Pontocerebellar hypoplasias (PCHs) are a genetically and clinically heterogeneous group of autosomal recessive inherited neurodevelopmental disorders. In the past decade, whole-exome sequencing (WES) has led to the identification of new genes, allowing the recognition of at least 10 different PCH types with broad and overlapping phenotypes.1
PCH type 9 (PCH9) (MIM 615809) is caused by homozygous mutations in the adenosine monophosphate deaminase 2 (AMPD2) gene.
AMPD2 plays an evolutionary conserved role in purine nucleotide metabolism by regulating the guanine nucleotide biosynthesis and protein translation.2 Patients typically present with severe developmental delay, microcephaly, axonal neuropathy, and epilepsy. Neuroimaging in patients with PCH9 shows a typical midbrain “figure of 8” appearance, callosal hypoplasia, and periventricular white matter involvement.3
Previous reports have demonstrated the pivotal role of AMPD2 during neurogenesis and showed a potentially rescue therapy in vitro by administration of purine precursors.2 Since the original report of 5 PCH9 families,2 only 6 additional patients from 2 families have been described.3,4 Furthermore, a homozygous frameshift mutation in the AMPD2 gene has been identified in 2 members of a consanguineous family affected by spastic paraplegia without PCH.5
We report a novel homozygous AMPD2 mutation in 3 siblings with severe PCH9 phenotype.
Clinical description.
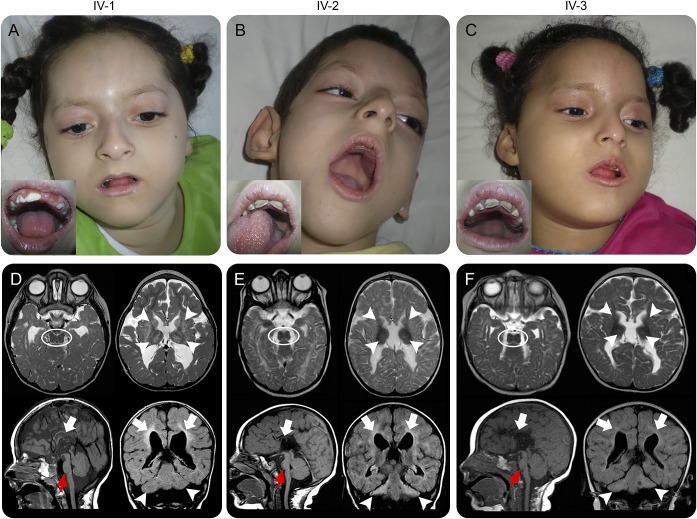
The probands are 2 girls (IV:1, aged 9 years and IV:3 aged 8 years) and 1 boy (IV:2, aged 7 years) born to first-cousin parents from the Middle East (figure e-1 at Neurology.org/ng). Family history was unremarkable. All individuals were born at term by normal delivery, following an uneventful pregnancy. Since the first months of life, developmental delay without any motor skills acquisition, drug-resistant focal and tonic-clonic seizures, and progressive spasticity were noticed. All patients shared microcephaly and common dysmorphic features, including sloping forehead, large and posterior rotated ears, upper lateral incisor agenesis, mottled and fragile teeth with multiple cavities, mandibular hypoplasia, and clinodactyly of the V fingers (figure 1, A–C). In all individuals, brain MRI showed PCH, severe callosal hypoplasia, leukoencephalopathy, and basal ganglia involvement (figure 1, D–F). Diffusion tensor imaging revealed marked thinning of the corticospinal tracts (figure e-2), and MR spectroscopy showed low N-acetyl aspartate in the thalamic region (data not shown) in patient II-3. Electrophysiologic studies performed in individual IV-2 did not reveal signs of axonal neuropathy. At the final follow-up, all patients showed cortical blindness and were using wheelchair or were bedridden. Their clinical and imaging details are summarized in table e-1.
Figure 1. Clinical and neuroradiologic features of patients carrying homozygous AMP2 mutations.
(A–C) Facial images demonstrate shared features of microcephaly, sloping forehead, large and posterior rotated ears, and mandibular hypoplasia. In insets, mottled teeth with multiple cavities. (D–F) Brain MRIs from each patient showing characteristic “figure of 8” midbrain appearance (dotted ovals) and small hyperintense basal ganglia and thalami (arrowheads) on axial T2-weighted images. Sagittal T1-weighted images show small pons (red arrows) and extremely severe callosal hypoplasia (empty arrows). Coronal FLAIR images reveal hypoplasia/atrophy of the cerebellar hemispheres (arrowheads) with relative sparing of the vermis. Leukoencephalopathy similar to periventricular leukomalacia present in all patients, with loss of white matter bulk, periventricular hyperintensity, and enlarged lateral ventricles (empty arrows). FLAIR = fluid-attenuated inversion recovery.
Genetic study.
