Abstract
Establishment of cell lines from primary mouse embryo fibroblasts depends on loss of either the Arf tumor suppressor or its downstream target, the p53 transcription factor. Mouse p19Arf is encoded by the Ink4a-Arf locus, which also specifies a second tumor suppressor protein, the cyclin D-dependent kinase inhibitor p16Ink4a. We surveyed bone marrow-derived cells from wild-type, Ink4a-Arf-null, or Arf-null mice for their ability to bypass senescence during continuous passage in culture. Unlike preB cells from wild-type mice, those from mice lacking Arf alone could be propagated indefinitely when placed onto stromal feeder layers engineered to produce IL-7. The preB cell lines remained diploid and IL-7-dependent and continued to express elevated levels of p16Ink4a. By contrast, Arf-null bone marrow-derived macrophages that depend on colony-stimulating factor-1 for proliferation and survival in culture initially grew at a slow rate but gave rise to rapidly and continuously growing, but still growth factor-dependent, variants that ceased to express p16Ink4a. Wild-type bone marrow-derived macrophages initially expressed both p16Ink4a and p19Arf but exhibited an extended life span when p16Ink4a expression was extinguished. In all cases, gene silencing was accompanied by methylation of the Ink4a promoter. Therefore, whereas Arf loss alone appears to be the major determinant of establishment of murine fibroblast and preB cell lines in culture, p16Ink4a provides an effective barrier to immortalization of bone marrow-derived macrophages.
Keywords: cell cycle checkpoints, cyclin-dependent kinase inhibitors
Primary mammalian cells in culture exhibit a limited proliferative capacity, after which they senesce (1, 2). In proliferating human somatic cells in which active telomerase is not normally expressed, telomere erosion from the ends of chromosomes serves as a “mitotic clock” that counts cell divisions. After a finite number of division cycles, ensuing telomere dysfunction activates cell cycle checkpoint controls to trigger replicative arrest (“senescence”). Abrogation of the tumor suppressive functions of the retinoblastoma protein (Rb) and p53 transcription factor bypasses this checkpoint arrest and endows cells with an extended life span. However, progressive telomere shortening ultimately leads to gross chromosomal instability and apoptosis (“crisis”) from which few if any cells survive. To escape crisis, cells must restabilize their telomeres, either by reactivating telomerase or by exploiting alternative recombinational mechanisms for telomere maintenance. The relative inability of normal human cells in culture to bypass both senescence and crisis likely accounts for their rare establishment as continuously proliferating cell lines (reviewed in refs. 3 and 4).
Cultured rodent cells behave differently. Laboratory mice are endowed with telomeres 5- to 10-fold longer than those of human chromosomes (5), and telomerase activity can be detected in many somatic cells (6). Indeed, mice lacking the gene encoding the telomerase RNA subunit are perfectly healthy and reveal consequences of telomere dysfunction only when interbred through multiple sequential generations (7). Yet, despite the apparent absence of a telomerase-based mitotic clock, explanted mouse embryo fibroblasts (MEFs) undergo senescence much more rapidly than human skin fibroblasts. In MEFs, the induction of inhibitors of cell cycle progression represents the major telomere-independent barrier to cellular immortalization. One hypothesis is that senescence of cultured rodent cells reflects the activation of p53- and Rb-dependent cell cycle checkpoints in response to nonphysiologic conditions of tissue culture per se (“culture shock”) (4, 8). Human cells likely respond in this manner as well, but as a longer-lived species, their cells may have evolved a more effective means of insulating themselves from stresses imposed by the culture environment. This idea has gained support from experiments in which specific alterations in culture conditions greatly extended the life span of both human and rodent cells and in some instances guaranteed seemingly continuous proliferation of explanted primary rodent cells (9–11).
The Ink4a-Arf locus encodes two unrelated proteins, p16Ink4a and p19Arf, encoded in part by alternative reading frames within exon 2 of the gene (12). These proteins are tumor suppressors that brake the cell cycle by modulating the activities of Rb and p53, respectively (13). The p16Ink4a protein inhibits the cyclin D-dependent kinases, Cdk-4 and Cdk-6, preventing them from phosphorylating and inactivating Rb (14). The unrelated p19Arf protein is induced in response to an elevated threshold of mitogenic signals that arise from overexpression of c-Myc, E2F-1, E1A, Ras, v-Abl, and other oncoproteins (15–19). p19Arf binds and inactivates functions of the p53 negative regulator, Mdm2, thereby potentiating p53 activity and inducing p53-dependent growth arrest or apoptosis, depending on the biologic context (20–23).
Although not detected during mouse embryonic development, p16Ink4a and p19Arf are induced and accumulate progressively as explanted MEFs are passaged in culture (24). Spontaneously immortalized cell lines derived from primary MEF cultures exhibit either p53 or Arf loss of function in a mutually exclusive manner (24). Moreover, MEFs derived from mice lacking Arf alone or Ink4a-Arf fail to undergo any detectable phase of replicative senescence (25, 26), although those from Arf-null mice continue to express high levels of p16Ink4a. Therefore, of the two products encoded by the Ink4a-Arf locus, p19Arf plays the dominant role in preventing MEF immortalization.
