We compared messenger RNA gene transcript profiles with standard wildlife diagnostic blood panels (e.g. hematology, biochemistry, serology and cytology) and physical examinations in order to improve interpretation of health and immune function in adult Agassiz’s desert tortoises.
Keywords: blood biochemistry, clinical status, desert tortoise, gene transcription, health, hematology, Mojave Desert, molecular, morphology, necropsy, reptile
Abstract
The analysis of blood constituents is a widely used tool to aid in monitoring of animal health and disease. However, classic blood diagnostics (i.e. hematologic and plasma biochemical values) often do not provide sufficient information to determine the state of an animal’s health. Field studies on wild tortoises and other reptiles have had limited success in drawing significant inferences between blood diagnostics and physiological and immunological condition. However, recent research using gene transcription profiling in the threatened Mojave desert tortoise (Gopherus agassizii) has proved useful in identifying immune or physiologic responses and overall health. To improve our understanding of health and immune function in tortoises, we evaluated both standard blood diagnostic (body condition, hematologic, plasma biochemistry values, trace elements, plasma proteins, vitamin A levels) and gene transcription profiles in 21 adult tortoises (11 clinically abnormal; 10 clinically normal) from Clark County, NV, USA. Necropsy and histology evaluations from clinically abnormal tortoises revealed multiple physiological complications, with moderate to severe rhinitis or pneumonia being the primary cause of morbidity in all but one of the examined animals. Clinically abnormal tortoises had increased transcription for four genes (SOD, MyD88, CL and Lep), increased lymphocyte production, biochemical enzymes and organics, trace elements of copper, and decreased numbers of leukocytes. We found significant positive correlations between increased transcription for SOD and increased trace elements for copper, as well as genes MyD88 and Lep with increased inflammation and microbial insults. Improved methods for health assessments are an important element of monitoring tortoise population recovery and can support the development of more robust diagnostic measures for ill animals, or individuals directly impacted by disturbance.
Introduction
Animal health plays an important role in the management and conservation of sensitive wildlife (Chabanet et al., 2016; Desforges et al., 2016; Rodriguez-Jorguera et al., 2016; Schoeman, 2016). Although a number of survey practices (i.e. physical examinations, hematological and biochemical blood panels, serology tests, and pathogen and toxicology screens) are used to assess wildlife health, these practices have had limited success in accurately tracking health status, particularly in free-ranging reptiles (Christopher et al., 2003; Madliger et al., 2016). Wildlife declines are generally the result of a combination of both anthropogenic (e.g. overharvest, habitat destruction, introductions of exotic species, contaminants) and natural (e.g. disease epidemics, drought, flood) stressors and rarely due to one single factor operating in isolation (Irwin and Irwin, 2006; Micheli et al., 2016). The response of an individual or population to environmental stressors often differs according to individual variability and the cumulative effects of the stressors present in their environment (Patyk et al., 2015). As a result, understanding animal health and the effects of environmental change requires the monitoring and assessments of these complex interactions, ideally using a range of innovative ecological, biomedical and behavioral indicators (Lloyd et al., 2016).
In reptiles, robust health assessments for disease status or physiological problems are further complicated by lack of research, challenging logistics of individual capture for repeated assessments, and their metabolic characteristics (Christopher et al., 2003; Keller et al., 2006; Allender et al., 2016; Buckley and Huey, 2016). As ectotherms, reptiles undergo strong seasonal shifts in behavior, physiology and metabolism (Porter and Gates, 1969; Huey, 1982; Zimmerman et al., 2010; Vitt, 2016), which can directly influence how reptiles partition resources to self-maintenance activities, including the immune system and other physiological functions (Zimmerman et al., 2010; Sandmeier and Tracy, 2014). Consequently, their physiological responses are highly variable and change with age, sex, nutritional, individual, seasonal and environmental conditions (Wright and Cooper, 1981; Zapata et al., 1992; Dickinson et al., 2002; Moore and Jessop, 2003, MacDonald et al., 2007; Nechaeva, 2011; Vitt, 2016).
Despite these drawbacks, standard wildlife diagnostic blood panels (e.g. hematology, biochemistry, serology and cytology) and physical examinations are still routinely used on reptiles to investigate conditions that may affect blood cells or cause a change in blood cell composition including anemia, inflammation, hematopoietic disorders and parasitemia (Christopher et al., 1999; Christopher, 1999; Jacobson and Origgi, 2002; Nardini et al., 2013; Sheldon et al., 2016). Comprehensive panels often require more blood or plasma than can be safely collected from wild or smaller reptiles, and the interpretation of these data is challenging because of the low number of studies and the lack of reference values for certain species or wild populations (Nardini et al., 2013; Lloyd et al., 2016). Furthermore, these laboratory tests are not designed for or capable of identifying specific intrinsic or extrinsic stressors impacting animal health.
Gene transcription profiling has been experimentally validated to improve the interpretation of health status and response of reptiles to environmental stressors (Krivoruchko and Storey, 2013, 2015; Drake et al., 2016). Using a small amount of tissue (e.g. 0.1 ml blood), ecologists and microbiologists can identify specific genes responding to perceived environmental stressors and measure their response by quantifying the amount of messenger RNA (mRNA) that is transcribed for targeted genes (Burcyznski et al., 2000; Bartosiewicz et al., 2001; Sitt et al., 2008; Bowen et al., 2012; Miles et al., 2012). This approach improves the assessment of physiological status by detecting the earliest observable signs of changes in health at the cellular level (Acevedo-Whitehouse and Duffus, 2009). For chelonians, this technology has proven useful for understanding the systemic health effects that occur with dietary changes and malnutrition such as the down-regulation of immune function and reductions in growth, calcium metabolism and shell calcification (Drake et al., 2016) as well as changes that occur in stressed or physiologically perturbed animals (Krivoruchko and Storey, 2013, 2015; Bowen et al., 2015). As reference genomes and primer sequences for targeted genes are made available for a wider range of non-mammalian species, our ability to understand molecular reactions to intrinsic and extrinsic stressors as well as assessments of animal and ecosystem health will improve.
Here, we provide an integrative health assessment approach for the Agassiz’s desert tortoise (Gopherus agassizii) by pairing molecular biomarkers with traditional blood panel and physical assessments. This threatened species is subject to a wide variety of environmental and human stressors (USFWS, 1994, 2011, 2016), and considerable attention has been focused on understanding aspects of health, nutrition, disease and general survivorship in this species (Brown et al., 1994; Christopher et al. 2003; Longshore et al. 2003; Tracy et al. 2006; Sandmeier et al. 2009; Esque et al. 2014). Most studies and field experiments have found it challenging to track, monitor and measure health in tortoises and other ectotherms (Christopher et al., 1999; Tracy et al., 2004; Hunter et al., 2008; Sandmeier et al., 2013, 2016).
Due to the reduced metabolic state and activity level of poikilothermic organisms, clinical signs of health or disease conditions may be slow to emerge (Christopher et al., 2003; McArthur, 2004; Allender et al., 2016). The use of diagnostic assays largely designed for mammals often lead to difficulties in interpretation (Llyod et al., 2016), as tortoises exert less control over their homeostatic mechanisms than birds and mammals and their ‘normal’ ranges are often wider and in many species subject to marked seasonal variation (Lillywhite, 1987; Peterson et al., 1993; Wilkinson, 2004). Furthermore, humoral immune reactions such as targeted antibody responses can be highly delayed (>18 months) in Agassiz’s desert tortoises following the appearance of clinical abnormalities (Maloney, 2011; Aiello et al., 2016; our unpublished work).
We evaluated both standard (hematologic and plasma biochemistry values, trace element screens, proteins, vitamin A levels) and gene-based (transcript profiling) diagnostic blood panels from clinically abnormal and normal adult tortoises from Clark County, NV, USA. In addition, we investigated aspects of both constitutive (e.g. white blood cell and leukocyte counts, gene transcription) and humoral (e.g. antibody production) immune responses to known pathogens (Mycoplasma agassizii and Testudinid herpesvirus 2; Brown et al., 2002; Wendland et al., 2007; Jacobson et al., 2012; Braun et al., 2014). Necropsy and histological evaluation were performed on clinically abnormal tortoises with chronic illness to provide additional evidence of health condition relative to blood panels. We investigated whether transcript profiles for genes involved in immune responses to pathogenic microbes (SAA, ATF, CD9, MX1, Myd88; Kibenge et al., 2005; Tumpey et al., 2007; Zhou et al., 2008, 2011; Li et al., 2011) and genes often correlated with malnutrition (Lep; Otero et al., 2005) and cellular stress superoxide dismutase (SOD; Walsh et al., 2010) would be higher in clinically abnormal tortoises suspected of having bacteria-related infection and disease (Bowen et al., 2015; Drake et al., 2016). By pairing molecular biomarkers with traditional hematologic and biochemical blood panels, and comparing data from necropsy and histological evaluation, our efforts consider how new screening methods can improve the diagnostic capacity of blood-based health assessments in reptiles.
Materials and Methods
Study animals
Captive adult tortoises (n = 11; 5 Female:6 Male) from the Desert Tortoise Conservation Center in Clark County, NV, USA that were classified as ‘clinically abnormal’ based on long-term health evaluations by specialized veterinarians were used in this study. Each tortoise in this category presented multiple clinical signs of potential illnesses associated with long-term weight loss and reduced or under-conditioned body condition. Due to their poor overall health, these tortoises were euthanized, and immediately necropsied to evaluate tissue conditions morphologically and histologically. ‘Clinically normal’ tortoises were selected from an in situ wild population that had been monitored since 2006 (n = 10; 6 Female:4 Male) in Hidden Valley, Clark County, NV, USA (Drake et al., 2015) to act as a healthy control for comparison. These tortoises were deemed clinically normal based on visual examination by specialized veterinarians and tortoise biologists and each tortoise had been evaluated and assessed as clinically normal for nine consecutive years (Drake et al., 2015). Clinically normal, wild tortoises were not euthanized. All tortoises were assessed and sampled in July between 06:00 and 08:00 h to minimize circadian influences on measured blood analytes. All handling and experiments using animals were conducted according to Institutional Animal Care and Use Committee guidelines (US Geological Survey WERC 2012-03, University of California-Davis WERC-2007-02) and covered under state (Nevada Division of Wildlife Permit #S33762) and federal (US Fish and Wildlife Service TE-030 659) permits.
Animal condition
All tortoises were assessed to characterize their general health and body condition. Assessments included an examination of the animal’s general posture, respiration, face (with specific attention to the eyes, periocular tissue, nares, mouth, tongue and oral mucosa), skin and shell for any clinical signs of disease, abnormalities, damage or discoloration (USFWS, 2016). We looked for discharge from the cloaca, eyes, nares and mouth and examined the skin for evidence of ulceration, erythema, swelling or discharge. We also palpated the coelomic cavity to confirm masses (e.g. urolith and egg) present during evaluation (USFWS, 2016). Clinical condition was quantified for each tortoise by summing the number of signs of disease, abnormalities, damage or discoloration present (Table 1).
