Abstract
Atrial fibrillation (AF) is the most common sustained arrhythmia and is associated with pronounced morbidity and mortality. Its prevalence, expected to further increase for the forthcoming years, and associated frequent hospitalizations turn AF into a major health problem. Structural and electrical atrial remodelling underlie the substrate for AF, but the exact mechanisms driving this remodelling remain incompletely understood. Recent studies have shown that microRNAs (miRNA), short non-coding RNAs that regulate gene expression, may be involved in the pathophysiology of AF. MiRNAs have been implicated in AF-induced ion channel remodelling and fibrosis. MiRNAs could therefore provide insight into AF pathophysiology or become novel targets for therapy with miRNA mimics or anti-miRNAs. Moreover, circulating miRNAs have been suggested as a new class of diagnostic and prognostic biomarkers of AF. However, the origin and function of miRNAs in tissue and plasma frequently remain unknown and studies investigating the role of miRNAs in AF vary in design and focus and even present contradicting results. Here, we provide a systematic review of the available clinical and functional studies investigating the tissue and plasma miRNAs in AF and will thereafter discuss the potential of miRNAs as biomarkers or novel therapeutic targets in AF.
Electronic supplementary material
The online version of this article (doi:10.1007/s10557-017-6736-z) contains supplementary material, which is available to authorized users.
Keywords: Atrial fibrillation, microRNA, Fibrosis, Electrical remodelling, Therapy
Introduction
Atrial fibrillation (AF) is the most common sustained arrhythmia, associated with a pronounced morbidity and mortality. AF is a major healthcare problem as it possesses a serious burden on socioeconomic budget due to its increasing prevalence and associated frequent hospitalizations. The most important risk factor for new-onset AF is ageing, and ageing of the population importantly contributes to the growing number of patients with AF. AF prevalence is expected to double by 2050 [1, 2]. The disease may present as lone AF or in association with other systemic or cardiovascular diseases such as hypertension, obesity, ischemic heart disease, or valvular disease [2, 3].
AF is a progressive disease that is classified on the basis of clinical presentation, duration and spontaneous termination of AF episodes. AF is subsequently categorized as ‘paroxysmal AF’ (parAF), defined by self-terminating or cardioverted AF within 7 days after onset, ‘persistent AF’ (persAF), defined by AF lasting longer than 7 days, ‘long-standing persAF’, which is AF lasting over 1 year and ‘permanent AF’ (permAF), when patient and physician have accepted that the rhythm will not return to normal sinus rhythm [4]. Recently, the concept of ‘atrial cardiomyopathy’ was introduced, which proposes a classification based on the pathological processes present in atrial disease in addition to the clinical classification [5]. However, we are insufficiently able to recognize the electrical and structural remodelling underlying the atrial cardiomyopathy present in AF patients. Moreover, AF remodelling may differ per aetiology or comorbidities present and does not necessarily parallel to the staged clinical classification [6]. Not surprisingly, treatment strategies that target atrial remodelling are lacking [4, 7]. A better understanding of the pathophysiological processes underlying AF and non-invasive methods to classify patients based on the underlying atrial cardiomyopathy would enable early and mechanism-specific diagnosis of AF followed by the development of mechanism-specific treatments [8].
Emerging studies have uncovered a role for microRNAs (miRNA, miR) in the initiation and maintenance of cardiovascular disease [9]. MiRNAs are highly conserved, small, non-coding RNAs that regulate physiological and disease processes at the post-transcriptional level. MiRNAs bind to a (partially) complementary sequence in the 3′-untranslated region (3’UTR) of the mRNA and can thereby induce degradation or inhibition of translation of this mRNA [10]. MiRNAs regulate approximately 30% of all protein-coding genes and have therefore been predicted to be involved in almost all cellular processes [11].
MiRNA regulation in cardiovascular disease was first described in 2006, when specific miRNAs were found to be up- or downregulated in mouse models of cardiac hypertrophy and heart failure (HF) [9]. Later, it was demonstrated that AF is associated with altered miRNA levels in atrial tissue and plasma [12, 13]. The regulatory function of specific miRNAs has been studied in the structural and electrical remodelling underlying AF, ischemic heart disease, cardiac hypertrophy, ion channel modification and extracellular matrix (ECM) formation [9, 14–19]. Furthermore, circulating miRNAs associated with AF could serve as potential biomarkers of the disease, whereas specific tissue miRNAs could become targets for therapy [20–22].
Currently available studies investigating the role of miRNAs in AF are diverse, with various designs and focuses, and show contradicting results. This systematic review aims to provide a structured overview of the available clinical studies exploring the tissue and plasma miRNA expression profiles in AF patients. Next, we will discuss the available experimental evidence for the functional role of miRNAs in pathophysiological processes underlying AF and discuss the future possible clinical applications of miRNAs in AF.
MiRNAS as Biomarkers of Atrial Fibrillation
MiRNAs may be suitable as biomarkers of disease, because of their tissue- and pathology-specific expression. They are stable in plasma because they are either incorporated in microparticles (exosomes, microvesicles and apoptotic bodies), or bound to proteins or high-density lipoproteins and are thereby protected from RNase activity. Moreover, miRNAs are detectable in plasma or serum with high sensitivity and specificity [20, 23, 24].
MiRNAs have been suggested as biomarkers of several cardiac diseases, including heart failure and coronary artery disease [25–27]. For example, plasma miR-208b demonstrated a high diagnostic accuracy for myocardial infarction similar to troponine T [28, 29]. This example underscores the potential of miRNAs to serve as clinical biomarkers of cardiovascular disease, while no such biomarker is currently available for AF. Once a diagnosis of AF has been established, biomarkers may give insight into the specific atrial cardiomyopathy underlying AF, which may have implications for prognosis and treatment and may enhance patient-tailored care [4].
MiRNAs Associated with Atrial Fibrillation Onset
McManus et al. [30] performed the only study to date that evaluated the prognostic value of circulating miRNAs for the occurrence of new-onset AF. In this study, which included 2292 participants from the Framingham Heart cohort without AF, 107 participants developed AF after a median follow-up of 5.4 years, but none of the 385 investigated miRNAs in whole blood were associated with new-onset AF.
Two prospective studies [31–34] investigated whether the occurrence of postoperative AF (POAF) was associated with miRNA plasma levels at the time of coronary bypass surgery (CABG) (Online Resource Table 2). POAF has a complex, but specific aetiology that involves considerable systemic and local inflammation following cardiac surgery [35]. Harling et al. [31] collected serum prior to surgery and concomitantly retrieved atrial tissue during cardiac surgery. They found 16 miRNAs that were differentially expressed in atrial tissue between 11 patients who developed POAF and 11 patients who did not. The cardiomyocyte-enriched miR-208a (further described in Tables 3 and 4) was most downregulated and miR-483-5p most upregulated. MiR-483-5p serum levels were also increased in these patients (ROC area 0.78) whereas miR-208a was undetectable in serum at any time point. Krogstad et al. [34] used qPCR to study the levels of over 30 miRNAs in plasma of 92 CABG patients, of whom 27 developed POAF. However, they found no miRNA associated with POAF onset.
Table 2.
microRNA discovery studies in tissue
| Ref | Tissue | Study Population | Technique | miRNA expression in AF patients |
|---|---|---|---|---|
| 67 | RAA | CABG, AVR and/or MVR: 4 parAF 4 matched no AF |
mRNA-seq miRNA-microarray qPCR |
Upregulated: 26a-2-3p, 27b-3p, 30b-3p, 30e-3p, 101-3p, 125a-5p, 125b-5p, 145-3p, 199a-3p, 199a-5p, 199b-5p, 222-3p, 223-3p, 1910-3p, 3135b, 3197, 381-5p, 3939, 4280, 4486, 6753-5p, 6820-5p, 7843-5p Downregulated: 1227-5p, 1273 g-3p, 1915-3p, 3196, 3656, 3665, 3960, 4281, 4466, 4497, 4516, 4530, 4690-5p, 4707-5p, 4734, 4787-5p, 4800-3p, 5096, 5787, 6090, 6125, 6775-5p, 6786-5p, 6791-5p, 7110-5p, 7704 |
| 43 | MVR: RAA, LAA CABG: RAA HTx: LAA |
~7 MVR and permAF ~6 MVR no AF 8 CABG no AF 5 HTx no AF |
Microarray qPCR |
Upregulated in RAA: 16, 21†, 21*, 142-3p, 142-5p, 146b-5p†‡, 198, 223, 224, 337-5p, 377, 483-5p, 1202, 1290, 1308 Downregulated in RAA: let-7c, 22*, 24–1*, 29c*,30‡, 30a, 30a*, 30b, 30c†, 30e, 99a, 99b, 125b-2*, 128, 133a†, 133b†, 139-5p, 143*, 145, 149, 181c, 181d, 197, 203, 331-3p, 367, 374b, 378, 378*, 484, 490-3p†‡, 490-5p, 628-5p No miRNAs were up- or downregulated in LAA. |
| 70 | RAA | CABG, AVR and/or MVR: 4 permAF 4 No AF |
Microarray qPCR miR-499 |
Upregulated:1, 7–1*, 24–1*, 145*, 187*, 208b, 301a, 302a, 302b, 375, 454, 499†, 885-3p, 1244 Downregulated:21*, 23a*, 27a*, 138, 193b*, 299-3p, 1270 |
| 55, 45¥ | LAA | 6 MVR and AF 6 MVR no AF |
Microarray qPCR |
Upregulated: 15b-5p, 21-5p, 466†, 574-3p†, 3178, 3196, 3613-3p†, 4492, 4497, 4707-5p Downregulated: 1†, let-7 g-5p, 24-3p, 26a-5p†, 26b-5p, 29a-3p, 151a-5p, 195-5p, 361-5p, 720, 4454, 5100 |
| RAA, LAA | 10 MVR and AF 8 MVR no AF |
Microarray qPCR |
Upregulated LAA: LAA: let-7d-3p, 15b-5p, 21-5p, 30a-5p, 149-3p, 181a-5p, 331-3p, 466, 494, 574-3p, 1307-3p, 1973, 3178, 3196, 3591-3p, 3613-3p, 3940-5p, 4485, 4492, 4497, 4534, 4707-5p Upregulated RAA: 149-3p, 574-5p, 762, 940, 1281, 1915-3p, 1973, 2861, 3141, 3656, 3940-5p, 4281, 4284, 4298, 4443, 4459, 4463, 4466, 4484†, 4485, 4488, 4497, 4505, 4508, 4530, 4534, 4707-5p, 4687-3p Downregulated LAA: let-7f-5p, 1†, 23b-3p†, 23c, 26a-5p†, 26b-5p, 143-3p†, 151a-5p, 151b, 195-5p, 361-5p, 378a-3p, 378d, 720, 2861, 4281, 4442, 4454†, 5100, 5190 Downregulated RAA: let-7a-5p, 16-5p, 21-5p, 22-3p, 25-3p, 26a-5p†, 26b-5p, 29a-3p, 30a-5p, 30b-5p, 30c-5p†, 30d-5p, 99a-5p, 107, 125a-5p, 125b-5p†, 133b†, 143-3p†, 145-5p†, 151a-5p, 151b, 152, 181a-5p, 191-5p, 195-5p, 221-3p, 222-3p, 331-3p, 378a-3p, 378d, 451a, 455-3p, 486-5p, 4286, 4324, 4454†, 5100 |
|
| 53 | RAA | CABG, aortic (valve) repair, MVR, maze, TVR, septal defects 9 persAF 11 no AF |
Microarray qPCR of miR-30 family |
Upregulated: 22, 24, 24–1*, 30a, 30b*†, 30d*†‡, 30e, 125a-3p, 185, 208b, 210, 324-5p, 499-5p‡, 505*, 574-5p, 602, 652, 671-5p, 1181, 1224-5p, 1290, 1305, 1972, 1973, 3125, 3195, 3610, 3648, 3679-5p, 4257, 4291, 4298, 4299, 4306 Downregulated: 10a, 31, 100 |
| 52 | RAA, plasma |
CABG, aortic (valve) repair, MVR, TVR: 16 AF (4 permAF; 12 maze; 10 succesful maze) 13 no AF (CABG/AVR) |
Microarray | Upregulated: let-7a, let-7d, let-7f, 20b, 21†‡, 22, 23b‡, 24, 27a, 27b, 28-5p, 32, 34a, 93, 95, 101, 103, 106b, 125b, 127-3p, 129-3p, 130a, 130b, 134, 140-5p, 142-3p, 146b,148b, 152, 15a, 15b, 181a, 181c, 184, 185, 187, 190, 193a-3p, 196b, 199a-5p, 199b-5p‡, 203, 208b†‡, 210, 215, 216a, 216b, 217, 320, 324-5p, 330-3p, 337-5p, 361-5p, 362-5p, 371-3p, 372, 423-5p, 424, 439, 449a, 450a, 455-5p, 487a, 487b, 494, 495, 499-5p, 500, 504, 505, 508-3p, 509-5p, 511, 517a, 517c, 518b, 518f, 520e, 522, 539, 542-5p, 545, 548d-5p, 579, 597, 618, 652, 660, 671-3p, 758, 874, 886-5p, 887, 888 Downregulated: 31, 200b, 429, 885-5p |
| 107, 99¥ | RAA, LAA | MVR, CABG and/or AVR: 21 AF (11 parAF; 10 permAF) 16 no AF |
Microarray qPCR validation of all selected miRNAs Mitochondrial respiration |
Upregulated: LAA: 18a, 18b, 19a, 19b, 23a, 25, 30a, 93, 106a, 106b, 144, 363, 451, 486-5p, 590-5p RAA: 15b, 106b, 144, 451 Downregulated in both LAA and RAA: 208a |
| 102, 101 | LAA | parAF, HTx: Discovery phase: 8 parAF; 5 HTx Validation phase: 30 parAF, 17HTx |
Microarray qPCR |
Upregulated: 19b†, 142-3p, 146b-5p†‡, 155†‡, 193b, 223, 301b, 486-5p, 519b-3p Downregulated: 193a-5p |
| 68 | RAA | 9 MVR and AF 4 MVR no AF 9 HTx |
Microarray qPCR |
Upregulated: 188-5p, 212, 335†, 630,1181, 1202†, 1207-5p, 1225-5p Downregulated: 95, 26b*, 125a-5p, 125b, 125b-2*, 143*, 145, 145*, 149, 181a†, 181a*, 181b, 181c†, 181d, 324-5p, 497, 500, 501-5p, 550, 874 |
* anti-sense miRNA; † validated with qPCR; ‡ most upregulated or downregulated miRNA
¥ There is an overlap of study population between the 2 studies. The first reference focused on the comparison between AF and no AF whereas the second reference focused on a comparison between RAA and LAA.
