Abstract
To better understand the phylogeny and evolution of mosquitoes, the complete mitochondrial genome (mitogenome) of Anopheles stephensi and An. dirus were sequenced and annotated, and a total of 50 mosquito mitogenomes were comparatively analyzed. The complete mitogenome of An. stephensi and An. dirus is 1,5371 bp and 1,5406 bp long, respectively. The main features of the 50 mosquito mitogenomes are conservative: 13 protein-coding genes (PCGs), two ribosomal RNA genes, 22 transfer RNA genes, positive AT-skew and negative GC-skew. The gene order trnA-trnR in ancestral insects is rearranged. All tRNA genes have the typical clover leaf secondary structure but tRNA Ser. The control regions are highly variable in size. PCGs show signals of purifying selection, but evidence for positive selection in ND2, ND4 and ND6 is found. Bayesian and Maximum Likelihood phylogenetic analyses based on all PCG nucleotides produce an identical tree topology and strongly support the monophyly of subgenera Cellia, Anopheles, Keterszia and Nyssorhynchus, the sister relationship of the subgenera Nyssorhynchus and Keterszia, and Cellia and Anopheles. The most recent ancestor of the genus Anopheles and Culicini + Aedini exited ~145 Mya ago. This is the first comprehensive study of mosquito mitogenomes, which are effective for mosquito phylogeny at various taxonomic levels.
Introduction
Mosquitoes (Diptera: Culicidae) are the most important group of medical and veterinary insects, and they are world-widely distributed in temperate and tropical regions1. The females of many mosquito species may transmit devastating pathogens and/or parasites through blood-sucking, including malaria, dengue, yellow fever, and encephalitis, which causes millions of human deaths every year2. Due to the medical importance, an accurate and reliable taxonomy and phylogeny construction of mosquito species are essential for our understanding of the ecology, life history strategy and disease transmission efficiency, and the design and implementation of effective control measures of mosquitoes.
Morphology-based classification is time-consuming and not always sufficient for identification of closely related species or cryptic species with overlapping geographical distributions. Therefore, multidisciplinary approaches, such as morphological characters, isoenzyme, and various molecular markers, have been adopted for mosquito taxonomy and phylogeny3–15. Currently, the family Culicidae is classified into two recognized subfamilies, Anophelinae and Culicinae. Subfamily Anophelinae harbors three genera (485 species) and Culicinae has 109 genera that are further divided into 11 tribes (3061 species)1. Though many efforts have been made on the phylogeny of genus Anopheles or more broadly across the family Culicidae, topological conflicts and/or discrepancies were frequently recovered from different datasets and inferring methods. Most previous studies supported that subfamily Anophelinae was monophyletic and formed the basal lineage of other Culicidae, and genus Chagasia presented a basal lineage of other Anophelinae species1, 16, 17. However, the phylogenetic relationships of genus Bironella, subgenera Lophopodomyia and Stethomyia within this subfamily were still problematic5. Within genus Nysorhynchus, the complete taxonomic picture for Albitarsis Complex is not yet clear, because there are conflicting tree topologies18. The An. punctulatus (AP) Group of mosquitoes, the major vectors of malaria and filariasis in the South West Pacific areas, currently contains at least 13 sibling species19. However, their phylogeny are not well resolved and sometimes are contradictory20, 21. Furthermore, a recent molecular phylogeny based on mitochondrial gene COI revealed that subgenera Anopheles and Pyretophorus were not monophyletic, because An. sinensis (Anopheles subgenus) and An. epiroticus (Pyretophorus series) were claded into subfamily Culicinae and Neomyzomyia series, respectively22. This result is a real challenge to the traditional taxonomy at the subgenus level. Although considerable advances in our understanding of phylogenetic relationships of mosquito lineages, the definition and identification of a specie or cryptic species complex are still problematic or remain limited.
Complete mitochondrial genome (mitogenome) has been widely used for molecular evolution, phylogenetics, phylogeography and population genetics due to its maternal inheritance, simple genome organization, and the ability to provide more phylogenetic information than individual genes23, 24. Using the complete mitogenome sequences in insect phylogeny has produced some remarkable results in Diptera25, Orthoptera26, Hymenoptera27 and Heteroptera28. However, it frequently produced high incongruence with the nuclear and morphological data to elucidate the relationships between orders or at higher levels in some insect groups26–28. Since the first mosquito mitogenome of An. gambiae was published29, the availability of mosquito mitogenome sequences is growing as a result of recent advances in sequencing strategies. However, only few studies focused on phylogeny using the complete genome sequences. Nevertheless, inconsistent phylogenies were also observed, largely due to different taxa sampling, dataset types and inferring methods. Phylogeny of 26 Anopheles species based on Maximum Likelihood analysis of the concatenated nine protein-coding genes recovered An. atroparvus + An. quadrimaculatus as a monophyletic clade30. More robust studies including more species will be really needed to better resolve this issue by using more reasonable molecular markers and rational inferring methods. Hua et al.31 recovered the monophyly of series Neomyzomia + (Pyretophorus + Myzomyia), whereas a more recent study supported the monophyly of series Pyretophorus + (Neomyzomia + Myzomyia)30. Therefore, how mitochondrial datasets and inferring methods affect the phylogenetic reconstruction of mosquitoes need to be further evaluated.
In the present study, we sequenced and annotated the complete mitogenomes of An. stephensi and An. dirus, the subgenus Cellia, which are of crucial importance in malaria transmission throughout the South-East Asian and Southern China. More importantly, we systematically compared 50 mosquito mitogenomes with aims to answer the following questions: i) does the positive selection act on protein-coding genes in different species or ecotypes of species at the mitogenomic level? ii) how do the mitochondrial datasets and inferring methods affect the phylogenetic reconstruction? iii) when did the major lineages in family Culicidae diverge during the evolution?
Results and Discussions
General features of An. stephensi and An. dirus mitogenomes
The complete mitogenome of An. stephensi (KM 899887) and An. dirus (KM 899888) is a typical circular, double-stranded molecule with the length of 15,371 bp and 15,406 bp, respectively. Each contains a conserved set of 37 genes, including 13 protein-coding genes (PCGs), large and small ribosomal genes (rrL and rrS), 22 transfer RNA (tRNA) genes and a control region (also known as the AT-rich region) (Fig. 1). 23 and 14 genes are located on the majority strand (J-strand) and the minority strand (N-strand), respectively (Fig. 1). The gene order of trnR-trnA is the same as in other mosquito mitogenomes, which is a reverse order of trnA-trnR in ancestral insect mitogenome32. The gene order is a feature of mitogenome that can provide important evidence to establish evolutionary relationships among taxa at low and/or high taxonomic level33–35. More mosquito mitogenome sequences will be helpful to clarify whether this inversion pattern represents an evolutionary event in family Culicidae.
Figure 1.
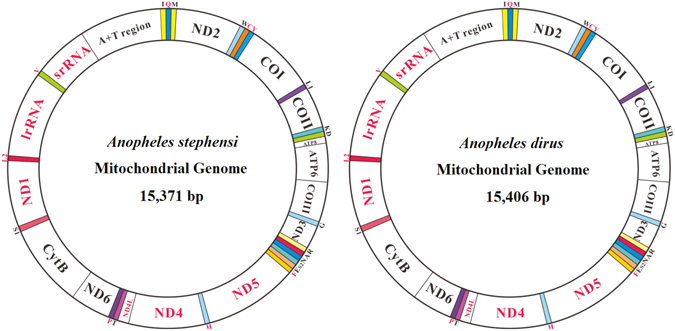
Mitochondrial genome structure of Anopheles stephensi and An. dirus. The color-filled blocks indicate tRNAs, while the un-filled white blocks denote protein-coding genes, rRNA and control regions. The protein-coding genes, rRNA and control regions with black name are located on majority strand, whereas those with red names are on minority strand.