Different genetic events may well govern the immortalization of other cell types in culture, and many lines of evidence argue that p16Ink4a plays a major role in imposing a proliferative barrier, particularly in various human cell strains (3, 4, 27). Here, we show that, whereas elimination of Arf is apparently sufficient to establish bone marrow (BM)-derived preB cells as continuously proliferating cell lines in culture, BM-derived macrophages preferentially silence Ink4a without losing expression of the closely linked Cdk inhibitor, p15Ink4b, or of p19Arf.
Materials and Methods
Cells and Culture Conditions.
Cells harvested from the femurs and tibias of mice were explanted into cultures onto NIH 3T3 feeder layers in RPMI 1640 medium (Mediatech, Herndon, VA) supplemented with 5% FCS/55 μM 2-mercaptoethanol/2 mM glutamine, penicillin, and streptomycin (GIBCO) (28, 29) and phenotyped as described (30). The NIH 3T3 subclone was engineered to secrete human recombinant interleukin-7 (DNAX) and provides efficient stromal cell support for culturing either primary preB cells or IL-7-dependent cell lines (ref. 30 and data not shown). Two-thirds of the preB cells were removed at 3-day intervals, medium was replenished, and residual cells were allowed to proliferate until growth ceased (1–2 months for cultures of wild-type cells) or until experiments with Arf-null cells were terminated. At day 10 after explantation, some preB cells were transferred from feeders to IL-7-conditioned liquid medium without stromal support (30). Cells were plated in triplicate at a density of 2.5 × 105/ml in 6-well plates, counted every 2 days, and rediluted to the starting density in fresh medium every fourth day. Under these conditions, IL-7 was not rate-limiting for cell proliferation.
Primary cultures of BM-derived macrophages were generated from 4- to 8-week-old Arf-null, Ink4a-Arf-null, and wild-type mice. Cells were seeded at 1 × 106 cells per ml in DMEM (BioWhittaker) supplemented with 15% FCS/5.5 μM 2-mercaptoethanol/2 mM glutamine, penicillin, and streptomycin, and L cell-derived colony-stimulating factor-1 (CSF-1) (31). Nonadherent cells were removed 24 h later, resuspended at the same cell density in fresh medium, replated, and then removed after a second 48-h incubation to separate progenitor cells from adherent phagocytes and fibroblasts. Nonadherent cells were seeded a final time at the same density and propagated for 3–4 days until confluent [Passage (P)-1]. Staining with antibodies to Mac-1 (M1/70 PharMingen) and F4/80 (32) revealed that all cells expressed the macrophage markers. Scraped cells were replated at 4 × 105 per ml, fed with L cell-conditioned-medium every 2 days, counted when confluent, diluted 1:5 in fresh medium before replating, and maintained on this protocol throughout the experiment. Growth kinetics were determined for six independent cultures of each genotype counted in triplicate. The mouse CSF-1-dependent macrophage cell line BAC1.2F5 was maintained as described (32); preB cells transformed by p210BCR-Abl were a gift from Owen Witte (University of California, Los Angeles). MEF (clone 10) containing mutant p53 and expressing high levels of both p19Arf and p16Ink4a was derived previously (24).
Irradiation and Apoptosis Assays.
PreB cells were suspended at 106 cells per ml in medium containing IL-7 and were irradiated with 4 Gy from a cesium source. Cell viability was determined 24 h later by analysis of cells stained with annexin V and propidium iodide (33). BM-derived macrophages were irradiated with 5 Gy, harvested, lysed, and analyzed by immunoblotting for induction of p53 and Mdm2.
Immunoblotting.
Frozen cell pellets were lysed in ice-cold EBC buffer (34). Nuclei and debris were sedimented, and protein quantified by using a BCA Protein Assay Kit (Pierce). Proteins (200 μg per lane) separated on 12.5% denaturing polyacrylamide gels containing SDS were transferred to nitrocellulose (Osmonics, Westborough, MA) and visualized (Western Blot Chemiluminescence Reagent, NEN Life Science, Boston) after blotting with affinity-purified rabbit polyclonal antisera specific to the C termini of p19Arf (12) or p16Ink4a (35), with antibody 2A10 to Mdm2 (15), or with commercial antibodies to p53 (Ab7, Calbiochem) or to α-tubulin (T-5168 Sigma). Rabbit polyclonal antiserum (RAE) to the p15Ink4b C terminus was used to detect the protein by sequential precipitation and blotting (24).
RNA Analysis.
Total RNA was extracted from cultured macrophages by using RNAzol B (Tel-Test, Friendswood, TX). RNA (50 μg/lane) was separated electrophoretically in formaldehyde-containing gels, blotted to Hybond-N+ nylon hybridization membranes (Amersham Pharmacia Biotech) and detected by using a 110-bp [32P]-labeled probe specific for exon 1α of the Ink4a-Arf locus (35).
Methylation-Specific PCR (MSP).