Both studies by Wang et al. describe the exact same study population and microarray results
Abbreviations: AF atrial fibrillation, AVR aortic valve repair, CABG coronary artery bypass grafting, HTx heart transplantation, LAA left atrial appendage, MVR mitral valve repair, parAF paroxysmal AF, permAF permanent/chronic AF, persAF persistent AF, RAA right atrial appendage, seq sequencing, qPCR quantitative polymerase chain reaction
Table 3.
MicroRNAs described for their role in electrical remodelling
| miRNA in AF | Target(s) in AF | Experimental Model | Function | Reporting studiesa | ||
|---|---|---|---|---|---|---|
| miR-31 | ↑ | nNOS, dystrophine [52] | ↓ | 51 AF, 165 SR patients, human cardiac myocytes, goat ATP model | Upregulation in AF correlated with decreased nNOS and unchanged Dystrophine mRNA levels. In vitro inhibition restored nNOS protein and normalized APD in myocytes from AF patients. MiR-31 directly targets both dystrophin and nNOS, and negatively reduce the respective protein by promoting nNOS mRNA decay and inhibiting the translation of dystrophin mRNA. | [30, 52–54] |
| miR-21 | ↑ | CACNA1C, CACNB2 [59] |
↓ | 10 permAF, 10 SR patients, atrial cadiomyocytes, HL-1 cells | An upregulationin AF was seen along with a decrease in CACNA1C, CACNB2 mRNA and ICaL in human atrial cardiomyocytes. Its overexpression in vitro decreased ICaL density and Cav1.2 protein levels. This miR directly targeted CACNA1C and CACNB2. | [40, 44, 46, 53, 56, 59, 74] |
| miR-208a/b | ↑ | CACNA1C, CACNB2 [58] | ↓ | 16 permAF, 15 SR patients, sheep ATP, human cardiac myocytes, HL-1 cells | Upregulation of miR-208b, but not of miR-208a in AF patients compared to controls correlated with decreased mRNA, protein levels and ICaL density. Overexpression of both miR208a/b in vitro reduced Cav1.2 protein levels. MiR-208a/b directly targeted CACNA1C and CACNB2. | [31, 53, 58, 62, 71, 74, 111, 114] |
| miR-328 | ↑ | CACNA1C, CACNB1 [45] | ↓ | 12 AF, 10 SR patients, canine ATP model, mice burst pacing, miR-328 TG mice, miR-328 sponge TG mice, in vivo forced expression in canines, neonatal rat cardiomyocytes | Upregulation in AF correlated with decreased human and canine mRNA and Cav1.2, Cavβ1 protein levels. In vivo overexpression of this miR promoted AF vulnerability, decreased APD, Cav1.2, Cavβ1 and ICaLdensity. Inhibition dampened AF vulnerability. MiR-328 directly targeted CACN1C and CACNB1. | [30, 37–39, 45, 56, 57] |
| miR-1 | ↑ | KCNE1, KCNB2 [63] | ↓ | Rabbit ATP model, in vivo forced expression and inhibition in rabbits | ATP was associated with increased miR-1 decreased KCNE1 and KCNB2 mRNA and protein levels, shortening of AERP and an increase in IKs and AF susceptibility. MiR-1 in vivo overexpression further enhanced these effects, while inhibition with antimiR-1 alleviated these results. MiR-1 directly targeted KCNE1 and KCNB2. | [15, 46, 63, 74, 135] |
| ↓ | KCNJ2 [15] | ↑ | 31 AF, 31 SR patients, in vitro TP of human atrial slices | Downregulation in tissue of AF patients correlated with increased mRNA, Kir2.1 levels and IK1 density. TP of human atrial slices induced a miR-1 decrease and Kir2.1 increase. | ||
| mir-26a/b | ↓ | KCNJ2 [16, 68] | ↑ | 12 AF, 10 SR patients, canine A/VTP model, mice A/VTP model, in vivo forced expression in mice, canine and mice fibroblasts, TG and KO mice, H9c2 cells | Downregulation of miR-26b and miR-26a in particular in AF patients or AF animal models correlated with an upregulation of mRNA, Kir2.1 levels and IK1 density. Both in vivo and in vitro inhibition of miR-26 increased IK1 and AF vulnerability, whereas overexpression of dampened AF vulnerability. MiR-26 directly targeted KCNJ2. | [16, 46, 68–70, 86] |
| miR-30d | ↑ | KCNJ3 [71] | ↓ | 14 AF, 19 SR patients, neonatal rat cardiomyocytes | Upregulation in cardiomyocytes from AF patients correlated with decreased mRNA and Kir3.1 levels. MiR-30d overexpression in vitro decreased KCNJ3, Kir3.1 and IKACh, while inhibition had the opposite effects. MiR-30d directly targeted KCNJ3. | [46, 71] |
| miR-499 | ↑ | KCNN3 [74] | ↓ | 4 permAF, 4 SR, HL-1 cells | Upregulation in tissue of AF patients correlated with decreased SK3 protein. MiR-499 in vitro overexpression suppressed KCNN3 levels and SK3 levels while inhibition enhanced SK3. MiR-499 directly targeted KCNN3. | [45, 53, 58, 71, 74] |
aStudies reporting about the specific miRNA in AF. These include both explorative and functional studies in tissue and plasma and may present conflicting data regarding upregulation or downregulation of the miRNA
Abbreviations: AF atrial fibrillation, APD action potential duration, ATP atrial tachypacing, cav1.2 L-type voltage-dependent calcium channel subunit α1C, Cavβ1 L-type voltage-dependent calcium channel subunit β1, I Ca,L, L-type voltage-dependent calcium channel current, KO knockout, LA left atrium, MVS mitral valve stenosis, ox-LDL oxidized low-density lipoprotein, permAF permanent/chronic AF, RA right atrium, SR sinus rhythm/controls, AERP atrial effective refractory period, AF atrial fibrillation, ATP atrial tachypacing, I K1 inward rectifier K+ current, I KACh acetylcholine regulated inward rectifier potassium current, IKs potassium currents, Kir2.1 inward rectifier potassium channel 2, Kir3.1 acetylcholine regulated inward rectifier potassium channel 3, LA left atrium, permAF permanent/chronic AF, RA right atrium, SR sinus rhythm/controls, SK3 small conductance calcium-activated potassium channel 3, TG transgenic, TP tachypacing, VTP ventricular tachypacing
Table 4.
MicroRNAs described for their role in structural remodelling
| MiRNA in AF | Target(s) in AF | Experimental Model | Function | Reporting Studiesa | ||
|---|---|---|---|---|---|---|
| miR-21 | ↑ | Pitx2c [136] | ↓ | Pig ATP model, HL-1 cells | Upregulation in AF was correlated with decreased PITX2C protein. Its overexpression in vitro decreased mRNA and protein while inhibition with antimiR-21 had opposite effects. | [17, 18, 40, 44, 53, 56, 59, 74, 96–98, 136] |
| SPRY1 [17] Spry1 [18] | ↓ | 5 valvular AF, 5 SR patients, TG mice expressing Rac1, neonatal rat fibroblasts, in vivo inhibition in an ischemic rat/mice model. | Upregulation in LAA from AF patients and in an ischemic mice model was seen and correlated with increased fibrotic content and a decrease of SPRY1. Administration of Ang-II induced an increase of CTGF and miR-21 in cardiac fibroblasts while Spry1 decreased.. In vivo inhibition with antagomir-21 or a 15-mer-LNA based antimiR-21 suppressed the fibrotic response and prevented increased AF susceptibility. | |||
| STAT3b [98] | ↑ | Sterile rat pericarditis model with ATP, in vivo inhibition in pericarditis rat, neonatal and adult rat atrial fibroblats | Pericarditis in rats increased AF susceptibility and fibrosis and upregulated IL1B, IL-6, TGFB, TNFa, STAT3 and miR-21. In vitro inhibition of miR-21 suppressed STAT3 phophorylation, Col1A1 and Col3A1 mRNA, while overexpression had opposite effects. In vivo inhibition with antagomir-21 decreased STAT3 phosphorylation, fibrosis and AF vulnerability, | |||
| Smad7 [97] | ↓ | Rabbit ATP model, in vivo forced expression in rabbits, rat cardiac fibroblasts | Upregulation of TGF-β1 mRNA and protein in ATP rabbits correlated with increased miR-21 and decreased Smad7. In vivo pre-treatment with miR-21 inhibitor restored Smad7 and prevented a decrease in collagen I/III mRNA and protein. MiR-21 directly targeted Smad7. | |||
| mir-26a | ↓ | TRPC3 [86] | ↑ | VTP canine model with CHF, ATP goat model, canine and rat cardiac fibroblasts | Downregulation in isolated LA fibroblast form AF dogs correlated with increased TRPC3 protein. Its overexpression in vitro suppressed TRPC3 protein and fibroblast number, while inhibition had opposite effects. Administration of NFAT-blocker increased miR-26a/b in vitro. MiR-26a directly targeted TRPC3. | [16, 46, 68–70, 86] |
| miR-29b | ↓ | COL1A1, COL3A1, FBN [103] | ↑ | RA from 17 AF, 19 SR patients, VTP canine model with CHF, canine fibroblasts, in vivo inhibition in mice, human AF plasma samples | Downregulation was seen in RA tissue of AF patients and LA tissue and fibroblasts from VTP dogs. Plasma levels of AF patients were also lower. VTP dogs demonstrated increased COL1A1, COL3A1 and FBN in fibroblasts. MiR-29b inhibition in vitro with miR-29b sponge increased mRNA levels and protein of those ECM components while overexpression had opposite effects. In vivo inhibition with miR-29b sponge in mice increased atrial COL1A1 and tissue collagen content. | [30, 103] |
| miR-30a | ↓ | Snail 1[104] | ↑ | Rabbit ATP model, cardiac rat fibroblasts | Downregulation in ATP rabbits correlated with increased Snail1 and Periostin mRNA and protein levels. MiR-30a overexpression xin vitro suppressed snail1 and periostin mRNA and protein, while inhibition increased their expression. MiR-30a directly targeted Snail1. | [104, 137] |
| miR-133 | ↓ | TGF-β1 [107] | ↑ | 19 AF patients, with or without nicotine abuses, canine model with nicotine administration, canine atrial fibroblasts | Downregulation in dogs and canine fibroblasts correlated with increased nicotine concentration. Nicotine usage was also associated with downregulation in human RA. MiR-133 overexpression in vitro decreased TGF-β1 protein and collagen content, while inhibition increased TGF-1 and collagen content. Nicotine administration in vitro decreased miR-133. MiR-133 directly targeted TGF-β1. | [44, 46, 107, 137] |
| miR-146b | ↑ | TIMP-4[108] | ↓ | 30 parAF, 17 SR patients, mice cardiac fibroblasts | Upregulation in AF correlated with decreased TIMP-4. MiR-146b in vitro overexpression decreased TIMP-4, which could be prevented by inhibition. A downregulation of TIMP-4 was associated with increased MMP-9 and collagen content. MiR-146b directly targeted TIMP-4. | [37, 44, 53, 108, 109] |
| miR-208a/b | ↑ | Thrap1, myostatin GATA4 [111] | ↓ | Mice pressure overload model, rat cardiomyocytes, TG and KO mice | MiR-208a KO mice developed spontaneous AF. TG mice overexpressing miR-208a had reduced Thrap1 and myostatin protein levels and hypertrophic growth while KO induced an increase. MiR-208a/b directly targeted Thrap1, myostatin and miR-208 targeted GATA4. | [31, 53, 58, 62, 71, 74, 111, 114] |
| miR-208a/b | ↑ | Sox5, Sox6 [58] | ↓ | 4 permAF, 2 SR patients, human cardiac myocytes, HL-1 cells | Upregulation in AF correlated with decreased Sox5, Sox6 and increased Myh7. Overexpression of miR-208b in vitro suppressed Sox6 and increased Myh7. Overexpression of miR-208a in vitro suppressed Sox5 and moderately increased Myh7. | [31, 53, 58, 62, 71, 74, 111, 114] |
| miR-590 | ↓ | TGF-βR2 [107] | ↑ | 19 AF patients, with or without nicotine abuses, canine model with nicotine administration, canine atrial fibroblasts | Downregulation in dogs and canine fibroblasts correlated with increased nicotine concentration. Nicotine usage was also associated with downregulation in human RA. MiR-590 overexpression in vitro decreased TGF-βR2 protein and collagen content, while inhibition increased TGF-β2 and collagen content. Nicotine administration in vitro decreased miR-590. MiR-590 directly targeted TGF-βR2. | [107, 114] |
aStudies reporting about the specific miRNA in AF. These include both explorative and functional studies in tissue and plasma and may present conflicting data regarding upregulation or downregulation of the miRNA
bNot a direct target of the miRNA
Abbreviations: AF atrial fibrillation, Ang-II angiotensin-II, APD action potential duration, ATP atrial tachypacing, CHF congestive heart failure, CTGF connective tissue growth factor, FBN fibrillin, FKBP5 FK506 binding protein 5, KO knockout, LA left atrium, MMP-9 Matrix metallopeptidase 9, MVS mitral valve stenosis, NFAT nuclear factor of activated T-cells, permAF permanent/chronic AF, PITX2 paired-like homeodomain transcription factor 2, RA right atrium, Smad7 decapentaplegic homologue 7, SPRY1 sprouty RTK signalling antagonist 1 gene, SR sinus rhythm/controls, STAT3 signal transducer and activator of transcription 3, TG transgenic, TGF-β1 transforming growth factor-β1, TGF-βR2 transforming growth factor β receptor type 2, THRAP1 thyroid hormone-associated protein 1, TIMP-4 Metalloproteinase inhibitor 4, TRPC3 transient receptor potential canonical-3, VTP ventricular tachypacing
Hence, the prognostic value of individual miRNAs has not been irrevocably demonstrated. MiR-483-5p was the only circulating miRNA associated with the occurrence of POAF and no circulating miRNA was associated with new-onset AF. Tissue miRNAs were more frequently associated with POAF than circulating miRNAs. However, as tissue is not standardly retrieved during (cardiothoracic) surgery, standardization of retrieval and processing is needed before tissue miRNAs can be considered for the prognostication of (PO)AF. Altogether, there is insufficient evidence to implement current findings into clinical practice. Future studies investigating the prognostic value of miRNAs for new-onset AF or POAF should focus on high risk patients and perform extensive clinical profiling to enable differentiating patients according to AF aetiology.