Table 1:
The clinical condition of adult captive tortoises (A1–A11; 6 F:5 M) and wild tortoises (N1–N10; 6 F:4 M) in Clark County, NV, USA
| ID | Sex | MCL (mm) | Mass (g) | Assessed status | Eyes | Nares | Oral cavity | Skin | Shell | Other | BCS |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A1 | F | 250 | 2411 | Abnormal | E | A, DS | − | − | − | − | 4 |
| A2 | F | 274 | 2862 | Abnormal | E, R | A, DM, Er | − | − | − | − | 4 |
| A3 | M | 270 | 3914 | Abnormal | E, R | A, DM, Er | L | − | − | − | 4 |
| A4 | M | na | 2507 | Abnormal | E, R | DS, Er | − | − | − | − | 4 |
| A5 | M | 274 | 2165 | Abnormal | − | A, DS, Er | − | − | − | CM | 3 |
| A6 | F | 283 | 4329 | Abnormal | DS, CR, E, R | A, DS, Er | − | − | − | − | 4 |
| A7 | F | 260 | 3037 | Abnormal | E, DM | A, DM, Er, O | − | − | − | LR | 4 |
| A8 | F | 246 | 2430 | Abnormal | E, DS | A, DM, Er, O | − | − | − | − | 4 |
| A9 | M | 213 | 1248 | Abnormal | E, R | − | − | L | − | − | 3 |
| A10 | M | 171 | 856 | Abnormal | − | A, DS, Er, O | − | − | − | − | 4 |
| A11 | F | 259 | 2764 | Abnormal | E, R | A, DS, Er | − | − | − | CM | 4 |
| N1 | F | 240 | 2415 | Normal | R | − | − | − | − | − | 4 |
| N2 | M | 285 | 4200 | Normal | R | − | − | − | − | − | 5 |
| N3 | M | 294 | 5035 | Normal | R | − | − | − | − | − | 4 |
| N4 | F | 244 | 2710 | Normal | R | − | − | − | − | − | 4 |
| N5 | M | 309 | 4305 | Normal | R | − | − | − | − | − | 4 |
| N6 | F | 224 | 2095 | Normal | R | − | − | − | − | − | 4 |
| N7 | F | 261 | 3370 | Normal | R | − | CP | − | − | − | 4 |
| N8 | F | 267 | 3498 | Normal | R | − | CP | − | − | − | 4 |
| N9 | M | 317 | 5525 | Normal | R | − | − | − | − | − | 4 |
| N10 | F | 256 | 3200 | Normal | R | − | − | − | − | − | 4 |
Tortoises were evaluated mid-summer (July) immediately before sampling of blood. The following codes indicate clinical anomalies observed during evaluation: A, asymmetrical; CM, coelomic mass; CP, coloration pale; CR, coloration red; DS, discharge serous; DM, discharge mucoid; E, edema; Er, eroded; L, lesions present; LR, labored respiration; O, occluded; R, recessed; ‘−’, clinically normal; MCL, maximum Carapace Length.
Numerical body condition scores (BCSs) were used to assess overall muscle condition and fat stores with respect to skeletal features of the head and limbs (USFWS, 2016). BCS scores were first categorized as ‘under,’ ‘adequate’ or ‘over’ condition, and then numerical values were assigned to provide a more precise and repeatable measurement (i.e. under: 1–3, adequate: 4–6, over: 7–9) (USFWS, 2016). The shell length for each tortoise was recorded to the nearest 1.0 mm using digital calipers.
Animal necropsy and histology
The clinically abnormal tortoises were in poor condition and were euthanized, necropsied, and evaluated for morphological and microscopic conditions at the Wildlife Disease Laboratories, San Diego Zoo Institute for Conservation (San Diego, CA, USA). All major organs and tissues including the eyes, palpebra, nasal cavity, trachea, lungs, oral cavity and tongue, esophagus, stomach, small intestine, large intestine, pancreas, liver, spleen, heart, kidney, bladder, adrenal glands, gonads, cloaca, brain, skeletal muscles and shell/skeletal structure were evaluated.
Blood collection
Blood (~3 ml) was extracted from all tortoises via jugular venipuncture (Jacobson et al., 1992) using a 1.91-cm, 25-gage needle-IV infusion set and 3 ml syringe (Fig. 1). Blood was collected from clinically abnormal tortoises prior to euthanasia. Aliquots of whole blood were placed immediately into an RNeasy® Animal Protect collection tube (0.5 ml blood for gene transcription analyses; Qiagen, Valencia, CA, USA) and BD Microtainer® tubes with lithium heparin (~2.5 ml blood for complete blood counts, hematological evaluations, chemistry panel screens, trace element screens and vitamin A analyses; Becton Dickinson and Company, Franklin Lakes, NJ, USA). Samples were stored on ice for no >2 h. A small droplet of blood (0.01 ml) was smeared onto a microscope slide for hematological evaluations. Aliquots of whole blood were shipped overnight for analysis of hematology. Plasma was separated from the remaining sample using centrifugation with a force of 1318×g and stored in an ultracold freezer (−70°C) until analysis. Aliquots of plasma (0.01 ml) were screened for antibodies to M. agassizii using an enzyme-linked immunosorbent assay (ELISA; Wendland et al., 2007). Additionally, sloughed epithelial cells were collected using sterile oral swabs, and screened for Testudinid herpesvirus 2 using polymerase chain reaction (PCR) (Jacobson et al., 2012; Braun et al., 2014).
Figure 1:
Photograph of blood (~3 ml) being extracted from an adult Agassiz desert tortoise (G. agassizii) in Clark County, NV, USA. Blood was collected via jugular venipuncture using a 1.91-cm, 25-gage needle-IV infusion set and 3 ml syringe. Photographs taken by D. Johnson.
Gene transcription
RNA extractions and cDNA synthesis were performed as described in Bowen et al. (2015) on each sample collected for gene transcription. PCR primers developed for G. agassizii were used to amplify 11 genes of interest and 1 ribosomal housekeeping gene within each sample (Supplementary Table 1, Bowen et al., 2015, Drake et al., 2016). Gene transcription cycle threshold values (CT) were measured for the housekeeping gene (18 S) and the genes of interest: CaM-Calmodulin, AHR-Arylhydrocarbon Receptor, Mx1, HSP70-Heat Shock Protein 70, SAA-Serum Amyloid A, MyD88-Myeloid Differentiation Factor 88, CD9, SOD, ATF, CL-Cathepsin L and Lep-Leptin (Supplementary Table 1) from each sample in duplicate using quantitative PCR. Amplifications were conducted on a StepOnePlus™ Real-Time PCR System (Thermo Fisher Scientific, Hanover Park, IL, USA). Gene transcription measures were normalized by subtracting the average 18 S housekeeping ribosomal gene CT value from the gene of interest CT for each tortoise.
We analyzed the qPCR gene transcript data using normalized CT values. These values are inversely proportional to the amount of subject mRNA in the sample such that the lower the normalized value, the more transcripts are present. A change in normalized value of two is approximately equivalent to a 4-fold change in the amount of the transcript. We evaluated normalized gene transcript profiles for each gene of interest to examine potential differences in immune function between animal sex and clinical status (Supplementary Table 1).
Blood analytes
Blood samples were evaluated for complete blood counts, leukocyte morphology, biochemical panels, trace elements and vitamin A levels at the San Diego Zoo Global Clinical Laboratory (Escondido, CA, USA) and the University of California, Davis William R. Pritchard Veterinary Medical Teaching Hospital, Clinical Diagnostic Laboratory (Davis, CA, USA).
‘Hematology’—Blood erythrocyte parameters (hematocrit, red blood cell (RBC) morphology), leukocyte morphology (white blood cell-WBC; WBC differentials including lymphocytes, heterophils, azurophils, eosinophils, basophils, heterophil to lymphocyte ratio) and other parameters (platelets, plasma protein, plasma fibrinogen, protein to fibrinogen ratio and icterus) (Table 2). Packed cell volume was determined by centrifugation of blood at 10 000 ×g in a Sorvall Legend Micro 17 Micro-centrifuge (Thermo Scientific Corp, Waltham MA, USA). Two hundred cell differential and leukocyte counts were done manually by certified clinical laboratory scientists on Wright-stained blood smears stained with an automatic stainer (Wescor Inc., Logan, UT, USA). Fibrinogen was measured by the standard micro heat-precipitation.
Table 2:
Geometric mean normalized CT transcription values for 11 genes of interest for 21 adult Mojave desert tortoises (G. agassizii) that were assessed as clinically normal (5 F:5 M) and clinically abnormal (6 F:4 M)
| Gene | Range | All torts | Clinically normal | Clinically abnormal |
|---|---|---|---|---|
| SAA | 13.80–20.17 | 15.82 | 16.04 | 15.63 |
| HSP70 | 11.41–14.14 | 12.86 | 12.57 | 13.13 |
| MX1 | 14.15–21.00 | 17.45 | 17.87 | 17.08 |
| CD9 | 10.92–14.38 | 12.52 | 12.81 | 12.27 |
| SOD* | 9.35–14.23 | 11.25 | 12.29 | 10.37 |
| AHR | 12.34–16.47 | 14.90 | 15.20 | 14.64 |
| MyD88* | 14.37–18.05 | 16.11 | 16.93 | 15.40 |
| CaM | 8.75–11.91 | 10.42 | 10.28 | 10.55 |
| ATF | 7.58–16.05 | 11.12 | 11.87 | 10.48 |
| CL* | 13.59–22.17 | 16.15 | 16.72 | 15.65 |
| Lep* | 11.35–15.44 | 13.43 | 14.14 | 12.82 |
The smaller the mean value, the higher the level of transcript for the 11 genes. ‘*’ Indicates significant difference (P ≤ 0.05) between clinical groups.
