Abstract
AIM
To investigate the miRNA expression in colonic mucosal biopsies from endoscopically inflamed and non inflamed regions of ulcerative colitis (UC) patients.
METHODS
Colonic mucosal pinch biopsies were analyzed from the inflamed and non inflamed regions of same UC patient. Total RNA was isolated and differential miRNA profiling was done using microarray platform. Quantitative Real Time PCR was performed in colonic biopsies from inflamed (n = 8) and non-inflamed (n = 8) regions of UC and controls (n = 8) to validate the differential expression of miRNA. Potential targets of dysregulated miRNA were identified by using in silico prediction tools and probable role of these miRNA in inflammatory pathways were predicted.
RESULTS
The miRNA profile of inflamed colonic mucosa differs significantly from the non-inflamed. Real time PCR analysis showed that some of the miRNA were differentially expressed in the inflamed mucosa as compared to non inflamed mucosa and controls (miR-125b, miR-223, miR-138, and miR-155), while (miR-200a) did not show any significant changes. In contrast to microarray, where miR-378d showed downregulation in the inflamed mucosa, qRT-PCR showed a significant upregulation in the inflamed mucosa as compared to the non inflamed. The in silico prediction analysis revealed that the genes targeted by these miRNAs play role in the major signaling pathways like MAPK pathway, NF-κB signaling pathway, cell adhesion molecules which are all assciated with UC.
CONCLUSION
The present study reports disease specific alteration in the expression of miR-125b, miR-155, miR-223 and miR-138 in UC patients and also predict their biological significance.
Keywords: Ulcerative colitis, Colon mucosa, MicroRNA, Microarray, qRT-PCR, In silico analysis
Core tip: In order to get an insight into the pathogenesis of ulcerative colitis (UC), we explored spatial expression of microRNA in the inflamed and non-inflamed region of mucosal tissue of patients. Profiling of differentially expressed miRNA was generated by microarray from three paired samples. Few significantly dysregulated microRNA were validated using qRT-PCR. The present study reports disease specific alteration in the expression of miR-125b, miR-155, miR-223 and miR-138 in UC patients and also analyzed their biological significance using in silico tools. MiR-223 exhibited elevated expression independent of disease activity therefore, could be a potential candidate as biomarker for UC.
INTRODUCTION
Inflammatory bowel disease (IBD) is a gastrointestinal disorder which is chronic and relapsing in nature and exists in two clinical forms ulcerative colitis (UC) and Crohn’s disease (CD)[1]. Over the past decade, the incidence and prevalence of IBD are continuously increasing worldwide[2]. The etiology of IBD is not defined yet, but several reports in the past have hypothesized that there is complex interplay of microbial, environmental and genetic factors which predisposes an individual to develop the disease[3]. Various systematic approaches like candidate gene identification or genome wide association studies have been made to identify the genes that get dysregulated and affect the downstream signaling pathways and subsequent gene expression during disease condition. Molecular changes in the expression of several genes have been found to be assciated with IBD pathogenesis[4], and some of these genes play important roles in the major inflammatory pathways.
MicroRNA are a class of small sized (about 18-22 nucleotides in length), non coding, single stranded, endogenous mRNA which modulates the expression of their target mRNA by binding to them in their 3’UTR by either carrying out their degradation or inhibiting the translation prcess[5-8]. However, target sites for miRNA have also been reported in the 5’UTR of many genes[9]. Around 2000 mature miRNA have been reported so far and they are known to modulate the expression of about one third of human genes[10]. Recent evidences suggest that miRNAs play key role in modulating the expression of target genes involved in the pathogenesis of various diseases. According to a recent estimate almost 60% of genes in a cell are regulated by miRNAs[11].
In the past few years miRNAs have emerged as the new epigenetic regulators in IBD[12,13] also differential expression of miRNAs have been assciated with several autoimmune diseases and cancer including IBD[14]. These studies have revealed differential profiling of miRNAs in mucosal tissues of UC and CD patients thus indicating their importance as potential biomarkers[15]. Our aim in this study had been to profile the miRNAs exhibiting differential expression in inflamed and non-inflamed region of the colon tissue in UC patients. Further we attempted to assess using bioinformatics tools, the functional significance of selected miRNAs involved in the regulation of target genes. Therefore, studying these miRNAs could provide better ways for disease diagnosis and therapeutics by establishing potential biomarkers.