Circulating miRNAs Associated with Prevalent Atrial Fibrillation
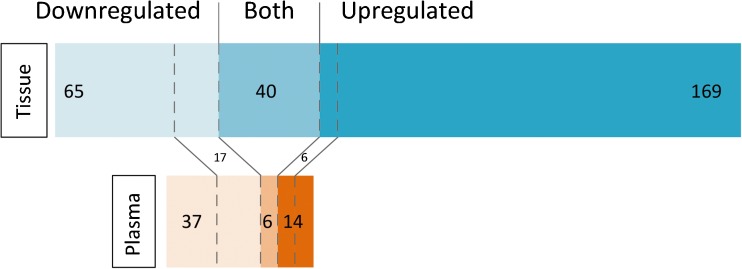
We identified 6 studies [30, 36–39] investigating miRNAs in plasma samples from patients with AF and controls without AF (Table 1). Twenty circulating miRNAs demonstrated higher levels in AF patients versus controls in one or more studies and 43 miRNAs demonstrated lower levels. (Fig. 1. Online Resource Table 3 gives a complete list of plasma miRNAs in AF)
Table 1.
MicroRNA discovery studies in plasma
| Ref | Study population | Technique | miRNA expression in AF patients |
|---|---|---|---|
| 35 | HF patients with EF <40% and healthy volunteers: 15 HF and AF 26 HF no AF 35 matched controls |
Platelets: microarray Serum: qPCR of 89 miRNAs |
Upregulated: None Downregulated: miR-150 (both plasma and platelets) |
| 36 | Discovery phase: 5 parAF, 5 persAF, 5 controls Validation phase: 30 parAF, 30 persAF, 30 controls |
MPSS qPCR (146a, 150, 19a, 375) |
Upregulated: 19a, 125a-5p, 146a, 146b-5p, 148b, 221, 342-3p, 409-3p, 421, 589, 598, 941 Downregulated: 99b, 100, 150b, 199a-5p, 199b-5p, 320b, 375 |
| 37 | Discovery phase: pooled samples 30 AF, 30 controls qPCR validation: pooled independent samples 30 AF, 30 controls qPCR validation: non-pooled independent samples 40 AF, 40 controls. |
Solexa sequencing qPCR |
Upregulated: 9, 152, 374a, 454, 664 Downregulated: 16–2a, 328b, 338-5p, 409-3pb, 432b, 478b, 486-5p, 493, 766, 874, 4732-3p |
| 29 | 153 prevalent AF at baseline 1017 new-onset AF after median FU 5.4y 2185 No AF at baseline or FU |
qPCR of 385 miRNAs | Upregulated prevalent AF: 31-3p, 182-5p, 196b-5p Downregulated prevalent AF: 28-5p, 99b-5p, 150-5p, 328, 331-3p, 339-5p Upregulated incident AF:29b-2-5p, 134, 151a-3p, 152, 193a-5p, 200c-3p, 221-3p, 375, 1274b, 720 Downregulated incident AF: None |
| 39 | 112 AF 99 no AF |
High throughput qPCR of 86 miRNAs LAA removed |
Upregulated: none Downregulated: let-7b-5p, let-7c-5p, 10b-5p, 21-5p, 24-3p, 29a-3p, 30c-5p, 99b-5p, 100-5p, 122-5p, 125a-5p, 125b-5p, 126-3p, 146a-5p, 148b-3p, 150-5p, 221-3p, 223-3p, 342-3p, 375, 411-5p |
| 38 | 122 AF (31 parAF, 91 pers. + permAF) 122 no AF |
Microarray (4 pooled groups) | Upregulated: 9, 19, 146, 152,374a, 454, 634, 664 Downregulated: 1, 145, 162, 222, 328, 432, 493b |
Abbreviations: AF atrial fibrillation, EF ventricular ejection fraction, FU follow-up, HF heart failure, LAA left atrial appendage, MPSS massively parallel signature sequencing, parAF paroxysmal AF, permAF permanent/chronic AF, persAF persistent AF, qPCR quantitative polymerase chain reaction
aAnti-sense miRNA
bValidated with qPCR
cMost upregulated or downregulated miRNA
Fig. 1.
microRNAs expressed in plasma and tissue. This figure illustrates the number of miRNAs differentially expressed in tissue and in plasma in AF patients. Note the high number of upregulated miRNAs in tissue, whereas miRNAs in plasma more often have lower levels in AF. Furthermore, there is little overlap between tissue and plasma expression of the miRNAs that have lower or higher levels in AF (between the dotted lines). There is a substantial proportion of miRNAs that present contradicting results (miRNAs described to be both up- and downregulated in tissue or plasma)
A limited number of miRNAs were consistently reported to have higher plasma levels in AF compared to controls. MiRNAs with higher levels in AF patients in two studies were miR-9, miR-152, miR-374a, miR-454 and miR-664. No miRNA was reported to be increased by three or more studies. More often, lower miRNA levels in AF were seen. MiRNAs with lower levels in AF described by three or more studies were miR-99b, miR-150 and miR-328. Interestingly, these three miRNAs were also reported by McManus et al. [30], who studied new-onset AF in the Framingham Heart cohort and used the same cohort to compare 2185 participants without AF with 153 patients with prevalent AF. In this study, miR-99b, miR-150 and miR-328 showed lower levels in AF, but only miR-328 levels remained significantly lower in patients with prevalent AF after correction for age, sex and technical covariates, like RNA quality and concentration.
MiR-150 was lower in plasma of AF patients in 4 out of 6 miRNA discovery studies [30, 36, 39, 40]. Goren et al. [36] included 41 patients with heart failure (HF), with or without AF, and found decreased levels of miR-150 in both plasma and platelets of AF patients. After a full separation of platelets and plasma, miR-150 was abundantly present in platelets. Platelet and plasma levels were significantly correlated, suggesting that platelets are the origin or transport mode of miR-150 [36]. The exact functional role of miR-150 in AF pathophysiology was not established by this study. Goren et al. [36] speculated that lower platelet miR-150 could be involved in many pathways leading to AF, including inflammation, platelet function and fibrosis [36, 41, 42]. Another study demonstrated that miR-150 promotes megakaryocytopoiesis in platelet progenitor cells [43].
Some of the miRNAs reported in explorative plasma studies were also described by studies exploring atrial tissue miRNA expression. For example, miR-99b was found to be decreased in plasma studies [30, 37, 40] and also in the left atrial appendage (LAA) of AF patients [44]. Overall, however, there was limited correlation between the levels of miRNAs discovered in plasma and tissue levels. For instance, miR-150 and miR-328, for which there is considerable evidence for lower plasma levels in AF patients, have not been described by any explorative study investigating atrial tissue. Conversely, miR-328 was actually upregulated in a study using a canine tachypacing model [45]. Interestingly, overall a larger proportion of miRNAs was found to be decreased in AF in plasma, whereas most differentially expressed miRNAs in tissue were found to be upregulated (Fig. 1).These findings illustrate that the differential expression in tissue does not necessarily give rise to different miRNA levels in plasma, and vice versa. However, higher miRNA levels in tissue and inverse lower levels in plasma suggest that tissue overexpression may at least partly result from an active process of miRNA retention in the cell or uptake of miRNAs from the circulation.
Myocardial miRNAs Associated with Prevalent Atrial Fibrillation
We identified 12 original case-control studies (including at least four cases per study group) in 9 non-overlapping study populations, which explored miRNA expression in atrial tissue. Studies were performed between 2011 and 2016 and used a diverse range of microarray platforms and cut-offs for the discovery of differentially expressed miRNAs. In general, studies validated their most significant results by qPCR (Table 2).
In total, 209 miRNAs were reported upregulated in AF in one or more studies and 105 miRNAs were downregulated. MiRNAs that were consistently upregulated in three or more studies were miR-15b, miR-21, miR-24, miR-30a, miR-142-3p, miR-146b, miR-208b, miR-223 and miR-499. Downregulated miRNAs described by at least three studies were miR-125b, miR-143 and miR-145. However, discrepancy exists concerning the up- or downregulation of five of these miRNAs (miR-21, miR-24, miR-30a, miR-125b, miR-145) (Online Resource Table 4 gives a complete list of tissue miRNAs in AF). The various origins of the tissue may in part explain this as exemplified by miR-21, which was downregulated in right atrial (RA) tissue, but upregulated in left atrial (LA) tissue [46]. However, tissue origin may not be the only explanation and an influence of study populations, comorbidities and techniques cannot be excluded.
Functional Implications of Tissue MiRNAS in Atrial Fibrillation
A predisposition to AF results from atrial remodelling processes that are generally thought to involve ion channel remodelling, Ca2+ overload, structural remodelling such as fibrosis and autonomic dysregulation (extensively reviewed earlier [41, 47–50]).
Numerous explorative studies have implicated miRNAs in these AF-induced remodelling processes. These findings comprise association, but the functional targets are often only predicted by bioinformatics analysis and proof for an arrhythmogenic mechanism in AF is often lacking (Tables 1 and 2). Below, we discuss miRNAs described in functional studies that provide evidence for a specific regulatory role in AF by gain- and loss of function models and/or with luciferase reporter assays (Tables 3 and 4).
Ion Channel Remodelling
Remodelling of ion channels occurs within hours after the initiation of AF and is characterized by a prominent downregulation of L-type Ca2+ (ICaL) current and transient outward current (Ito) and by an upregulation of inward rectifier K+ current(IK1) and acetylcholine-dependent K+ current (IKACh). These changes shorten action potential duration (APD) and the effective refractory period (ERP). Subsequent shortening of the wavelength (the mathematical product of ERP and conduction velocity) then facilitates AF induction and perpetuation [50].
For example, neuronal nitric oxide synthase (nNOS), which is an upstream regulator of several ion channels, was described to be suppressed by miR-31 [51]. Reilley et al. [52] found LA-specific upregulation of miR-31 in AF patients, and a goat model of AF. MiR-31 promoted the decay of nNOS and altered the localization of nNOS by repression of dystrophin translation. MiR-31-5p hairpin inhibitor in human atrial myocytes restored nNOS and dystrophin protein levels and APD, which may contribute to the termination of AF. MiR-31 expression in AF has not been reported to be upregulated in the explorative studies so far, but a downregulation in AF patients was described by two studies [53, 54].
