells. Proteins in the periplasmic fraction were extracted with 10 mM Tris·HCl (pH 8) by using three cycles of freeze-and-thaw and centrifuging at 16,000 × g to remove debris. The supernatant was loaded on a 20-ml DEAE HR 16/10 column (Toyopearl 650S, Tosoh, Tokyo). Unless otherwise mentioned, all separations by ion-exchange chromatography were done with a linear gradient of 0–500 mM NaCl in 10 mM Tris·HCl (pH 8). Fractions containing the target protein were further purified on a 20-ml Q-Sepharose HR 16/10 column (Amersham Pharmacia Biosciences) and, subsequently, on a Mono-Q HR 10/10 column (Amersham Pharmacia Biosciences). PBP fractions were concentrated by using Centriprep-10 (Millipore) and loaded on a Superdex-75 26/60 gel-filtration column (Amersham Pharmacia Biosciences) preequilibrated with 200 mM NaCl and 20 mM Tris·HCl (pH 8). Fractions were analyzed by SDS/PAGE and MS. Fractions containing even traces of nontarget proteins were repurified by Mono-Q by using a shallower gradient of 0–300 mM. Highly purified protein fractions [>98% by liquid chromotography electrospray ionization (LC-ESI)/MS] were concentrated by Centricon-10, desalted on four 5-ml HiTrap desalting columns (Amersham Pharmacia Biosciences) in tandem and by using water as mobile phase, analyzed by LC-ESI/MS, lyophilized, and stored at –80°C until use. The concentrations of the recombinant proteins were measured by UV radiation at 280 nm in 20 mM sodium phosphate (pH 6.5) and 6 M guanidine HCl by using the theoretical extinction coefficients calculated with expasy software (http://us.expasy.org/tools/protparam.html). For fluorescence-emission experiments, the concentrations of the native and mutated BmorPBP samples were adjusted to give identical detector responses in MS analysis (see below). The peptide MDVAVGEILAEV, corresponding to the C terminus of BmorPBP, was synthesized and purified (98%) by SynPep (Dublin, CA). The molecular mass determined by LC-ESI/MS (observed, 1,245.53 Da; see below) confirmed the purity and identity of the unprotected peptide.
Cold Binding Assay. To 50 μl of protein solution (6.2 μM) in 100 mM buffer [ammonium acetate (pH 7) or sodium acetate (pH 5)] in a glass insert deactivated by Silcote CL7 treatment (Kimble Glass, Vineland, NJ), 1 μl of a 3.2 mM ethanol solution of the ligand was added. The sample was incubated in a gel shaker (100 rpm) at 25°C ± 2°C for 5, 30, or 60 min. The reaction mixture was transferred to a washed Microcon YM-10 centrifugal filter unit (Millipore) and centrifuged (12,000 × g) at 4°C for 5 min. The filtrate was either used to analyze the amount of free ligand or was discarded. The retentate was transferred to a 100-μl V-vial (Wheaton Scientific), and 10 μl of the reaction buffer was added to wash the centrifugal device and was then pooled in the V-vial. To extract the bound ligand, 50 μl of a low-pH buffer [1 M sodium formate (pH 3)] was added to the pooled retentate. After shaking the vial gently, 20 μl of a hexane solution (2.5 ppm) of the internal standard (eicosyl acetate, Fuji Flavor, Tokyo) was added. The vial was capped, vortexed for 1 min, and centrifuged (2,500 × g) at 4°C for 5 min. The hexane fraction was recovered and analyzed by GC-MS and GC. Each experiment was performed with five replicates and was repeated at least four times. After the identity of the ligand was confirmed by MS, quantification was routinely done by GC. As determined by LC-ESI/MS (see method below), retentate recovery for BmorPBP was 95%. Binding could be measured with a wide range of protein concentrations (0.62 μM to 62 μM) and 0.5–50 equivalents of ligand. Data presented in this article were obtained with 6.2 μM protein and 10× ligand. GC and GC-MS were done on a 6890 Series GC and a 5973 Network Mass Selective Detector (Agilent Technologies, Palo Alto, CA), respectively. Both instruments were equipped with the same type of capillary column (HP-5MS, 25 m × 0.25 mm; 0.25 μm; Agilent Technologies) operated under the same temperature program (100°C for 1 min, increased to 250°C at a rate of 10°C/min, and held at the final temperature for 10 min). Each sample was analyzed three times.