MATERIALS AND METHODS
Patients and tissue samples
The study included 8 UC patients and 8 non IBD controls. Colonic pinch biopsies were collected from the endoscopically inflamed as well as non inflamed regions of UC patients from the Department of Gastroenterology, All India Institute of Medical Sciences, New Delhi, India. The disease activities of the samples were measured on the basis of SCCAI score. Demographic features of study subjects are enlisted in Table 1. For obtaining inflamed mucosal samples, pinch biopsies were withdrawn from the colonic regions which showed clear symptoms of UC such as loss of vascular pattern, erythema, spontaneous bleeding or ulceration while in case of non inflamed mucosal samples, biopsies were collected from colonic regions of same UC patients with no disease activity. In case of control subjects biopsies were collected from rectosigmoid area. All the controls were age and sex matched with patients. Controls included were individual attending the clinic for routine colonoscopy and were without any inflammatory disorder of intestine and without any IBD symptoms. Mucosal biopsy samples were collected in RNA later solution. Diagnosis of UC was confirmed through endoscopic and histological examination by following ECCO guidelines[16].
Table 1.
Demographic features of study subject n (%)
| Characteristics | UC patients | Non IBD control |
| No. of patients (total) | 8 | 8 |
| Sex (M/F) | 3/5 | 2/6 |
| Age (mean ± SD, range, yr) | 39.75 ± 10.29 (25-62) | 40.75 ± 11.86 (24-60) |
| Disease duration (mean ± SD, Range, yr) | 6.35 ± 6.52 (1.5-21) | NA |
| Disease extent | ||
| Proctitis | 3 (37.5) | |
| Left sided colitis | 5 (62.5) | |
| Medication | ||
| Mesalamine | 5 (62.5) | 0 |
| Azathioprine | 3 (37.5) | 0 |
| Steroids | 0 | 0 |
UC: Ulcerative colitis; IBD: Inflammatory bowel disease.
Ethical approval
The study has been ethically approved from the Institute Ethics Committee, All India Institute of Medical Sciences (Ref. No. T-290/23.06.2015, RT-7/27.01.2016) and Institutional Ethics Review Board, JNU (IERB Ref. No.2016/Student/93). Informed consent was taken from all the subjects included in study.
RNA isolation and quality check
Total RNA was extracted from the mucosal biopsies using mirVana miRNA isolation kit (Ambion INC, TX, United States) according to manufacturer’s protcol and quantified by NanoDrop 2000 Spectrophotometer (Thermo Scientific, Waltham, United States). The 260/280 values were above 1.9. Quality check for RNA was assayed by Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, United States).
miRNA microarray profiling
The array included three pairs of inflamed and non inflamed samples from UC patients. Samples were submitted to Affymetrix for miRNA profiling using GeneChip® miRNA 4.0 Array which is designed to interrogate all mature miRNA sequences in miRBase Release 20. The differentially expressed miRNA were statistically assessed by one way ANOVA paired method. P value of 0.05 was considered significant and fold change of 1.5 was taken as cut off for upregulation and -1.5 for downregulation of miRNA in a given sample.
Reverse transcription and quantitative real time PCR
Reverse transcription for each miRNA (800 ng) was carried out using gene specific looped primers[17], using revert aid cDNA synthesis kit (Fermentas St. Leon Rot, Germany). Sequence of primers used for reverse transcription and qRT-PCR are enlisted in Table 2. Differential expression of six selected miRNAs identified by microarray was performed using BioRad CFX96 Real Time System with C1000 Touch Thermal cylcer. (BioRad, Hercules, CA, United States) by SYBR Green method, and the cycles were as follows: initial denaturation - 94 °C for 2 min, denaturation - 94 °C for 30 s, annealing - 60 °C for 1 min for 40 cycles. The relative expression differences of miRNAs were normalized to internal reference U6 snoRNA and analyzed using 2-ΔΔct method. The statistical analysis was done using unpaired, two way student’s t-test and a P value < 0.05 were considered significant.