Calcium Channels
Decreased ICaL, which shortens action potential duration, may result from a decreased expression of the L-type voltage-dependent calcium channel subunit α1C (Cav1.2; CACNA1C) and the L-type voltage-dependent calcium channel subunit β1 and β1 (Cavβ1/2; CACNB1/2) [50]. Several miRNAs have been implicated to regulate Ca2+-channels (Table 3).
miR-328
MiR-328 was the highest upregulated miRNA after microarray analysis of LA tissue of AF patients and in a canine atrial tachypacing (ATP) model [45]. In vivo adenoviral overexpression of miR-328 in dogs and transgenic mice overexpressing miR-328, resulted in decreased Cav1.2, Cavβ1 and ICaL., shortening of the APD and enhanced AF susceptibility. Consequently, antagomir-328 reversed the condition in dogs and genetic knockdown with a miR-328 sponge in mice decreased AF susceptibility. Interestingly, miR-328 was consistently found to be lower in plasma of both AF and heart failure patients [30, 38, 39, 55]. However, other AF studies specifically mentioned no difference in miR-328 in plasma [37] or atrial tissue [56] or even described an increase in plasma [57].
miR-208a/b
Canon et al. [58] performed a microarray screen in 4 patients with AF and 2 without AF and identified miR-208a and miR-208b in particular as the most significantly increased miRNA in AF. In this study, miR-208a was not significantly associated with reduced CACNA1C mRNA levels or decreased ICaL density. Moreover, transfection with either miR-208a or miR-208b in HL-1 cells decreased Cav1.2 protein levels. Luciferase assay confirmed CACNA1C and CACNB2 as direct targets of miR-208a/b. The miR-208 family is cardiomyocyte specific and encoded by introns of the cardiac myosin heavy chain genes (MYH6 and MYH7). Besides their role in electrical remodelling, Canon et al. [58] and others suggested that miR-208a and miR-208b play a role in structural remodelling, as described below and in Table 4.
miR-21
MiR-21 was upregulated in RA cardiomyocytes from AF patients and correlated with decreased CACNA1C and CACNB2 levels [59]. In vitro overexpression of miR-21 decreased ICaL density and luciferase assays confirmed miR-21 to target CACNA1C and CACNB2. This miRNA has extensively been studied for its involvement in the structural remodelling underlying AF, described below and in Table 4.
Potassium Channels
An increase in inward rectifier current IK1, is a prominent feature of AF electrical remodelling. Increased IK1 results from increased expression of Kir2.1 protein encoded by KCNJ2 and causes shortening of APD and hyperpolarization of the membrane potential, which may promote re-entry and stabilize atrial rotors [60, 61](Table 3).
miR-1
MiR-1 is a muscle-specific miRNA and the most abundantly expressed miRNA in both ventricles and atria [62]. MiR-1 was upregulated in a rabbit ATP model along with a decrease of KCNE1 and KCNB2 mRNA and protein, shortening of the atrial effective refractory period (AERP) and increase of AF susceptibility. In vivo upregulation of miR-1 through atrial injection of a recombinant lentivirus carrying miR-1, resulted in enhanced downregulation of KCNE1 and KCNB2 and, unlike what may be expected, caused an increase in delayed rectifier potassium current (IKs) and AF susceptibility. Conversely, anti-miR-1 attenuated the decrease of KCNE1 and KCNB2, and decreased AF vulnerability. KCNE1 and KCNB2 were confirmed as direct targets of miR-1 with luciferase assays [63]. However, because human atrial myocytes have different electrophysiological properties compared to rabbits, upregulation of IKs is less likely to contribute to AF pathophysiology in patients [64] and others have reported contradicting results about the regulation of miR-1 in AF (Table 3). Most importantly, Girmatsion et al. [15] found greatly reduced levels of miR-1 in atrial tissue of AF patients, which corresponded to an upregulation of KCNJ2, the inward-rectifier potassium ion channel protein (Kir2.1) and IK1. Furthermore, reduced miR-1 levels and increased Kir2.1 could be induced by ex vivo tachypacing of human atrial slices [15].
MiR-1 has extensively been studied in the context of ventricular arrhythmogenesis and interestingly, an upregulation of ventricular miR-1 was reported in patients with coronary artery disease and in a mouse model of ischemia, where it also had pro-arrhythmogenic effects [65]. Moreover, Zhao et al. [66] suggested miR-1 in the ventricle to target the transcription factor Irx5, which regulates several ion channels and gap junction proteins, and Terentyev et al. [67] related miR-1 to calcium handling abnormalities in the failing heart. As electrical remodelling in AF is also characterized by a dysregulation of ion channels and calcium handling, this suggests that the effects of miR-1 in AF are much broader than currently described.
miR-26a/b
A downregulation of miR-26 in atria of AF patients was associated with increased KCNJ2 and Kir2.1 [68]. Two subsequent studies of the same study group including both canine and mice tachypacing models resulted in a decreased expression of miR-26b and an even stronger decrease of miR-26a in atrial tissue [68] and cardiac fibroblasts [16]. In vitro knockdown of miR-26a with an LNA-based antimiR in canine fibroblasts induced an increase in Ik1, hyperpolarized the resting membrane potential and increased fibroblast proliferation [16]. In vivo inhibition of miR-26a in mice with antimiR-26a increased IK1 and promoted AF, whereas in vivo adenoviral overexpression of miR-26a decreased IK1 and damped AF vulnerability [68]. Luciferase assays confirmed miR-26 to directly target KCNJ2 [68]. Of note, besides these two reports from the same study group, other discovery studies are inconsistent about the up- or downregulation of miR-26 isoforms in atrial tissue [56, 69, 70].
miR-30d
Morishima et al. [71] performed a microarray screen to identify miRNAs involved in electrical remodelling and found miR-30d to be highly expressed in patients with persAF, corresponding to downregulation of KCNJ3 and Kir3.1. MiR-30d was found to directly target KCNJ3 by luciferase assays and in vitro transfection of miR-30d was associated with a downregulation of KCNJ3, Kir3.1 and IKACh. In patients with sustained AF, the acetylcholine-regulated K+-current (IKACh), which is an inward rectifier current carried by the Kir3.1 and Kir3.4 subunits, was found downregulated [72]. However, another study revealed constitutive activity of IKACh alongside an increase in IK1, which may contribute to APD shortening and AF [73].
miR-499
MiR-499 was upregulated in a microarray screen of RA tissue of 4 AF patients with and 4 patients without AF. Correspondingly, there was a decrease of the small conductance Ca2+-activated potassium channel (SK3) [74]. Luciferase assay confirmed the encoding KCNN3 as a direct target of miR-499 and in vitro overexpression of miR-499 downregulated KCNN3 and SK3 while antimiR-499 upregulated SK3 levels. MiR-499 is a cardiomyocyte-enriched miRNA and circulating miR-499 has extensively been studied as a biomarker of myocardial infarction and HF [75]. Three explorative studies reported miR-499 to be upregulated in tissue of AF patients [53, 71, 74], but in other studies, miR-499 was downregulated [45, 58].
Sodium Channels
Aside from potassium current remodelling, sodium channel (INa) density may also be reduced in AF [76, 77]. This is supported by the fact that loss-of-function mutations in the SCN5A gene, encoding a subunit of the cardiac voltage gated sodium channel Nav1.5, have been associated with familial AF [78]. Zhao et al. [79] reported upregulation of miR-192-5p in AF patients which corresponded to downregulation of SCN5A and Nav1.5 protein. In vitro overexpression of miR-192-5p decreased INa density. MiR-192-5p is thereby the only miRNA that has been associated with sodium channel remodelling in AF.
Calcium Handling
High atrial rates during AF cause a Ca2+-overload and an imbalance in intracellular Ca2+ homeostasis which contributes to AF perpetuation. Increased diastolic leak of Ca2+ from the sarcoplasmic reticulum through the ryanodine receptor 2 (RYR2) promotes increased Na+/Ca2+ exchange through the Na+/Ca2+exchanger (NCX) [80–82]. This, in turn, depolarizes the cell membrane and thereby facilitates triggered activity [83–85]. In addition, abnormalities in cellular Ca2+ homeostasis may indirectly affect structural and electrical remodelling. For example, Ca2+-overload may activate the Ca2+-dependent calcineurin/nuclear factor of activated T cells (NFAT) system, which results in hypertrophy and fibrosis [16, 86].
miR-208b
Ca2+ from the cytosol is transported back into the sarcoplasmic reticulum through the sarcoplasmic reticulum Ca2+adenosine triphosphates type 2a (SERCA2a), which thereby influences sarcoplasmic and cytosolic Ca2+ concentration. Canon et al. [58] found an inverse correlation between an upregulation of miR-208b, but not of miR-208a, and a decrease in SERCA2 mRNA in atrial myocytes from AF and control patients. In vitro overexpression of miR-208b also decreased SERCA2 protein expression.
miR-106b-25 Cluster
Downregulation of the miR-106b-25 cluster was seen in parAF patients compared to controls and was associated with increased RYR2 protein expression. RyR2 mRNA expression was unchanged, suggestive of an inhibition of RYR2 translation by miR-106b-25. Luciferase assay confirmed miR-93, belonging to the cluster, to directly target RyR2. Moreover, spontaneous local and global sarcoplasmic reticulum Ca2+-releases were increased in miR-106b-25 knockout mice, resulting in higher AF susceptibility [87].
Extracellular Matrix Remodelling
Atrial fibrosis is considered the hallmark of atrial structural remodelling in AF. Fibrosis may promote re-entry by conduction slowing, increased anisotropy or unidirectional conduction block [88, 89]. Increased collagen content was found in AF patients compared to controls [90, 91] and an altered composition of ECM proteins was related to AF progression [89, 92]. Fibroblasts form the most abundant cell type in cardiac tissue and may, under pathophysiological conditions, provide pro-fibrotic signalling, or differentiate into myofibroblasts which secrete ECM proteins. Fibroblasts may furthermore by interactions with cardiomyocytes affect excitability and thereby conduction velocity [88, 93]. Various pro-fibrotic signalling pathways are involved in atrial fibrosis, such as the renin-angiotensin-aldosterone pathway, transforming growth factor-β1 (TGF-β1) [94], connective tissue growth factor (CTGF) and platelet-derived growth factor (PDGF) [88]. Here, we present the most important miRNAs that have been implicated as regulators of atrial fibrosis in AF.
miR-21
Upregulation of miR-21, highly expressed in fibroblasts, has been associated with increased cardiac fibrosis, not limited to AF [17, 18, 95–98]. Several mechanisms have been proposed for the potential effect of miR-21 on fibrosis, but most attention has been paid to miR-21 repression of SPRY1 (Sprouty 1, RTK signalling antagonist 1). Sprouty-1 inhibits the extracellular signal-regulated kinases (ERK) signalling pathway, which promotes fibrosis [17, 18, 95]. Upregulation of miR-21 was associated with downregulation of SPRY1 in patients with valvular AF and in rats with myocardial ischemia [17, 18]. In vivo inhibition with antagomir-21 in mice or a 15-mer LNA-based antimiR-21 in rats suppressed fibrosis and AF [18]. However, in the heart failure field contradicting results about the role of miR-21 in fibrosis have been reported. Thum et al. [99] showed that inhibition of miR-21 by antagomir injection protected mice against cardiac fibrosis and attenuated cardiac dysfunction in response to thoracic aorta constriction (TAC). On the other hand, Patrick et al. [100] reported that neither genetic deletion of miR-21 nor inhibition with tiny LNA-based antimiRs altered cardiac fibrosis in response to various stresses, like TAC and MI. Contradicting findings may results from different effectiveness of the antimiR-21 and antagomir-21 chemistries, used in the different studies [101].
MiR-21 may also promote inflammation-associated atrial fibrosis through the phosphorylation of the transcription factor signal transducer and activator of transcription 3 (STAT3). Inhibition with antagomir-21 in rats with pericarditis and AF suppressed STAT3 phosphorylation, the expression of fibrosis-related genes and AF vulnerability. MiR-21 promotes STAT3 phosphorylation through targeting the protein inhibitor of activated STAT3 (PIAS3) in multiple myeloma cells [102]. Expression of miR-21 itself was also found to be positively regulated by phosphorylated STAT3 and may thus form a feedback loop. Moreover, cardiac fibroblasts stimulated with the cytokine interleukin-6 increased STAT3 phosphorylation and miR-21 expression. This pathway may therefore link atrial inflammation to fibrosis formation [98].