Other Analytical Procedures. ESI-MS was performed with a LCMS-2010 (Shimadzu). HPLC separations were done on a ZorbaxCB C8 column (150 × 2.1 mm; 5 μm; Agilent Technologies) with a gradient of water and acetonitrile plus 2% acetic acid as a modifier. The detector was operated with the nebulizer gas flow at 1.5 liters/min and the curved desolvation line and heat block at 250°C. Stopped-flow fluorometry was recorded on a KineAsyst SF-61SX2 instrument (Hi-Tech Scientific, Salisbury, U.K.) having a dead time of 1.6 ms (excitation, 280 nm; emission, 330 nm). To analyze the basic to acidic (B → A) pH-mediated conformational change, 14 μM BmorPBP in 20 mM ammonium acetate (pH 7) was mixed in a ratio of 1:5 with 200 mM sodium acetate (pH 4) to give a final protein concentration of 2.33 μM. Aliquots of these mixtures were collected to measure the final pH (4.7 ± 0.1). The reverse reaction (A → B) was analyzed by a similar procedure, with the protein in sodium acetate (pH 5) mixed with ammonium acetate buffer (pH 7) for a final pH of 6.3 ± 0.1. These experiments were repeated at least 10 times for each pH jump. To measure the kinetics of the bombykol–BmorPBP association, 0.6 μM BmorPBP in 50 mM buffer [ammonium acetate (pH 7) or sodium acetate (pH 5)] was mixed with a solution of the test ligand (bombykol and cetyl alcohol) in the same test buffer plus ethanol, to a final concentration of 4.5%. The concentration of bombykol in solution (65.6 ± 9.5 μM) was measured directly by GC of extracts from similar mixtures but devoid of protein. Data were analyzed by nonlinear regression. The reduced χ2 is given in figure legends. Steady-state fluorescence measurements were done on a spectrofluorophotometer (RF-5301, Shimadzu) at 25°C ± 1°C. The proteins were excited at 280 nm, and the emission spectra were recorded between 295 and 420 nm. Emission and excitation slit widths were 1.5 and 10 nm, respectively. Protein solutions (0.6 μM) were prepared in 20 mM ammonium acetate (pH 7, pH 6.5, and pH 6), or sodium acetate (pH 5.5, pH 5, and pH 4). CD spectra were recorded by using a J-810 spectropolarimeter (Jasco, Easton, MD) with 21 μM BmorPBPΔP129-V142 and 80 mM synthetic peptide MDVAVGEILAEV in either 20 mM ammonium acetate (pH 7) or 20 mM sodium acetate (pH 5).
Results
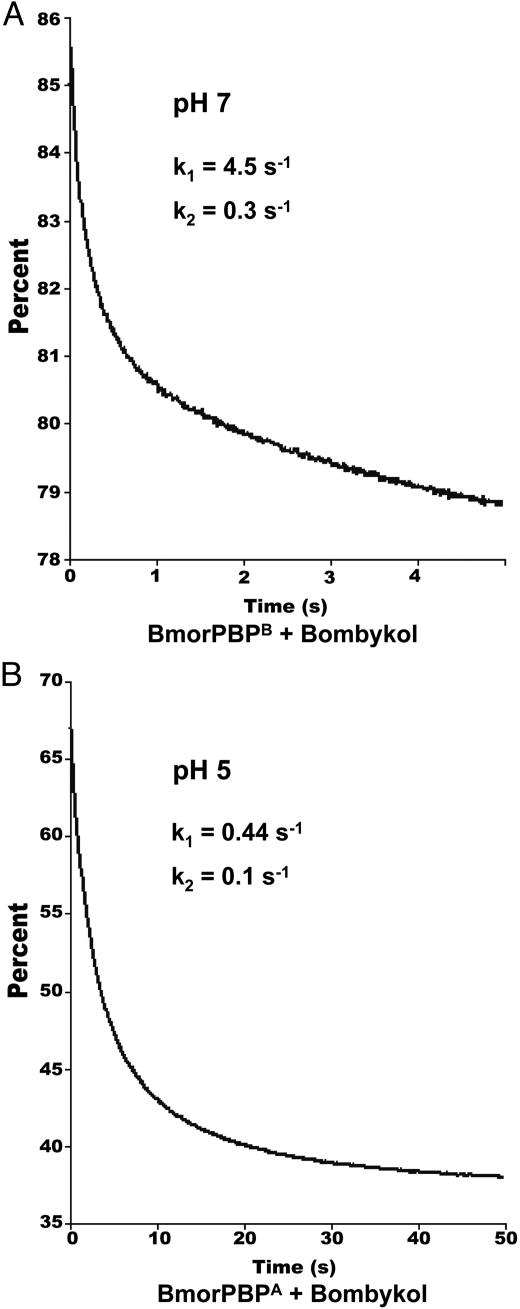
Kinetics of Pheromone Binding. To determine the rate constants for the binding of bombykol to BmorPBP, kinetic fluorescence traces were obtained by mixing protein and ligand at both high and low pH. The curves were fitted to a two-exponential process with rate constants k1 = 4.5 ± 0.06 s–1 and k2 = 0.3 ± 0.008 s–1 at pH 7 (Fig. 1A). As a negative control, we measured the kinetic parameters for the interaction of cetyl alcohol and BmorPBP at pH7(k1 = 0.76 ± 0.02 s–1 and k2 = 0.09 ± 0.002 s–1). The slower interaction