Table 2.
List of primers used for reverse transcription and qRT-PCR
| Name | Primer | Sequence (5’-3’) |
| Reverse Universal Primer | GTGCAGGGTCCGAGGT | |
| hsa-miR-125b-5p | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGTCACAAGT |
| Forward | TCCCTGAGACCCTAACTTG | |
| hsa-miR-223-3p | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGTGGGGTAT |
| Forward | TGTCAGTTTGTCAAATACCC | |
| hsa-miR-155-5p | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCCTAT |
| Forward | TTAATGCTAATCGTGATAGG | |
| hsa-miR-138-5p | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGCGGCCTGA |
| Forward | AGCTGGTGTTGTGAATCAG | |
| hsa-miR-200a-3p | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACATCGTT |
| Forward | TAACACTGTCTGGTAACGAT | |
| hsa-miR-378d | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGTTTCTGTC |
| Forward | ACTGGACTTGGAGTCAGAAA | |
| Sno RNA U6 | RT | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAAAATATG |
| Forward | CAAATTCGTGAAGCGTTCCA |
RT primers were used to sythesize miRNA specific cDNA. For real time PCR miRNA specific forward primer and reverse universal primers were used.
Target prediction for differentially expressed miRNAs
Target prediction for differentially expressed miRNAs was carried out by miRWalk database (http://www.ma.uniheidelberg.de/apps/zmf/mirwalk/). This database simultaneously searches several other databases[18]. Additionally, TargetScan, miRBD, RNA22, DIANA, Pictar were also selected for target prediction and genes picked at least by three tools were selected for our list of potential targets. Further, involvement of these miRNAs in different biological pathways was investigated using mirPath v.3: DIANA TOOLS.
RESULTS
MiRNA microarray profiling of inflamed colonic mucosa of UC patients
To investigate the disease specific changes in the miRNA profiling, mucosal biopsy tissues from endoscopically inflamed and non inflamed region of UC patients were submitted for microarray analysis. Microarray analysis revealed that miRNAs are differentially expressed in the inflamed colonic region of UC patients as compared to the non inflamed ones Supplementary Figure 1. We had selected three matched pairs for miRNA microarray profiling, out of which two pairs (Sample NI1, I1 and NI6, I6) exhibited dysregulated expression of several miRNAs in the inflamed mucosa which indicates the disease specific behavior of miRNAs. In the third matched pair (NI5, I5) the changes in miRNA expression were not comparable probably due to the mild disease activity in the patient.
Figure 1.
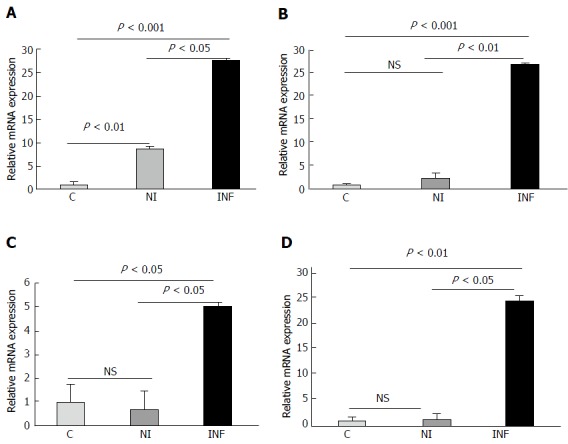
Differential expression of miRNAs investigated by performing real time PCR. A: miR-223; B: miR-125b; C: miR-138; D: miR-155. C denotes Control, NI denotes non-inflamed and INF denotes inflamed. NS: Not significant.