Another suggested signalling pathway of miR-21 in AF involves the downregulation of Smad7, which is an inhibitory Smad of the TGFβ-pathway. Loss of Smad7 upregulates collagen I and III. MiR-21 expression was significantly increased in ATP rabbits along with a decrease of Smad7. In vivo inhibition of miR-21 suppressed the decrease of Smad7 and the increase of collagen I/III. Luciferase assays validated Smad7 as a direct target of miR-21 [97].
miR-26a
MiR-26 may, aside from potassium channel regulation, also play a role in ECM formation. MiR-26a was downregulated in the LA from dogs with heart failure and AF and corresponded to an increase of the Ca2+-permeable transient receptor potential canonical-3 (TRPC3) protein. Increased TRPC3 in turn stimulated fibroblast proliferation, differentiation and activation [86]. Luciferase assays validated miR-26a to directly target TRPC3 and in vitro inhibition of miR-26a with an LNA-based antimiR increased TRPC3 protein expression and promoted fibroblast proliferation. Moreover, increased TRPC3 expression was positively correlated with ERK phosphorylation and the expression of several ECM-related genes. Expression of miR-26 itself may be under the control of the NFAT system. The NFAT system downregulates miR-26 in response to Ca2+-loading and promotes fibroblast proliferation [86].
miR-29b
MiR-29b was downregulated in the atrium and in fibroblasts from dogs after ventricular tachypacing [103]. Both AF duration (after burst pacing) and atrial fibroblast COL1A1, COL3A1 and FBN mRNA increased significantly after prolonged ventricular tachypacing in these dogs. Moreover, overexpression of miR-29b in canine fibroblasts decreased COL1A1, COL3A1 and FBN expression, along with a decrease of Col1a1 protein in the supernatant. Adeno-associated virus (AAV) mediated knockdown with a miR-29b sponge had the opposite effect in fibroblasts and increased atrial Col1a1 mRNA and ventricular collagen content (Masson trichrome) in AAV-mediated knockdown mice [103]. Interestingly, this study also found miR-29b levels to be lower in tissue and plasma of patients with heart failure and concomitant AF.
miR-30a
In a rabbit ATP model, miR-30a was decreased in atrial tissue along with an increase of the transcription factor Snail 1, the matricellular protein Periostin and fibrotic tissue [104]. A functional role of miR-30a in the regulation of Snail1 and Periostin was revealed by overexpression and inhibition of miR-30a in rat cardiac fibroblasts [104]. Luciferase assays confirmed Snail 1 as a direct target of miR-30a. However, the mechanisms by which Snail 1 regulates Periostin, remains unclear.
Moreover, Li et al. [105] also associated the downregulation of miR-30 and miR-133 with an increase in fibrosis (Masson’s trichrome) in dogs with AF induced by tachypacing. However, they did not elaborate on potential targets of these miRNAs. In the heart failure field, Duisters et al. [19] concluded that miR-30 and miR-133 directly target CTGF and thereby control collagen content. Of note, miR-30a was described upregulated in AF patients in two studies investigating the LAA [46, 106] and in one study in the RAA [71]. Downregulation was seen in two studies investigating the RAA [44, 46].
miR-133 and miR-590
The expression levels of miR-133 and miR-590 were decreased in human atrial tissue of AF patients with nicotine abuses compared to non-users, and in a canine model with nicotine administration [107]. Transfection of miR-133 and miR-590 in canine atrial fibroblasts decreased TGF-β1, TGF-βRII and collagen content, which was reversed by inhibition by antagomirs respectively. Nicotine administration induced a downregulation of miR-133 and miR-590 in fibroblasts in a dose-dependent manner. TGF-β1 and TGF-βRII were established as direct targets of miR-133 and miR-590, respectively. MiR-133 was previously described to directly targeted CTGF, an important pro-fibrotic protein [19]. Moreover, CTGF is induced by TGF-β1 [19]. In explorative studies, miR-133 was downregulated in RAA in AF patients [44, 46].
miR-146b
Wang et al. [108] used LAAs from patients with and without AF and performed an integrated analysis of miRNA and mRNA expression profiles using microarray discovery followed by qPCR validation. This resulted in the miRNA-mRNA pair: miR-146-5p and tissue inhibitor of metalloproteinase 4 (TIMP-4). TIMP-4 is thought to inhibit matrix metallopeptidase 9 (MMP-9), involved in the degradation of extracellular matrix and formation of fibrosis. MiR-146b-5p was upregulated in AF patients along with an increase in MMP-9 and collagen content, but downregulation of TIMP-4. TIMP-4 was established as a direct target of miR-146b-5p by luciferase assays and transfection of miR-146b-5p in cardiac fibroblasts reduced TIMP-4 and increased collagen content. MiR-146b expression was furthermore correlated with LA diameter, AF duration and high-sensitivity CRP plasma levels [109]. In one of the explorative studies, miR-146b was actually the most upregulated miRNA in AF [44].
miR-208a/b
MiR-208a and miR-208b have frequently been implicated in AF pathophysiology and were described above for their role in Ca2+-handling and calcium channel regulation. However, these miRNAs are much better known for their role in structural remodelling in cardiovascular disease. MiR-208a and miR-208b are located within an intron of the α-cardiac muscle myosin heavy chain (MYH6) and the β-cardiac muscle myosin heavy chain (MYH7) gene, respectively [110]. Callis et al. [111] found that transgenic mice overexpressing miR-208a developed cardiac hypertrophy with suppressed expression of the targets thyroid hormone-associated protein (Thrap1, a known repressor of MYH7 transcription) and myostatin (a known repressor of muscle growth) [112]. MiR-208a knockout mice, on the other hand, developed spontaneous AF, accompanied by decreased Connexin 40 and elevated GATA4 protein levels [111]. However, the mechanism of arrhyhtmogenesis induced by miR-208 depletion remained unclear.
Canon et al. [58] found increased expression of miR-208a and especially of miR-208b in tissue of AF patients. Computational analysis predicted Sox5 and Sox6, negative factors of Myh7 transcription, as putative targets of miR-208. In vitro overexpression of miR-208a and miR-208b suppressed the expression of Sox5 and Sox6, respectively. Because the atrial tissue of AF patients showed a drastically increase of MYH7 protein levels, it is suggested that the increased expression of miR-208a/b in AF contributes to high MYH7 protein levels via inhibiting the expression of Sox5/6. Given that the healthy adult heart mainly expresses miR-208a and not miR-208b, miR-208a may initially target Thrap1 in AF pathophysiology. This in turn promotes MYH7 and simultaneously miR-208b transcription, which targets Sox5/6 and ultimately reinforces MYH7 transcription. MYH7 is a hallmark of cardiac hypertrophy, and the switch in MYH6:MYH7 expression ratio is linked to cardiac hypertrophy and heart failure. However, the mechanistic implications of MYH7 in AF remain unclear. MiR-208 has extensively been studied and was suggested as diagnostic biomarker of acute myocardial infarction or as therapeutic target in HF. For example, inhibition of this miRNA in several HF models has successfully prevented the formation of both cardiomyocyte hypertrophy and fibrosis, as discussed below [28, 110, 113]. Altogether, numerous studies associated miR-208a and miR-208b expression levels to cardiovascular disease and AF and proposed pathophysiological mechanisms involved in AF electrical and structural remodelling [31, 53, 58, 71, 74, 111, 114]. MiR-208 should therefore be considered as a potential target for AF therapy.
Autonomic Nervous System
Autonomic dysregulation plays an important role in AF onset and maintenance [115]. This involves the upregulation of the acetylcholine dependent Ik+ current (IKACh) which shortens APD [116]. Furthermore, structural remodelling of the autonomic nerves consists of sympathic hyperinnervation of the atria and an imbalance between sympathetic and parasympathetic nerves [117, 118].
No miRNA has directly been associated with autonomic dysregulation in AF. As described above, miR-30d was found upregulated in patients with persAF and associated with downregulation of IKACh [71]. Another study in ATP dogs demonstrated highly increased miR-206 levels, associated with structural remodelling of the autonomic nerves. In vivo lentiviral mediated overexpression of miR-206 in ATP dogs was associated with increased reactive oxygen species (ROS), nerve density and shortened AERP [119]. Luciferase assays confirmed that miR-206 directly regulated the anti-oxidant superoxide dismutase 1 (SOD1). These results suggest that miR-206 may induce autonomic nerve remodelling through a decrease of SOD1 and an increase of ROS.
Other miRNAs in Atrial Fibrillation
Some miRNAs found to be involved in AF pathophysiology have not been associated to the regulation of a specific ion channel or extracellular matrix genes. For example, the upregulation of miR-199a in AF was found to suppress and target FKBP5, also known as the FK506 binding protein 5, an immunoregulative protein. Chiang et al. [69] performed a miRNA-mRNA interaction study in atrial biopsies from patients with AF and matched controls. The upregulation of miR-199a correlated with a downregulation of FKBP5. Luciferase assays revealed a direct interaction between miR-199a-5p and FKBP5. FKBP5 may interact with heat shock protein and may be involved in stabilizing microtubules and intracellular trafficking. However, FKBP5 may also affect Ca2+-regulation, but its function in AF pathogenesis has not yet been demonstrated [120]. MiR-199a has been associated with POAF by Yamac et al. [32], who suggested miR-199a to target SIRT1, a protein with antioxidant activity.
Discussion
Limitations of microRNA Studies
Circulating and tissue miRNAs regulate determinants of AF pathophysiology and have emerged as biomarkers of this disease. In this review, we present an extensive list of supporting evidence for the role of miRNAs in AF, but the inconsistencies among the explorative and functional studies cannot be denied. So far, no distinct miRNA has been identified as a clinically useful biomarker or as target for AF treatment. The inconsistencies between studies might be the result of the variation between studies in biological and technical design and more standardized comparisons of different disease models and technical approaches for modulation of miRNA levels are needed before miRNAs can be used in clinical practice.
Many of the discrepancies between plasma and tissue studies may be the result of the biological variation between the populations studied. Study populations used for explorative studies were small and usually did not include over 10 samples per group whereas some studies were even based on pooled samples for the discovery phase. In general, study designs did not allow to take the progression of AF into account, although study populations ranged from parAF to permAF. Furthermore, study populations consisted of patients with various, but relevant comorbidities such as CABG or mitral valve disease, but frequently, studies provided inadequate clinical details or were insufficient in size to correct for these comorbidities. Finally, miRNA expression can be tissue specific and miRNA differences are likely to depend on the origin of the tissue which can be RA or LA. Indeed, studies investigating both RA and LA found significant differences in miRNA expression between these two atria [44, 46, 56, 106, 114].
From a technical point of view, variation between studies can be introduced at several levels. For example, at the time of sample retrieval, patients may have received heparin for cardiothoracic surgery or plasma may have been stored in heparin holding tubes. Heparin interferes with enzymes in the RT-PCR and thus may affect its results [121]. Furthermore, most studies used a diverse range of microarray platforms for the discovery phase. The limiting aspects of microarray technology such as low comparability and sensitivity may be in part responsible for the discrepancies between studies [122]. Data-analysis pipelines also showed clear variation in the thresholds used for miRNA selection and studies rarely performed multivariate analyses.
Studies usually validated the microarray discoveries with qPCR amplification. qPCR is the most sensitive technique to detect miRNAs, but reliability may be hampered by low miRNA levels, especially in plasma. Because of low miRNA levels, normalization is particularly important for plasma studies. This requires standardized techniques and an adequate correction for differences in starting material with reference genes (tissue) or reference miRNAs or miRNA spike-ins. The optimal endogenous reference panel may vary with the present clinical characteristics of each study population in both plasma and tissue. Tissue miRNA levels are generally high and normalization has not been problematic in other cardiovascular disease, but this may not be the case for AF as a recent report indeed demonstrated that the commonly used reference gene U6 was the worst normalizer of a panel of five for human atrial tissue [123]. To date, there is no standardized protocol for the use of endogenous normalizers in AF. Alternatively, synthetic miRNAs such as Caenorhabditis elegans-derived miRNAs have successfully been spiked-in as exogenous normalizers, but only when added after full inactivation of RNase activity [23].
The discovery of miRNA biomarkers in animal models may not directly be extrapolated to the diagnosis or prognosis of clinical AF. Firstly, human heart physiology differs from the animal heart and artificial AF models simplify the complexity and multifactorial character of the disease [64]. Animal models of AF used for miRNA studies enable functional analysis, but studies rarely started with an explorative miRNA expression screen to select the most differentially expressed miRNAs. MiRNAs studied for their function were usually selected based on an established mechanism in other (cardiovascular) diseases, or were derived from previous studies in AF.
Altogether, we can conclude that there are many factors contributing to the inconsistencies between both functional and explorative studies.
Future Perspective of miRNAs as Circulating Biomarkers
Despite the inconsistencies of miRNA expression among the current studies, circulating miRNAs remain promising biomarkers of AF. However, the specificity, the origin and function of miRNAs in the circulation are largely unknown. MiRNAs in the circulation have been demonstrated in conjunction with apoptotic bodies, microvesicles, exosomes or the so called protein-protected protein-miRNA complexes [20, 23, 24]. Circulating miRNAs are suggested to play a role in cell-to-cell signalling [124, 125]. For example, Bang et al. [126] demonstrated that cardiac fibroblasts excreted miR-21-3p in exosomes, which induced hypertrophy in cardiomyocytes. However, most miRNAs are thought to be excreted with protein complexes and to have limited function in cell-to-cell signalling [127, 128].
Circulating miRNAs do not necessarily reflect miRNA tissue levels. In this study, we demonstrated many contradictions in atrial tissue and plasma levels of specific miRNAs. Furthermore, our results imply that in tissue of AF patients, miRNAs were more frequently upregulated, whereas miRNAs levels in plasma were more often lower in AF patients. For example, plasma miR-328 levels were described to be lower in prevalent AF in multiple studies [30, 38, 39], whereas tissue miR-328 expression increased in dogs with AF and in LA tissue of AF patients [45]. A possible explanation for the contradicting increases and decreases in tissue and plasma could be retaining of cellular miRNA at the expense of miRNA secretion into the circulation. Alternatively, it is intriguing to speculate that the uptake by affected cells of circulating regulator miRNAs to restore intracellular levels might contribute to the difference between tissue and plasma levels.