‘Biochemical Panels’—Plasma parameters were evaluated for biochemical panels including: ‘enzymes’ (alkaline phosphatase—ALK, aspartate aminotransferase—AST, creatine kinase—CK, glutamate dehydrogenase—GLUD and lactic dehydrogenase—LDH), ‘organics’ (blood urea nitrogen-BUN, cholesterol-Chol, glucose-Gluc, triglycerides-Trig, total protein-TP and uric acid-UA), ‘proteins’ (prealbumin, albumin-ALB, α1 globulins, α2 globulins, β-globulins, ϒ-globulins, albumin to globulin ratio (A:G) and total protein-TP), ‘minerals’ (calcium-Ca, phosphorus-P), and ‘electrolytes’ (anion gap, chloride-CL, sodium-Na, potassium-K, total carbon dioxide-CO2) (Table 3; Taylor and Jacobson, 1982; O’Connor et al., 1994; Christopher et al., 1999; Christopher et al., 2003). Frozen plasma samples were thawed at room temperature and analyzed using a Roche cobas c501 system (Roche Diagnostics, Indianapolis, IN, USA). Analytes were measured using commercially available kits for albumin (ALB, Roche-ALB2), ALP (Roche-ALP2), anion gap (calculated), AST (AST, Roche-AST-L), bicarbonate (Roche-CO2-L), calcium (CA-Roche-CA2), cholesterol (CHOL-Roche = CHOL2), CK (Roche-CKL), electrolytes-sodium (Na), potassium (K) and calcium (Ca, Roche-ISE indirect Gen 2), globulin (Glob, calculated), glucose (GLU, Roche-GLUC3), glutamate dehydrogenase (Randox-GL441), inorganic phosphate (P, Roche-PHOS2), total protein (TP, Roche-TP2), urea nitrogen (BUN, Roche-UREAL) and uric acid (UA, Roche-UA2) (Table 3).
Table 3.
Blood hematology values for 21 adult Mojave desert tortoises (G. agassizii) that were assessed as captive clinically abnormal (5 F:6 M) and wild clinically normal (6 F:4 M) in July at Clark County, NV, USA
| Parameters | PS | Abnormal range | Normal range | Reference range | Abnormal geometric mean | Normal geometric mean |
|---|---|---|---|---|---|---|
| RBC (%) | All | 19−26 | 13−43.5 | 19.5−37.1 | 23 | 25 |
| WBC (/μl)* | All | 3500−21 700 | 6900−10 000 | 1496−10 924 | 6421 | 8321 |
| Heterophils (%)* | All | 9−68 | 20−58 | NA | 21 | 36 |
| Heterophils (/μl)* | All | 520−4556 | 1500−5320 | 719−7159 | 1326 | 2900 |
| Lymphocytes (%)* | All | 14−67 | 12−47 | NA | 47 | 23 |
| Lymphocytes (/μl) | All | 938−14 322 | 962−3700 | 63−2746 | 3027 | 1878 |
| Hetero:Lympho* | All | 0.14−4.86 | 0.43−4.67 | NA | 0.44 | 1.5 |
| Azurophils (%)* | All | 0−5 | 3−13 | NA | 1 | 7 |
| Azurophils (/μl)* | All | 0−335 | 258−949 | 0−557 | 22 | 570 |
| Eosinophils (%) | All | 0−26 | 4−23 | NA | 6 | 12 |
| Eosinophils (/μl)* | All | 0−1312 | 300−1980 | 0−950 | 292 | 1005 |
| Basophils (%) | All | 6−22 | 6−27 | NA | 10 | 15 |
| Basophils (/μl)* | All | 228−2387 | 438−2300 | 62−3574 | 625 | 1258 |
Reference values were taken from published research wild adult Mojave desert tortoises in the summer season (Christopher et al., 1999). Population Specifications (PS): All, all tortoises. ‘*’ indicates significant difference (P ≤ 0.05) between clinical groups. Hetero: Lympho, heterophils: lymphocytes ratio.
‘Trace Element Screens’—Plasma was evaluated for trace elements of calcium (Ca), copper (Cu), iron (Fe), magnesium (Mg), potassium (K), sodium (Na) and zinc (Zn). A 0.5 ml sample of plasma was placed into a test tube with 4 ml of protein precipitating internal standard solution. The sample was mixed and centrifuged to produce a clear supernatant free of proteins. The sample was then analyzed for elements by inductively coupled plasma-atomic emission spectroscopy (ICP-AES; Thermo iCAP 6500 Radial instrument). Each element was reported in parts per million (ppm), except Na and K which were reported as milli-equivalents per liter (mEq/l; Table 3).
‘Vitamin A Analysis’—Vitamin A (retinol) was extracted from 0.5 ml plasma with petroleum ether after precipitation of proteins with ethanol. Each sample was mixed and centrifuged to produce a clear supernatant free of protein. After concentration to dryness, samples were exchanged into methanol and filtered through a 0.45 μm syringe into an auto-sampler vial. Quantitation was performed by high-performance liquid chromatography (Agilent 1200 series) using fluorescence detection (Waters model 2475 fluorescence detector).
Statistical analyses
Most blood analytes were not normally distributed even after log transformations; therefore, we used non-parametric tests for analyses using R statistical software (R Development Core Team, 2016). The geometric means and 95% confidence intervals were calculated for all blood analytes in each clinical group. We used conventional mean responses per clinical group (clinically abnormal or normal) and animal sex (male or female) with data assessed for statistical significance between classification ranks using Wilcoxon Signed Rank Tests (Hollander et al., 2014; R package stats v3.2.2). Analyte responses between clinical groups and animal sex were compared using a non-parametric Multivariate Analysis of Variance (permutation MANOVA; R package vegan v2.3–1). We also performed a nonmetric, multivariate, multidimensional scaling (NMDS; R package vegan v2.3–1; Oksanen et al., 2011) ordination with the Bray–Curtis similarity measure in conjunction with cluster analysis for statistical and graphical representation of individual tortoises clustered by similarity in transcription, hematology, biochemistry and trace element screens and not by pre-defined groups such as clinical status. Statistical significance was based on P-values ≤ 0.05. Spearman’s rank-order correlation was used to measure the strength and direction of association between variables in our data set, using a statistical significance based on P-values ≤ 0.01.
Results
Physical condition
All clinically normal tortoises had optimal ranged BCSs (range 4–5), indicating adequate muscle and fat deposits relative to skeletal features such as the sagittal crest. Recessed eyes were consistently observed in clinically normal tortoises, which is a typical finding for desert tortoises during the dry summer season and likely indicates a period of temporary dehydration (Table 1). Two clinically normal tortoises were found to have pale oral and tongue muscosa. Each tortoise classified as clinically abnormal was found to have multiple significant physical anomalies. Anomalies included periocular edema, conjunctival edema and hyperemia due to inflammation, recession of periocular tissue, ocular and nasal discharge (both serous and mucoid), occluded and eroded nares, labored respiration, pale and reddened oral muscoa and tongue, coelomic masses indicative of urolithiasis, skin lesions and associated lethargy (Table 1). General signs of upper respiratory tract disease (URTD) (e.g. nasal discharge, occluded and eroded nares, periocular edema) were observed in all abnormal tortoises. Two out of 11 clinically abnormal tortoises had BCSs classified as a ‘3’ indicating a palpable or visible sagittal crest and atrophied forelimbs (Table 1).
Necropsy
Necropsy and histological evaluation was performed on 9 of the 11 clinically abnormal tortoises to provide more comprehensive information on physical condition and health. The primary findings were moderate to severe rhinitis in seven of the tortoises and moderate to severe pneumonia in the other two tortoises. Additional significant findings were renal gout in one tortoise and a chronic liver hematoma in another. Secondary inflammatory findings were mild glossitis, mild to moderate tracheitis, mild gastritis and mild enteritis in several of the animals. Two tortoises also showed degenerative changes in the kidney including glomerular atrophy. Incidental findings were skeletal muscle sarcocysts in one tortoise and urolithiasis in three tortoises. These impairments likely contributed to the observed poor health and body condition each animal exhibited (Supplementary Table 2).
Pathogens
ELISA test results for antibodies specific to M. agassizii for all clinically normal tortoises were negative. In contrast, ELISA test results were positive for all clinically abnormal tortoises, potentially indicating prior or current mycoplasmid infections. PCR test results for Testudinid herpesvirus 2 were negative for all tortoises.
Gene transcription
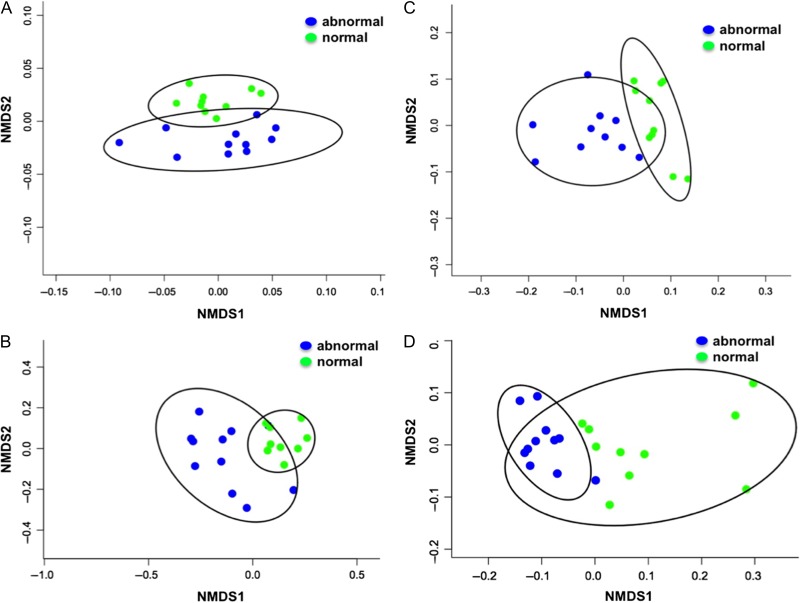
Gene transcript (CT) profiles indicative of immune and physiological function were statistically different between clinically abnormal and normal individuals (perMANOVA F1,20 = 4.95, P < 0.01). When analyzed without a priori structure of clinical status, tortoises separated into two well-defined groups as depicted by NMDS and cluster analysis (Fig. 2). Transcription levels of most genes were higher in clinically abnormal than normal tortoises (Table 2). When grouping tortoises by clinical status, we found significant differences in the genes SOD (W = 12.0, P < 0.01), MyD88 (W = 5.0, P < 0.01), CL (W = 27.0, P = 0.05) and Lep (W = 12.5, P < 0.01) (Table 2). These genes were indicative of molecular reactions for enzymatic protection from superoxide radicals (SOD; Walsh et al., 2010), signaling of innate immunity against microbial infection (Myd88; Li et al., 2011), protein synthesis (CL; Zhou et al., 2008), and overall nutritional condition and neuroendocrine and immune functions (Lep; Otero et al., 2005). Transcript levels for SOD and MyD88 in abnormal tortoises represented a fourfold increase in transcription compared to normal animals. Other genes important for innate and adaptive immune defenses against bacterial and microbial infection and inflammation (i.e. SAA, MX1, CD9 and ATF), environmental toxicants (AHR), detoxification (HSP70), calcium metabolism and cellular regulation (CaM) did not differ between normal and abnormal tortoises (Table 2). We found no evidence of differences in gene transcript profiles between males and females (perMANOVA F1,20 = 1.51, P = 0.20).