We first excluded copy number and structural DNA variations by karyotyping and array–comparative genomic hybridization. We then performed WES on DNA samples of all family members, using a standard Illumina pipeline (e-Methods, tables e-2 and e-3) and identified a novel homozygous frameshift mutation c.495delG (p.R165fs*21) in the gene encoding for the adenosine monophosphate deaminase 2 enzyme (AMPD2, NM_001257360.1). Sanger sequencing showed that the mutation segregates according to a recessive model of inheritance (figure e-2). The institutional review board approved the use of human samples for this study.
Discussion.
AMPD2 encodes 1 of 3 adenosine monophosphate (AMP) deaminase enzyme homologs, which convert AMP to IMP. AMPD2 deficiency results in accumulation of adenosine nucleotides and depletion of guanine nucleotide, impairing the guanosine-5'-triphosphate–dependent initiation of protein translation, similarly to what was observed in transfer RNA splicing endonuclease complex genes, linked to other PCH types.2,6 Recently, AMPD2 mutant mice and Ampd2−/− mice have been associated with nephrotic syndrome and proteinuria in the absence of any brain abnormality.7 A neurodegenerative phenotype has been observed when both Ampd2 and Ampd3 are knocked out, suggesting a functional redundancy among AMP deaminase homologs. However, it remains to be elucidated whether humans carrying AMPD2 mutations may have renal involvement.
To date, 14 patients with PCH9 have been reported. All but 1 AMPD2 mutation reside within the conserved catalytic AMP deaminase domain2–4 (table e-1). The novel identified homozygous AMPD2 mutation (c.495delG p.R165RfsX21) lies outside the catalytic domain and is associated with a very severe phenotype as described for c.751C>T (p.R251W).4 Neuroimaging revealed the involvement of the basal ganglia and thalami and marked hypoplasia/atrophy of the corticospinal tracts, unraveling the anatomical basis of the characteristic midbrain “figure of 8” appearance and pontine flattening observed in PCH9 individuals. However, our patients showed peculiar facial dysmorphisms and teeth abnormalities that might be an additional feature associated with PCH9. Genetic studies did not show any additional homozygous variant that could readily explain these findings (table e-3). Further studies will clarify the phenotypic spectrum associated with AMPD2 mutations.
Supplementary Material
Footnotes
Supplemental data at Neurology.org/ng
Author contributions: A.A., P.S., V.C., and F.Z. contributed to the conception and design of the study and the acquisition, analysis, and interpretation of data. M.I., A.T., and V.N. performed NGS data analysis. F.P., A.O., M.S.V., R.S., C.M., and M.S. contributed to the acquisition, analysis, and interpretation of data. All authors contributed to the critical revision of the final version of the manuscript for important intellectual content.
Disclosure: A. Accogli, M. Iacomino, F. Pinto, A. Orsini, M. S. Vari, R. Selmi, and A. Torella report no disclosures. V. Nigro has served on the editorial board of Neuromuscular Disorders. C. Minetti and M. Severino report no disclosures. P. Striano has served on the scientific advisory board of the Italian Agency of the Drug (AIFA) and has served on the editorial board of Epilepsia. V. Capra reports no disclosures. F. Zara has received research support from the Italian Ministry of Health. Go to Neurology.org/ng for full disclosure forms.
Study funding: F.Z. has been supported by the Italian Ministry of Health (RF-2010-2314356). P.S. has been supported by the Italian Ministry of Health (GR-2011-02346749). A.A., V.C., M.I., A.T., V.N., F.P., A.O., M.S.V., R.S., C.M., and M.S. report no disclosures. The Article Processing Charge was funded by the authors.
References
- 1.Rudnik-Schoneborn S, Barth PG, Zerres K. Pontocerebellar hypoplasia. Am J Med Genet C Semin Med Genet 2014;166C:173–183. [DOI] [PubMed] [Google Scholar]
- 2.Akizu N, Cantagrel V, Schroth J, et al. . AMPD2 regulates GTP synthesis and is mutated in a potentially treatable neurodegenerative brainstem disorder. Cell 2013;154:505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marsh AP, Lukic V, Pope K, et al. . Complete callosal agenesis, pontocerebellar hypoplasia, and axonal neuropathy due to AMPD2 loss. Neurol Genet 2015;1:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marsh AP, Yap P, Tan T, et al. . A novel AMPD2 mutation outside the AMP deaminase domain causes pontocerebellar hypoplasia type 9. Am J Med Genet A 2017;173:820–823. [DOI] [PubMed] [Google Scholar]
- 5.Novarino G, Fenstermaker AG, Zaki MS, et al. . Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science 2014;343:506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Budde BS, Namavar Y, Barth PG, et al. . tRNA splicing endonuclease mutations cause pontocerebellar hypoplasia. Nat Genet 2008;40:1113–1118. [DOI] [PubMed] [Google Scholar]
- 7.Helmering J, Juan T, Li CM, et al. . A mutation in Ampd2 is associated with nephrotic syndrome and hypercholesterolemia in mice. Lipids Health Dis 2014;13:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.