If possible, miRNA expression in AF should be studied in the different compartments of the blood separately and tissue and plasma levels should be studied in parallel for a better understanding of their relation and indications for the specificity of circulating miRNAs. Because AF is such a complex disease, future studies should ideally hold larger study populations and perform more detailed clinical profiling to enable analysis of comorbidities. Ideally, studies should not only focus on the diagnostic value of miRNAs for AF presence, but also prospectively investigate the prognostic value of a single miRNA or a panel of miRNAs, for the occurrence of new-onset AF, AF recurrence or AF progression. In addition, miRNAs may be studied for monitoring of the disease or response to AF therapy.
Future Perspective of miRNAs as Therapeutics
MiRNAs hold a promise for the development of a new class of therapeutics as expression and function can be enhanced or repressed by the systemic or local delivery of synthetic miRNA mimics and inhibitors respectively [129]. MiRNA mimics are double-stranded oligonucleotides that resemble the miRNA-duplex. The ‘guide strand' from the miRNA mimic is then incorporated in the RNA-induced silencing complex (RISC) to become functional and bind to the target mRNA. As the ‘guide strand’ from the miRNA mimic has to be recognized properly and undergo processing as an endogenous pre-miRNA, chemical modification for the delivery of miRNA mimics to the site of function is more challenging compared to antimiR chemistry. MiRNA overexpression may therefore be established using different serotypes of AAVs to increase organ and cell specificity [130]. AAVs may also be used for knockdown with the delivery of a miRNA sponge. AntimiRs are antisense singe-stranded oligonucleotides that are complementary to a full or a part of the mature miRNA and block its function after hybridization. AntimiRs can be chemically modified at their sugar backbone to increase their stability, prevent against degradation and improve their cellular uptake (e.g. 2’-O-methoyethyl, 2′-fluoro, LNA modifications and cholesterol particles) [21, 131].
MiRNA research in AF and their role in atrial cardiomyopathies lags behind research in the field of ventricular cardiomyopathies since the first human explorative studies in AF were performed in 2011 and most functional studies, besides a few exceptions, date from 2012 onwards. Meanwhile, progress has been made on miRNA-based therapies in pre-clinical trials for other cardiac diseases. As an example, miR-208 is one of the most important miRNAs studied as a potential target for miRNA inhibition in ventricular cardiomyopathies [113]. It was also designated as an important regulator of atrial remodelling in this review and could thus also play a role in atrial cardiomyopathies. MiR-208a knockout mice exhibited reduced fibrosis in response to cardiac stress and failed to upregulate Myh7 [110]. Montgomery et al. [113] demonstrated that the delivery of an LNA-based antimiR against miR-208a suppressed fibrosis, improved cardiac function and improved survival in a hypertension induced heart-failure model in rats. Of note, caution with the inhibition of miR-208 should be taken as miR-208a knockout mice displayed spontaneous AF [111]. Meanwhile, no clinical trials have been performed with miRNA therapeutics in cardiovascular pathology, but in other fields, miRNA therapeutics have successfully shown to suppress hepatitis C virus replication in phase 2a clinical trials without long-term relevant side-effects [132].
Functional studies with in vivo manipulation of miRNAs in AF suggest that also for atrial cardiomyopathies, a specific miRNA based therapeutic might be developed [17, 45, 63]. For example, Lu et al. [45] found that after in vivo adenoviral mediated forced expression of miR-328, the antagomir-328 successfully reversed AF susceptibility in ATP dogs. Jia et al. [63] demonstrated the potential of inhibiting miR-1 by administration of LNA-based antimiR-1, which prolonged AERP and reduced AF susceptibility and duration in ATP rabbits.
Before miRNA-based therapy is implemented in clinical practice, concerns about the safety of miRNA therapeutics in humans need to be overcome [21, 131, 133]. The most important concern about miRNA therapy arises from its potential to target multiple pathways. MiRNAs may interfere with physiological pathways by the delivery of (a high load of) miRNA mimics in non-targeted organs, or non-targeted pathways in the targeted tissue. MiRNAs mimics could also interfere with normal gene regulation through competition with endogenous uptake of double-stranded RNA or compete with incorporation in the RISC complex. Conversely, miRNA inhibitors may also have off-target effects. Using tissue-specific miRNA inhibitors or targeted delivery of miRNA mimics or inhibitors, which should be the focus of future research, could overcome these issues. AAVs could be used for targeted delivery, but as these rely on the delivery of genetic material, the effects may be permanent and long-term negative effects need to be established [130, 134]. On the other hand, anti-miRs inhibit miRNA function for a prolonged period of time, but they are eventually degraded and thus their pharmacokinetic properties may be improved by chemical modifications. [113]. The first clinical trials with the systemic delivery of liver specific miRNAs did not show long-term adverse effects and miRNA therapeutics remain promising [132]. Meanwhile, future studies should focus on in vivo effects of cardiovascular miRNA therapeutics in order to demonstrate safety and firmly determine therapeutic potentials [21].
Conclusion
In this systematic review, we present up-to-date evidence on the role of miRNAs in AF pathophysiology. Explorative studies have indicated tissue and plasma miRNAs to be differentially expressed in patients with and without AF and functional studies implicated miRNAs in several pathophysiological pathways. However, the controversy among the studies was striking and careful attention needs to be paid when interpreting previous studies about the discovery or function of miRNA(s) in AF. Despite the explicit variation among the studies investigating miRNAs in AF, they may help to uncover the mechanisms underlying AF, have the potential to form a new class of biomarkers and promote the development of innovative therapies.
Electronic supplementary material
(DOC 57 kb)
Compliance with Ethical Standards
J.R. de Groot received a grant from NWO/ZonMW(106.146.310). A.J. Tijsen received a grant from NWO/ZonMW(016.166.150). To the best of our knowledge, no conflict of interest, financial or other, exists for the co-authors. This article does not contain any original studies with human participants or animals performed by any of the authors. There were no human cases included and thus no informed consent was needed.
Footnotes
SEARCH
A systematic search was performed in OVID MEDLINE (31–10-2016) to identify relevant articles. We excluded articles in Chinese or poor quality articles. We included human and animal studies. The search terms included synonyms for “atrium”, “atrial fibrillation” and “microRNAs”. (Details Online Resource)
Electronic supplementary material
The online version of this article (doi:10.1007/s10557-017-6736-z) contains supplementary material, which is available to authorized users.
References
- 1.Ball J, Carrington MJ, McMurray JJV, et al. Atrial fibrillation: profile and burden of an evolving epidemic in the 21st century. Int J Cardiol. 2013;167:1807–1824. doi: 10.1016/j.ijcard.2012.12.093. [DOI] [PubMed] [Google Scholar]
- 2.Chen LY, Shen W-K. Epidemiology of atrial fibrillation: a current perspective. Hear Rhythm Off J Hear Rhythm Soc. 2007;4:S1–S6. doi: 10.1016/j.hrthm.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 3.Andrade J, Khairy P, Dobrev D, et al. The clinical profile and pathophysiology of atrial fibrillation: relationships among clinical features, epidemiology, and mechanisms. Circ Res. 2014;114:1453–1468. doi: 10.1161/CIRCRESAHA.114.303211. [DOI] [PubMed] [Google Scholar]
- 4.Kirchhof P, Benussi S, Kotecha D, et al. 2016 ESC guidelines for the management of atrial fibrillation developed in collaboration with EACTS: the task force for the management of atrial fibrillation of the European Society of Cardiology (ESC)developed with the special contribution of the Europea. Eur Heart J 2016;:ehw210.
- 5.Goette A, Kalman JM, Aguinaga L, et al. EHRA/HRS/APHRS/SOLAECE expert consensus on atrial cardiomyopathies: definition, characterisation, and clinical implication. 2016. [DOI] [PMC free article] [PubMed]
- 6.Singh JP, Morady F. Patient selection and classification for atrial fibrillation ablation: thinking beyond duration. Hear Rhythm. 2009;6:1522–1525. doi: 10.1016/j.hrthm.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 7.Woods CE, Olgin J. Atrial fibrillation therapy now and in the future: drugs, biologicals, and ablation. Circ Res. 2014;114:1532–1546. doi: 10.1161/CIRCRESAHA.114.302362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smit MD, Van Gelder IC. New treatment options for atrial fibrillation: towards patient tailored therapy. Heart. 2011;97:1796–1802. doi: 10.1136/heartjnl-2011-300276. [DOI] [PubMed] [Google Scholar]
- 9.van Rooij E, Sutherland LB, Liu N, et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci U S A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 11.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;2008:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 12.Luo X, Yang B, Nattel S. MicroRNAs and atrial fibrillation: mechanisms and translational potential. Nat Rev Cardiol. 2015;12:80–90. doi: 10.1038/nrcardio.2014.178. [DOI] [PubMed] [Google Scholar]
- 13.Arora P, Wu C, Khan AM, et al. Atrial natriuretic peptide is negatively regulated by microRNA-425. J Clin Invest. 2013;123:3378–3382. doi: 10.1172/JCI67383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Rooij E, Sutherland LB, Thatcher JE, et al. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc Natl Acad Sci U S A. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girmatsion Z, Biliczki P, Bonauer A, et al. Changes in microRNA-1 expression and IK1 up-regulation in human atrial fibrillation. Hear Rhythm Off J Hear Rhythm Soc. 2009;6:1802–1809. doi: 10.1016/j.hrthm.2009.08.035. [DOI] [PubMed] [Google Scholar]
- 16.Qi X-Y, Huang H, Ordog B, et al. Fibroblast inward-rectifier potassium current upregulation in profibrillatory atrial remodeling. Circ Res. 2015;116:836–845. doi: 10.1161/CIRCRESAHA.116.305326. [DOI] [PubMed] [Google Scholar]
- 17.Adam O, Löhfelm B, Thum T, et al. Role of miR-21 in the pathogenesis of atrial fibrosis. Basic Res Cardiol. 2012;107:278. doi: 10.1007/s00395-012-0278-0. [DOI] [PubMed] [Google Scholar]
- 18.Cardin S, Guasch E, Luo X, et al. Role for MicroRNA-21 in atrial profibrillatory fibrotic remodeling associated with experimental postinfarction heart failure. Circ Arrhythm Electrophysiol. 2012;5:1027–1035. doi: 10.1161/CIRCEP.112.973214. [DOI] [PubMed] [Google Scholar]
- 19.Duisters RF, Tijsen AJ, Schroen B, et al. miR-133 and miR-30 Regulate Connective Tissue Growth Factor. Circ Res 2009;104. [DOI] [PubMed]
- 20.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18282. [DOI] [PubMed]
- 21.van Rooij E, Olson EN. MicroRNA therapeutics for cardiovascular disease: opportunities and obstacles. Nat Rev Drug Discov. 2012;11. [DOI] [PMC free article] [PubMed]
- 22.Weckbach LT, Grabmaier U, Clauss S, et al. MicroRNAs as a diagnostic tool for heart failure and atrial fibrillation. Curr Opin Pharmacol. 2016;27:24–30. doi: 10.1016/j.coph.2016.01.001. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber JA, Baxter DH, Zhang S, et al. The MicroRNA Spectrum in 12 body fluids. Clin Chem. 2010;56. [DOI] [PMC free article] [PubMed]
- 25.Tijsen AJ, Creemers EE, Moerland PD, et al. MiR423-5p as a circulating biomarker for heart failure. Circ Res. 2010;106:1035–1039. doi: 10.1161/CIRCRESAHA.110.218297. [DOI] [PubMed] [Google Scholar]
- 26.Fichtlscherer S, De Rosa S, Fox H, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107:677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 27.D’Alessandra Y, Devanna P, Limana F, et al. Circulating microRNAs are new and sensitive biomarkers of myocardial infarction. Eur Heart J. 2010;31:2765–2773. doi: 10.1093/eurheartj/ehq167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devaux Y, Mueller M, Haaf P, et al. Diagnostic and prognostic value of circulating microRNAs in patients with acute chest pain. J Intern Med. 2015;277:260–271. doi: 10.1111/joim.12183. [DOI] [PubMed] [Google Scholar]