Comparative analyses of 50 mosquito mitogenomes
Like other insect mitogenome sequences, base composition of mosquito mitogenomes is heterogeneous among species (Fig. 2; Supplementary Table 3). The AT content of the complete sequences excluding the control regions ranges from 76.7% for An. christyi to 78.7% for Ae. notoscriptus. All mitogenomes exhibit a positive AT-skew with the range of variation from 0.01 for subgenus Culex and Ar. subalbatus to 0.044 for An. christyi. However, all mitogenomes display negative GC-skews ranging from −0.2 for Ae. aegypti to −0.13 for An. punctulatus in Papua New Guinea (PNG)(Fig. 2). Insect mitogenomes exhibit a distinct strand-based nucleotide composition bias, which was thought to be due to either replication-mutation or transcription-associated mutation24.
Figure 2.
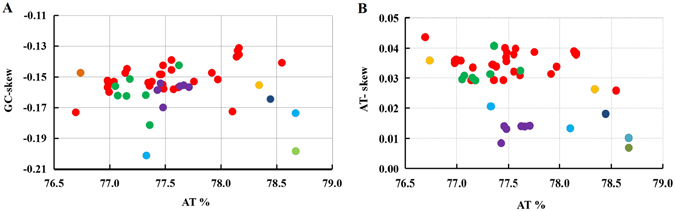
Comparative analysis of AT content, AT- and GC-skew in 50 mosquito mitogenomes. Red, orange, green, light blue, purple, blue and light green dots represent mitogenomes from the subgenus Cellia, Anopheles, Nyssorhynchus, Aedes, Culex, Kertezia and Armigeres, respectively.
AT content for all PCGs ranges from 75.3% for An. christyi and 79.1% for An. minimus. ATN, TTG, GTT and GTG are used as canonical start codons for invertebrate mitogenomes36. Most mosquito PCGs use ATN as the start codon, while gene ND5 in 39 species uses GTG as the start codon, which has been considered as a common feature across various organisms37. All COI genes use a TCG start codon, which is non-canonical start codon for mitochondrial genes. Non-canonical start codon TCG in COI gene was previously observed in Polystoechotes punctatus and Ascaloptynx appendiculatus 38. Stop codons for almost all PCGs are invariable complete and incomplete stop codon T or TA, which was observed in all metazoan mitogenomes39 and can be completed to the full stop codon TAA through post-transcriptional modification40. The third codon position has a higher AT content than the first or the second position. Purifying selection against deleterious mutations is expected to be less severe on third codon position, thus higher AT content is possibly associated with the bias usage of synonymous codon.
Codon usage bias can be caused by many factors, including gene function, recombination, mutation bias, GC composition, gene length, codon position, environmental stress, population size and others41. Codons ending with A or U are used more frequently than those ending with CG or GC, which is a common feature in many dipteran insects. Effective number of codon (ENC) values for all PCGs range from 24.4 to 43.9, reflecting a strong codon bias. ENC-GC3 plot showed that most of the values are not close to the standard curve (Fig. 3A; Supplementary Fig. 1), indicating that not only mutation but also other factors, such as natural selection and/or translational selection, are likely to be involved in shaping the codon bias in mosquito mitogenomes. The neutrality plot revealed that all PCGs have a narrow GC3 distribution and there is no significant correlation between GC12 and GC3 (Y = −0.028x + 0.155, R 2 = 0.001, P = 0.739) (Fig. 3B). Our result addresses an important selection process that the codon bias is mainly dominated by natural selection, and mutational pressure only lightly affects the usage bias.
Figure 3.
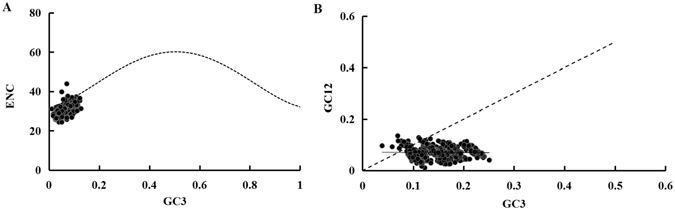
Evaluation of codon bias in 50 mosquito mitogenomes. (A) ENC-GC3 plot. The standard curve shows the functional relation between ENC and GC3 under mutation pressure without selection. Points on or close to the curve indicate codon use bias caused by mutation pressure; otherwise, they are affected by natural selection or other factors. (B) Neutrality plot of GC12 versus GC3. GC12 indicates the average value of GC content in the first and second position of the codons (GC1 and GC2); GC3 refers to the GC content in the third position.
Hierarchical clustering of codon frequencies showed that about 50% codons are used in a bias pattern (Supplementary Fig. 2). However, the clustering result does not accurately reflect the phylogeny of mosquitoes inferred from data PCG123 below. This result will contribute to our understanding of codon usage patterns and provide better insights into their evolutionary relationships.
Most tRNAs can be folded into the typical clover-leaf structure except tRNASer, in which the dihydrouracil (DHU) arm is absent. The lack of a DHU arm in this gene has been commonly observed across metazoan mitogenomes42 (Supplementary Fig. 3). Notably, tRNALeu(CUN) uses UUR as the anticodon rather than CUN, which might be driven by the natural selection to adapt to the codon use bias. It is worthy to note that the terminal CCA is absent in all tRNAs, which is speculated to be edited by CCA-adding enzymes. Pairwise genetic distance (p-distance, pDis and maximum- likelihood distance, MLdis) and base difference (BDps) allows the level of conservation and the possible understanding of the substitution patterns in mosquito tRNAs. Our results showed that the tRNA Val is the most conserved one (BDps = 0.0945, MLdis = 0.007 ± 0.001 and pDis = 0.009 ± 0.003), whereas tRNA Leu(CUN) is the most variable one (BDps = 9.6, MLdis = 0.18 ± 0.124 and pDis = 0.13 ± 0.0083) (Supplementary Table 4). A total of 1472 base substitutions, ranging from 20 in tRNA Val to 245 in tRNA Leu(CUN), were detected and mapped on their secondary structures (Supplementary Fig. 3). Most variations including base substitutions and indels were found in loops, the TΨC (thymidine pseudouridine cytidine) and DHU arms. Due to the substitution heterogeneity, a perfect correspondence does not exist between the percentage of stem positions involved in the base change and the global percentage of base substitution in a single tRNA. A total of 645 BCNS (base change number in a stem) including 347 FCBC (fully compensatory base change), 51HCBC (hemi-compensatory base change), and 247 mismatches were identified in all tRNAs (Supplementary Fig. 3). Due to suffering greater mutation pressures, mitochondrial tRNAs accumulated more deleterious mutation relative to their nuclear tRNA counterparts43. Although mitochondrial mRNA editing was observed in some eukaryote taxa44, it has not been reported in mosquito mitogenomes. Therefore, we speculated that the mismatched stems could be edited by template-dependent RNA editing mechanism.