with the nonnatural ligand was similar to that of the cognate ligand (bombykol) at pH 5 (k1 = 0.44 ± 0.003 s–1 and k2 = 0.1 ± 0.001 s–1) (Fig. 1B). The dependence of the observed rate (kobs) on ligand concentration follows a simple linear function (kobs = kon[L] + koff), where kon and koff represent the binding and dissociation constants, respectively (10). A plot of kobs (i.e., k1) vs. [L] gives the “on” rate (kon = 0.068 ± 0.01 μM–1·s–1) for the binding of bombykol to BmorPBP at pH 7. This value is smaller than the calculated value (kon = 0.154 μM–1·s–1) for the interaction of the wild silkmoth Antheraea polyphemus PBP (ApolPBP) with its cognate ligand (11). Because bombykol is released from the BmorPBP–pheromone complex by a pH-dependent conformational change, the relevant “off” rate (for the release of the pheromone) is directly related to the B → A conformational change rather than the “off” rate of BmorPBPB at the bulk pH of the sensillar lymph.
Fig. 1.
Association kinetics of bombykol and BmorPBP. (A) Fluorescence decrease upon mixing bombykol and BmorPBPB at pH 7. The curve was fitted (reduced χ2, 0.76) by using two-exponential terms and used to calculate the association rate (kon). (B) At low pH, the association of bombykol and BmorPBPA is 10 times slower as indicated by the time course and the kinetic parameters obtained by curve fitting (reduced χ2, 0.15) with two-exponential terms.
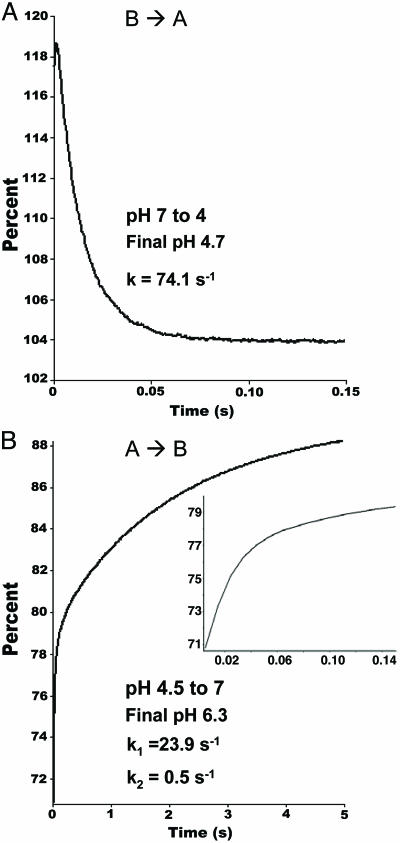
Kinetics of Conformational Change. The kinetics of the pH-dependent conformational change in BmorPBP were measured by mixing a solution of BmorPBP with a buffer solution to change the pH from 7 to 4.7, and the reaction was followed by stopped-flow fluorescence. The reaction BmorPBPB → Bmor-PBPA proceeded under first-order conditions (Fig. 2A), with an observed rate constant of 74.1 ± 0.32 s–1. Under these conditions, the t1/2 of the conversion of BmorPBP from basic to acidic form was then ≈9 ms, which is also the expected t1/2 of the BmorPBP–bombykol complex when the pH is dropped from 7 to ≈5, given that BmorPBPA does not retain the ligand at low pH (see below). On the other hand, the fluorescence transients collected for the reverse reaction (BmorPBPA → BmorPBPB) were fitted by a double-exponential equation, with the faster process (k1 = 23.9 ± 0.39 s–1) contributing to nearly half of the total fluorescence signal amplitude. It might be that the unwinding of the C-terminal α-helix in BmorPBPA is a slower process than the “reconstitution” of the N-terminal helical segment. These data suggest that the two conformations of BmorPBP exist in equilibrium, probably favoring the formation of BmorPBPB at the bulk pH of the sensillar lymph. Interactions with negatively charged membranes or a lowering of the pH shifts this equilibrium toward the formation of BmorPBPA.
Fig. 2.