Figure 2:
Multivariate, nonmetric multidimensional scaling (NMDS) 2D plots for (A) normalized cycle threshold values for 11 genes of interest, (B) counts and percentages for 13 blood hematology analytes, (C) 19 plasma biochemical analytes and (D) seven trace element analytes from collected blood samples. Adult Agassiz’s desert tortoises (G. agassizii; n = 21) were sampled and categorized based on clinical condition (11 abnormal and 10 normal) in July.
Hematology
Hematological blood parameters were also statistically different between individuals assessed as clinically abnormal or normal tortoises (perMANOVA F1,20 = 4.40, P < 0.01). As with the gene transcription, we found no statistical differences by sex (perMANOVA F1,20 = 1.02, P = 0.37). Multivariate analyses (nMDS and cluster analysis) of hematological data revealed two distinct groups of tortoises (Fig. 2). Most hematological values including counts of white blood cells, heterophils, azurophils, eosionphils and basophils as well as percentages of hematocrit, heterophils, azurophils, eosinophils and basophils were lower in abnormal tortoises (Table 3). Direct counts and percentages of lymphocytes were markedly higher in abnormal tortoises, indicating increased humoral immune reactions (antibody production; Table 3). Statistical analyses using a priori clinical groupings indicated significant differences in WBCs (#/ul, W = 18, P = 0.01), heterophils (%, W = 24.5, P = 0.03; #/μl, W = 22.5, P = 0.02), lymphocytes (%, W = 97, P < 0.01), azurophils (%, W = 4, P < 0.01; #/μl, W = 2, P < 0.01), eosinophils (#/μl, W = 22, P = 0.02) and basophils (#/μl, W = 27, P = 0.05) (Table 3). Counts of WBCs were lower in clinically abnormal tortoises, despite noted tissue inflammation and infection in clinically abnormal tortoises (Supplementary Table 2). Heterophil:lymphocyte ratios were higher in clinically normal tortoises than clinically abnormal tortoises (W = 18, P < 0.01; Table 3).
Plasma biochemical panels
We found no significant overall differences in plasma biochemical values between clinically normal or abnormal individuals (perMANOVA F1,19 = 1.24 P = 0.26). However, differences were found between animal sex (perMANOVA F1,19 = 3.74, P = 0.04), as levels of calcium (W = 99, P < 0.01) and cholesterol (W = 84, P = 0.03) were higher in female than male tortoises (Table 4). Most biochemical values including enzymes (AST, CPK), organics (Alb, BUN, Chol, Globulins, Gluc, Fibrogen, total protein and UA), minerals (Ca, P) and electrolytes (Cl, Na) were increased but not statistically different in abnormal tortoises compared to clinically normal tortoises (Table 4). Measurements of the enzyme ALK, electrolyes K and C02, and vitamin A were higher in clinically normal tortoises. We found statistical differences in ALK (W = 17.5, P < 0.01), AST (W = 95.5, P < 0.02), Alb (W = 92, P < 0.01), BUN (W = 94, P < 0.01), Chol (W = 90, P = 0.01), Globulins (W = 98, P < 0.01), Total Protein (W = 87, P = 0.03) and UA (W = 91.5, P = 0.01) (Table 4) between clinically abnormal and normal tortoises.
Table 4.
Plasma biochemical results for adult Mojave desert tortoises (G. agassizii) that were assessed as captive clinically abnormal (5 F:6 M) and wild clinically normal (5 F:5 M) in July at Clark County, NV, USA
| Biochemical parameters | Abbreviations | PS | Abnormal range | Normal range | Reference range | Abnormal geometric mean | Normal geometric mean |
|---|---|---|---|---|---|---|---|
| Enzymes | |||||||
| Alkaline phosphatase (U/l) | ALK* | All | 19–116 | 26–134 | 25–114 | 32.86 | 65.34 |
| Aspartate Animotransferase (U/l) | AST* | All | 25–369 | 19–46 | NA | 58.46 | 29.97 |
| AST | M | – | – | 24–123 | – | – | |
| AST | F | – | – | 15–78 | – | – | |
| Creatine kinase (U/l) | CK | All | 604–108 520 | 955–2774 | NA | 2081.74 | 1343.67 |
| Glutamate dehydrogenase (U/L) | GLDH | All | NM | 0–4 | NA | NM | 0.80 |
| Lactic dehydrogenase (U/l) | LDH | All | 159–4270 | NM | 25–250 | 315.89 | NM |
| Organics | |||||||
| Albumin (g/dl) | Alb* | All | 1.2–2.0 | 0.4–1.6 | 0.08–1.9 | 1.61 | 1.01 |
| PreAlbumin | PreAlb | All | 0.54–1.16 | NM | 0.01–0.19 | 0.79 | NM |
| Albumin:Globulins | Alb:Glob | All | 0.27–0.56 | 0.31–0.64 | 0.41–0.97 | 0.38 | 0.49 |
| Blood urea nitrogen (mg/dl) | BUN* | All | 3–45.0 | 0.9–6.0 | 1–37 | 8.32 | 2.42 |
| Cholesterol (mg/dl) | Chol* | All | 79–361 | 23–164 | NA | 162.67 | 76.70 |
| Chol* | M | 23–56 | – | 60–381 | – | – | |
| Chol* | F | 94–164 | – | 33–217 | – | – | |
| Globulins (g/dl) | Glob* | All | 1.91–4.46 | 0.9–3.0 | 1.33–3.9 | 3.20 | 2.06 |
| α1 Globulins | α1 Glob | All | 0.1–0.26 | NM | 0.03–1.22 | 0.18 | NM |
| α2 Globulins | α2 Glob | All | 0.38–0.89 | NM | 0.03–1.15 | 0.62 | NM |
| β Globulins | β Glob | All | 0.92–2.76 | NM | 0.05–2.82 | 1.79 | NM |
| ϒ Globulins | ϒ Glob | All | 0.34–0.73 | NM | 0.02–0.50 | 0.57 | NM |
| Glucose (mg/dl) | Gluc | All | 67–182 | 27–85 | 65–186 | 90.08 | 63.26 |
| Plasma fibrinogen (mg/dl) | Fib | All | 3.5–6.0 | 2.6–5.4 | NA | 4.7 | 3.9 |
| Plasma protein (mg/dl) | Pro | All | NM | 100–300 | NA | NM | 120 |
| Triglycerides (mg/dl) | Trig | All | 5.0–546 | NM | NA | 52.16 | NM |
| Trig | M | – | – | 7–32 | – | – | |
| Trig | F | – | – | 14–603 | – | – | |
| Total Protein (g/dl) | TP* | All | 3–6.0 | 1.4–4.6 | 2.3–5.3 | 4.45 | 3.09 |
| Uric acid (mg/dl) | UA* | All | 1.9–12.5 | 0.9–5.4 | 1.7–9.2 | 4.90 | 2.41 |
| Minerals | |||||||
| Calcium (mg/dl) | Ca | All | 9.2–20.4 | 0–20.6 | NA | 12.48 | 9.30 |
| Ca* | M | 8.1–11.0 | 8.6–12.5 | 10.24 | 9.18 | ||
| Ca* | F | 0.0–20.6 | 11.3–23.9 | 14.66 | 9.38 | ||
| Phosphorus (mg/dl) | P | All | 12–70 | 14–52 | NA | 29.71 | 26.85 |
| P | M | – | – | 1.1–3.3 | – | – | |
| P | F | – | – | 2.0–6.5 | – | – | |
| Electrolytes | |||||||
| Anion gap (mmol/l) | AnionG | All | NA | 5–18 | 2–29 | NM | 9.22 |
| Chloride (mmol/l) | Cl | All | 107–161 | 103–145 | 101–138 | 124.44 | 118.78 |
| Sodium (mmol/l) | Na | All | 139–190 | 136–172 | 127–176 | 153.50 | 146.33 |
| Potassium (mmol/l) | K | All | 3–5.5 | 3–6.9 | 3.7–7.5 | 4.21 | 4.64 |
| Total CO2 (mmol/l) | CO2 | All | 15–27 | 17–26 | 14–32 | 21.22 | 22.24 |
| Other | |||||||
| Vitamin A (ppm) | Vit A | All | 0–0.23 | 0.06–0.28 | NA | 0.10 | 0.16 |
| Trace Elements | |||||||
| Calcium (ppm) | Ca | All | 52–230 | 66–170 | NA | 106.56 | 116.15 |
| Ca | M | – | – | NA | – | – | |
| Ca | F | – | – | NA | – | – | |
| Copper (ppm) | Cu* | All | 0.4–0.8 | 0.0–0.3 | – | 0.58 | 0.16 |
| Cu | M | – | – | 0.5–0.7 | – | – | |
| Cu | F | – | – | 0.3–0.7 | – | – | |
| Iron (ppm) | Fe | All | NA | 0.2–2.5 | NA | NM | 0.69 |
| Fe | M | – | – | NA | – | – | |
| Fe | F | – | – | NA | – | – | |
| Magnesium (ppm) | Mg | All | 19–51 | 29–62 | NA | 32.38 | 37.56 |
| Mg | M | – | – | NA | – | – | |
| Mg | F | – | – | NA | – | – | |
| Phosphorus (ppm) | P | All | 12–70 | 14–52 | NA | 29.71 | 26.85 |
| P | M | – | – | NA | – | – | |
| P | F | – | – | NA | – | – | |
| Potassium (mEq/l) | K | All | 2.1–5.0 | NM | NA | 3.5 | NV |
| K | M | – | – | 3.5–4.7 | – | – | |
| K | F | – | – | 3.7–4.7 | – | – | |
| Sodium (mEq/l) | Na | All | 77–160 | 110–130 | NA | 124.13 | 113.82 |
| Na | M | – | – | 122.4–136 | – | – | |
| Na | F | – | – | 122.3–138.5 | – | – | |
| Zinc (ppm) | Zn | All | 1.0–3.5 | 0.6–2.4 | NA | 1.68 | 1.70 |
| Zn | M | – | – | 0.4–3.4 | – | – | |
| Zn | F | – | – | 0.7–3.7 | – | – | |
Reference values were taken from published research wild adult Mojave desert tortoises (Christopher et al.,1994; Christopher et al.,1999; Christopher et al., 2003). NM = not measured, NA = information not available, and ‘–’ = information not calculated. ‘*’ indicates significant difference (P ≤ 0.05) between clinical abnormal and normal tortoises (All) or animal sex (M, F). Population specifications (PSs): All, all tortoises, M, male, F, female.