The control region, known for the initiation of replication in vertebrates and invertebrates45, is located between rrnS and tRNA Ile with a varying size. The conserved motif 5′CCCCTA3′ followed by a 15–27 bp poly-T stretch was identified in the control regions of 31 mosquito mitogenomes. This motif was putatively involved in the origin of the light strand replication. Ae. notoscriptus, An. dirus China, An. cracens, and An. dirus A use the motif 5′ATTGTA3′, whereas Ae. albopictus uses the motif 5′TTACTA3′. In 24 mosquito mitogenomes, tandem repeat sequences harbor two repeat unit types and are interrupted by a non-coding region (Fig. 4). Overall, control regions of mosquito mitogenomes showed a distinct sequence and structural characteristic, which may be taxon-specific and can be potentially used as a genetic marker for evolutionary and/or population genetic studies of mosquito species.
Figure 4.

The structural organization of the control region of 30 fully completed mosquito mitogenomes. The location and copy number of tandem repeats are shown by colored circle. Non-repeat regions are indicated by colored box with sequence size inside.
Evidence of positive selection on protein-coding genes
To evaluate the selective pressures acting on mosquito mitogenomes, pairwise analyses of the non-synonymous (Ka) and synonymous (Ks) substitution ratio (Ka/Ks) were performed. The Ka/Ks ratio ranges from 0.039 ± 0.011 in COI to 0.295 ± 0.016 in ND6, and displays the following order: COI < ATP6 = COIII < CtyB < COII < ND1 < ND3 < ND4l < ND5 < ND4 < ND2 < ATP8 < ND6 (Fig. 5A). This result indicated that 13 PCGs in mosquito mitogenomes were globally evolving under negative constraints. Purifying selection seems to be particularly strong in Complex IV (COI, COII and COII) and Complex III (CtyB), in which Ka/Ks ratios are low and narrow (Fig. 5A). Indeed, due to their crucial roles in cellular respiration, PCGs have been expected to be mostly under purifying selection. Our results further support that COI is a suitable barcoding marker for mosquito phylogeny at some taxonomic levels. Subunits of NADPH complex I yielded higher average Ka/Ks values than other PCGs, which can be explained by relaxed purifying selection (here defined by Ka/Ks below one but significantly higher than other genes due to less conservative evolutionary constraints) or positive selection on them. The fixed effects likelihood (FEL) analyses (P < 0.05) revealed that the subunits with the highest percentage of codons under the negative selection presented as the following order: COI > CytB > COIII > COII > ND2 > ND4 > ND5 > ND4l > ND3 > ND6 > ND1 > ATP6 > ATP8 (Fig. 5B). Both analyses confirmed that purifying selection might be the major selection constraint for the maintenance of the complete mitogenome.
Figure 5.

Positive selection estimated by FEL site-by-site analyses and pairwise divergence Ka/Ks ratio across mitochondrial protein-coding genes in 50 mosquito mitogenomes. (A) Ratios estimation. Box plot for pairwise divergence of Ka/Ks ratio for each one of the mitochondrial subunits. (B) Percentage of sites under positive selection.
Given strong evolutionary constraints, it does not exclude the possibility for positive selection acting on single gene or few codon positions. From branch-site model test, the mixed effects model of evolution (MEME) analysis (posterior probability ≥ 95%) was able to identify a few positive selected codons in gene ND1, ND4L, ND5, ND6 and COX1 (Fig. 6). Therefore, changes in mitochondrial energy transduction system may play a crucial role in the evolution of mosquitoes. Since the mosquitoes occupy a variety of climates and habitants, the polymorphisms of some of OXPHO genes may facilitate climatic adaptions, long distance host-seeking and the host-parasites co-evolution. However, no positive selection acting on the protein-coding genes of four Anopheles mosquito mitogenomes was detected46. This result is largely due to few sequences available, very low sequence divergence, or lack of power of the analysis.
Figure 6.
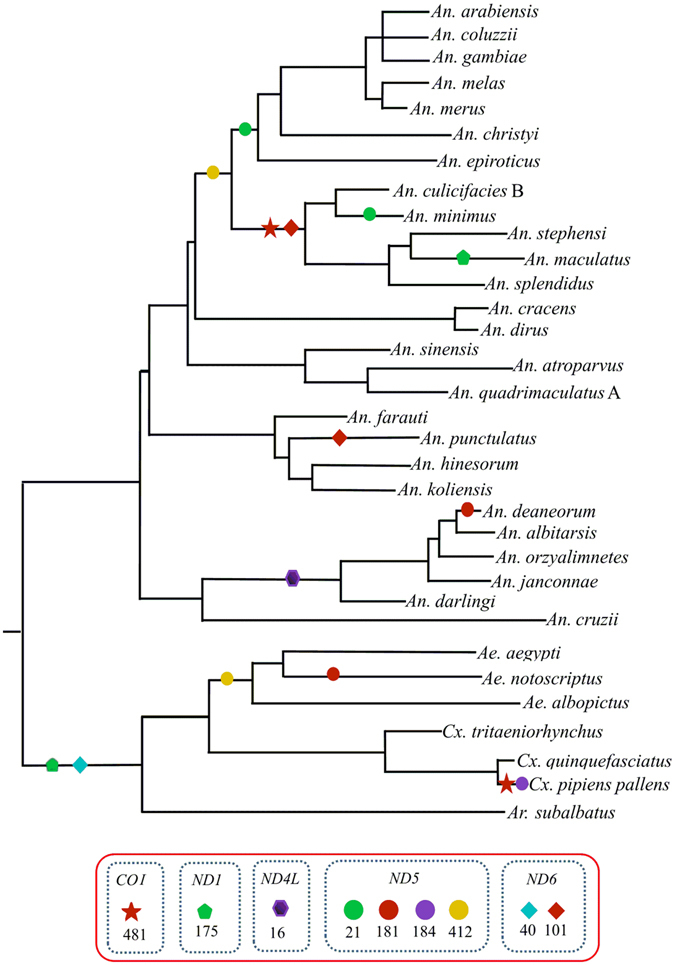
Adaptive evolution analyses based on MEME branch-site selection model. Identification of the maximum likelihood tree branches to test the adaptive evolution of each of the 13 PCGs in 34 representative mosquito species. The putatively positive selection sites with posterior probabilities ≥95% (p-value < 0.05) were marked with different symbols on the represented neighbor-joining tree of ATP6.
Substitution saturation analysis
Plots of transversion and transition against the genetic distance showed a linear relationship for all datasets (Supplementary Fig. 4). The index of substitution saturation (Iss for PCG1 = 0.153, Iss for PCG2 = 0.071, and Iss for PCG3 = 0.176) was significantly lower (P = 0.000) than the critical value of the index of saturation (Iss.c for PCG1 = 0.831, Iss.c for PCG2 = 0.831 and Iss.c for PCG3 = 0.843), implying that the nucleotides of 50 mosquito mitogenomes experienced little substitution saturation, but they are qualified for the phylogenetic reconstruction.
Phylogeny of 50 mosquito mitogenomes
The phylogenetic analysis based on Bayesian inference (BI) and Maximum- likelihood (ML)-PCG123 produced an identical tree topology (Fig. 7). However, disparities between different datasets and inferring approaches were also evident (Supplementary Figs 5–8). The third codon position has proved to be less restricted by purifying selection and easily saturated with substitutions47, and therefore it was usually excluded in the phylogenetic analysis48. In the present study, however, the removing of the third codon position did not generate a better reliable tree topology as well as the concatenated amino acid sequence. In addition, the phylogenetic analyses based each individual gene also failed to provide identical tree topologies with these based on the concatenated 13 PCG123 (data not shown).
Figure 7.