Some of the miRNAs were upregulated with a fold change more than 10 such as miR-138, miR-708-5p, miR-212-3p, miR-4521, miR-17,-3p, miR-223,-3p, miR-21. The top 44 differentially expressed miRNAs have been enlisted in Table 3 with their respective fold changes. MiR-138 and miR-223 were upregulated more than 10 folds in the inflamed tissues and they have also been reported to be dysregulated in UC. miR-125b and miR-155 were upregulated with a fold change of 2.56 and 2.33 respectively in our study. Additionally, miR-200a and miR-378d were downregulated with a fold change of -2.14 and -4.06 respectively. We selected these six miRNAs as candidate miRNAs for further analysis because these miRNAs were reported to be dysregulated during autoimmune diseases and in various cancers. Some of these miRNAs were reported to be differentially regulated during UC but whether these miRNAs show any spatial expression within the endoscopically inflamed and non inflamed regions was not known. Also the gene targeted by these miRNAs play role in major inflammatory pathways.
Table 3.
List of top 44 differentially expressed miRNAs in the inflamed colonic mucosal biopsies of ulcerative colitis patients determined by microarray
| MicroRNA | F.C1 | Expression | MicroRNA | F.C | Expression |
| hsa-miR-138-5p | 35.16 | Upregulated | hsa-miR-148a-5p | 2.42 | Upregulated |
| hsa-miR-708-5p | 34.7 | Upregulated | hsa-miR-155-5p | 2.33 | Upregulated |
| hsa-miR-212-3p | 24.06 | Upregulated | hsa-miR-21-5p | 1.81 | Upregulated |
| hsa-miR-4538 | 26.12 | Upregulated | hsa-miR-196b-3p | 1.50 | Upregulated |
| hsa-miR-4521 | 17.69 | Upregulated | hsa-miR-552-3p | -7.82 | Downregulated |
| hsa-miR-4417 | 16.63 | Upregulated | hsa-miR-196b-5p | -7.60 | Downregulated |
| hsa-miR-17-3p | 15.28 | Upregulated | hsa-miR-378d-5p | -4.06 | Downregulated |
| hsa-miR-424-3p | 14.17 | Upregulated | hsa-miR-141-3p | -3.03 | Downregulated |
| hsa-miR-874-3p | 13.47 | Upregulated | hsa-miR-10b-5p | -2.85 | Downregulated |
| hsa-miR-25-5p | 13.41 | Upregulated | hsa-miR-215-5p | -2.56 | Downregulated |
| hsa-miR-223-3p | 13.01 | Upregulated | hsa-miR-192-5p | -2.52 | Downregulated |
| hsa-miR-1271-5p | 11.96 | Upregulated | hsa-miR-194-3p | -2.31 | Downregulated |
| hsa-miR-148b-3p | 11.31 | Upregulated | hsa-miR-422a | -2.22 | Downregulated |
| hsa-miR-501-5p | 11.26 | Upregulated | hsa-miR-200a-3p | -2.14 | Downregulated |
| hsa-miR-4486 | 10.86 | Upregulated | hsa-miR-378a-3p | -2.04 | Downregulated |
| hsa-miR-224-3p | 10.46 | Upregulated | hsa-miR-6732-5p | -1.94 | Downregulated |
| hsa-miR-21-3p | 8.46 | Upregulated | hsa-miR-147b | -1.78 | Downregulated |
| hsa-miR-146b-5p | 6.83 | Upregulated | hsa-miR-200b-3p | -1.7 | Downregulated |
| hsa-miR-149-5p | 6.24 | Upregulated | hsa-miR-572 | -1.63 | Downregulated |
| hsa-miR-31-5p | 4.04 | Upregulated | hsa-miR-4649-5p | -1.61 | Downregulated |
| hsa-miR-491-5p | 3.92 | Upregulated | hsa-miR-638 | -1.6 | Downregulated |
| hsa-miR-125b-5p | 2.56 | Upregulated | hsa-miR-299-5p | -1.59 | Downregulated |
F.C denoted the fold change in the expression of respective miRNA in the inflamed biopsies compared to the non inflamed biopsy.