Bisulfite modification of genomic DNA and MSP were performed (36) with primer sets designed to amplify PCR products from the Ink4a promoter region and exon 1α. Primers that detected methylated DNA yielded a 199-bp PCR product (nucleotide positions 8–207 according to GenBank accession no. L76150), whereas those that hybridized to unmethylated templates amplified a 197-bp product (nucleotide positions 10–207). Retention of exon 1α was confirmed by amplifying untreated genomic DNA with wild-type primers to produce a 201-bp fragment (nucleotide positions 6–207). Primer sequences were: (methylated) 5′-ATACGATTGGGCGATTGGGCG-3′ (sense) and 5′-CACCTAAATCGAAATACGACCGA-3′ (antisense) (unmethylated); 5′-ATGATTGGGTGA- TTGGGTG-3′ (sense) and 5′-CATCACCTAAATCAAAATACAA-3′ (antisense); and (wild-type) 5′-TCACACGACTGGGCGATTGG-3′ (sense) and 5′-CACCTGAATCGGGGTACGAC-3′ (antisense). Reactions were initiated at 95°C for 7.5 min, Taq polymerase (Promega) was added, and DNA was amplified for 30 cycles of denaturation (95°C, 30 s), annealing (61°C, methylated; 51°C, unmethylated, and 56°C, wild-type, 30 s) and extension (72°C, 45 s) by using a thermal cycler (5331 Mastercycler, Brinkmann). PCR products were visualized on 2.0% agarose gels in Tris-acetate/EDTA electrophoresis buffer containing ethidium bromide (0.5 ng/ml).
Results
p19Arf Deficiency Prevents Senescence of Primary preB Cells.
We derived long-term IL-7-dependent B lymphocyte cultures from bone marrow of age-matched wild-type and Arf-null mice. When cultured on NIH 3T3 feeder cells engineered to express IL-7, myeloid progenitors cannot survive, and a homogeneous population of preB cells (>90%) grows out after 10 days. PreB cells from wild-type mice could be continuously expanded on feeder layers, initially doubling every 2–3 days, but they underwent replicative senescence within 1–2 months (28). In contrast, Arf-null preB cells proliferated continuously and at significantly faster rates doubling every 24 h, even after 3 months in culture when many such experiments were terminated (data not shown). When wild-type preB cells were removed from feeder layers after 10 days of culture and propagated under more stringent liquid culture conditions in the presence of saturating quantities of IL-7 but without other stromal factors, the cells proliferated more slowly, doubling every 5 days. After 2 weeks in liquid medium, their proliferation ceased and cells gradually died (Fig. 1A). In contrast, Arf-null preB cells doubled every 1.5 days and proliferated continuously throughout the duration of the experiment (Fig. 1A). Indeed, the generation time of Arf-null preB cells was indistinguishable from that of an established, transformed and tumorigenic preB cell line expressing the p210BCR-Abl oncoprotein.
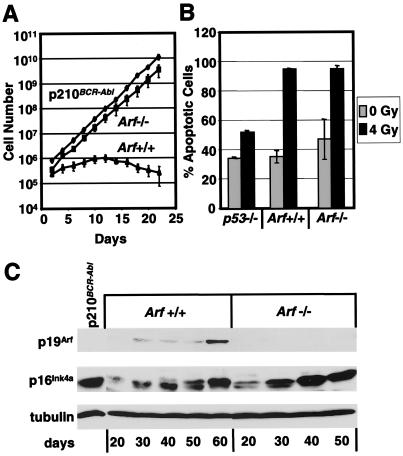
Figure 1.
p19Arf deficiency bypasses senescence of preB cells. (A) Kinetics of cell growth were determined for five independently derived lines of Arf-null (■) and wild-type (▴) preB cells in IL-7-containing medium and compared with a previously immortalized preB cell line transformed by BCR-Abl (⧫). (B) After exposure to 4 Gy γ-irradiation, preB cells of the indicated genotypes (Bottom) were cultured for 24 h and stained with annexin V and propidium iodide to determine cell viability. (C) PreB cells maintained on IL-7-expressing feeder layers were harvested and lysed at 10-day intervals (days, Bottom) over a 2-month period. Results are compared with those obtained with cells transformed by BCR-Abl (p210). Proteins were detected by direct immunoblotting by using antibodies raised against p19Arf and p16Ink4a, as indicated in the left margin. Detection of α-tubulin was used to confirm equal protein loading.
Arf-null MEFs become established immediately after explantation into culture, even though they accumulate high levels of p16Ink4a on further passage (26). Similarly, both wild-type and Arf-null preB cells accumulated p16Ink4a protein as they were passaged (Fig. 1C), even when maintained on IL-7-producing feeder layers. Late passage Arf-null preB cells continued to proliferate at the same high rate, despite robust p16Ink4a expression. Increased p19Arf protein expression was observed as wild-type preB cells senesced (Fig. 1C). Cells immortalized by BCR-Abl also expressed high levels of p16Ink4a, although selectively eliminating expression of p19Arf (Fig. 1C), consistent with previous observations that v-Abl expression in preB cells selects for loss of either p19Arf or p53 function (19). Therefore, of the two proteins encoded by the Ink4a-Arf locus, only p19Arf is required to impose replicative arrest in preB cells.