Fluorescence transients of BmorPBP upon changing the pH. (A) The time dependence of the basic to acidic (B → A) conformational change is fitted by one exponential term to generate the observed rate constant (k) (reduced χ2, 1.95). (B) The time course of the reverse reaction (A → B) was fitted by two-exponential terms (k1 and k2). Note the faster step followed by a second, slower process (reduced χ2, 0.59). (Inset) A steep increase in fluorescence intensity in the first 150 ms of the reaction is shown.
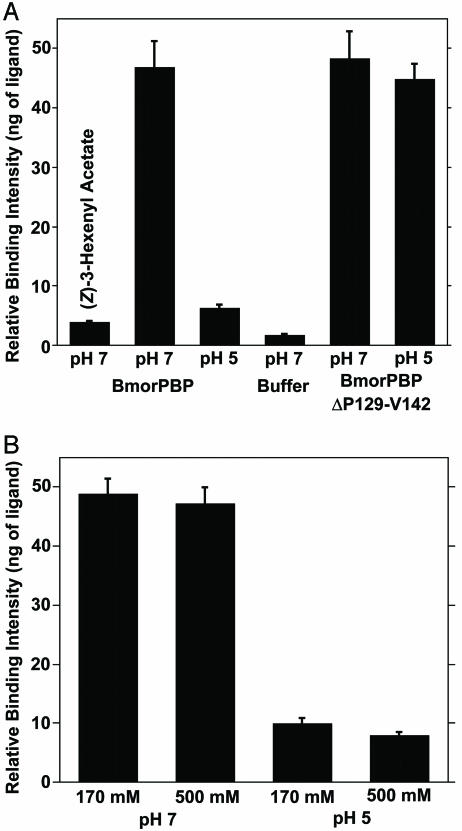
Binding of Bombykol to BmorPBP. We have developed a binding assay based on the separation of bound and free ligands by a centrifugal filter device. After incubation of a test compound with a PBP (under the desired conditions of temperature, pH, ionic strength, etc.), the free ligand is filtered out of the mixture, whereas the ligated protein is retained by the filter's membrane. Then, the ligand is released from the protein by lowering the pH, extracted with an organic solvent by using an internal standard, and analyzed by GC-MS (for identification) and/or by GC for quantification. This “cold binding assay” does not require radiolabeled compounds. The procedure allows the evaluation of binding by direct detection of the ligand but requires larger amounts of protein than the quantitative native gel assay (12). Our cold binding assay was extensively evaluated by using BmorPBP and bombykol, whose binding ability at high pH (and lack of binding at low pH) has been well documented (8, 13, 14). Results of the cold binding assay (Fig. 3A) were consistent with high and low binding affinities reported at pH 7 and pH 5, respectively. A small background was detected when the ligand was incubated in buffer (Fig. 3A). This “unspecific binding” can be decreased by additional centrifugation of the reaction mixture after adding 50 μl of 20 mM buffer (same pH as for the reaction). However, the extended residence time led to a decrease (≈30%) in the recovery of bound ligand at pH 7 (data not shown). On the other hand, there was no significant difference in the detected binding when the reaction was incubated for 5, 30, and 60 min. Data presented were obtained with a 60-min incubation. Two lines of evidence suggest that the cold binding assay allows the determination of specific binding. We detected no binding of bombykol to a nonolfactory protein, β-lactoglobulin (pH 7, 5.8 ± 0.5; pH 5, 7.4 ± 0.7; control, 3 ± 0.8 ng of ligand) and no affinity of a plant-derived semiochemical, (Z)-3-hexenyl acetate, to BmorPBP (Fig. 3A).
Fig. 3.
Bombykol binding to native and truncated PBPs. (A) pH-dependent binding ability. As a negative control, the green leaf volatile (Z)-3-hexenyl acetate did not show significant binding to BmorPBP. The effect of pH on the binding ability of the native conformation of BmorPBP and rescue of binding affinity at low pH in a truncated protein (BmorPBPPΔP129-V142) are shown. (B) Ionic strength and binding ability. Physiologically relevant or extremely high salt concentrations did not affect the binding affinity at high pH and did not rescue binding ability at low pH.
The dissociation constants for bombykol at pH 7 and pH 5, determined from these binding assays, were 105 and 1,600 nM, respectively. A higher value (KD 1,100 nM) at pH 8, determined by a tryptophan fluorescence-based binding assay, has been reported (14). In contrast to fluorescence assays, the cold binding assay allows accurate measurements of the concentrations of free and bound ligand (by GC) and protein (by LC-ESI/MS), thus permitting a more accurate determination of dissociation constants. Discrepancies of one order of magnitude have been previously documented for the interaction of the A. polyphemus major pheromone constituent with ApolPBP. Whereas Kaissling et al. (15) determined a value of 60 nM using isolated native PBP, Du and Prestwich (16) reported a KD of 640 nM with a recombinant protein and a different assay.