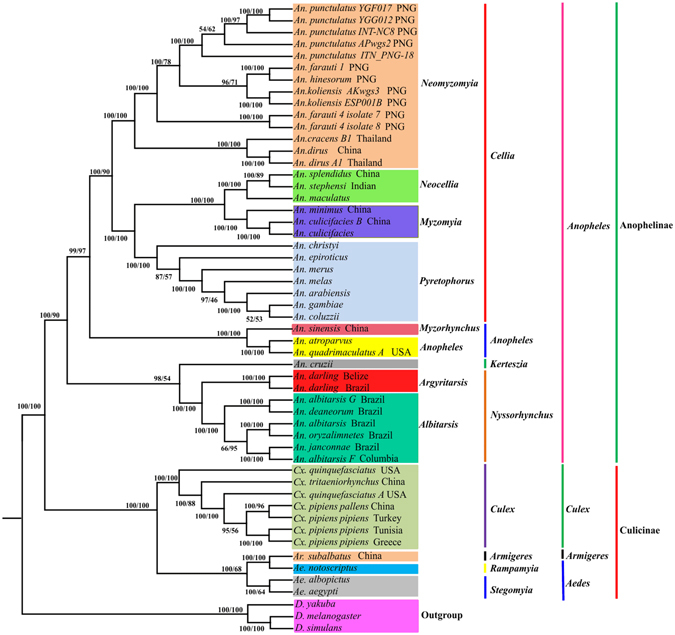
Reconstruction of phylogenetic tree determined by RAxML rapid bootstrap and Bayesian inference methods based on all sites (PCG123) of the concatenated protein-coding genes of 50 mosquito mitogenomes. Bayesian posterior probabilities (BPP) (left) and bootstrap support values (BP) are shown at relevant branches of the BI tree.
Anophelinae Subfamily
The monophyly of Anophelinae subfamily containing four Anopheles subgenera (Cellia, Anopheles, Keterszia and Nyssorhynchus), and the sister relationships of two subgenera Cellia and Anopheles were recovered with a high statistical support, which is consistent with the traditional taxonomic views based on morphology and molecular phylogenetic studies1, 5, 49.
Subgenus Cellia
Two major clades (Neomyzomyia, and Pyretophorous + (Myzomyia + Neocellia)) were clearly recognized in BI-tree and ML-PCG123 tree. As shown in other studies, Neomyzomyia, Pyretophorous, Myzomyia and Neocellia were shown to be monophyletic groups based on ribosomal (18S RNA and 28S RNA) and individual mitochondrial genes (COI and COII)5. Contrary to our findings, the relationships within Cellia were poorly resolved by the morphological analysis1.
Phylogenetic analysis of morphological and molecular data indicated that the AP Group contains at least 13 sibling species19. However, their evolutionary relationships were not well resolved and sometimes were contradictory20, 21. A phylogenetic study using 18S RNA and maximum parsimony inferring method recovered An. koliensis as the basal lineage of Farauti clade, and An. farauti 4 as the basal lineage of Punctulatus clade50. Logue et al.51 extended molecular characterizations by mitogenome analysis and concluded that An. farauti 4 was the most divergent, while An. farauti.s.s and An. hinesorum were most closely related to An. punculatus. The monophyly of the An. punctulatus Group was also recovered in BI-PCG123 and ML-PCG123 trees (Fig. 7) and formed the basal lineage of An. farauti 4, which further supported that AP group in PNG was colonized through a single migration event followed by speciation51. However, not all members of An. farauti-like species were clustered together because An. koliensis was consistently grouped with An. farauti and An. hinesorum.
Phylogenetic construction of An. gambiae Complex based on molecular markers was complicated, largely due to the degree of genetic similarity caused by the ancestral polymorphism and introgression. The sister relationship between An. gambiae and An. arabiensis was also confirmed in PCG123 data analysis. However, inversion phylogeny recovered An. gambiae and An. merus as sister taxa52. Our results revealed that An. christyi branched out earlier than An. epiroticus in BI- and ML-PG123, BI- and ML-AA (amino acid) trees, but in BI- and ML-PCG12 trees An. epiroticus branched out prior to An. christyi (Fig. 7; Supplementary Figs 5–8)
Subgenera Nyssorhynchus and Keterszia
The monophyly and sister relationship of the subgenera Nyssorhynchus and Keterszia was in agreement with previous studies based on morphological or molecular data (Fig. 7; Supplementary Fig. 7). However, it was worthy to note that Nyssorhynchus was recovered as the sister to the clade including subgenera Kerteszia and Stethomyia based on mitochondrial gene COI and COII, and 5.8S rRNA53. In the present study, An. cruzii was recovered as the basal lineage of genus Anopheles in BI-PCG12 tree, ML- and BI-AA trees (Supplementary Figs 5, 6 and 8), suggesting that these datasets were less phylogenetically informative. Only one Keterszia species was included in the present study, more mitogenomic sequences will be helpful to resolve the monophyletic status of Keterszia. The sister relationship between subgenus Cellia and Nyssorhynchus + Kerteszia was recovered by morphological data analysis47, but this relationship was not supported in their recent study1. Our BI- and ML-PCG123 phylogenetic trees strongly supported the sister relationship of (Nyssorhynchus + Kerteszia) and (Cellia + Anopheles), which was consistent with the phylogeny inferred from 1,085 orthologs in 18 mosquito genomes54. The discrepancies between different analyses are likely due to the number and selection of samples, different molecular markers and computerized methods, which varied significantly among previous studies.
For the subgenus Nyssorhynchus, many approaches have been tried to resolve the phylogeny of Albitarsis Complex, including morphology, behavior, alloenzyme and molecular analysis9, 18, 55–58. However, the complete taxonomic picture for Albitarsis Complex is not yet clear, because there are conflicting tree topologies. The monophyly of Albitarsis Complex was previously recovered based on gene ND6 and combined data sets58, ribosomal internal transcribed spacer 2 (ITS2) and rDNA59. However, this group was less well resolved in the white gene tree, which placed An. marajoara as a basal lineage to {An. albitarsis B (An. albitarsis [An. deaneorum 1, An. deaneorum 2])}58. An. albitarsis was recovered as a sister species to An. deaneorum in the white gene tree, but it was recovered as a sister to the remaining Albitarsis species in ND6 gene tree. Another study based on the concatenated sequences of COI + ND4 + ITS2 + D2 supported the relationship of An. albitarsis + [An. albitarsis B (formerly An. oryzalimnetes) + (An. marajoara + An. deaneorum)]56. Our BI- and ML-PCG123 trees supported the monophyly of Albitarsis Complex, but An. albitarsis G was recovered as a sister specie of An. deaneorum, and (An. albitarsis G + An. deaneorum) and formed the basal lineage of [(An. albitarsis + An. albitarsis) + (An. Janconnae (formerly An. albitarsis E) + An. albitarsis F].
Culicinae Subfamily
Morphological and molecular evidence indicated that Aedini was a monophyletic taxon60, 61. However, Aedini was not recovered as a monophyletic group by Wilerson et al.62. The monophyly of Aedini was supported in BI- and ML-PCG123 trees, and Ar. subalbatus (Armigeres subgenus) was recovered as a sister of Aedini genus. Our results also found that Pipiens Complex was paraphyletic due to the inclusion of Cx. quinquefasciatus A USA.