Five separate Arf-null preB cell lines maintained on IL-7-producing feeders for at least 3 months remained dependent on IL-7 for growth and survival. G1 phase cells remained diploid, which is characteristic of immortalized Arf-null MEFs and distinguishes them from established fibroblast cell lines that lack p53 function and rapidly become tetraploid (24, 26). PreB cell lines expressed very low levels of wild-type p53, as determined with antibodies that distinguish normal from mutant isoforms (data not shown). Because mutant p53 is unable to transcriptionally activate its negative regulator Mdm2, mutant forms of p53 are stable. The failure to accumulate p53 protein also implied that p53 mutations had not occurred. When preB cells were exposed to a low dose of γ-irradiation (4 Gy), and apoptotic cells were stained 24 h later with annexin V, Arf-null cells, like their wild-type counterparts, were highly sensitive and rapidly underwent apoptosis (Fig. 1B), whereas p53-null preB cells were markedly resistant (Fig. 1B) (37). Collectively, these data imply that Arf-null preB cells can continue to proliferate at a high rate in culture without sustaining p53 or p16Ink4a loss of function.
p16Ink4a Down-Regulation Accompanies Establishment of Bone Marrow-Derived Macrophage Cell Lines.
Murine macrophage cell lines have previously been derived after infection of peritoneal macrophages or myeloid progenitors with replication-defective simian virus (SV)40 and propagation of infected cells in CSF-1-containing medium. Established macrophage cell lines emerged 1–2 months after viral infection but were aneuploid and no longer depended on CSF-1 for proliferation or survival (38, 39). To date, only one CSF-1-dependent, diploid murine macrophage cell line has been derived after infection of myeloid progenitors with SV40 virus (32, 40). These BAC1.2F5 cells sustained bi-allelic deletion of the Ink4a-Arf locus, but lack detectable SV40 viral DNA (35).
To determine whether primary Arf-null macrophages, like Arf-null murine preB cells and MEFs, would exhibit an extended life span in culture, we explanted BM-derived macrophages into medium containing CSF-1. Under these conditions, established Ink4a-Arf-null BAC1.2F5 macrophages grew with a constant doubling time (Td) of 1.3 days throughout serial passage (Fig. 2A and Table 1). We emphasize that these cells have been propagated in culture for many years and have been periodically subcloned to retain CSF-1 dependence; they therefore have had ample opportunity to acquire additional growth-promoting mutations during their long passage history. Strikingly, Ink4a-Arf-null macrophages explanted directly from mice also maintained a constant doubling time throughout the duration of the experiment (Td = 1.9 days) that approached that of BAC1.2F5 cells. Wild-type macrophages proliferated much more slowly (Td = 7.1 days) during their first month in culture (Fig. 2A and Table 1). Macrophages lacking Arf alone proliferated faster than wild-type cells during this period (Td = 3.3 days) but grew slower than their Ink4a-Arf-null counterparts (Table 1). Eventually, however, more rapidly proliferating cells emerged from both the Arf-null and wild-type populations, and these exhibited faster doubling times for the remainder of the experiment. The latter cultures were maintained for an additional 3 months without any further change in proliferative rate. Late passage cells of all genotypes remained growth factor-dependent, retained macrophage cell surface markers, and demonstrated phagocytosis of antibody-coated avian erythrocytes (data not shown). Together, these data suggested (i) that both p16Ink4a and p19Arf cooperate in preventing establishment of murine macrophage cell lines, and (ii) that wild-type and Arf-null macrophages likely sustained additional mutations that facilitated their adaptation to culture.
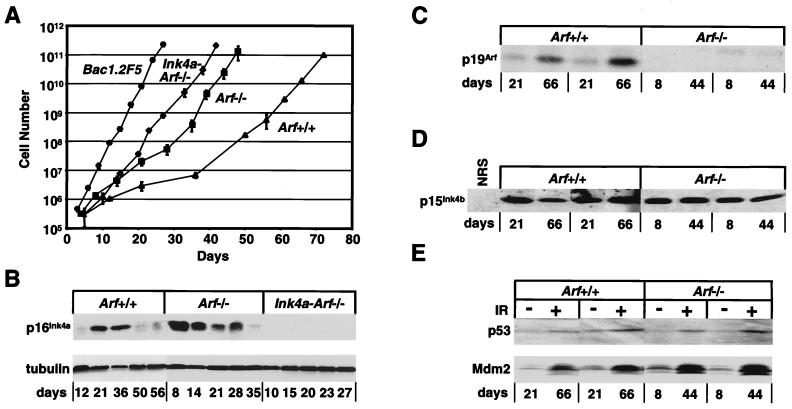
Figure 2.
Proliferation kinetics and protein expression in macrophages of different genotypes. (A) BM-derived macrophages cultured in 60-mm diameter dishes were counted when confluent and rediluted 1:5 for the next passage. Data were plotted for six independent cultures derived from Arf-null (■), Ink4a-Arf-null (⧫) and wild-type (▴) mice, and compared with those obtained with the established macrophage cell line, BAC1.2F5 (●). (B) Cells taken at indicated times were assayed for expression of p16Ink4a and α-tubulin by immunoblotting. Macrophages grown for the designated times (days) were lysed and assayed by direct immunoblotting for p19Arf (C) or by sequential immunoprecipitation and blotting for p15Ink4b (D). (E) Macrophages of the indicated genotypes (Top) after different times in continuous culture (Bottom) were exposed to 5 Gy γ-irradiation (+) or left untreated (−) and analyzed by immunoblotting for p53 and Mdm2 protein induction. Representative results with several independently derived populations are shown in C–E.