To test the hypothesis that physiological K+ concentration counterbalances the effect of low pH (17), we have evaluated pheromone binding in the presence of KCl. Binding of bombykol to BmorPBP at pH 7 was not affected by salt (Fig. 3B). Physiological and extremely high concentrations of KCl did not rescue binding at low pH (Fig. 3B).
Effect of C-Terminal α-Helix on Binding Affinity at Low pH. Previously, we suggested that the loss of binding affinity at low pH is due to the occupation of the binding cavity by a C-terminal α-helix in Bmor-PBPA (Fig. 3A) (18, 19). To test this hypothesis, we expressed a truncated form of BmorPBPB by removing the segment P129-SMDVAVGEILAE-V142 (see Fig. 5, which is published as supporting information on the PNAS web site), which is involved in the formation of the C-terminal α-helix (MDVAVGEILAE) (18). The molecular mass of BmorPBPΔP129-V142 obtained by LC-ESI/MS (observed, 14,466 Da; calculated, 14,472 Da) indicated that all Cys residues were linked to form three disulfide bridges, as in the native conformation (18–22). CD data (see Fig. 6, which is published as supporting information on the PNAS web site) showed that the helical contents of the truncated protein at high and low pH are nearly identical, thus implying that, as opposed to the intact BmorPBP, there is no unwinding of the N-terminal α-helix at low pH. This observation is further supported by steady-state fluorescence measurements of BmorPBPΔP129-V142, which showed little difference in the range of pH 6.5 to pH 4 (data not shown). On the other hand, a synthetic peptide with the same primary sequence as the C-terminal α-helix in BmorPBPA showed typical CD spectra of unordered peptides at high and low pH values (Fig. 6 Inset). Although not entirely surprising, given the size of this peptide (23), this observation indicates that the formation of the C-terminal α-helix might be stabilized by interactions of the terminal residues with residues forming the binding cavity (19, 22). Except for three acidic residues, the terminal fragment is composed almost entirely of hydrophobic amino acid residues.
Binding assays demonstrated that the binding affinity at pH 7 was not affected by the removal of the C-terminal α-helix (Fig. 3A). Binding of bombykol to the native conformation and to truncated forms of BmorPBP were indistinguishable. As opposed to the native conformation, however, the truncated protein showed high binding affinity at low pH (Fig. 3A). Attempts to prevent binding of bombykol to BmorPBPΔP129-V142 by the addition of the terminal peptide to the reaction mixture were unsuccessful. The formation of the helical segment MDVAVGEILA might be possible only in the protein context and, possibly, synchronized with the unfolding of the N-terminal segment. When isolated from the protein, the peptide has larger entropy and an additional N-terminal charge. The PBP N-terminal helix seems to be preserved in BmorPBPΔP129-V142 at low pH (Fig. 6). In other words, the peptide by itself did not fold or occupy the binding pocket.
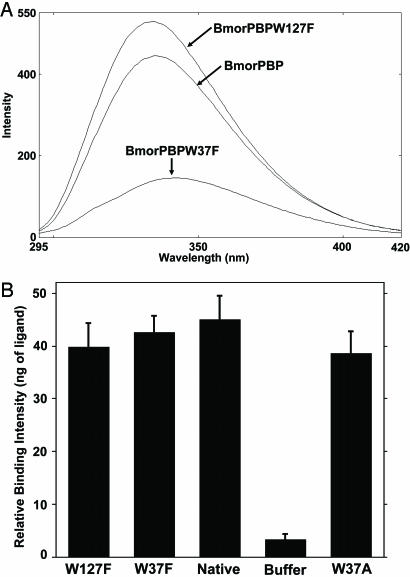
Effects of Trp-37 and Trp-137 on Fluorescence Emissions and Binding. The pH-dependent conformational change in BmorPBP is associated with a dramatic change in tryptophan fluorescence (8), thereby allowing the measurement of transient kinetics by stopped-flow fluorometry. Comparison of the two forms of BmorPBP indicates that the environment of Trp-110 remains unchanged in the acidic and basic forms. On the other hand, the environments of Trp-127 and Trp-37 are remarkably different (see Fig. 7, which is published as supporting information on the PNAS web site), possibly accounting for the decrease in intrinsic fluorescence at low pH (8). Comparison of the steady-state fluorescence of mutated and native BmorPBP indicated that the intrinsic fluorescence of these proteins decreases in the order BmorPBPW127F > BmorPBP > BmorPBPW37F at pH 7 (Fig. 4A). These experiments were carried out with normalized concentrations of proteins. The higher emission amplitude of the protein with Trp-127 replaced by Phe and low emission of the Trp-37 mutation are consistent with findings in A. polyphemus PBP (24). BmorPBPW127F displayed a similar pH-dependence of the tryptophan fluorescence to that observed with the native protein (data not shown), i.e., larger emission amplitude at high pH and smaller amplitude at low pH. By contrast, the intrinsic fluorescence of BmorPBPW37F increased as the pH decreased, i.e., BmorPBPW37F showed the highest intensity at pH 4 and the lowest at pH 7 (data not shown). BmorPBPW127F gave a larger fluorescence emission than the native protein because Trp-127 somewhat quenches the emission associated with Trp-37 in BmorPBP.