Trace element screens
We found no significant overall differences in trace elements between clinically abnormal and normal individuals (perMANOVA F1,20 = 1.07, P < 0.31) or by sex (perMANOVA F1,20 = 1.63, P = 0.20), although quantitatively levels of copper (cu), phosphorus (p) and sodium (na) were numerically higher in clinically abnormal tortoises, and calcium (ca), magnesium (mg) and zinc (zn) were higher in clinically normal tortoises (Table 4). Statistical analyses using a priori clinical groupings indicated significant differences only in trace elements of cu (W = 110, P < 0.01; Table 4).
Correlations
We found 74 significant (P ≤ 0.01) spearman’s rank variable correlations within evaluated blood variables including gene transcript, hematological, plasma biochemical and trace element panels (Supplementary Table 3).
Discussion
Here, we present the first comprehensive comparative study investigating molecular, constitutive and humoral aspects of the immune system along with other physiological and histological characteristics in tortoises. We found Agassiz’s desert tortoises with long-term illness (classified as clinically abnormal) had general signs of URTD (Brown et al., 1994; Berry and Christopher, 2001; Sandmeier et al., 2009; Jacobson et al., 2014) whereas all clinically normal animals exhibited recessed eyes, a potential sign of dehydration (USFWS, 2016). Two tortoises previously determined to be clinically normal were found to have pale oral and tongue muscosa, which may indicate anemia associated with limited food availability during the sampling period. URTD was confirmed in clinically abnormal tortoises by necropsy and lesions in other tissues, particularly in the lung, kidney and liver. Notable differences in molecular (gene transcript) and hematological blood profiles were found between animals assessed as clinically abnormal or normal; yet, profiles for most plasma biochemistry, trace element screens and vitamin A levels were similar for all tortoises evaluated. Most variables evaluated did not differ between male and female tortoises, except for plasma biochemical panels, as females had higher levels of calcium and cholesterol (Christopher et al., 1999). Transcription levels were increased in clinically ill tortoises including genes responding to defenses against microbial pathogens (MyD88; Li et al., 2011), cellular and oxidative stress (SOD; Walsh et al., 2010; Sarma and Sharma, 2016), protein synthesis (CL; Zhou et al., 2008) and malnutrition (Lep; Otero et al., 2005). Ill tortoises also showed increases in lymphocyte production, antibodies to pathogenic bacteria (M. agassizii), biochemical enzymes (AST), organics (BUN, chol, globulins, total protein and UA), and trace elements of copper. Hematological biomarkers (e.g. white blood cells, heterophils, azurophils, eosinophils, basophils and heterophil to lymphocyte ratios) routinely used to evaluate infection and inflammation in reptiles (Aguirre et al., 1995; Christopher et al., 1999, 2003; Keller et al., 2006; Zimmerman et al., 2010; Eshar et al., 2014; Sandmeier et al., 2016) were similar or lower in clinically abnormal animals than tortoises presumed healthy in our study and within reference ranges for wild Agassiz’s desert tortoises during the summer season (Christopher et al., 1994, 1999, 2003). Collectively, these findings emphasize the complexities involved in assessing, diagnosing, and interpreting fitness, immune function, and general health in tortoises.
Assessment of Tortoise Health
Hematological and plasma biochemical patterns, clinical disease and laboratory abnormalities in adult Agassiz’s desert tortoises have been well studied (Nagy and Medica, 1986; Peterson et al., 1993; Christopher et al., 1994, 1999, 2003; O’Connor et al., 1994; Peterson, 1996a, b; Homer et al., 1998; Christopher, 1999; Berry and Christopher, 2001; Sandmeier et al., 2016). However, many blood analytes in these studies were likely strongly influenced by sampling time, season and year, animal condition such as age, sex, hydration state and nutritional condition, environmental variables such as rainfall and plant food availability, and the geographic location of the populations evaluated, making it difficult to compare those diagnostic findings with current and future health studies. In addition, puncture site and levels of blood and lymph mixtures can strongly affect counts and concentrations of blood solutes (Bonnet et al., 2016), making it important to compare results from precise sampling techniques. To avoid some of these pitfalls, we sampled tortoises during discrete time periods (e.g. within 1 week between 06:00 and 08:00 h) to capture similar environmental conditions and used jugular venipuncture to minimize lymph contamination.
Findings from the literature suggest that comparisons among individuals with and without clinical abnormalities were helpful to identify potential disease cues or immune responses within populations, but typically do not indicate direct associations between laboratory data and specific disorders (Christopher et al., 2003). For example, previous research on plasma biochemistry in clinically abnormal tortoises documented marked azotemia (BUN > 100 mg/dl), mild hyperuricemia, moderate cholesterolemia, hypophosphatemia and increased AST activity (Christopher et al., 1999, 2003; Christopher, 1999). Although Christopher et al. (2003) found that tortoises thought to be responding to inflammation and infection had marked increases in heterophilia, leukocytosis and lymphocytosis, we found no evidence of this in our data.
Transcript Profiling
By analyzing transcript profiles in tortoises with clinical and pathophysiological impairments, our study expands the diagnostic tool kit being applied to domestic and wild tortoise populations. Gene transcript profiling has the ability to expedite and expand the detection of physiological changes and immune reactions at the cellular level, often before other changes in hematology, biochemistry or clinical conditions have occurred (McLoughlin et al., 2006; Acevedo-Whitehouse and Duffus, 2009; Miles et al., 2012). Previous studies in ectothermic organisms used transcript profiling with small suites of genes to identify specific molecular signatures to chemical contaminants (Connon et al., 2012; Hirakawa et al., 2012), malnutrition (Drake et al., 2016), cellular and environmental stressors (Ju et al., 2002; Connon et al., 2012; Bowen et al., 2015), and other physiological perturbations (Costanzo and Lee, 2013; Krivoruchko and Storey, 2013, 2015; Sujiwattanarat et al., 2016), highlighting environmental and anthropogenic stressors negatively impacting animal health and survival (McLoughlin et al., 2006; Acevedo-Whitehouse and Duffus, 2009; Connon et al., 2012).
We investigated if transcription for genes responding to innate and adaptive immune defenses against microbial pathogens (e.g. SAA, ATF, CD9, CL, MX1, MyD88; Supplementary Table 1) would increase in tortoises with lethargy, clinical abnormalities and presumed disease. URTD in tortoises is caused by exposure to bacterial pathogens (M. agassizii and M. testidenum; Brown et al., 1994) and subsequent infection, and is arguably one of the more important chronic infectious diseases of wild and captive North American and European tortoises (Jacobson et al., 2014). Infection and inflammation associated with Mycoplasma spp. and other pathogenic microbes such as Testudinid herpesvirus 2 are often subclinical and hide in tissues for years within the host without showing clinical signs (e.g. nasal exudate, periocular edema) or physical impairment (Jacobson et al., 2014). Therefore, we assumed that most clinically ill tortoises likely had multiple morphological, histological and physiological impairments in various organs, and that transcripts responding to cellular and oxidative stress (SOD), antigen processing and protein synthesis (CL), and malnutrition (Lep) would also be increased in ill animals.
We found significant differences in transcript profiles between tortoises classified as clinically abnormal or normal. These genetic differences likely reflect long-term suboptimal nutritional and environmental conditions endured prior to this study as well as repeated bouts with microbial infection, inflammation, disease and improper organ function. Clinically, abnormal tortoises in our study had access to food and water for two years prior to the experiment; however, their nutritional and disease conditions may reflect prior years in captivity or physiological and behavioral conditions that prevented them from foraging or processing important nutrients. Genes transcribed at significantly higher levels (SOD, Myd88, CL and Lep) in clinically abnormal tortoises may indicate a response to increased environmental toxicants, oxidative stress, microbial and bacterial infections, and malnutrition respectively. Increased transcription for SOD and Lep were previously described in malnourished juvenile tortoises used in a controlled food trial (Drake et al., 2016) and other wild adult tortoises presumed to be environmentally stressed (Bowen et al., 2015).
Oxidative Stress
Micro-molecules such as reactive oxygen species (ROS) are released by phagocytic cells during pathogen attack, and are generically cytotoxic because they do not differentiate between cells and tissues of the host and infective agent (Cherry and Silverman, 2006; Sorci and Faivre, 2009). Oxidative stress resulting from ROS is regularly cited as a major cause of immunopathology in humans and non-human animals (Costantini, 2008; Pursall and Rolff, 2012). Previous studies found elevated levels of copper and oxidative stress or superoxide (O2−) radicals in blood were linked to diseases such as cancer, diabetes, premature aging and hyptertension (Strausak et al., 2001). In our study, both trace elements of copper and transcription for the gene regulating oxidative stress (SOD; Walsh et al., 2010; Sarma and Sharma, 2016) were elevated and highly correlated in clinically ill tortoises. Copper is an essential element for the activity of physiological important enzymes and is also a cofactor of the enzyme Cu/Zn-SOD, which plays a key role in the cellular response to oxidative stress by scavenging ROS (Harman, 1956, 1994; Fridovich, 1989; Strausak et al., 2001). Free radicals such as superoxide are a type of ROS that can strip electrons from proteins, lipids or nucleic acids, thereby destroying their function and resulting in cell dysfunction or death. In chelonians, two SOD enzymes have been described (Willmore and Storey, 1997; Sarma and Sharma, 2016), with SOD1 (Cu/Zn-SOD) occurring in the cytoplasm and outer mitochondrial space protecting the cells against any lethal effects of radiation, drugs or toxicity of ROS (Epstein et al., 1987) and SOD2 (Mn SOD) being found in the inner mitochondrial space, promoting cellular differentiation, apoptosis, tumorgenesis and hypoxia induced pulmonary disease (Scott et al., 1989; Wispe et al., 1992; St. Clair et al., 1994).
Conservation Implications
Improving our understanding of wildlife health is essential to inform management and policy decisions. However, ‘health’ remains a difficult concept to define and assess (Deem et al., 2008; Nordenfelt, 2011; Hanisch et al., 2012; Stephen and Karesh, 2014; Stephen, 2014). Research on tortoises and other wildlife health is limited, largely reactive and disease-centric, with an emphasis on responding to existing or imminent animal health events instead of understanding environmental and physiological conditions in the context of vulnerability and resilience (Stephens, 2014). Most wildlife and ecosystems face an assault of threats from continued habitat disturbance or loss (Hughes et al., 2003; Cox et al., 2006; Irwin and Irwin, 2006; Patz et al., 2008; Jarvis et al., 2009; Gioria and Pysek, 2016) and chemical contamination and pollution (Hughes et al., 2003; Keller et al., 2004) from increased human use. Long-lived species such as the desert tortoise may be at particular risk from the effects of degraded habitats and accumulated contaminants (Tracy et al., 2004; Gardner, 2006; Keller et al., 2006). Conservation efforts are especially challenged by a paucity of basic knowledge on how reptiles respond, tolerate or adapt to environmental stressors (Anderson et al., 1997; Gibbons et al., 2000; Hayes et al., 2016; Masters et al., 2016; Wise et al., 2016). In addition, abnormalities and subclinical infection (e.g. recent exposure to bacterial pathogens) often go undetected in tortoises and other reptiles (Aiello et al. 2016), potentially impacting accurate assessments of health. Incorporating new technologies and studying targeted physiological changes may be key to understanding how species cope with environmental variability and increasing anthropogenic stressors.