Table 1.
Generation times of primary bone marrow-derived macrophages
| Genotype | Passage | Days in culture | Doubling time, days |
|---|---|---|---|
| Wild type | P2–P5 | 1–36 | 7.1 |
| P5–P10 | 36–72 | 2.6 | |
| Arf-null | P2–P6 | 1–28 | 3.3 |
| P6–P10 | 28–48 | 1.7 | |
| Ink4a-Arf-null | P2–P10 | 1–42 | 1.9 |
| BAC1.2F5 | P2–P10 | 1–27 | 1.3 |
Macrophages were passaged as described in Fig. 2, and their doubling times were determined at the designated intervals. Because cells were passaged just after they became confluent, passage numbers for cells of different genotypes correspond to different numbers of days in culture. Immortal CSF-1-dependent BAC1.2F5 cells have sustained bi-allelic deletion of the Ink4a-Arf locus.
Given the initial differences in generation times of explanted Ink4a-Arf-null and Arf-null macrophages, an obvious possibility was that additional mutations accelerating the proliferation of Arf-null cells might reflect subsequent loss of expression of p16Ink4a. Immunoblotting analyses of cell lysates revealed that the emergence of faster growing variants from cultures of either wild-type or Arf-null macrophages correlated with down-regulation of p16Ink4a protein expression (Fig. 2B). Loss of p16Ink4a from wild-type cells between 36 and 50 days of culture (Fig. 2B) was accompanied by an abrupt shortening of cell generation time during this interval (Fig. 2A; Table 1), whereas similar changes in Arf-null cells occurred more rapidly between days 28–35 (Fig. 2 A and B; Table 1). The p19Arf protein continued to accumulate in wild-type macrophages even after faster growing cells emerged (Fig. 2C), and the latter variants proliferated more slowly than cells lacking both p19Arf and p16Ink4a (Table 1). Expression of the 15 kDa product of the closely linked Ink4b gene was similarly maintained in late passage wild-type and Arf-null cells, as determined by sequential immunoprecipitation and blotting (Fig. 2D). All cell populations accumulated p53 and its transcriptional target Mdm2 after γ-irradiation, arguing for retention of functional wild-type p53 (Fig. 2E). Therefore, p16Ink4a expression was eliminated during propagation of both Arf-null and wild-type BM-derived macrophages, whereas the Ink4b coding region was retained and expressed.
Methylation-Associated Suppression of Ink4a Expression in BM-Derived Macrophages.
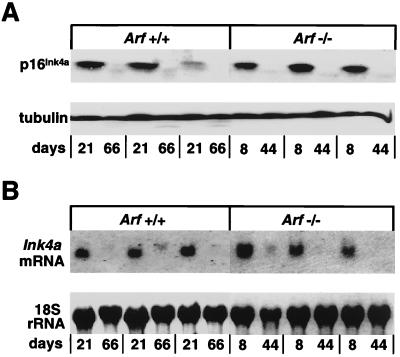
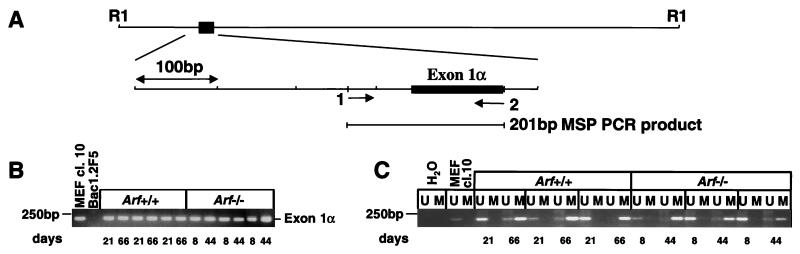
Although initially observed, p16Ink4a protein could no longer be detected in later passage wild-type or Arf-null macrophages (Fig. 3A). Similarly, Northern blot analysis performed with a 110-bp exon 1α cDNA probe that specifically detects Ink4a but not Arf transcripts (12, 35) revealed no Ink4a mRNA at later times (Fig. 3B). We therefore carried out methylation-specific PCR analysis of bisulfite-modified genomic DNA (36). Although each of the three Ink4a coding exons contains a CpG island, only methylation of exon 1α and the 5′ promoter region occurs in cell lines and tumors derived from both humans and mice (41, 42). We designed oligonucleotide primers to amplify a ≈200-bp region including 75 bp from the 5′ proximal promoter region and extending through exon 1α, as shown in Fig. 4A. We first used primers specific for unmodified DNA to confirm that exon 1α was not selectively deleted in established cell lines. PCR amplification yielded a 200-bp product from all late passage macrophage cell lines, except Ink4a-Arf-null BAC1.2F5 cells (Fig. 4B). Analysis of bisulfite-modified genomic DNA by using primer sets designed to distinguish between methylated and unmethylated DNA revealed that in each early-passage cell line in which high levels of p16Ink4a protein are detectable (Figs. 2B and 3A), only primers to unmethylated DNA gave rise to PCR products (Fig. 4C). In contrast, later passage cells, which lacked p16Ink4a protein (Figs. 2B and 3A), yielded PCR products that arose almost exclusively from primers to methylated DNA (Fig. 4C). Therefore, elimination of the p16Ink4a protein product in late passage macrophages correlated with methylation of exon 1α and transcriptional repression.