- 29.Wang G-K, Zhu J-Q, Zhang J-T, et al. Circulating microRNA: a novel potential biomarker for early diagnosis of acute myocardial infarction in humans. Eur Heart J. 2010;31:659–666. doi: 10.1093/eurheartj/ehq013. [DOI] [PubMed] [Google Scholar]
- 30.McManus DD, Lin H, Tanriverdi K, et al. Relations between circulating microRNAs and atrial fibrillation: data from the Framingham Offspring Study. Hear Rhythm Off J Hear Rhythm Soc. 2014;11:663–669. doi: 10.1016/j.hrthm.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harling L, Lambert J, Ashrafian H, et al. Elevated serum microRNA 483-5p levels may predict patients at risk of post-operative atrial fibrillation. Eur J Cardio-Thoracic Surg Off J Eur Assoc Cardio-Thoracic Surg Published Online First: 2016. [DOI] [PMC free article] [PubMed]
- 32.Yamac AH, Kucukbuzcu S, Ozansoy M, et al. Altered expression of micro-RNA 199a and increased levels of cardiac SIRT1 protein are associated with the occurrence of atrial fibrillation after coronary artery bypass graft surgery. Cardiovasc Pathol Off J Soc Cardiovasc Pathol. 2016;25:232–236. doi: 10.1016/j.carpath.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 33.Slagsvold KH, Rognmo O, Høydal M, et al. Remote ischemic preconditioning preserves mitochondrial function and influences myocardial microRNA expression in atrial myocardium during coronary bypass surgery. Circ Res. 2014;114:851–859. doi: 10.1161/CIRCRESAHA.114.302751. [DOI] [PubMed] [Google Scholar]
- 34.Krogstad LEB, Slagsvold KH, Wahba A. Remote ischemic preconditioning and incidence of postoperative atrial fibrillation. Scand Cardiovasc J SCJ. 2015;49:117–122. doi: 10.3109/14017431.2015.1010565. [DOI] [PubMed] [Google Scholar]
- 35.Maesen B, Nijs J, Maessen J, et al. Post-operative atrial fibrillation: a maze of mechanisms. Europace. 2012;14. [DOI] [PMC free article] [PubMed]
- 36.Goren Y, Meiri E, Hogan C, et al. Relation of reduced expression of MiR-150 in platelets to atrial fibrillation in patients with chronic systolic heart failure. Am J Cardiol. 2014;113:976–981. doi: 10.1016/j.amjcard.2013.11.060. [DOI] [PubMed] [Google Scholar]
- 37.Liu Z, Zhou C, Liu Y, et al. The expression levels of plasma micoRNAs in atrial fibrillation patients. PLoS One. 2012;7:1–9. doi: 10.1371/journal.pone.0044906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu T, Zhong S, Rao F, et al. Catheter ablation restores decreased plasma miR-409-3p and miR-432 in atrial fibrillation patients. Eur Eur Pacing, Arrhythmias, Card Electrophysiol J Work Groups Card Pacing, Arrhythmias, Card Cell Electrophysiol Eur Soc Cardiol. 2016;18:92–99. doi: 10.1093/europace/euu366. [DOI] [PubMed] [Google Scholar]
- 39.Lu Y, Hou S, Huang D, et al. Expression profile analysis of circulating microRNAs and their effects on ion channels in Chinese atrial fibrillation patients. Int J Clin Exp Med. 2015;8:845–853. [PMC free article] [PubMed] [Google Scholar]
- 40.McManus DD, Tanriverdi K, Lin H, et al. Plasma microRNAs are associated with atrial fibrillation and change after catheter ablation (the miRhythm study) Hear Rhythm Off J Hear Rhythm Soc. 2015;12:3–10. doi: 10.1016/j.hrthm.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schotten U, Verheule S, Kirchhof P, et al. Pathophysiological mechanisms of atrial fibrillation: a translational appraisal. Physiol Rev. 2011;91. [DOI] [PubMed]
- 42.Choudhury A, Chung I, Blann AD, et al. Platelet surface CD62P and CD63, mean platelet volume, and soluble/platelet P-selectin as indexes of platelet function in atrial fibrillation. J Am Coll Cardiol. 2007;49:1957–1964. doi: 10.1016/j.jacc.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 43.Lu J, Guo S, Ebert BL, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cooley N, Cowley MJ, Lin RCY, et al. Influence of atrial fibrillation on microRNA expression profiles in left and right atria from patients with valvular heart disease. Physiol Genomics. 2012;44:211–219. doi: 10.1152/physiolgenomics.00111.2011. [DOI] [PubMed] [Google Scholar]
- 45.Lu Y, Zhang Y, Wang N, et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122:2378–2387. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]
- 46.Liu H, Qin H, Chen G, et al. Comparative expression profiles of microRNA in left and right atrial appendages from patients with rheumatic mitral valve disease exhibiting sinus rhythm or atrial fibrillation. J Transl Med. 2014;12:90. doi: 10.1186/1479-5876-12-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schotten U, Dobrev D, Platonov PG, et al. Current controversies in determining the main mechanisms of atrial fibrillation. J Intern Med. 2016;279:428–438. doi: 10.1111/joim.12492. [DOI] [PubMed] [Google Scholar]
- 48.Andrade J, Khairy P, Dobrev D, et al. The clinical profile and pathophysiology of atrial fibrillation: relationships among clinical features, epidemiology, and mechanisms. Circ Res. 2014;114:1453–1468. doi: 10.1161/CIRCRESAHA.114.303211. [DOI] [PubMed] [Google Scholar]
- 49.Nattel S, Harada M. Atrial remodeling and atrial fibrillation: recent advances and translational perspectives. J Am Coll Cardiol. 2014;63:2335–2345. doi: 10.1016/j.jacc.2014.02.555. [DOI] [PubMed] [Google Scholar]
- 50.Heijman J, Voigt N, Nattel S, et al. Cellular and molecular electrophysiology of atrial fibrillation initiation, maintenance, and progression. Circ Res. 2014;114 [DOI] [PubMed]
- 51.Tamargo J, Caballero R, Gomez R, et al. Cardiac electrophysiological effects of nitric oxide. Cardiovasc Res. 2010;87:593–600. doi: 10.1093/cvr/cvq214. [DOI] [PubMed] [Google Scholar]
- 52.Reilly SN, Liu X, Carnicer R, et al. Up-regulation of miR-31 in human atrial fibrillation begets the arrhythmia by depleting dystrophin and neuronal nitric oxide synthase. Sci Transl Med. 2016;8:340ra74. doi: 10.1126/scitranslmed.aac4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishi H, Sakaguchi T, Miyagawa S, et al. Impact of microRNA expression in human atrial tissue in patients with atrial fibrillation undergoing cardiac surgery. PLoS One. 2013;8 doi: 10.1371/journal.pone.0073397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Morishima M, Iwata E, Nakada C, et al. Atrial fibrillation-mediated upregulation of miR-30d regulates myocardial electrical remodeling of the G-protein-gated K(+) channel, IK.ACh. Circ J Off J Japanese Circ Soc. 2016;80:1346–1355. doi: 10.1253/circj.CJ-15-1276. [DOI] [PubMed] [Google Scholar]
- 55.Watson CJ, Gupta SK, O’Connell E, et al. MicroRNA signatures differentiate preserved from reduced ejection fraction heart failure. Eur J Heart Fail. 2015;17:405–415. doi: 10.1002/ejhf.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu H, Chen G, Liang M, et al. Atrial fibrillation alters the microRNA expression profiles of the left atria of patients with mitral stenosis. BMC Cardiovasc Disord. 2014;14:10. doi: 10.1186/1471-2261-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soeki T, Matsuura T, Bando S, et al. Relationship between local production of microRNA-328 and atrial substrate remodeling in atrial fibrillation: J Cardiol Published Online First; 2016. [DOI] [PubMed]
- 58.Cañón S, Caballero R, Herraiz-Martínez A, et al. miR-208b upregulation interferes with calcium handling in HL-1 atrial myocytes: implications win human chronic atrial fibrillation. J Mol Cell Cardiol. 2016;99:162–173. doi: 10.1016/j.yjmcc.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 59.Barana A, Matamoros M, Dolz-Gaitón P, et al. Chronic atrial fibrillation increases microRNA-21 in human atrial myocytes decreasing L-type calcium current. Circ Arrhythm Electrophysiol. 2014;7:861–868. doi: 10.1161/CIRCEP.114.001709. [DOI] [PubMed] [Google Scholar]
- 60.Pandit SV, Berenfeld O, Anumonwo JMB, et al. Ionic determinants of functional reentry in a 2-D model of human atrial cells during simulated chronic atrial fibrillation. Biophys J. 2005;88:3806–3821. doi: 10.1529/biophysj.105.060459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobrev D, Graf E, Wettwer E, et al. Molecular basis of downregulation of G-protein-coupled inward rectifying K+ current (IK,ACh) in chronic human atrial fibrillation: decrease in GIRK4 mRNA correlates with reduced IK,ACh and muscarinic receptor-mediated shortening of action potentials. Circulation. 2001;104:2551–2557. doi: 10.1161/hc4601.099466. [DOI] [PubMed] [Google Scholar]
- 62.Kakimoto Y, Tanaka M, Kamiguchi H, et al. MicroRNA deep sequencing reveals chamber-specific miR-208 family expression patterns in the human heart. Int J Cardiol. 2016;211:43–48. doi: 10.1016/j.ijcard.2016.02.145. [DOI] [PubMed] [Google Scholar]
- 63.Jia X, Zheng S, Xie X, et al. MicroRNA-1 accelerates the shortening of atrial effective refractory period by regulating KCNE1 and KCNB2 expression: an atrial tachypacing rabbit model. PLoS One. 2013;8:e85639. doi: 10.1371/journal.pone.0085639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Christ T, Wettwer E, Ravens U. Letter by Christ et al regarding article, "Angiotensin II potentiates the slow component of delayed rectifier K+ current via the AT1 receptor in Guinea pig atrial Myocytes". Circulation. 2006;114:e565. doi: 10.1161/CIRCULATIONAHA.106.639377. [DOI] [PubMed] [Google Scholar]
- 65.Yang B, Lin H, Xiao J, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 66.Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 67.Terentyev D, Belevych AE, Terentyeva R, et al. miR-1 overexpression enhances Ca2+ release and promotes cardiac Arrhythmogenesis by targeting PP2A regulatory subunit B56 and causing CaMKII-dependent hyperphosphorylation of RyR2. Circ Res. 2009;104:514–521. doi: 10.1161/CIRCRESAHA.108.181651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Luo X, Pan Z, Shan H, et al. MicroRNA-26 governs profibrillatory inward-rectifier potassium current changes in atrial fibrillation. J Clin Invest. 2013;123:1939–1951. doi: 10.1172/JCI62185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chiang DY, Zhang M, Voigt N, et al. Identification of microRNA-mRNA dysregulations in paroxysmal atrial fibrillation. Int J Cardiol. 2015;184:190–197. doi: 10.1016/j.ijcard.2015.01.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xiao J, Liang D, Zhang Y, et al. MicroRNA expression signature in atrial fibrillation with mitral stenosis. Physiol Genomics. 2011;43:655–664. doi: 10.1152/physiolgenomics.00139.2010. [DOI] [PubMed] [Google Scholar]
- 71.Morishima M, Iwata E, Nakada C, et al. Atrial fibrillation-mediated upregulation of miR-30d regulates myocardial electrical remodeling of the G-protein-gated K(+) channel, IK.ACh. Circ J. 2016;80:1346–1355. doi: 10.1253/circj.CJ-15-1276. [DOI] [PubMed] [Google Scholar]
- 72.Voigt N, Trausch A, Knaut M, et al. Left-to-right atrial inward rectifier potassium current gradients in patients with paroxysmal versus chronic atrial fibrillation. Circ Arrhythmia Electrophysiol. 2010;3:472–480. doi: 10.1161/CIRCEP.110.954636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dobrev D, Friedrich A, Voigt N, et al. The G protein-gated potassium current IK,ACh Is constitutively active in patients with chronic atrial fibrillation. Circulation. 2005;112:3697–3706. doi: 10.1161/CIRCULATIONAHA.105.575332. [DOI] [PubMed] [Google Scholar]
- 74.Ling T-Y, Wang X-L, Chai Q, et al. Regulation of the SK3 channel by microRNA-499--potential role in atrial fibrillation. Hear Rhythm Off J Hear Rhythm Soc. 2013;10:1001–1009. doi: 10.1016/j.hrthm.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Corsten MF, Dennert R, Jochems S, et al. Circulating MicroRNA-208b and MicroRNA-499 reflect myocardial damage in cardiovascular disease. Circ Cardiovasc Genet. 2010;3:499–506. doi: 10.1161/CIRCGENETICS.110.957415. [DOI] [PubMed] [Google Scholar]
- 76.Sossalla S, Kallmeyer B, Wagner S, et al. Altered Na(+) currents in atrial fibrillation effects of ranolazine on arrhythmias and contractility in human atrial myocardium. J Am Coll Cardiol. 2010;55:2330–2342. doi: 10.1016/j.jacc.2009.12.055. [DOI] [PubMed] [Google Scholar]
- 77.Gaspo R, Bosch RF, Bou-Abboud E, et al. Tachycardia-induced changes in Na+ current in a chronic dog model of atrial fibrillation. Circ Res. 1997;81:1045–1052. doi: 10.1161/01.RES.81.6.1045. [DOI] [PubMed] [Google Scholar]
- 78.Ellinor PT, Nam EG, Shea MA, et al. Cardiac sodium channel mutation in atrial fibrillation. Hear Rhythm. 2008;5:99–105. doi: 10.1016/j.hrthm.2007.09.015. [DOI] [PubMed] [Google Scholar]