Supplementary Material
Acknowledgements
We thank many people for their contributions in this study, especially M. Walden, R. Foster and S. Kelly, L. Dang, K. Holder, N. Nourn M. Bechtel, R. Poppenga, L. Aston, D. Carrade Holt and L. Keener. In addition, we thank A. Walsh, N. Lamberski, A. Fesnock, K. Field and R. Averill-Murray for supporting this research. Any use of trade, product or firm names in this publication is for descriptive purposes only and does not imply endorsement by the US government.
Supplementary material
Supplementary material is available at Conservation Physiology online.
Funding
The work was supported in part by the US Geological Survey, Western Ecological Research Center [GX16ZC00BQAP2, GX16ZC00BQAP4] and Coyote Springs Investment LLC. Equipment and support staff were provided by the San Diego Zoo Global. In addition, funds from the Harold and June Grant Memorial and James and Mary Crouch Memorial Scholarships through the Department of Biology, San Diego State University also contributed to this work.
References
- Acevedo-Whitehouse K, Duffus ALJ (2009) Effects of environmental change on wildlife health. Philos T Roy Soc B 364:3429–3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre AA, Balazs GH, Spraker TR, Gross TS (1995) Adrenal and hematological responses to stress in juvenile green turtles (Chelonia mydas) with and without fibropapillomas. Physiol Zool 68:831–854. [Google Scholar]
- Aiello CA, Nussear KE, Esque TC, Emblidge PG, Sah P, Bansal S, Hudson PJ (2016) Host contact and shedding patterns clarify variation in pathogen exposure and transmission in threatened tortoise Gopherus agassizii: implications for disease modeling and management. J Animal Ecol 85(3):829–842. [DOI] [PubMed] [Google Scholar]
- Allender MC, Philips CA, Baker SJ, Wylie DB, Narotsky A, Dreslik MJ (2016) Hematology in an eastern Massasauga (Sistrurus catenatus) population and the emergence of ophidiomyces in Illinois, USA. J Wildlife Dis 52(2):258–269. [DOI] [PubMed] [Google Scholar]
- Anderson NL, Wack RF, Hatcher R (1997) Hematology and clinical chemistry reference ranges for clinically normal, captive New Guinea snapping turtle (Elseya novaeguineae) and the effects of temperature, sex, and sample type. J Zoo Wildlife Med 28:394–403. [PubMed] [Google Scholar]
- Bartosiewicz M, Penn S, Buckpitt A (2001) Applications of gene arrays in environmental toxicology: fingerprints of gene regulation associated with cadmium chloride, benzo(a)pyrene, and trichloroethylene. Environ Health Persp 109:71–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry KH, Christopher MM (2001) Guidelines for the field evaluation of desert tortoise health and disease. J Wildlife Dis 37:427–450. [DOI] [PubMed] [Google Scholar]
- Bonnet X, El Hassani MS, Lecq S, Michel CL, El Mouden EH, Michaud B, Slimani T (2016). Blood mixtures: impact of puncture site on blood parameters. J Comp Physiol B 186(6):787–800. [DOI] [PubMed] [Google Scholar]
- Bowen L, Miles AK, Drake KK, Waters SC, Nussear KE, Esque TC (2015) Integrating gene transcription-based biomarkers to understand desert tortoise and ecosystem health. Ecohealth 12(3):501–512. [DOI] [PubMed] [Google Scholar]
- Bowen L, Miles AK, Murray M, Haulena M, Tuttle J, Van Bonn W, Adam L, Bodkin JL, Estes J, Tinker MT, et al. (2012) Gene transcription in sea otters (Enhydra lutris); development of a diagnostic tool for sea otter and ecosystem health. Mol Ecol Resour 12:67–74. [DOI] [PubMed] [Google Scholar]
- Braun J, Schrenzel M, Witte C, GoKool L, Burchell J, Rideous BA (2014) Molecular methods to detect Mycoplasma spp. and Testudinid herpesvirus 2 in desert tortoises (Gopherus agassizii) and implications for disease management. J Wildlife Dis 50(4):757–766. [DOI] [PubMed] [Google Scholar]
- Brown MB, Schumacher IM, Klein PA, Harris K, Correll T, Jacobson ER (1994) Mycoplasma agassizii causes upper respiratory tract disease in the desert tortoise. Infect Immun 62:4580–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DR, Schumacher IM, McLaughlin GS, Wendland LD, Brown MB, Klein PA, Jacobson ER (2002) Application of diagnostic tests for mycoplasmal infections of desert and gopher tortoises, with management recommendations. Chelonian Conserv Biol 4:497–507. [Google Scholar]
- Buckley LB, Huey RB (2016) How extreme temperatures impact organisms and the evolution of their thermal tolerance. Integr Comp Biol 56(1):98–109. [DOI] [PubMed] [Google Scholar]
- Burczynski ME, McMillian M, Ciervo J, Li L, Parker JB, Dunn RT II, Hicken S, Farr S, and Johnson MD (2000) Toxicogenomics-based discrimination of toxic mechanism in HepG2 human hepatoma cells. Toxicol Sci 58:399–415. [DOI] [PubMed] [Google Scholar]
- Chabanet P, Bigot L, Nicet JB, Durvill P, Masse L, Mulochau T, Russo C, Tessier E, Obura D (2016) Coral reef monitoring in the Iles Eparses, Mozambigue Channel (2011–2013). Acta Oecol 72:62–71. [Google Scholar]
- Cherry S, Silverman M (2006) Host-pathogenn interactions in drosophilia: new tricks from an old friend. Nat Immunol 7:911–917. [DOI] [PubMed] [Google Scholar]
- Christopher MM. (1999) Physical and biochemical abnormalities associated with prolonged entrapment in a desert tortoise. J Wildl Dis 35(2):361–366. [DOI] [PubMed] [Google Scholar]
- Christopher MM, Berry KH, Henen BT, Nagy KA (2003) Clinical disease and laboratory abnormalities in free-ranging desert tortoises in California (1990–1995). J Wildlife Dis 39(1):35–56. [DOI] [PubMed] [Google Scholar]
- Christopher MM, Berry KH, Wallis IR, Nagy KA, Henen BT, Peterson CC (1999) Reference intervals and physiologic alterations in hematologic and biochemical values of free-ranging desert tortoises in the Mojave Desert. J Wildlife Dis 35(2):212–238. [DOI] [PubMed] [Google Scholar]
- Christopher MM, Brigmon R, Jacobson E (1994) Seasonal alterations in plasma β-hydroxybutyrate and related biochemical parameters in the desert tortoise (Gopherus agassizii). Comp Biochem Physiol, Part A Physiol 108 A(2/3):303–310. [Google Scholar]
- Connon RE, D’Abronzo LS, Hostetter NJ, Javidmehr A, Roby DD, Evans AJ, Loge FJ, Werner I (2012) Transcription profiling in environmental diagnostic: health assessments in Columbia River basin steelhead (Oncorhynchus mykiss). Environ Sci Technol 46:6081–6087. [DOI] [PubMed] [Google Scholar]
- Constantini D. (2008) Oxidative stress in ecology and evolution: lessons from avian studies. Ecol Lett 11(11):1238–1251. [DOI] [PubMed] [Google Scholar]
- Costanzo JP, Lee RE Jr (2013) Avoidance and tolerance of freezing in ectothermic vertebrates. J Exp Biol 216:1961–1967. [DOI] [PubMed] [Google Scholar]
- Cox N, Chanson J, Stuart S (Compilers) (2006) The status and distribution of reptiles and amphibians of the Mediterranean Basin. IUCN, Gland, Switzerland and Cambridge, UK, v + 42 pp. [Google Scholar]
- Deem S, Parker P, Miller E (2008) Building bridges: connecting the health and conservation professions. Biotopica 40:662–665. [Google Scholar]
- Desforges JW, Sonne C, Levin M, Siebert U, De Guise S, Dietz R (2016) Immunotoxic effects of environmental pollutants in marine mammals. Environ Int 86:126–139. [DOI] [PubMed] [Google Scholar]
- Dickinson VM, Jarchow JL, Trueblood MH (2002) Hematology and plasma biochemistry reference range values for free-ranging desert tortoises in Arizona. J Wildlife Dis 38(1):143–153. [DOI] [PubMed] [Google Scholar]
- Drake KK, Esque TC, Nussear KE, DeFalco LA, Modlin AT, Medica PA (2015) Desert tortoise use of burned habitat in the eastern Mojave Desert. J Wildlife Mgmt 79(4):618–629. [Google Scholar]
- Drake KK, Bowen L, Nussear KE, Esque TC, Berger AJ, Custer NA, Waters SC, Johnson JD, Miles AK, Lewison RL (2016) Negative impacts of invasive plants on conservation of sensitive desert wildlife. Ecosphere 00(0):e01531.10.1002/esc2.1531. [Google Scholar]
- Epstein CJ, Avraham KB, Lovett M, Smith S, Elroy-Stein O, Rotman G, Bry C, Groner Y (1987) Transgenic mice with increased Cu/Zn-superoxide dismutase activity: animal model of dosage effects in Down syndrome. Proc Natl Acad Sci 84(22):8044–8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshar D, Ganez AY, Avni-Magen N, King R, Beaufrere H (2014) Hematologic, plasma biochemistry, and acid-base analysis of adult negev desert tortoises (Testudo werneri) in Isreal. J Zoo Wildlife Med 45(4):979–983. [DOI] [PubMed] [Google Scholar]
- Esque TC, Drake KK, Nussear KE (2014) Water and food acquisition and their consequences on life history and metabolism of North American tortoises In: Rostal DC, McCoy ED, Mushinsky HR, eds. Biology and Conservation of North American Tortoises. John Hopkins Press, Baltimore, pp 85–95. [Google Scholar]
- Fridovich I. (1989) Superoxide dismutase. An adaptation to a paramagnetic gas. J Biol Chem 264(14):7761–7764. [PubMed] [Google Scholar]
- Gardner SC. (2006) Introduction to reptilian toxicology In: Gardner SG, Oberdorster E, eds. Toxicology of Reptiles. CRC Press, Boca Raton, pp 1–8. [Google Scholar]
- Gibbons JW, Scott DE, Ryan TJ, Buhlmann KA, Tuberville TD, Metts BS, Greene JL, Mills T, Leiden Y, Poppy S, et al. (2000) The global decline of reptiles, déjà vu amphibians. Biosci 50(8):653–666. [Google Scholar]