Fig. 4.
Fluorescence emission spectra and binding affinity of native and tryptophan-mutated PBPs at pH 7. (A) Intrinsic fluorescence of BmorPBP, BmorPBPW127F, and BmorPBPW37F with excitation at 280 nm and maximal emission at 335, 334, and 341 nm, respectively. The concentrations of the four proteins were adjusted by LC-ESI/MS. (B) Effect of tryptophan on binding ability. The three tryptophan mutants (BmorPBPW127F, BmorPBPW37F, and BmorPBPW37A) showed binding ability to bombykol nearly identical to that of the native conformation (BmorPBP).
It has been suggested that an unsaturated aliphatic pheromone chain interacts with the well conserved aromatic side chain of Trp-37 before entering the internal cavity of PBPs (25, 26). Our binding data (Fig. 4B) with BmorPBPB, BmorPBPW37F, and BmorPBPW37A did not support this hypothesis. Mutated and native conformations of BmorPBP bound bombykol with nearly identical affinity. In conclusion, Trp-37 and Trp-127 are directly involved in the change in fluorescence associated with the pH-dependent conformational change. However, Trp-37 is unlikely to be involved in the uptake of bombykol by BmorPBP.
Discussion
Rapid pheromone binding and release is essential for the orientation of insect flight toward a pheromone source (5). Here, we have studied the kinetics of pheromone binding and release in the bombykol/BmorPBP system. The “on” rate of bombykol binding to BmorPBP is very small when compared, for example, to the “on” rates for the binding of fatty acids to their binding proteins (kon ≈ 10 μM–1·s–1) (27). However, the small rate is compensated for by high concentrations (10 mM) of PBP in the sensillar lymph (28). Under these physiological conditions, the t1/2 of the unbound bombykol is only ≈1 ms. A fast uptake of pheromone is important for the fast rise of the receptor potential and a fast onset of the nerve impulse (11). On the other hand, the dissociation rate (based on a KD of 100 nM) is very slow (“off” rate, ≈0.007 s–1). This corresponds to a t1/2 of the ligand–protein complex of ≈100 s, an extremely slow process given the dynamics of the insect olfactory system. However, it has been strongly suggested that the release of pheromone is triggered by a pH-dependent conformational change (8, 18, 19, 22, 29), and this is a very fast process (t1/2 ≈ 9 ms). The data presented here thus support the hypothesis that PBPs are essential for the dynamics of the insect olfactory system. This notion is supported by voltage-clamp recordings of a sexpheromone receptor from the silkmoth expressed in Xenopus laevis oocytes (9). The latency of response and the steepness of the rise (and the decay) of potential was very slow in this heterologous system, which was most likely devoid of PBPs.
We have developed a facile binding assay that does not require the use of radiolabeled ligands, permits the evaluation of binding under various conditions of ionic strength, pH, and temperature, and can even be used in competitive assays. The assay can be used as a low-throughput protocol for ligand fishing, i.e., the screening of potential insect attractants and/or repellents.
Previously, we demonstrated that model anionic membranes mimic the pH-dependent conformational changes in BmorPBP that lead to low binding affinity at low pH (8). In other words, interaction of BmorPBP at high bulk pH with negatively charged membranes (i.e., localized low pH) significantly decreases binding affinity (8). In addition, negatively charged surface coats have been found on the pore tubules and dendritic membranes of male moth olfactory hairs by application of cation markers (30–32). This finding led to the hypothesis that interactions of PBP–pheromone complexes with dendritic membrane surfaces are physiologically relevant for the release of the pheromone (3–5, 33). Kowcun and collaborators (17) have suggested that the calculated K+ concentration near a membrane counterbalances the decrease in pheromone-binding affinity caused by the decrease in pH (17). We have already demonstrated that the presence of physiological concentrations of KCl (34, 35) reduces the effect of model membranes (see figure 7 of ref. 8), but the interaction with negatively charged membranes in the presence of 170 mM KCl was still comparable to the effect of lowering the pH to 5. We have now tested Kowcun's hypothesis with the newly developed cold binding assay. BmorPBP showed high and low binding affinities at high and low pH values (Fig. 3B), respectively, when the binding was measured at either a physiological (170 mM) or an extremely high (500 mM) concentration of KCl. These data further support the finding that, under physiological conditions, a decrease in pH (such as that caused by a negatively charged dendritic membrane) leads to the release of bombykol from the BmorPBP–pheromone complex.