Divergence time estimation of mosquito species
Divergence time analysis based on PCG123 data revealed that the split date between subfamily Culicinae and Anophelinae was ~145.03 Mya, in the late Jurassic (Fig. 8). This estimation is in reasonable agreement with the report by Krzywinski et al.63, who speculated the divergence time between Aedes (Stegomyia) and Anopheles was ~146 Mya based on mitochondrial DNA sequences. However, Zhou et al.64 and Bertone et al.65 pushed backward the split date to ~122 Mya, and the time estimated by Moreno M et al.46 and Chen et al.66 was ~190 Mya (Early Jurassic) and 217.5 Mya (Late Triassic), respectively. The different evolutionary patterns of the molecular marker (nuclear DNA vs mitochondrial DNA), besides the algorithm and incomplete species sampling, partially explain the apparent incongruence between the estimations. Both Zhou et al.64 and Chen et al.66 estimated the divergence time using gene orthologous at genome level, whereas the study by Bertone et al.65 was aimed at deeper divergence within lower Diptera not within Culicidae and only a single molecular marker (28S rDNA) was used. Our estimation is also consistent with other evidences suggesting that mosquitoes likely originated in the Jurassic. The Anopheles radiation occurred during the early Cretaceous, and the split date between subgenera Anopheles and Cellia was estimated at ~90.41 Mya. This timescale inferred here is in agreement with the previous report ~90 Mya12, and ~93.6 Mya46. However, the last common ancestor of Anopheles was inferred in the Early Cretaceous (~113 Mya), and ~100 Mya based on 16 Anopheles genome54. Kerteszia and Nyssorhynchus are both distributed in South America, and was regarded as sister taxon. The divergence date of subgenera Nyssorhynchus was estimated at ~79 Mya46 or ~ 94 Mya51. Our estimation (~89.23 Mya) for Nyssorhynchus was comparable with those studies. Within the subgenus Cellia, the split date between Myzomyia (An. minimus + An. culicifacies) and Neocellia (An. stephensis) was ~61.94 Mya. The most recent ancestor of An. dirus from China and An. dirus from India was estimated at ~2.99 Mya.
Figure 8.
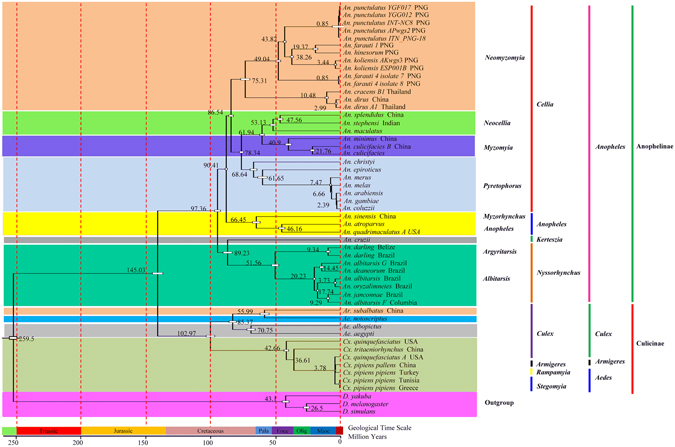
Evolutionary timescale for subfamily Anophelinae and Culicinae inferred from mtgenome PCG123 datasets based on BI tree. Numbers near the nodes indicate the average divergence time estimated (million years, Mya). In the geological time scale: Pala, Eoce, Olig and Mioc indicate Palaeocene, Eocene, Oligocene and Miocene, respectively.
Materials and Methods
Sample collection and DNA extraction
An. stephensi strain Hor (originally collected from India) and An. dirus (Hainan, China) were reared in the Third Military Medical University, China. Adult individuals were collected and stored at −80 °C. Total genomic DNA from a single adult was extracted using the modified sodium dodecyl-sulphate (SDS)/proteinase K method as described before67. The DNA was examined on 1% agarose gel and then used for PCR amplifications.
PCR amplification and sequencing
To amplify the entire mitogenome of An. stephensi and An. dirus, 18 pairs of primers were designed based on the known mitogenomes of mosquito species (Supplementary Table 1), and PCR amplifications were conducted in 50 μl reaction mixture, including 5 μL of 25 mM MgCl2, 5 μL of 10 × PCR buffer (Mg2+ free), 8 μL of dNTP (2.5 mM each), 2 μL of each primer (10 mM), 2.5 U of Taq DNA polymerase (Takara, Japan) and 2 μL template DNA. The amplification conditions were as follows: an initial denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 1 min, 48–55 °C (depend on different primer pairs) for 45 s, and 68 °C for 1 min, and a final extension at 72 °C for 10 min. All PCR products were separated on 1% agarose gel by electrophoresis, purified using a QIAquick Gel Extraction Kit (Qiagen, China) and then sequenced. The control region was cloned into pMD-19T vector and then transformed into chemical competent Escherichia coli DH5α cells. The positive clone was sequenced at least three times.
Mitogenome sequence assembly and analysis
Raw nucleotide sequences were trimmed to remove the low quality bases and assembled using DNAMAN V4.0 (Lynnon Biosoft). Annotations of the protein-coding genes, rrnL and rrnS gene were performed by a combination of BLAST searching and ORF Finder in GenBank, and MITOS68. Transfer RNA genes were identified using MITOS68, tRNAscan-SE69, and DOGMA70. The identification of tRNAArg and tRNASer, and the secondary structures of all tRNA genes were analyzed by comparison with the nucleotide sequences of other well-known mosquito tRNAs. Sequence motifs in the control region were identified using the Tandem Repeats Finder program71. AT- and GC-skew were calculated with the formula: AT skew = (A − T)/(A + T) and GC skew = (G − C)/(G + C)72. Codon usage bias was evaluated by calculating of effective number of codon (ENC) with the CodonW73. The relative synonymous codon usage (RSCU) was calculated by the DNAStar74, clustered and displayed using Cluster3.0 and Java TreeView1.2375, respectively.
Tracking positive selection events on protein-coding genes
To detect evidence of selection on each protein-coding gene, amino acid sequence alignments were independently conducted by MAFFET program76. Nucleotide sequences were aligned based on the alignment of amino acid sequences to maintain the reading frame using the RevTrans77. The pair-wise comparison of the ratios of non-synonymous substitutions (Ka) and synonymous (Ks) substitutions was conducted using DnaSP5.078. Further analyses were performed using the MEME (Mixed Effects Model of Evolution) method on Datamonkey server (www.datamonkey.org), which allows the rate of Ka/Ks to vary from site to site (fixed effect) or from branch to branch at a site (random effect). For the analysis, the best-fitting nucleotide substitution model for ND2 (model 012034), ND4 (model TrN93) and ND6 (model 011120) were predicted on Datamonkey server.
Sequence alignment, saturation and phylogenetic analyses
Two complete mitogenome sequences obtained in the present study, and 48 complete or nearly complete mosquito mitogenome sequences from NCBI or our unpublished data were used for phylogenetic analyses (Supplementary Table 2). Sequence data from three Drosophila species (D. melanogaster NC_001709, D. yakuba NC_001322, and D. simulans NC_005781) were included as the outgroup. Nucleotide and amino acid sequences of 13 protein-coding genes were separately aligned using MAFFT76. After removing the poorly aligned and divergent regions using Gblocks79, the individual alignment was then concatenated following their orders in the mitogenome with Sequence Matrix v1.7.680. Substitution saturation was tested by plotting the number of transitions and transversions against genetic divergence (GTR) using DAMBE81.