Figure 3.
Transcriptional repression of Ink4a in established BM-derived macrophages. (A) Lysates from macrophages after different times in continuous culture were immunoblotted for p16Ink4a and α-tubulin; three independent clones of wild-type and Arf-null cells were studied. (B) Northern blot analysis was carried out with RNA extracted from parallel cultures of the same cells described in A. RNA was hybridized with an exon 1α probe specific for Ink4a; 18S rRNA levels were determined by methylene blue staining.
Figure 4.
Transcriptional silencing and methylation of Ink4a in late passage macrophages. (A) Schematic map of exon 1α and the 5′ proximal promoter region indicating the location of sense (1→) and antisense (←2) oligonucleotides used to amplify a 201-bp product after bisulfite modification of genomic DNA from BM-derived macrophages. (B) PCR analysis of unmodified genomic DNA from macrophages harvested at various times (days in culture), together with DNA from cells of known p16Ink4a status (MEF clone 10 positive control; BAC1.2F5, negative control) by using oligonucleotides to the wild-type genetic sequence. (C) PCR analysis of bisulfite-modified DNA from the same cells described in B by using oligonucleotides that distinguish between unmethylated (U) and methylated (M) sequences. A PCR reaction lacking template (H2O) was used as a negative control. Several independently derived populations of each genotype were compared.
Discussion
Disabling the Arf–Mdm2–p53 surveillance pathway is sufficient to permit rapid establishment of continuously growing MEF cell lines, whereas disruption of the p16Ink4a–cyclin D/Cdk–Rb signaling pathway is not a prerequisite. To evaluate whether Arf deficiency might be sufficient to yield immortalized cell types other than MEFs, we generated primary cultures of BM-derived preB cells and macrophages from Arf-null mice. Like MEFs from these animals, primary Arf-null preB cells grew continuously and at a constant rate after explantation into culture, despite continued maintenance of p16Ink4a protein expression. Arf-null preB cells were dependent on IL-7 for growth and survival, retained functional p53, remained diploid after extended passage, and showed little evidence of karyotypic abnormalities (data not shown). These data are consistent with the finding that infection of preB cells with Abelson murine leukemia virus can select for immortalized clones that have sustained Arf deletions (19). Astrocytes derived from mice lacking both Ink4a and Arf are also immortal in culture (43), and we have made similar observations with astrocytes explanted from animals lacking Arf alone (J. Alan Diehl and C.J.S., unpublished data). However, other cell types from Arf-null mice, such as keratinocytes, endothelial cells, and cardiac myocytes have proven difficult to immortalize. Therefore, genes affecting signaling through p19Arf-independent pathways must prevent various cell types from adapting to continuous growth in culture.
On the basis of studies with human cells, the most likely contributing candidates in enabling bypass of cellular senescence in these other cell types are genes in the Rb pathway (3, 4, 8, 27). In support of this concept, BM-derived CSF-1-dependent macrophages from Arf-null mice, although initially able to proliferate faster than wild-type cells, spontaneously yielded even faster growing populations that exhibited doubling times comparable to cells lacking both Arf and Ink4a. Strikingly, these later passage Arf-null macrophages no longer expressed Ink4a mRNA but continued to produce the related Cdk inhibitor, p15Ink4b, which is encoded by a locus very closely linked to Ink4a-Arf. These data implied that deletion or rearrangement of chromosomal DNA had not occurred in the region that contains both Ink4a-Arf and Ink4b and instead suggested that Ink4a was transcriptionally silenced. Use of a methylation-specific PCR assay (36) revealed that down-regulation of p16Ink4a expression in late passage cells temporally correlated with methylation within the first coding exon and immediate 5′ promoter sequences of the Ink4a gene. Like BAC1.2F5 cells, which lack the Ink4a-Arf locus, and unlike macrophage cell lines immortalized by simian virus 40, macrophage populations derived from Arf-null mice that no longer expressed Ink4a remained diploid and dependent on CSF-1 for growth and survival throughout many months in culture.
After a delay of several weeks, rapidly proliferating variant populations also arose spontaneously in macrophages cultured from normal bone marrow. At early passages, these cells proliferated much more slowly than their counterparts from Arf-null mice. However, at later passages, methylation and silencing of the Ink4a gene again correlated with accelerated growth. The latter cells continued to express p19Arf and could be maintained for an additional 3 months in culture without further changes in their generation time, suggesting that loss of p16Ink4a may, at least in part, alleviate selective pressure for mutations in the Arf–Mdm2–p53 pathway. It remains unclear whether the populations derived from wild-type macrophage progenitors are fully established, in the sense that their doubling times remained much slower than those of immortal BAC1.2F5 cells or of cells explanted from animals deficient in both Ink4a and Arf, which grew rapidly immediately after explantation. Nonetheless, it is evident that Ink4a strongly contributes in providing a significant barrier to continuous macrophage proliferation in culture. It will therefore be of some interest to determine the behavior of BM-derived macrophages from mice that retain Arf and lack Ink4a alone.