- 79.Zhao Y, Huang Y, Li W, et al. Post-transcriptional regulation of cardiac sodium channel gene SCN5A expression and function by miR-192-5p. Biochim Biophys Acta. 1852;2015:2024–2034. doi: 10.1016/j.bbadis.2015.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hove-Madsen L, Llach A, Bayes-Genís A, et al. Atrial fibrillation is associated with increased spontaneous calcium release from the sarcoplasmic reticulum in human atrial myocytes. Circulation. 2004;110:1358–1363. doi: 10.1161/01.CIR.0000141296.59876.87. [DOI] [PubMed] [Google Scholar]
- 81.Voigt N, Li N, Wang Q, et al. Enhanced sarcoplasmic reticulum Ca2+ leak and increased Na+−Ca2+ exchanger function underlie delayed afterdepolarizations in patients with chronic atrial fibrillation. Circulation. 2012;125:2059–2070. doi: 10.1161/CIRCULATIONAHA.111.067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Li N, Chiang DY, Wang S, et al. Ryanodine receptor-mediated calcium leak drives progressive development of an atrial fibrillation substrate in a transgenic mouse model. Circulation. 2014;129:1276–1285. doi: 10.1161/CIRCULATIONAHA.113.006611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Harada M, Luo X, Murohara T, et al. MicroRNA regulation and cardiac calcium signaling: role in cardiac disease and therapeutic potential. Circ Res. 2014;114:689–705. doi: 10.1161/CIRCRESAHA.114.301798. [DOI] [PubMed] [Google Scholar]
- 84.Nattel S, Dobrev D. The multidimensional role of calcium in atrial fibrillation pathophysiology: mechanistic insights and therapeutic opportunities. Eur Heart J. 2012;33. [DOI] [PubMed]
- 85.Vest JA, Wehrens XHT, Reiken SR, et al. Defective cardiac ryanodine receptor regulation during atrial fibrillation. Circulation. 2005;111. [DOI] [PubMed]
- 86.Harada M, Luo X, Qi XY, et al. Transient receptor potential canonical-3 channel-dependent fibroblast regulation in atrial fibrillation. Circulation. 2012;126:2051–2064. doi: 10.1161/CIRCULATIONAHA.112.121830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Chiang DY, Kongchan N, Beavers DL, et al. Loss of microRNA-106b-25 cluster promotes atrial fibrillation by enhancing ryanodine receptor type-2 expression and calcium release. Circ Arrhythm Electrophysiol. 2014;7:1214–1222. doi: 10.1161/CIRCEP.114.001973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Burstein B, Nattel S. Atrial fibrosis: mechanisms and clinical relevance in atrial fibrillation. J Am Coll Cardiol. 2008;51:802–809. doi: 10.1016/j.jacc.2007.09.064. [DOI] [PubMed] [Google Scholar]
- 89.Krul SPJ, Berger WR, Smit NW, et al. Atrial fibrosis and conduction slowing in the left atrial appendage of patients undergoing thoracoscopic surgical pulmonary vein isolation for atrial fibrillation. Circ Arrhythmia Electrophysiol. 2015;8. [DOI] [PubMed]
- 90.Frustaci A, Chimenti C, Bellocci F, et al. Histological substrate of atrial biopsies in patients with lone atrial fibrillation. Circulation. 1997;96. [DOI] [PubMed]
- 91.Boldt A, Wetzel U, Lauschke J, et al. Fibrosis in left atrial tissue of patients with atrial fibrillation with and without underlying mitral valve disease. Heart. 2004;90:400–405. doi: 10.1136/hrt.2003.015347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xu J, Cui G, Esmailian F, et al. Atrial extracellular matrix remodeling and the maintenance of atrial fibrillation. Circulation. 2004;109. [DOI] [PubMed]
- 93.Souders CA, Bowers SLK, Baudino TA. Cardiac fibroblast: the renaissance cell. Circ Res. 2009;105:1164–1176. doi: 10.1161/CIRCRESAHA.109.209809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lin CS, Pan CH. Regulatory mechanisms of atrial fibrotic remodeling in atrial fibrillation. Cell Mol Life Sci. 2008;65:1489–1508. doi: 10.1007/s00018-008-7408-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 96.Adam O, Zimmer C, Hanke N, et al. Inhibition of aldosterone synthase (CYP11B2) by torasemide prevents atrial fibrosis and atrial fibrillation in mice. J Mol Cell Cardiol. 2015;85:140–150. doi: 10.1016/j.yjmcc.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 97.He X, Zhang K, Gao X, et al. Rapid atrial pacing induces myocardial fibrosis by down-regulating Smad7 via microRNA-21 in rabbit. Heart Vessels Published Online First: 2016. [DOI] [PMC free article] [PubMed]
- 98.Huang Z, Chen X-J, Qian C, et al. Signal transducer and activator of transcription 3/MicroRNA-21 feedback loop contributes to atrial fibrillation by promoting atrial fibrosis in a rat sterile pericarditis model. Circ Arrhythm Electrophysiol. 2016;9. [DOI] [PMC free article] [PubMed]
- 99.Thum T, Gross C, Fiedler J, et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature. 2008;456:980–984. doi: 10.1038/nature07511. [DOI] [PubMed] [Google Scholar]
- 100.Patrick DM, Montgomery RL, Qi X, et al. Stress-dependent cardiac remodeling occurs in the absence of microRNA-21 in mice. J Clin Invest. 2010;120:3912–3916. doi: 10.1172/JCI43604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tijsen AJ, Pinto YM, Creemers EE. Non-cardiomyocyte microRNAs in heart failure. Cardiovasc Res. 2012;93:573–582. doi: 10.1093/cvr/cvr344. [DOI] [PubMed] [Google Scholar]
- 102.Xiong Q, Zhong Q, Zhang J, et al. Identification of novel miR-21 target proteins in multiple myeloma cells by quantitative proteomics. J Proteome Res. 2012;11:2078–2090. doi: 10.1021/pr201079y. [DOI] [PubMed] [Google Scholar]
- 103.Dawson K, Wakili R, Ordög B, et al. MicroRNA29: a mechanistic contributor and potential biomarker in atrial fibrillation. Circulation. 2013;127:1466–1475. doi: 10.1161/CIRCULATIONAHA.112.001207. [DOI] [PubMed] [Google Scholar]
- 104.Yuan C-T, Li X-X, Cheng Q-J, et al. MiR-30a regulates the atrial fibrillation-induced myocardial fibrosis by targeting snail 1. Int J Clin Exp Pathol. 2015;8:15527–15536. [PMC free article] [PubMed] [Google Scholar]
- 105.Li H, Li S, Yu B, et al. Expression of miR-133 and miR-30 in chronic atrial fibrillation in canines. Mol Med Rep. 2012;5:1457–1460. doi: 10.3892/mmr.2012.831. [DOI] [PubMed] [Google Scholar]
- 106.Slagsvold KH, Johnsen AB, Rognmo O, et al. Comparison of left versus right atrial myocardium in patients with sinus rhythm or atrial fibrillation - an assessment of mitochondrial function and microRNA expression. Physiol Rep 2014;2. [DOI] [PMC free article] [PubMed]
- 107.Shan H, Zhang Y, Lu Y, et al. Downregulation of miR-133 and miR-590 contributes to nicotine-induced atrial remodelling in canines. Cardiovasc Res. 2009;83:465–472. doi: 10.1093/cvr/cvp130. [DOI] [PubMed] [Google Scholar]
- 108.Wang J, Wang Y, Han J, et al. Integrated analysis of microRNA and mRNA expression profiles in the left atrium of patients with nonvalvular paroxysmal atrial fibrillation: role of miR-146b-5p in atrial fibrosis. Hear Rhythm Off J Hear Rhythm Soc. 2015;12:1018–1026. doi: 10.1016/j.hrthm.2015.01.026. [DOI] [PubMed] [Google Scholar]
- 109.Wang J, Song S, Xie C, et al. MicroRNA profiling in the left atrium in patients with non-valvular paroxysmal atrial fibrillation. BMC Cardiovasc Disord. 2015;15:97. doi: 10.1186/s12872-015-0085-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van Rooij E, Sutherland LB, Qi X, et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 111.Callis TE, Pandya K, Seok HY, et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J Clin Invest. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Morissette MR, Cook SA, Foo S, et al. Myostatin regulates cardiomyocyte growth through modulation of Akt signaling. Circ Res. 2006;99:15–24. doi: 10.1161/01.RES.0000231290.45676.d4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Montgomery RL, Hullinger TG, Semus HM, et al. Therapeutic inhibition of miR-208a improves cardiac function and survival during heart failure. Circulation. 2011;124:1537–1547. doi: 10.1161/CIRCULATIONAHA.111.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Slagsvold KH, Johnsen AB, Rognmo O, et al. Mitochondrial respiration and microRNA expression in right and left atrium of patients with atrial fibrillation. Physiol Genomics. 2014;46:505–511. doi: 10.1152/physiolgenomics.00042.2014. [DOI] [PubMed] [Google Scholar]
- 115.Shen MJ, Choi E-K, Tan AY, et al. Neural mechanisms of atrial arrhythmias. Nat Rev Cardiol. 2012;9:30–39. doi: 10.1038/nrcardio.2011.139. [DOI] [PubMed] [Google Scholar]
- 116.Kovoor P, Wickman K, Maguire CT, et al. Evaluation of the role of I(KACh) in atrial fibrillation using a mouse knockout model. J Am Coll Cardiol. 2001;37:2136–2143. doi: 10.1016/S0735-1097(01)01304-3. [DOI] [PubMed] [Google Scholar]
- 117.Chen P-S, Chen LS, Fishbein MC, et al. Role of the autonomic nervous system in atrial fibrillation: pathophysiology and therapy. Circ Res. 2014;114:1500–1515. doi: 10.1161/CIRCRESAHA.114.303772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Deneke T, Chaar H, de Groot JR, et al. Shift in the pattern of autonomic atrial innervation in subjects with persistent atrial fibrillation. Hear Rhythm. 2011;8:1357–1363. doi: 10.1016/j.hrthm.2011.04.013. [DOI] [PubMed] [Google Scholar]
- 119.Zhang Y, Zheng S, Geng Y, et al. MicroRNA profiling of atrial fibrillation in canines: miR-206 modulates intrinsic cardiac autonomic nerve remodeling by regulating SOD1. PLoS One. 2015;10:e0122674. doi: 10.1371/journal.pone.0122674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cioffi DL, Hubler TR, Scammell JG. Organization and function of the FKBP52 and FKBP51 genes. Curr Opin Pharmacol. 2011;11:308–313. doi: 10.1016/j.coph.2011.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kim D-J, Linnstaedt S, Palma J, et al. Plasma components affect accuracy of circulating cancer-related microRNA quantitation. J Mol Diagn. 2012;14:71–80. doi: 10.1016/j.jmoldx.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006;25:6220–6227. doi: 10.1038/sj.onc.1209914. [DOI] [PubMed] [Google Scholar]
- 123.Masè M, Grasso M, Avogaro L, et al. Selection of reference genes is critical for miRNA expression analysis in human cardiac tissue. A focus on atrial fibrillation. Sci Rep. 2017;7:41127. doi: 10.1038/srep41127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Creemers EE, Tijsen AJ, Pinto YM. Circulating MicroRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 125.Rayner KJ, Hennessy EJ. Extracellular communication via microRNA: lipid particles have a new message. J Lipid Res. 2013;54:1174–1181. doi: 10.1194/jlr.R034991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bang C, Batkai S, Dangwal S, et al. Cardiac fibroblast–derived microRNA passenger strand-enriched exosomes mediate cardiomyocyte hypertrophy. J Clin Invest. 2014;124:2136–2146. doi: 10.1172/JCI70577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Turchinovich A, Tonevitsky AG, Cho WC, et al. Check and mate to exosomal extracellular miRNA: new lesson from a new approach. Front Mol Biosci. 2015;2:11. doi: 10.3389/fmolb.2015.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.van Rooij E, Purcell AL, Levin AA. Developing microRNA therapeutics. Circ Res. 2012;110:496–507. doi: 10.1161/CIRCRESAHA.111.247916. [DOI] [PubMed] [Google Scholar]
- 130.Asokan A, Samulski RJ. An emerging adeno-associated viral vector pipeline for cardiac Gene therapy. Hum Gene Ther. 2013;24:906–913. doi: 10.1089/hum.2013.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Laurence J, Van Beusekom M. Translating MicroRNAs to the clinic. Academic Press 2017. https://books.google.nl/books?id=bGklBgAAQBAJ&printsec=frontcover&dq=translating+microRNAs+to+the+clinic&hl=nl&sa=X&redir_esc=y#v=onepage&q=several chemical modifications are applied&f=false. Accessed 15 Mar 2017.
- 132.Janssen HLA, Reesink HW, Lawitz EJ, et al. Treatment of HCV infection by targeting MicroRNA. N Engl J Med. 2013;368:1685–1694. doi: 10.1056/NEJMoa1209026. [DOI] [PubMed] [Google Scholar]
- 133.Elmén J, Lindow M, Schütz S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 134.Grimm D, Streetz KL, Jopling CL, et al. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 135.Li Y-D, Hong Y-F, Yusufuaji Y, et al. Altered expression of hyperpolarization-activated cyclic nucleotide-gated channels and microRNA-1 and -133 in patients with age-associated atrial fibrillation. Mol Med Rep. 2015;12:3243–3248. doi: 10.3892/mmr.2015.3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Torrado M, Franco D, Lozano-Velasco E, et al. A MicroRNA-transcription factor blueprint for early atrial Arrhythmogenic remodeling. Biomed Res Int. 2015;2015:263151. doi: 10.1155/2015/263151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li H, Li S, Yu B, et al. Expression of miR-133 and miR-30 in chronic atrial fibrillation in canines. Mol Med Rep. 2012;5:1457–1460. doi: 10.3892/mmr.2012.831. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 57 kb)