For phylogenetic analyses, three data types were used: 1) concatenated amino acid sequences of 13 protein coding genes; 2) all three nucleotides in codons of 13 protein-coding genes (PCG123 with 11192 nucleotides); 3) the 1st and 2nd nucleotide in codons of 13 protein-coding genes (PCG12 with 7462 nucleotides). The General Time Reversible (GTR) model incorporating invariant sites and a gamma distribution (GTR + I + G) for nucleotide datasets was selected by Modeltest82 using the Akaike Information Criterion (AIC), and the site-heterogeneous model GTR + CAT (CAT model, named due to classifying sites into categories) was used for amino acid sequences. Maximum likelihood analyses were conducted under the best model using RAxML83. Node support was estimated by analyzing 1000 bootstrap replicates. Bayesian inference was conducted using MrBayes84 and two independent runs with four chains (three heat and one cold) were performed simultaneously for 1,000,000 generations. The runs were stopped till the average deviation of split frequencies fall below 0.01. Stationarity is considered to be reached when estimated samples size (ESS) value is above 100 and potential scale reduction factor (PSRF) approach 1.0. The chains were sampled every 100 generations with the discard of the first 25% as burn-in. Posterior probabilities (PP) were calculated in a consensus tree. The topology of the best-scoring trees were visualized and edited in FigTree V1.4 (http://www.molecularevolution.org).
Divergence time Estimation
Estimation of divergence times among mosquitoes were calculated in BEAST1.8.385. The GTR + I + G substitution model, empirical base frequencies and speciation Yule model were applied as Tree prior. Two independent MCMC runs were each performed for a total of 1,000,000 generations with the first 25% burned-in under the uncorrelated lognormal relaxed clock and sampled every 1000 generations to estimate the divergence time. The split date (259.9 Mya) between Anopheles and Drosophila was used as the calibration63, 86.
Electronic supplementary material
Acknowledgements
This research was supported by the following, Par-Eu Scholars Program (20136666), The National Natural Science Foundation of China (31672363, 31372265), Coordinated Research Project of the International Atomic Energy Agency (18268), National Key Program of Science and Technology Foundation Work of China (2015FY210300), and Scientific and Technological Research Program of Chongqing Municipal Education Commission (KJ1600338).
Author Contributions
Bin Chen conceived and designed the research and experiments. Yi-Li Zou, Yi-Ran Ding, Wen-Yue Xu, Zhen-Tian Yan, Xu-Dong Li, Wen-Bo Fu and Ting-Jing Li performed mitochondrial sequence amplifications, assembly and annotation and/or collected samples used in this study. You-Jin Hao, Bin Chen and Yi-Li Zou performed the analysis and writing of the paper. All authors have read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
You-Jin Hao, Yi-Lin Zou and Bin Chen contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at doi:10.1038/s41598-017-07977-0
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Harbach RE, Kitching IJ. The phylogeny of Anophelinae revisited: inferences about the origin and classification of Anopheles (Diptera: Culicidae) Zoologica Scripta. 2015;45:34–47. doi: 10.1111/zsc.12137. [DOI] [Google Scholar]
- 2.Benelli G. Research in mosquito control: current challenges for a brighter future. Parasitology Research. 2015;114:2801–2805. doi: 10.1007/s00436-015-4586-9. [DOI] [PubMed] [Google Scholar]
- 3.Beebe NW, Cooper RD. Distribution and evolution of the Anopheles punctulatus group (Diptera: Culicidae) in Australia and Papua New Guinea. International Journal for Parasitology. 2002;32:563–574. doi: 10.1016/S0020-7519(01)00359-9. [DOI] [PubMed] [Google Scholar]
- 4.Krzywinski J, Wilkerson RC, Besansky NJ. Toward understanding Anophelinae (Diptera, Culicidae) phylogeny: Insights from nuclear single-copy genes and the weight of evidence. Systematic Biology. 2001;50:540–556. doi: 10.1080/106351501750435095. [DOI] [PubMed] [Google Scholar]
- 5.Sallum MAM, et al. Phylogeny of Anophelinae (Diptera: Culicidae) based on nuclear ribosomal and mitochondrial DNA sequences. Systematic Entomology. 2002;27:361–382. doi: 10.1046/j.1365-3113.2002.00182.x. [DOI] [Google Scholar]
- 6.Collucci E, Sallum MA. Cladistic analysis of the subgenus Anopheles (Anopheles) Meigen (Diptera: Culicidae) based on morphological characters. Memorias do Instituto Oswaldo Cruz. 2007;102:277–271. doi: 10.1590/S0074-02762007005000026. [DOI] [PubMed] [Google Scholar]
- 7.Wilkerson RC, Reinert JF, Li C. Ribosomal DNA ITS2 sequences differentiate six species in the Anopheles crucians complex (Diptera: Culicidae) Journal of Medical Entomology. 2015;41:392–401. doi: 10.1603/0022-2585-41.3.392. [DOI] [PubMed] [Google Scholar]
- 8.Garros C, Harbach RE, Manguin S. Morphological assessment and molecular phylogenetics of the Funestus and Minimus groups of Anopheles (Cellia) Journal of Medical Entomology. 2005;42:522–536. doi: 10.1093/jmedent/42.1.7. [DOI] [PubMed] [Google Scholar]
- 9.Sukowati S, et al. Isozyme evidence for three sibling species in the Anopheles sundaicus complex from Indonesia. Medical & Veterinary Entomology. 1999;13:408–414. doi: 10.1046/j.1365-2915.1999.00197.x. [DOI] [PubMed] [Google Scholar]
- 10.Reidenbach KR, et al. Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC Evolutionary Biology. 2009;9:298. doi: 10.1186/1471-2148-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Besansky NJ, Fahey GT. Utility of the white gene in estimating phylogenetic relationships among mosquitoes (Diptera: Culicidae) Molecular Biology & Evolution. 1997;14:442–454. doi: 10.1093/oxfordjournals.molbev.a025780. [DOI] [PubMed] [Google Scholar]
- 12.Krzywinski J, Wilkerson RC, Besansky NJ. Evolution of mitochondrial and ribosomal gene sequences in anophelinae (Diptera: Culicidae): implications for phylogeny reconstruction. Molecular Phylogenetics and Evolution. 2001;18:479–487. doi: 10.1006/mpev.2000.0894. [DOI] [PubMed] [Google Scholar]
- 13.Chen B, Harbach RE, Butlin RK. Molecular and morphological studies on the Anopheles minimus group of mosquitoes in southern China: taxonomic review, distribution and malaria vector status. Medical and Veterinary Entomology. 2002;16:253–265. doi: 10.1046/j.1365-2915.2002.00373.x. [DOI] [PubMed] [Google Scholar]
- 14.Chen B, et al. Mitochondrial DNA variation in the malaria vector Anopheles minimus across China, Thailand and Vietnam: evolutionary hypothesis, population structure and population history. Heredity. 2011;106:241–252. doi: 10.1038/hdy.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B, Butlin RK, Harbach RE. Molecular phylogenetics of the oriental members of the Myzomyia series of Anopheles subgenus Cellia (Diptera: Culicidae) inferred from nuclear and mitochondrial DNA sequences. Systematic Entomology. 2003;28:57–69. doi: 10.1046/j.1365-3113.2003.00200.x. [DOI] [Google Scholar]
- 16.Rueda LM. Global diversity of mosquitoes (Insecta: Diptera: Culicidae) in freshwater. Hydrobiologia. 1970;595:477–487. doi: 10.1007/s10750-007-9037-x. [DOI] [Google Scholar]
- 17.Harbach RE, Kitching IJ. Phylogeny and classification of the Culicidae (Diptera) Systematic Entomology. 1998;23:327–370. doi: 10.1046/j.1365-3113.1998.00072.x. [DOI] [Google Scholar]