Transcriptional repression of Ink4a associated with hypermethylation of the proximal promoter and first coding exon has been previously reported to occur as a telomere-independent mechanism to overcome early senescence in human mammary epithelial cells and keratinocytes (44, 45). Although we believe that such telomere-independent barriers to proliferation occur as a checkpoint response to stress imposed by artificial culture conditions (3, 4), hypermethylation of p16Ink4a occurs in many forms of human cancer (46, 47). We might surmise that in mice, as well as in humans, the loss of Ink4a alone (with retention of Arf) can contribute to tumor formation in those cell types that rely strongly on p16Ink4a function for checkpoint control. A deeper understanding of this lineage specificity may help explain why Ink4a and Arf are differentially targeted in human cancers.
Acknowledgments
We thank Dr. Richard Ashmun for performing flow cytometric analyses, Dr. Tal Teitz for assistance in setting up the MSP analysis, Dr. William Walker for characterization of BM-derived macrophages, Dr. Ronald A. DePinho for generously providing Ink4a-Arf-null mice, and Dr. Owen Witte for transformed preB cells. We also gratefully acknowledge the excellent technical assistance of Rose Matthew, Camulous Hornsby, and Esther van de Kamp. This work was supported by National Institutes of Health Grants CA56819 and CA71907 (to M.F.R.), Cancer Core Grant CA21765, and the American Lebanese Syrian Associated Charities of St. Jude Children's Research Hospital. C.J.S. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- MEFs
murine embryonic fibroblasts
- MSP
methylation-specific PCR
- CSF-1
colony-stimulating factor 1
- BM
bone marrow
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hayflick L, Moorhead P S. Exp Cell Res. 1961;25:585–621. doi: 10.1016/0014-4827(61)90192-6. [DOI] [PubMed] [Google Scholar]
- 2.Todaro G J, Green H. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wright W E, Shay J W. Nat Med. 2000;6:849–851. doi: 10.1038/78592. [DOI] [PubMed] [Google Scholar]
- 4.Sherr C J, DePinho R A. Cell. 2000;102:407–410. doi: 10.1016/s0092-8674(00)00046-5. [DOI] [PubMed] [Google Scholar]
- 5.Kipling D, Cooke H J. Nature (London) 1990;347:400–402. doi: 10.1038/347400a0. [DOI] [PubMed] [Google Scholar]
- 6.Prowse K R, Greider C W. Proc Natl Acad Sci USA. 1995;92:4818–4822. doi: 10.1073/pnas.92.11.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blasco M A, Lee H-W, Hande M P, Samper E, Lansdorp P M, DePinho R A, Greider C W. Cell. 1997;91:25–34. doi: 10.1016/s0092-8674(01)80006-4. [DOI] [PubMed] [Google Scholar]
- 8.Wright W E, Shay J W. Science. 2001;291:839–840. doi: 10.1126/science.1058546. [DOI] [PubMed] [Google Scholar]
- 9.Ramirez R D, Morales C P, Herbert B-S, Rohde J M, Passons C, Shay J W, Wright W E. Genes Dev. 2001;15:398–403. doi: 10.1101/gad.859201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang D G, Tokumoto Y M, Apperly J A, Lloyd A C, Raff M C. Science. 2001;291:868–871. doi: 10.1126/science.1056780. [DOI] [PubMed] [Google Scholar]
- 11.Mathon N F, Malcolm D S, Harrisingh M C, Cheng L, Lloyd A C. Science. 2001;291:872–875. doi: 10.1126/science.1056782. [DOI] [PubMed] [Google Scholar]
- 12.Quelle D E, Zindy F, Ashmun R A, Sherr C J. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 13.Sherr C J. Genes Dev. 1998;12:2984–2991. doi: 10.1101/gad.12.19.2984. [DOI] [PubMed] [Google Scholar]
- 14.Serrano M, Hannon G J, Beach D. Nature (London) 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 15.Zindy F, Eischen C M, Randle D, Kamijo T, Cleveland J L, Sherr C J, Roussel M F. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Stanchina E, McCurrach M E, Zindy F, Shieh S-Y, Ferbeyre G, Samuelson A V, Prives C, Roussel M F, Sherr C J, Lowe S W. Genes Dev. 1998;12:2434–2442. doi: 10.1101/gad.12.15.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bates S, Phillips A C, Clarke P, Stott F, Peters G, Ludwig R L, Vousden K H. Nature (London) 1998;395:124–125. doi: 10.1038/25867. [DOI] [PubMed] [Google Scholar]
- 18.Palmero I, Pantoja C, Serrano M. Nature (London) 1998;395:125–126. doi: 10.1038/25870. [DOI] [PubMed] [Google Scholar]