Validation of differentially expressed miRNA by qRT-PCR
The differential expression of six candidate miRNAs (miR-155, miR-138, miR-125b, miR-223, miR-378d and miR-200a) was validated by performing qRT-PCR in matched pair samples from UC patients (n = 8) and non IBD controls (n = 8). The expression of miR-155, miR-138, miR-125b and miR-223 was found to be significantly upregulated in inflamed colonic mucosa of UC patients as compared to non inflamed mucosa and controls Figure 1. We found that miR-223 was significantly upregulated in the inflamed vs non inflamed tissues (P < 0.05) and in inflamed vs control (P < 0.001). Its expression was also significantly higher in the non inflamed vs controls (P < 0.01) as well. Similarly, miR-125b, was significantly higher in inflamed vs non inflamed (P < 0.01) as seen in microarray and inflamed vs controls (P < 0.001). In accordance with microarray, miR-138 exhibited increased expression in inflamed vs non inflamed and also in inflamed vs controls (P < 0.05) and mir-155 was also upregulated in inflamed vs non inflamed (P < 0.05) as well as in inflamed vs control (P < 0.01). In contrast to microarray results, the expression of miR-200a did not change significantly in the inflamed samples when compared with non inflamed and controls Figure 2. But interestingly, the expression of miR-200a in non inflamed mucosa was significantly lower in the non inflamed vs controls. As seen in microarray miRNA-378d was downregulated in the inflamed mucosa of UC patients but in qRT-PCR, it showed a significant upregulation in the inflamed mucosa as compared to the non inflamed (P < 0.01) and controls (P < 0.05). Interestingly, miR-378d showed less expression in the non inflamed mucosa as compared to the control.
Figure 2.

Real time analysis for miR-378d and miR-200a. A: miR-378d; B: miR-378d matched individual samples; C: miR-200a; D: miR-200a matched individual samples. C: Control; NI: Non inflamed; INF: Inflamed; NS: Not significant.
The qRT-PCR results were further reanalyzed individually for each matched pair and it showed that although inter sample variation was there in each case, the expression of miR-155, miR-138, miR-223, miR-125b was upregulated in most of the inflamed samples as compared to the non inflamed ones Figure 3 and these changes were found to be statistically significant. In contrast to microarray, miR-378d showed significant upregulation in the inflamed samples as compared to the non inflamed although inter sample variation was seen in this case also. In case of miR-200a, the inter sample variation was quite high, although the changes in expression were not significant it showed upregulation in the expression in four pairs whereas downregulation in the rest four pairs Figure 2. Therefore, we could not draw a conclusive trend for this miRNA.
Figure 3.
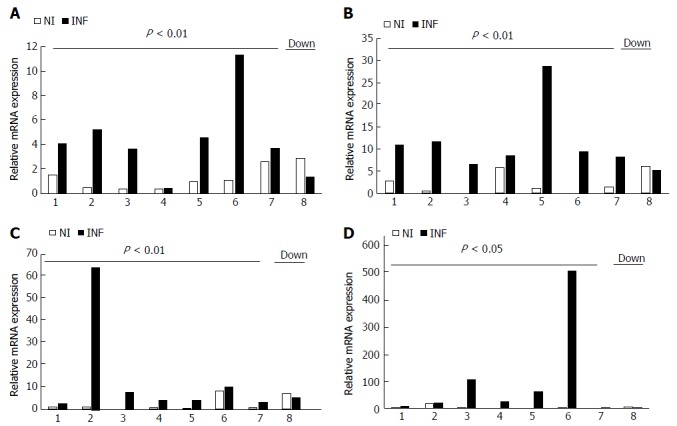
Differentially expressed miRNAs in the matched colonic biopsies of each ulcerative colitis patient determined by qRT-PCR analysis. A: miR- 223; B: miR-125-b; C: miR-138; D: miR-155. NI denotes non-inflamed and INF denotes inflamed. NS: Not significant.