The existence of a C-terminal α-helix occupying the binding cavity in the acidic form of BmorPBP (18) suggested that the ligand release is triggered by an intramolecular rearrangement. That bombykol binds to a truncated mutant (BmorPBPΔP129-V142) at both high and low pH suggests that the protein folds and forms a similar or identical binding cavity, even without the extended C-terminal segment. More importantly, this binding confirms that, indeed, the C-terminal α-helix is essential for preventing binding at low pH.
There is growing evidence that bombykol is ejected from the binding cavity by the molecular rearrangement leading to occupation of the binding cavity. However, it is not yet known how bombykol enters the binding pocket. Because the binding ability of BmorPBP was not affected by replacing Trp-37 by Phe or Ala, this residue is unlikely to be involved in the uptake of pheromone, as previously suggested. Tryptophan mutations showed, however, that both Trp-37 and Trp-127 contribute to the total fluorescence emission, with the contribution of Trp-127 being smaller and having the opposite pH-dependence to that of Trp-37.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institute of Allergy and Infectious Diseases, National Institutes of Health (1U01AI058267-01); the National Science Foundation (0234769); the National Research Initiative of the U.S. Department of Agriculture (USDA) Cooperative State Research, Education, and Extension Service (2003-35302-13468); the USDA Agricultural Research Service Chemical Affecting Behavior Laboratory (58-1275-1-042); the University of California Systemwide Mosquito Research Program; the California Pistachio Commission; and the Almond Board of California.
Author contributions: W.S.L. designed research; W.S.L., A.M.C., Y.I., V.P.C., M.L.E., T.I.M., and J.M.T. performed research; W.S.L. analyzed data; and W.S.L., Y.I., M.L.E., and T.I.M. wrote the paper.
Abbreviations: PBPs, pheromone-binding proteins; BmorPBP, PBP from the silkmoth; BmorPBPA, acidic form of BmorPBP; BmorPBPB, basic form of BmorPBP; LC-ESI, liquid chromotography electrospray ionization.
References
- 1.Murlis, J., Willis, M. A. & Cardé, R. T. (2000) Physiol. Entomol. 25, 211–222. [Google Scholar]
- 2.Vogt, R. G., Riddiford, L. M. & Prestwich, G. D. (1985) Proc. Natl. Acad. Sci. USA 82, 8827–8831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leal, W. S. (2003) in Insect Pheromone Biochemistry and Molecular Biology, The Biosynthesis and Detection of Pheromones and Plant Volatiles, eds. Blomquist, G. J. & Vogt, R. G. (Elsevier, London), pp. 447–476.
- 4.Leal, W. S. (2005) in Semiochemicals in Pest Management and Alternative Agriculture, eds. Petroski, R. J., Tellez, M. R. & Behle, R. W. (Oxford Univ. Press, New York), in press.
- 5.Leal, W. S. (2005) Top. Curr. Chem. 240, 1–36. [Google Scholar]
- 6.Vogt, R. G. & Riddiford, L. M. (1986) in Mechanisms in Insect Olfaction, eds. Payne, T. L., Birch, M. C. & Kennedy, C. E. J. (Oxford Univ. Press, New York), pp. 201–208.
- 7.Maïbèche-Coisne, M., Nikonov, A. A., Ishida, Y., Jacquin-Joly, E. & Leal, W. S. (2004) Proc. Natl. Acad. Sci. USA 101, 11459–11464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wojtasek, H. & Leal, W. S. (1999) J. Biol. Chem. 274, 30950–30956. [DOI] [PubMed] [Google Scholar]
- 9.Sakurai, T., Nakagawa, T., Mitsuno, H., Mori, H., Endo, Y., Tanoue, S., Yasukochi, Y., Touhara, K. & Nishioka, T. (2004) Proc. Natl. Acad. Sci. USA 101, 16653–16658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johnson, K. A. (2003) in Kinetic Analysis of Macromolecules, ed. Johnson, K. A. (Oxford Univ. Press, New York).