- 18.Marrelli MT, Sallum MA, Marinotti O. The second internal transcribed spacer of nuclear ribosomal DNA as a tool for Latin American anopheline taxonomy: a critical review. Memorias do Instituto Oswaldo Cruz. 2006;101:817–832. doi: 10.1590/S0074-02762006000800002. [DOI] [PubMed] [Google Scholar]
- 19.Bangs MJ, Taai K, Howard TM, Cook S, Harbach RE. The mosquito Anopheles (Cellia) oreios sp. n., formerly species 6 of the Australasian Anopheles farauti complex, and a critical review of its biology and relation to disease. Medical & Veterinary Entomology. 2015;29:68–81. doi: 10.1111/mve.12092. [DOI] [PubMed] [Google Scholar]
- 20.Beebe NW, Ellis JT, Cooper RD, Saul A. DNA sequence analysis of the ribosomal DNA ITS2 region for the Anopheles punctulatus group of mosquitoes. Insect Molecular Biology. 1999;8:381–390. doi: 10.1046/j.1365-2583.1999.83127.x. [DOI] [PubMed] [Google Scholar]
- 21.Foley DH, Bryan JH, Yeates D, Saul A. Evolution and systematics of Anopheles: insights from a molecular phylogeny of Australasian mosquitoes. Molecular Phylogenetics & Evolution. 1998;9:262–275. doi: 10.1006/mpev.1997.0457. [DOI] [PubMed] [Google Scholar]
- 22.Chu, H. et al. The phylogenetic relationships of known mosquito (Diptera: Culicidae) mitogenomes. Mitochondrial DNA 1–5 (2016). [DOI] [PubMed]
- 23.Talavera G, Vila R. What is the phylogenetic signal limit from mitogenomes? The reconciliation between mitochondrial and nuclear data in the Insecta class phylogeny. BMC Evolutionary Biology. 2011;11:498–503. doi: 10.1186/1471-2148-11-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cameron SL. Insect mitochondrial genomics: implications for evolution and phylogeny. Annual Review of Entomology. 2013;59:95–117. doi: 10.1146/annurev-ento-011613-162007. [DOI] [PubMed] [Google Scholar]
- 25.Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Molecular Biology & Evolution. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 26.Fenn JD, Song H, Cameron SL, Whiting MF. A preliminary mitochondrial genome phylogeny of Orthoptera (Insecta) and approaches to maximizing phylogenetic signal found within mitochondrial genome data. Molecular Phylogenetics & Evolution. 2008;49:59–68. doi: 10.1016/j.ympev.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 27.Cameron SL, et al. Mitochondrial genome organization and phylogeny of two vespid wasps. Genome. 2008;51:800–808. doi: 10.1139/G08-066. [DOI] [PubMed] [Google Scholar]
- 28.Hua J, et al. Phylogenetic analysis of the true water bugs (Insecta: Hemiptera: Heteroptera: Nepomorpha): evidence from mitochondrial genomes. BMC Evolutionary Biology. 2009;9:134. doi: 10.1186/1471-2148-9-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beard CB, Hamm DM, Collins FH. The mitochondrial genome of the mosquito Anopheles gambiae: DNA sequence, genome organization, and comparisons with mitochondrial sequences of other insects. Insect Molecular Biology. 1993;2:103–124. doi: 10.1111/j.1365-2583.1993.tb00131.x. [DOI] [PubMed] [Google Scholar]
- 30.Peng XY, Zhou P, Duan XY, Qian ZQ. The mitochondrial genomes of twelve Anopheles mosquitoes (Diptera: Culicidae) and their phylogenetic implications. Conservation Genetics Resources. 2016;8:1–4. doi: 10.1007/s12686-016-0565-4. [DOI] [Google Scholar]
- 31.Hua Y. Q. et al. Sequencing and analysis of the complete mitochondrial genome in Anopheles culicifacies species B (Diptera: Culicidae). Mitochondrial DNA 1–2 (2015). [DOI] [PMC free article] [PubMed]
- 32.Clary DO, Wolstenholme DR. The mitochondrial DNA molecular of Drosophila yakuba: nucleotide sequence, gene organization, and genetic code. Journal of Molecular Evolution. 1985;22:252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- 33.Boore JL, Collins TM, Stanton D, Daehler LL, Brown WM. Deducing the pattern of arthropod phylogeny from mitochondrial DNA rearrangements. Nature. 1995;376:163–165. doi: 10.1038/376163a0. [DOI] [PubMed] [Google Scholar]
- 34.Boore JL, Lavrov DV, Brown WM. Gene translocation links insects and crustaceans. Nature. 1998;392:667–668. doi: 10.1038/33577. [DOI] [PubMed] [Google Scholar]
- 35.Negrisolo E, Babbucci M, Patarnello T. The mitochondrial genome of the ascalaphid owlfly Libelloides macaronius and comparative evolutionary mitochondriomics of neuropterid insects. BMC genomics. 2010;12:1–26. doi: 10.1186/1471-2164-12-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. International Review of Cytology. 1992;141:173–216. doi: 10.1016/S0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- 37.Wolstenholme DR. Genetic novelties in mitochondrial genomes of multicellular animals. Current Opinion in Genetics & Development. 1992;2:918–925. doi: 10.1016/S0959-437X(05)80116-9. [DOI] [PubMed] [Google Scholar]
- 38.Beckenbach AT, Stewart JB. Insect mitochondrial genomics 3: the complete mitochondrial genome sequences of representatives from two neuropteroid orders: a dobsonfly (order Megaloptera) and a giant lacewing and an owlfly (order Neuroptera) Genome. 2008;52:31–38. doi: 10.1139/G08-098. [DOI] [PubMed] [Google Scholar]
- 39.Wolstenholme DR. Animal mitochondrial DNA: structure and evolution. International Review of Cytology. 1992;141:173–216. doi: 10.1016/S0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]
- 40.Ojala D, Montoya J, Attardi G. tRNA punctuation model of RNA processing in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 41.Behura SK, Severson DW. Codon usage bias: causative factors, quantification methods and genome-wide patterns: With emphasis on insect genomes. Biological Reviews of the Cambridge Philosophical Society. 2013;88:49–61. doi: 10.1111/j.1469-185X.2012.00242.x. [DOI] [PubMed] [Google Scholar]
- 42.Li, X., Wang, Y., Su, S. & Yang, D. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA 1–2 (2014). [DOI] [PubMed]
- 43.Lynch M. Mutation accumulation in transfer RNAs: molecular evidence for Muller’s ratchet in mitochondrial genomes. Molecular Biology & Evolution. 1996;13:209–220. doi: 10.1093/oxfordjournals.molbev.a025557. [DOI] [PubMed] [Google Scholar]
- 44.Lavrov DV, Adamski M, Chevaldonne P, Adamska M. Extensive mitochondrial mRNA editing and unusual mitochondrial genome organization in calcaronean sponges. Current Biology. 2016;26:86–92. doi: 10.1016/j.cub.2015.11.043. [DOI] [PubMed] [Google Scholar]
- 45.Fernández-Silva P, Enriquez JA, Montoya J. Replication and transcription of mammalian mitochondrial DNA. Experimental Physiology. 2003;88:41–56. doi: 10.1113/eph8802514. [DOI] [PubMed] [Google Scholar]
- 46.Moreno M, et al. Complete mtDNA genomes of Anopheles darlingi and an approach to anopheline divergence time. Malar. Journal. 2010;9:127. doi: 10.1186/1475-2875-9-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jermiin LS, Dan G, Lowe RM, Crozier RH. Analysis of directional mutation pressure and nucleotide content in mitochondrial cytochrome b genes. Journal of Molecular Evolution. 1994;39:160–173. doi: 10.1007/BF00163805. [DOI] [PubMed] [Google Scholar]
- 48.Castro LR, Dowton M. Mitochondrial genomes in the Hymenoptera and their utility as phylogenetic markers. Systematic Entomology. 2006;32:60–69. doi: 10.1111/j.1365-3113.2006.00356.x. [DOI] [Google Scholar]
- 49.Harbach RE, Kitching IJ. Reconsideration of anopheline mosquito phylogeny (Diptera: Culicidae: Anophelinae) based on morphological data. Systematics & Biodiversity. 2005;3:345–374. doi: 10.1017/S147720000500174X. [DOI] [Google Scholar]