Prediction of potential targets by in silico prediction tool
miRNAs (miR-155, miR-138, miR-125b andmiR-223) showing significant changes in qRT-PCR analysis were examined for their biological relevance by studying their respective potential gene targets and the signaling pathways involved. In silico target prediction analysis revealed several putative targets which play important role in major signaling pathways. Target prediction analysis indicated biological relevance of these dysregulated miRNAs as the genes targeted by these miRNAs are involved in the major inflammatory pathways such as NF-κB signaling pathways and MAPK pathway. Potential targets of all four miRNAs have been enlisted in Table 4. Potential targets of miR-125b included TNF receptor assciated factor 6 (TRAF6), TNF alpha induced protein 3 (TNFAIP3), IL6R of NF-κB signaling pathway and Signal transducer and activator of transcription (STAT3), JUN, AKT1, IRF6 and IRAK1 of TLR signaling. In addition to this, miR-125b also targets SMAD4 and cAMP responsive element binding protein1 (CREB1), SIRT5, DICER1, acetylcholine esterase (ACHE), CDK16, CCR2 and MUC1.Putative targets of miR-155 includes signaling molecules of NF-κB signaling such as TGF beta activated kinase 2 (TAB2), CARD6, RelA and TRAF3, suppressor of cytokine signaling (SCS1), KRAS, PAK2, BDNF of MAPK signaling, CTLA4, IL6R, nuclear factor of activated T cells 5 (NFAT5), IL17RB, Myd88, IKBKE, CTLA4, CLCN3 and APC.
Table 4.
List of potential targets and the pathway involved
| MicroRNA | Target gene | Pathway involved |
| hsa-miR-155-5p | TAB2, CFLAR, CARD11, | NF-κB signaling. |
| ReLA, TRAF3 | ||
| hsa-miR-155-5p | CARD6, ERBB2IP, MAPK10 | Nod like receptor signaling |
| hsa-miR-155-5p | S °CS1, KRAS, PAK2, RAP1B, | MAPK signaling |
| RFLA, TAOK1, CACNB4, BDNF | ||
| hsa-miR-155-5p | FOS, S °CS1, IKBKE, TRAF3, | TLR signaling |
| MAPK10, TAB2 | ||
| has-miR-155 | NFAT5, GSK3B, INPP5D, VAV3, PIK3CA | B cell receptor signaling |
| hsa-miR-138-5p | CSNK2A2, RELA | NF-κB signaling |
| hsa-miR-138-5p | CXCL8, MAP2K7 | TLR signaling |
| hsa-miR-138-5p | CLDN19, CD274, ITGAL | Cell Adhesion Molecules. |
| hsa-miR-125b-5p | MAPK14, JUN.AKT1, IRAK1, TRAF6, IRF5 | TLR signaling |
| hsa-miR-125b | TNFAIP3, CSNK2A1, IRAK1 | NF-κB signaling |
| hsa-miR-223-3p | STAT1, RELA | TLR signaling |
| hsa-miR-223-3p | TAB3, ERC1, PARP1, RELA | NF-κB signaling |
| hsa-miR-378d | MAPK1 | MAPK signaling |
| hsa-miR-378d | TRAF3 | NF-κB signaling |
| hsa-miR-200a | TAB2, TNFAIP3 | NF-κB signaling |
Similarly, miR-138 targets genes such as CXCL8, RELA of NF-κB signaling pathway and other genes like CASP3, HDAC4, SIRT1, MAST4, SMAD4, MyD88 and CDK6. miR-223 targets STAT1, RELA and TAB3 of NF-κB signaling. The putative targets of miR-378d include TRAF3, SCS2 and MAPK1. Putative target genes of miR-200a involved in inflammatory pathways were TAB2, CXCL12, TNFAIP3, SIRT1 and TGFB2.
DISCUSSION
In the past few years, miRNAs have been reported to be dysregulated during UC, there are several studies which claimed that miRNA profile in the blood and tissue of UC patients differs from that of control individuals[19,20]. MiRNA were also reported to play critical role in mediating cross talk between CD, UC and CRC[21].