- Gioria M, Pyšek P (2016) The legacy of plant invasions: changes in the soil seek bank of invaded plant communities. Biosci 66:40–53. [Google Scholar]
- Hanisch S, Riley S, Nelson M (2012) Promoting wildlife health or fighting wildlife disease: insights from history, philosophy and science. Wildl Soc Bull 36:477–482. [Google Scholar]
- Harman D. (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol 11(3):298–300. [DOI] [PubMed] [Google Scholar]
- Harman D. (1994) Aging: prospects for further increases in functional life span. Age 17(4):119–146. [Google Scholar]
- Hayes WK, Escobarr RA III, Fry SK, Fortune EM, Wasilewski JA, Tuttle DM, West KS, Iverson JB, Buckner SD, Carter RL (2016) Conservation of the endangered sandy cay rock iguanas (Cyclura rileyi cristata): Invasive species control, population response, priates, poaching, and translocation. Herpetol Conserv Biol 11(Monogr 6):106–120. [Google Scholar]
- Hirakawa I, Miyagawa S, Mitsui N, Miyahara M, Onishi Y, Kagami Y, Kusano T, Takeuchi T, Ohta Y, Iguchi T (2012) Developmental disorders and altered gene expression in the tropical clawed frog (Silurana tropicalis) exposed to 17 alpha-ethinylestradiol. J Appl Toxicol 33:1001–1010. [DOI] [PubMed] [Google Scholar]
- Hollander M, Wolfe DA, Chicken E (2014) Nonparametric statistical methods. John Wiley & Sons, New York: pp 1–848. [Google Scholar]
- Homer BL, Berry KH, Brown MB, Ellis G, Jacobson ER (1998) Pathology of diseases in wild desert tortoises from California. J Wildlife Dis 34(3):508–523. [DOI] [PubMed] [Google Scholar]
- Huey RB. (1982) Temperature, physiology, and the ecology of reptiles In: Gans C, Pough FH, eds. Biology of the Reptilia, Vol 12 Academic Press, New York, pp 25–91. [Google Scholar]
- Hughes TP, Baird AH, Bellwood DR, Card M, Connolly SR, Folke C, Grosberg R, Hoegh-Guldberg O, Jackson JBC, Kleypas J, et al. (2003) Climate change, human impacts, and the resilience of coral reefs. Science 301(5635):929–933. [DOI] [PubMed] [Google Scholar]
- Hunter KW Jr, duPre SA, Sharp T, Sandmeier FC, Tracy CR (2008) Western blot can distinguish natural and acquired antibodies to Mycoplasma agassizii in the desert tortoise (Gopherus agassizii). J Microbiol Methods 75(3):464–471. [DOI] [PubMed] [Google Scholar]
- Irwin L, Irwin K (2006) Global threats affecting the status of reptile populations In: Gardner SG, Oberdorster E, eds. Toxicology of Reptiles. CRC Press, Boca Raton, pp 9–34. [Google Scholar]
- Jacobson ER, Origgi F (2002) Use of serology in reptile medicine. Semin Avian Exot Pet Med 11(1):33–45. [Google Scholar]
- Jacobson ER, Schumacher J, and Green M (1992) Field and clinical techniques for sampling and handling blood for hematological and selected biochemical determinations in the desert tortoise, Xerobates agassizii. Copeia 1992:237–241. [Google Scholar]
- Jacobson ER, Berry KH, Wellehan JF Jr, Origgi F, Childress AL, Braun J, Schrenzel M, Yee J, Rideout B (2012) Serological and molecular evidence for Testudinin Herpesvirus 2 in wild Agassiz’s desert tortoises, Gopherus agassizii. J Wildlife Dis 48(3):747–757. [DOI] [PubMed] [Google Scholar]
- Jacobson ER, Brown MB, Wendland LD, Brown DR, Klein PA, Christopher MM, Berry KH (2014) Mycoplasmosis and upper respiratory tract disease of tortoises: a review and update. Vet J 201:257–264. [DOI] [PubMed] [Google Scholar]
- Jarvis A, Touval JL, Schmitz MC, Sotomayor L, Hyman GG (2009) Assessment of threats to ecosystems in South America. J Conserv Nature 18:180–188. [Google Scholar]
- Ju Z, Dunham RA, Liu Z (2002) Differential gene expression in the brain of channel catfish (Ictalurus punctatus) in response to cold acclimation. Mol Genet Genomics 268:87–95. [DOI] [PubMed] [Google Scholar]
- Keller JM, Kucklick JR, Stamper MA, Harms CA, McClellan-Green PD (2004) Associations between organochlorine contaminant concentrations and clinical health parameters in loggerhead sea turtles from North Carolina, USA. Environ Health Persp 112(10):1074–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller JM, McClellen-Green PD, Kucklick JR, Keil DE, Peden-Adams MM (2006) Effects of organochlorine contaminants on loggerhead sea turtle immunity: comparison of a correlative field study and in vitro exposure experiments. Environ Health Persp 114:70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibenge MJT, Munir K, Kibenge FSB (2005) Constitutive expression of Atlantic salmon Mx1 protein in CHSE-214 cells confers resistance to infectious salmon anaemia virus. Virol J 2:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivoruchko A, Storey KB (2013) Activation of the unfolded protein response during anoxia exposure in the turtle Trachemys scripta elegans. Mol Cell Biochem 374:91–103. [DOI] [PubMed] [Google Scholar]
- Krivoruchko A, Storey KB (2015) Turtle anoxia tolerance: biochemistry and gene regulation. Biochim Biophys Acta 1850(6):1188–1196. [DOI] [PubMed] [Google Scholar]
- Li X, Zhu B, Chen N, Hu H, Chen J, Zhang X, Li J, Fang W (2011) Molecular characterization and functional analysis of MyD88 in Chinese soft-shelled turtle Trionyx sinensis. Fish Shellfish Immun 30:33–38. [DOI] [PubMed] [Google Scholar]
- Lillywhite HN. (1987) Temperature, energetics, and physiological ecology In: Seigel RA, Collins JT, Novak S, eds. Snakes: Ecology and evolutionary biology. MacMillan Publishing Company, New York, pp 422–477. [Google Scholar]
- Lloyd TC, Allender MC, Archer G, Phillips CA, Byrd J, Moore AR (2016) Modeling hematologic and biochemical parameters with spatiotemporal analysis for the free-ranging eastern box turtle (Terrapene carolina carolina) in Illinois and Tennessee, a potential biosentinel. Ecohealth. doi:10.1007/s10393-016-1142-8. [DOI] [PubMed] [Google Scholar]
- Longshore KM, Jaeger JR, Sappington JM (2003) Desert tortoise (Gopherus agassizii) survival at two eastern Mojave Desert sites: death by short-term drought? J Herpetol 37(1):169–177. [Google Scholar]
- MacDonald EA, Czekala NM, Gerber GP (2007) Diurnal and seasonal patterns in corticosterone in the Turks and Caicos Iguana (Cyclura carinata carinata). Caribb J Sci 43(2):266–272. [Google Scholar]
- Madliger CL, Cooke SJ, Crespi EJ, Funk JL, Hultine KR, Hunt KE, Rohr JR, Sinclair BJ, Suski CD Willis CD et al. (2016) Success stories and emerging themes in conservation physiology. Conserv Physiol 4(1):cov057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloney NK. (2011) Transmission of microplasmal upper respiratory disease in the desert tortoise (Gopherus agassizii). MS Thesis, University of Nevada, Reno, NV, USA.
- Masters NJ, Alexander S, Jackson B, Sigler L, Chatterton J, Harvey C, Gibson R, Humphrey S, Rawdon TG, Science RP, et al. (2016) Dermatomycosis causes by Paranannizziopsis australasiensis in five tuatara (Sphenodon punctatus) and a coastal bearded dragon (Pogona barbata) in a zoological collection in New Zealand. N Z Vet J 64(5):301–307. [DOI] [PubMed] [Google Scholar]
- McArthur S. (2004) Interpretation of presenting signs In: McArthur S, Wilkinson R, Meyer J, Innis C, Hernandez-Divers S, eds. Medicine and Surgery of Tortoises and Turtles. Blackwell Publishing Ltd, Oxford, pp 273–300. [Google Scholar]
- McLoughlin K, Turteltaub K, Bankaitis-Davis D, Gerren R, Siconolfi L, Storm K, Cheronis J, Trollinger D, Macejak D, Tryon V, et al. (2006) Limited dynamic range of immune response gene expression observed in healthy blood donors using RT-PCR. Mol Med 12:185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micheli F, Heiman KW, Kappel CV, Martone RG, Sethi SA, Osio GC, Fraschetti S, Shelton AO, Tanner JM (2016) Combined impacts of natural and human disturbances on rocky shore communities. Ocean Coast Manage 126:42–50. [Google Scholar]
- Miles AK, Bowen L, Ballachey BE, Bodkin JL, Murray M, Estes JA, Keister RA, Stott JL (2012) Variations of transcript profiles between sea otters Enhydra lutris from Prince William Sound, Alaska, and clinically normal reference otters. Mar Ecol-Prog Ser 451:201–212. [Google Scholar]
- Moore IT, Jessop TS (2003) Stress, reproduction, and adrenocortical modulation in amphibians and reptiles. Horm Behav 43:39–47. [DOI] [PubMed] [Google Scholar]
- Nagy KA, Medica PA (1986) Physiological ecology of desert tortoises in southern Nevada. Herpetologica 42:73–92. [Google Scholar]
- Nardini G, Leopardi S, Bielli M (2013) Clinical hematology of reptilian species. Vet Clin N AM: Exot 16(1):1–30. [DOI] [PubMed] [Google Scholar]
- Nechaeva MV. (2011) Physiological responses to acute changes in temperature and oxygenation in bird and reptile embryos. Respir Physiol Neurobiol 178:108–117. [DOI] [PubMed] [Google Scholar]
- Nordenfelt L. (2011) Health and welfare in animals and humans. Acta Biotheor 59:139–152. [DOI] [PubMed] [Google Scholar]
- O’Connor MP, Grumbles JS, George RH, Zimmerman LC, Spotila JR (1994) Potential hematological and biochemical indicators of stress in free-ranging desert tortoises and captive tortoises exposed to a hydric stress gradient. Herpetol Monogr 8:5–26. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendrew P, O’Hara RB, Simpson GL, Stevens MHH, Wagner H (2011) Vegan: community ecology package. Version 1.17–11.