- 50.Beebe NW, Cooper RD, Morrison DA, Ellis JT. Subset partitioning of the ribosomal DNA small subunit and its effects on the phylogeny of the Anopheles punctulatus group. Insect Molecular Biology. 2000;9:515–520. doi: 10.1046/j.1365-2583.2000.00211.x. [DOI] [PubMed] [Google Scholar]
- 51.Logue K, et al. Mitochondrial genome sequences reveal deep divergences among Anopheles punctulatus sibling species in Papua New Guinea. Malar. Journal. 2013;12:64. doi: 10.1186/1475-2875-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Green CA. Cladistic analysis of mosquito chromosome data (Anopheles (Cellia) Myzomyia. Journal of Heredity. 1982;73:2–11. doi: 10.1093/oxfordjournals.jhered.a109571. [DOI] [PubMed] [Google Scholar]
- 53.Freitas, L. A. et al. Diversification of the genus Anopheles and a neotropical clade from the late Cretaceous. PloSone10, e0134462, doi:10.1371/journal.pone.0134462 (2015). [DOI] [PMC free article] [PubMed]
- 54.Neafsey DE, et al. Mosquito genomics. Highly evolvable malaria vectors: the genomes of 16 Anopheles mosquitoes. Science. 2015;347:1258522. doi: 10.1126/science.1258522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li C, Wilkerson RC. Identification of Anopheles (Nyssorhynchus) albitarsis complex species (Diptera: Culicidae) using rDNA internal transcribed spacer 2-based polymerase chain reaction primes. Memorias do Instituto Oswaldo Cruz. 2005;100:495–500. doi: 10.1590/S0074-02762005000500009. [DOI] [PubMed] [Google Scholar]
- 56.Wilkerson RC, Foster PG, Cong LI, Sallum MAM. Molecular phylogeny of neotropical Anopheles (Nyssorhynchus) albitarsis species complex (Diptera: Culicidae) Annals of the Entomological Society of America. 2005;98:918–925. doi: 10.1603/0013-8746(2005)098[0918:MPONAN]2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gómez G, Jaramillo L, Correa MM. Wing geometric morphometrics and molecular assessment of members in the Albitarsis Complex from Colombia. Molecular Ecology Resources. 2013;13:1082–1092. doi: 10.1111/1755-0998.12126. [DOI] [PubMed] [Google Scholar]
- 58.Bourke BP, Foster PG, Bergo ES, Calado DC, Sallum MA. Phylogenetic relationships among species of Anopheles (Nyssorhynchus) (Diptera, Culicidae) based on nuclear and mitochondrial gene sequences. Acta tropica. 2010;114:88–96. doi: 10.1016/j.actatropica.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 59.Marrelli MT, Sallum MA, Marinotti O. The second internal transcribed spacer of nuclear ribosomal DNA as a tool for Latin American Anopheline taxonomy‒a critical review. Memorias do Instituto Oswaldo Cruz. 2006;101:817–832. doi: 10.1590/S0074-02762006000800002. [DOI] [PubMed] [Google Scholar]
- 60.Harbach RE. The Culicidae (Diptera): A review of taxonomy, classification and phylogeny. Zootaxa. 2007;1668:591–638. [Google Scholar]
- 61.Reidenbach KR, et al. Phylogenetic analysis and temporal diversification of mosquitoes (Diptera: Culicidae) based on nuclear genes and morphology. BMC Evolutionary Biology. 2009;9:1–14. doi: 10.1186/1471-2148-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilkerson, R. C. et al. Making mosquito taxonomy useful: a stable classification of tribe Aedini that balances utility with current knowledge of evolutionary relationships. PloSone10 (2014). [DOI] [PMC free article] [PubMed]
- 63.Krzywinski J, Grushko OG, Besansky NJ. Analysis of the complete mitochondrial DNA from Anopheles funestus: an improved dipteran mitochondrial genome annotation and a temporal dimension of mosquito evolution. Molecular Phylogenetics and Evolution. 2006;39:417–423. doi: 10.1016/j.ympev.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 64.Zhou D, et al. Genome sequence of Anopheles sinensis provides insight into genetics basis of mosquito competence for malaria parasites. BMC Genomics. 2014;15:73–78. doi: 10.1186/1471-2164-15-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bertone MA, Courtney GW, Wiegmann BM. Phylogenetics and temporal diversification of the earliest true flies (Insecta: Diptera) based on multiple nuclear genes. Systematic Entomology. 2008;33:668–687. doi: 10.1111/j.1365-3113.2008.00437.x. [DOI] [Google Scholar]
- 66.Chen XG, et al. Genome sequence of the asian tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E5907–5915. doi: 10.1073/pnas.1516410112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yi-Lin Z, Yi-Ran D, Qian-Chun L, Bin C. The extraction method of mosquito mitochondrial genome. Chinese Journal of Vector Biology and Control. 2015;26:333–336. [Google Scholar]
- 68.Bernt M, et al. MITOS: Improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics & Evolution. 2013;69:313–319. doi: 10.1016/j.ympev.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 69.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Research. 1997;25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 71.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Research. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Perna NT, Kocher TD. Patterns of nueleotide composition at fourfold degenerate sites of animal mitochondrial genomes. Journal of Molecular Evolution. 1995;41:353–358. doi: 10.1007/BF01215182. [DOI] [PubMed] [Google Scholar]
- 73.Peden JF. Analysis of codon usage. University of Nottingham. 2000;90:73–74. [Google Scholar]
- 74.Burland TG. DNASTAR’s Lasergene sequence analysis software. Methods in Molecular Biology. 2000;132:71–91. doi: 10.1385/1-59259-192-2:71. [DOI] [PubMed] [Google Scholar]
- 75.Eisen MB, et al. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences. 1999;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Katoh, K. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology & Evolution30, 772–780 (2013).btm466 (2007). [DOI] [PMC free article] [PubMed]
- 77.Wernersson R, Pedersen AG. RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Research. 2003;31:3537–3539. doi: 10.1093/nar/gkg609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rozas J, Rozas R. DnaSP, DNA sequence polymorphism: an interactive program for estimating population genetics parameters from DNA sequence data. Computer Applications in the Biosciences Cabios. 1996;11:621–625. doi: 10.1093/bioinformatics/11.6.621. [DOI] [PubMed] [Google Scholar]
- 79.Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Systematic Biology. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- 80.Vaidya, G., David, J. L. & Meier, R. Sequence Matrix: concatenation software for the fast assembly of multigene datasets with character set and codon information (Blackwell Publishing Ltd, 2011). [DOI] [PubMed]
- 81.Xia X, Xie Z. DAMBE: software package for data analysis in molecular biology and evolution. Journal of Heredity. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- 82.Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- 83.Stamatakis A. RAxML Version 8: A tool for phylogenetic analysis and post-snalysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 85.Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Molecular Biology & Evolution. 2012;29:1969–1973. doi: 10.1093/molbev/mss075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gaunt MW, Miles MA. An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Molecular Biology & Evolution. 2002;19:748–761. doi: 10.1093/oxfordjournals.molbev.a004133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