Our microarray analysis included three matched pairs from the endoscopically inflamed and non inflamed regions of UC patients. One of the pair didn’t show much changes in the miRNA transcriptome of inflamed region when compared with the non inflamed one (NI5 and I5), which is more certainly due to the mild activity of disease in the patient. Other two pairs showed significant changes in the miRNA profile in the inflamed regions as compared to the non inflamed ones. We found that miR-223 and miR-138 were upregulated with a fold change of more than 10 folds in the inflamed mucosa and these miRNAs are also reported earlier to be dysregulated during UC[22-24] however the spatial expression of these miRNAs in the endoscopically inflamed and non inflamed regions are reported here for the first time. miR-125b and miR-155 mediates cross talk between UC and CRC[21] as reported earlier and also modulates the expression of genes playing role in inflammatory pathways[25]. miR-200a is earlier reported to get downregulated in UC[22]. Keeping the role of these miRNAs in mind we selected these six as our candidate miRNAs for further analysis, so that we can elucidate the disease specific behavior of these miRNA and correlate their expression with disease activity.
qRT-PCR analysis revealed that miR-223 was upregulated in inflamed tissues of UC patients, also this was the only miRNA in our study, which showed significantly higher expression in both the inflamed as well as non inflamed tissues of UC patients as compared to controls. Therefore, this miRNA could be established as a potential biomarker to differentiate between UC and controls as its expression does not depend upon disease activity. Moreover, previous studies have shown miR-223 plays a role in gastric cancer and also known to target the tumor suppressor gene EPB41L3[26,27]. Wang et al[28] showed that miR-223 targets claudin-8 (CLDN8) which is a tight junction protein and maintains the intestinal barrier. miR-223 targets CLDN8 thereby mediating cross talk between IL23 pathway during IBD.
In accordance with the microarray studies, miR-155 expression was also significantly higher in the inflamed vs non inflamed as well as inflamed vs controls. Its upregulation is also reported in breast cancer where it acts as an OncomiR by targeting suppressor of cytokine signaling 1 (SCS1)[29,30]. In addition to this Kong et al[31] claimed that miR-155 modulated the expression of tumor suppressor gene von Hippel-Lindau (VHL) and promotes tumor invasion, proliferation, angiogenesis and recruitment of proinflamatory cells in breast cancer. Expression of miR-125b was also found significantly higher in the inflamed mucosa as compared to non inflamed and controls. Since miRNAs (miR125b and miR-155) are responsible for mediating cross talk between UC and CRC[21], therefore, studying these miRNAs in detail could provide new insights in disease diagnosis and therapeutics.
In the present study, miR-138 was also highly expressed in the inflamed mucosa of UC patient. Elevated level of miR-138 expression reported here has not been shown earlier from mucosal tissue of UC patients but only from peripheral blood samples[32]. Some of the previous reports suggest tumor suppressive role of miR-138 in nasopharyngeal cancers by targeting oncogene CCDN1 and controlling tumor proliferation[33] and also downregulation of miR-138 was reported by Liu et al[34] in head and neck squamous cell carcinomas where it played role in promoting apoptosis and suppressing tumor invasion. Role of this microRNA, regulating the target gene of inflammatory pathway during UC is yet to be validated. In contrast with the microarray results, qRT-PCR analysis did not show significant changes in expression of miR-200a in the inflamed mucosa vs non inflamed mucosa of UC patients. Interestingly, miR-378d showed a significant upregulation in the inflamed mucosa as compared to the non inflamed and controls in paired samples, while our microarray data a reverse trend in the inflamed mucosa. We also investigated the expression of miR-200a and miR-378d in UC (n = 30) vs controls (n = 20) by qRT-PCR, where we again observed significant downregulation in miR-200a and significant upregulation in miR-378d (data not shown).
The next approach in our study was to find out the putative targets of candidate miRNAs using in silico target prediction tools. The potential targets of our candidate miRNAs included signaling molecules of inflammatory pathways such as MAPK signaling, NF-κB signaling and cytokine signaling pathways, which are known to be directly assciated with pathogenesis of IBD[35].
In conclusion, our study elucidated the spatial relationship of miR-125b, miR-155, miR-138 and miR-223 in the endoscopically involved and non involved pockets of UC patients. miR-223 showed differential expression independent of disease activity therefore, could be a potential target as biomarker for UC.