- Otero M, Lago R, Lago F, Casanueva FF, Dieguez C, Gomez-Reino JJ, Gualillo O (2005) Leptin, from fat to inflammation: old questions and new insights. FEBS Lett 579:295–301. [DOI] [PubMed] [Google Scholar]
- Patyk KA, Duncan C, Nol P, Sonne C, Laidre K, Obbard M, Wiig O, Aars J, Regehr E, Gustafson LL, et al. (2015) Establishing a definition of polar bear (Ursus maritimus) health: a guide to research and management activities. Sci Total Environ 514:371–378. [DOI] [PubMed] [Google Scholar]
- Patz JA, Olson SH, Uejio CK, Gibbs HK (2008) Disease emergence from global climate and land use change. Med Clin North AM 92:1473–1491. [DOI] [PubMed] [Google Scholar]
- Peterson CC, Gibson AR, Dorcas ME (1993) Snake thermal ecology: the causes and consequences of body-temperature variation In: Seigel RA, Collins JT, eds. Snakes: Ecology and Behavior. McGraw-Hill Inc, New York, pp 241–314. [Google Scholar]
- Peterson CC. (1996. a) Ecological energetics of the desert tortoise, Gopherus agassizii: effects of rainfall and drought. Ecology 77:1831–1844. [Google Scholar]
- Peterson CC. (1996. b) Anhomeostasis: seasonal water and solute relations in two populations of the desert tortoise (Gopherus agassizii) during chronic drought. Physiol Zool 69:1324–1358. [Google Scholar]
- Porter WP, Gates DM (1969) Thermodynamic equilibria of animals with environment. Ecol Monograph 39:227–244. [Google Scholar]
- Pursall ER, Rolff J (2012) Immunopathology in ecological immunology In: Demes GE, Nelson RJ, eds. Ecoimmunology. Oxford University Press Inc, New York, pp 530–547. [Google Scholar]
- R Development Core Team (2016) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org (last accessed 14 July 2016).
- Rodriguez-Jorquera IA, Silva-Sanchez C, Strynar M, Denslow ND, Toor GS (2016) Footprints or urban micro-pollution in protected areas: investigating the longitudinal distribution of perfluoroalkyl acids in wildlife preserves. PLoS One 11(2):e0148654 doi:10.1371/journal.pone.0148654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmeier FC, Horn KR, Tracy CR (2016) Temperature-independent, seasonal fluctuations in immune function in a reptile, the Mojave desert tortoise (Gopherus agassizii). Can J Zool 94(8):583–590. [Google Scholar]
- Sandmeier FC, Tracy RC (2014) The metabolic pace-of-life model: incorporating ectothermic organisms into the theory of vertebrate ecoimmunology. Integ Comp Biol 54(3):387–395. [DOI] [PubMed] [Google Scholar]
- Sandmeier FC, Tracy CR, duPré S, Hunter K (2009) Upper respiratory tract disease (URTD) as a threat to desert tortoise populations: a reevaluation. Biol Con 142:1255–1268. [Google Scholar]
- Sandmeier FC, Tracy CR, Hagerty BE, DuPre S, Mohammadpour H, Hunter K Jr (2013) Mycoplasmal upper respiratory tract disease across the range of the threatened Mojave desert tortoise: associations with thermal regime and natural antibodies. EcoHealth 10:63–71. [DOI] [PubMed] [Google Scholar]
- Sarma R, Sharma K (2016) Sequence to structure analysis of SOD1 and SOD2 from fresh water turtles. J Bioinform Proteomics Rev 1(2):1–6. [Google Scholar]
- Schoeman MC. (2016) Light pollution at stadiums favors urban exploiter bats. Anim Con 19(2):120–130. [Google Scholar]
- Scott MD, Meshnick SR, Eaton JW (1989) Superoxide dismutase amplifies organismal sensitivity to ionizing radiation. J Biol Chem 264(5):2498–2501. [PubMed] [Google Scholar]
- Sheldon JD, Stacy NI, Blake S, Cabrera F, Deem SL (2016) Comparison of total leukocyte quantification methods in free-living Galapagos tortoises (Chelonidis spp.). J Zoo Wildlife Med 47(1):196–205. [DOI] [PubMed] [Google Scholar]
- Sitt T, Bowen L, Blanchard MT, Smith BR, Gershwin LJ, Byrne BA, Stott JL (2008) Quantitation of leukocyte gene expression in cetaceans. Dev Comp Immunol 32:1253–1259. [DOI] [PubMed] [Google Scholar]
- Sorci G, Faivre B (2009) Inflammation and oxidate stress in vertebrate host-parasite systems. Phil Trans R Soc B 364(1513):71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St. Clair DK, Oberley TD, Muse KE, St. Clair WH (1994) Expression of manganese superoxide dismutase promotes cellular differentiation. Free Radic Biol Med 16(2): 275–282. [DOI] [PubMed] [Google Scholar]
- Stephen C. (2014) Toward a modernized definition of wildlife health. J Wildlife Dis 50(3):427–430. [DOI] [PubMed] [Google Scholar]
- Stephen C, Karesh WB (2014) Is one health delivering results? Introduction. Rev Sci Tech 33(2):375–392. [DOI] [PubMed] [Google Scholar]
- Strausak D, Mercer JFB, Dieter HH, Stemmel W, Multhaup G (2001) Copper in disorders with neurological symptoms: Alzheimer’s Menkes, and Wilson diseases. Brain Res Bull 55(2):175–185. [DOI] [PubMed] [Google Scholar]
- Sujiwattanarat P, Pongsanarakul P, Temsiripong Y, Temsiripong T, Thawarnkuno C, Uno Y, Unajak S, Matsuda Y, Choowongkomon K, Srikulnath K (2016) Molecular cloning and characterization of Siamese crocodile (Crocodylus siamensis) copper, zinc superoxide dismutase (CSI-CU,Zn-SOD) gene. Comp Biochem Physiol A 191:187–195. [DOI] [PubMed] [Google Scholar]
- Taylor RW, Jacobson ER (1982) Hematology and serum chemistry of the gopher tortoise, Gopherus polyphemus. Comp Biochem Physiol Part A—Physiol 72(2):425–428. [DOI] [PubMed] [Google Scholar]
- Tracy CR, Averill-Murray R, Boarman WI, Delehanty D, Heaton J, McCoy E, Morafka D, Nussear KE, Hagerty B, Medica P (2004) Desert Tortoise Recovery Plan Assessment. http://www.fws.gov/nevada/desert_tortoise/documents/dtrpac/dtrpac_report.pdf (last accessed 15 August 2016).
- Tracy CR, Nussear KE, Esque TE, Dean-Bradley K, Tracy CR, DeFalco LA, Castle KT, Zimmerman LC, Espinoza RE, Barber AM (2006) The importance of physiological ecology in conservation biology. Integr Comp Biol 46(6):1191–1205. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Szretter KJ, Van Hoeven N, Katz JM, Kochs G, Haller O, Garcia-Sastre A, Staeheli P (2007) The Mx1 gene protects mice against the pandemic 1918 and highly lethal human H5N1 influenza viruses. J Virol 81(19):10818–10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [USFWS] Fish and Wildlife Service (1994) Desert tortoise (Mojave population) recovery plan. U.S. Fish and Wildlife Service, Portland, Oregon, USA. [Google Scholar]
- [USFWS] Fish and Wildlife Service (2011) Revised recovery plan for the Mojave population of the desert tortoise (Gopherus agassizii). U.S. Fish and Wildlife Service, California and Nevada Region, Sacramento, California, USA. [Google Scholar]
- [USFWS] Fish and Wildlife Service (2016) Health assessment procedures for the desert tortoise (Gopherus agassizii): A handbook pertinent to translocation. U.S. Fish and Wildlife Service, Desert Tortoise Recovery Office, Reno, Nevada, USA. http://www.fws.gov/nevada/desert_tortoise/documents/reports/2016/may-2016-desert-tortoise-health-eval-handbook.pdf (last accessed 15 August 2016).
- Vitt LJ. (2016) Reptile diversity and life history In: Dodd Jr. CK , ed. Reptile Ecology and Conservation. A Handbook of Techniques. Oxford University Press, Oxford, pp 1–15. [Google Scholar]
- Walsh CJ, Leggett SR, Carter BJ, Colle C (2010) Effects of brevetoxin exposure on the immune system of loggerhead sea turtles. Aquat Toxicol 97:293–303. [DOI] [PubMed] [Google Scholar]
- Wendland LD, Zacher LA, Klein PA, Brown DR, Demcovitz D, Littell R, Brown MB (2007) Improved enzyme-linked immunosorbent assay to reveal Mycoplasma agassizii exposure: a valuable tool in the management of environmentally sensitive tortoise populations. Clin Vaccine Immunol 14:1190–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson R. (2004) Clinical pathology In: McArthur S, Wilkinson R, Meyer J, Innis C, Hernandez-Divers S, eds. Medicine and Surgery of Tortoises and Turtles. Blackwell Publishing, West Sussex, pp 141–186. [Google Scholar]
- Wilmore WG, Storey KB (1997) Antioxidant systems and anoxia tolerance in a freshwater turtle Trachemys scripta elegans. Mol Cell Biochem 170(1-2):177–185. [DOI] [PubMed] [Google Scholar]
- Wise SS, Wise C, Xie H, Guillette LJ Jr, Zhu C, Wise JP (2016) Hexavalent chromium is cytotoxic and genotoxic to American alligator cells. Aquat Toxicol 171:30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wispe JR, Warner BB, Clark JC, Dey CR, Neuman J, Glasser SW, Crapo JD, Chang LY, Whitsett JA (1992) Human Mn-superoxide dismutase in pulmonary epithelial cells of transgenic mice confers protection from oxygen injury. J Biol Chem 267(33):23937–23941. [PubMed] [Google Scholar]
- Wright RK, Cooper EL (1981) Temperature effects on ectotherm immune responses. Dev Comp Immunol 5(1):117–122. [Google Scholar]
- Zapata AG, Varas A, Torroba M (1992). Seasonal variations in the immune system of lower vertebrates. Immunol Today 13(4):142–147. [DOI] [PubMed] [Google Scholar]
- Zhou X, Guo QL, Dai HP (2008) Identification of differentially expressed immune-relevant genes in Chinese soft-shelled turtle (Trionyx sinensis) infected with Aeromonas hydrophila. Vet Immunol Immunopathol 125:82–91. [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang L, Feng H, Guo Q, Dai H (2011) Acute phase response in Chinese soft-shelled turtle (Trionyx sinensis) with Aeromonas hydrophila infection. Dev Comp Immunol 35:441–445. [DOI] [PubMed] [Google Scholar]
- Zimmerman LM, Vogel LA, Bowden RM (2010) Commentary understanding of the vertebrate immune system: insights from the reptilian perspective. J Exp Biol 213:661–671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.