- 11.Kaissling, K.-E. (2001) Chem. Senses 26, 125–150. [DOI] [PubMed] [Google Scholar]
- 12.Vogt, R. G. & Riddiford, L. M. (1981) Nature 293, 161–163. [DOI] [PubMed] [Google Scholar]
- 13.Maida, R., Steinbrecht, A., Ziegelberger, G. & Pelosi, P. (1993) Insect Biochem. Mol. Biol. 23, 243–253. [Google Scholar]
- 14.Campanacci, V., Krieger, J., Bette, S., Sturgis, J. N., Lartigue, A., Cambillau, C., Breer, H. & Tegoni, M. (2001) J. Biol. Chem. 276, 20078–20084. [DOI] [PubMed] [Google Scholar]
- 15.Kaissling, K.-E., Klein, U., de Kramer, J. J., Keil, T. A., Kanaujia, S. & Hemberger, J. (1985) in Molecular Basis of Nerve Activity. Proceedings of the International Symposium in Memory of D. Nachmansohn, eds. Changeux, J. P. & Hucho, F. (de Gruyter, Berlin), pp. 173–183.
- 16.Du, G. & Prestwich, G. D. (1995) Biochemistry 34, 8726–8732. [DOI] [PubMed] [Google Scholar]
- 17.Kowcun, A., Honson, N. & Plettner, E. (2001) J. Biol. Chem. 276, 44770–44776. [DOI] [PubMed] [Google Scholar]
- 18.Horst, R., Damberger, F., Luginbuhl, P., Guntert, P., Peng, G., Nikonova, L., Leal, W. S. & Wüthrich, K. (2001) Proc. Natl. Acad. Sci. USA 98, 14374–14379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, D., Damberger, F. F., Peng, G., Horst, R., Guntert, P., Nikonova, L., Leal, W. S. & Wüthrich, K. (2002) FEBS Lett. 531, 314–318. [DOI] [PubMed] [Google Scholar]
- 20.Leal, W. S., Nikonova, L. & Peng, G. (1999) FEBS Lett. 464, 85–90. [DOI] [PubMed] [Google Scholar]
- 21.Scaloni, A., Monti, M., Angeli, S. & Pelosi, P. (1999) Biochem. Biophys. Res. Commun. 266, 386–391. [DOI] [PubMed] [Google Scholar]
- 22.Sandler, B. H., Nikonova, L., Leal, W. S. & Clardy, J. (2000) Chem. Biol. 7, 143–151. [DOI] [PubMed] [Google Scholar]
- 23.Honig, B. & Yan, A.-S. (1995) Adv. Protein Chem. 46, 27–58. [DOI] [PubMed] [Google Scholar]
- 24.Bette, S., Breer, H. & Krieger, J. (2002) Insect Biochem. Mol. Biol. 32, 241–246. [DOI] [PubMed] [Google Scholar]
- 25.Mohanty, S., Zubkov, S. & Groneborn, A. M. (2004) J. Mol. Biol. 337, 443–451. [DOI] [PubMed] [Google Scholar]
- 26.Mohanty, S., Zubkov, S. & Groneborn, A. M. (2004) J. Mol. Biol. 338, 1037. [DOI] [PubMed] [Google Scholar]
- 27.Richieri, G. V., Ogata, R. T. & Kleinfeld, A. M. (1996) J. Biol. Chem. 271, 11291–11300. [DOI] [PubMed] [Google Scholar]
- 28.Klein, U. (1987) Insect Biochem. 17, 1193–1204. [Google Scholar]
- 29.Damberger, F., Nikonova, L., Horst, R., Peng, G., Leal, W. S. & Wüthrich, K. (2000) Protein Sci. 9, 1038–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keil, T. A. (1987) Ann. N.Y. Acad. Sci. 510, 403–405. [Google Scholar]
- 31.Keil, T. A. (1984) Zoomorphologie 104, 147–151. [Google Scholar]
- 32.Keil, T. A. (1984) Tissue Cell 16, 705–717. [DOI] [PubMed] [Google Scholar]
- 33.Leal, W. S. (2000) Biochem. Biophys. Res. Commun. 268, 521–529. [DOI] [PubMed] [Google Scholar]
- 34.Kaissling, K.-E. & Thorson, J. (1980) in Receptors for Neurotransmitters, Hormones and Pheromones in Insects, eds. Sattelle, D. B., Hall, L. M. & Hildebrand, J. G. (Elsevier Biomedical, Amsterdam), pp. 261–282.
- 35.Kaissling, K.-E. (1995) in Experimental Cell Biology of Taste and Olfaction. Current Techniques and Protocols, eds. Spielman, A. I. & Brand, J. G. (CRC, Boca Raton, FL), pp. 361–377.
- 36.Koradi, R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graphics 14, 51–55. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.