- 19.Radfar A, Unnikrishnan I, Lee H-W, DePinho R A, Rosenberg N. Proc Natl Acad Sci USA. 1998;95:13194–13199. doi: 10.1073/pnas.95.22.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pomerantz J, Schreiber-Agus N, Liégeois N J, Silverman A, Alland L, Chin L, Potes J, Chen K, Orlow I, Lee H-W, Cordon-Cardo C, DePinho R. Cell. 1998;92:713–723. doi: 10.1016/s0092-8674(00)81400-2. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y, Xiong Y, Yarbrough W G. Cell. 1998;92:725–734. doi: 10.1016/s0092-8674(00)81401-4. [DOI] [PubMed] [Google Scholar]
- 22.Kamijo T, Weber J D, Zambetti G, Zindy F, Roussel M F, Sherr C J. Proc Natl Acad Sci USA. 1998;95:8292–8297. doi: 10.1073/pnas.95.14.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stott F J, Bates S, James M C, McConnell B B, Starborg M, Brookes S, Palmero I, Ryan K, Hara E, Vousden K H, Peters G. EMBO J. 1998;17:5001–5014. doi: 10.1093/emboj/17.17.5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zindy F, Quelle D E, Roussel M F, Sherr C J. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 25.Serrano M, Lee H-W, Chin L, Cordon-Cardo C, Beach D, DePinho R A. Cell. 1996;85:27–37. doi: 10.1016/s0092-8674(00)81079-x. [DOI] [PubMed] [Google Scholar]
- 26.Kamijo T, Zindy F, Roussel M F, Quelle D E, Downing J R, Ashmun R A, Grosveld G, Sherr C J. Cell. 1997;91:649–659. doi: 10.1016/s0092-8674(00)80452-3. [DOI] [PubMed] [Google Scholar]
- 27.Ruas M, Peters G. BBA Rev Cancer. 1998;1378:F115–F177. doi: 10.1016/s0304-419x(98)00017-1. [DOI] [PubMed] [Google Scholar]
- 28.Whitlock C A, Witte O N. Methods Enzymol. 1987;150:275–286. doi: 10.1016/0076-6879(87)50085-4. [DOI] [PubMed] [Google Scholar]
- 29.Borzillo G V, Sherr C J. Mol Cell Biol. 1989;9:3973–3981. doi: 10.1128/mcb.9.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eischen C M, Weber J D, Roussel M F, Sherr C J, Cleveland J L. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanley E R. Methods Enzymol. 1986;116:564–587. doi: 10.1016/s0076-6879(85)16044-1. [DOI] [PubMed] [Google Scholar]
- 32.Morgan C, Pollard J W, Stanley E R. J Cell Physiol. 1987;130:420–427. doi: 10.1002/jcp.1041300316. [DOI] [PubMed] [Google Scholar]
- 33.Vermes I, Haanen C, Steffens-Nakken H, Reutelingsperger C. J Immunol Methods. 1995;184:39–51. doi: 10.1016/0022-1759(95)00072-i. [DOI] [PubMed] [Google Scholar]
- 34.Inoue K, Wren R, Rehg J E, Adachi M, Cleveland J L, Roussel M F, Sherr C J. Genes Dev. 2000;14:1797–1809. [PMC free article] [PubMed] [Google Scholar]
- 35.Quelle D E, Ashmun R A, Hannon G J, Rehberger P A, Trono D, Richter H, Walker C, Beach D, Sherr C J, Serrano M. Oncogene. 1995;11:635–645. [PubMed] [Google Scholar]
- 36.Herman J G, Graff J R, Myohanen S, Nelkin B D, Baylin S B. Proc Natl Acad Sci USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strasser A, Harris A W, Jacks T, Cory S. Cell. 1994;79:329–339. doi: 10.1016/0092-8674(94)90201-1. [DOI] [PubMed] [Google Scholar]
- 38.Stone L B, Takemoto K K. J Virol. 1970;6:621–627. doi: 10.1128/jvi.6.5.621-627.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mauel J, Defendi V. J Exp Med. 1971;134:335–350. doi: 10.1084/jem.134.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwarzbaum S, Halpern R, Diamond B. J Immunol. 1984;132:1158–1162. [PubMed] [Google Scholar]
- 41.Jones P A, Laird P W. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 42.Malumbres M, de Castro I P, Santos J, Melendez B, Mangues R, Serrano M, Pellicer A, Fernandez-Piqueras J. Oncogene. 1997;14:1361–1370. doi: 10.1038/sj.onc.1200969. [DOI] [PubMed] [Google Scholar]
- 43.Holland E C, Hively W P, Gallo V, Varmus H E. Genes Dev. 1998;12:3644–3649. doi: 10.1101/gad.12.23.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kiyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Nature (London) 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 45.Brenner A J, Stampfer M R, Aldaz C M. Oncogene. 1998;17:199–205. doi: 10.1038/sj.onc.1201919. [DOI] [PubMed] [Google Scholar]
- 46.Baylin S B, Herman J G, Graff J R, Vertino P M, Issa J-P. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 47.Esteller M, Corn P G, Baylin S B, Herman J G. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]