ACKNOWLEDGMENTS
The authors acknowledge the subjects who participated in the study. Valmiki S acknowledges the fellowship received from University Grants Commission, New Delhi India. The authors are grateful to Mr. Raju Ranjha for helping us with troubleshoots during experiments.
COMMENTS
Background
MicroRNA (miRNAs) are endogenous, single stranded, short length (18-22 nucleotides), non coding RNA which modulates the expression of their target mRNA by binding to them in the 3’UTR and causing RNA degradation or translational repression. The differential expression of miRNA has been reported in different diseases including psoriasis, multiple sclerosis, and inflammatory bowel disease.
Research frontiers
Expression profile of different miRNA was investigated in the colonic mucosal samples of endoscopically inflamed and non inflamed regions of the same ulcerative colitis (UC) patient to get an insight in to the status of these miRNA in the inflamed and non inflamed pockets of UC patients. Microarray and qRT-PCR showed a spatial distribution in expression of miR-125b, miR-223, miR-155 and miR-138 in the inflamed colonic samples. Putative targets of these miRNAs included the genes involved in the major inflammatory pathways such as NF-κB pathway and MAPK signaling which showed that these miRNA might be one of the contributing factors in disease pathogenesis.
Innovations and breakthroughs
In order to get an insight into the pathogenesis of UC, the authors explored spatial distribution in expression of microRNA in the inflamed and non-inflamed region of mucosal tissue of patients. Profiling of differentially expressed miRNA was generated by microarray from three paired samples. Few significantly dysregulated miRNA were validated using qRT-PCR. The present study reports disease specific alteration in the expression of miR-125b, miR-155, miR-223 and miR-138 in UC patients and also analyzed their biological significance using in silico tools. miR-223 exhibited elevated expression independent of disease activity therefore, could be a potential candidate as biomarker for UC.
Applications
miR-223 altered expression during UC, irrespective of inflammation, therefore it could be developed as a biomarker for UC. Expression of this miRNA in colonic biopsy of UC patient could indicate toward disease status.
Terminology
SCCAI (Simple Clinical Colitis Activity Index) score is a symptoms based diagnostic tool and questionnaire to estimate the Ulcerative colitis disease severity. It takes into account parameters such as general well-being, bowel frequency, urgency, abdominal pain, rectal bleeding and extra intestinal features.
Peer-review
The authors reported miRNA exhibit altered expression in the inflamed colonic mucosa of UC patients. The article is informative and well-presented.
Footnotes
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: India
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
Institutional review board statement: The study has been ethically approved from the Institute Ethics Committee, All India Institute of Medical Sciences (Ref. No. T-290/23.06.2015, RT-7/27.01.2016) and Institutional Ethics Review Board, JNU (IERB Ref. No.2016/Student/93).
Conflict-of-interest statement: The authors declare no conflict of interest in the present study.
Peer-review started: February 8, 2017
First decision: March 30, 2017
Article in press: June 1, 2017
P- Reviewer: Chiba T, Lakatos PL S- Editor: Ma YJ L- Editor: A E- Editor: Li D
References
- 1.Bouma G, Strober W. The immunological and genetic basis of inflammatory bowel disease. Nat Rev Immunol. 2003;3:521–533. doi: 10.1038/nri1132. [DOI] [PubMed] [Google Scholar]
- 2.Hisamatsu T, Kanai T, Mikami Y, Yoneno K, Matsuoka K, Hibi T. Immune aspects of the pathogenesis of inflammatory bowel disease. Pharmacol Ther. 2013;137:283–297. doi: 10.1016/j.pharmthera.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 3.Chen WX, Ren LH, Shi RH. Implication of miRNA for inflammatory bowel disease treatmernt: Systemic review. World J Gastrointest Pathophysiol. 2014;5:63–70. doi: 10.4291/wjgp.v5.i2.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 5.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 8.Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 9.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci USA. 2007;104:9667–9672. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loddo I, Romano C. Inflammatory Bowel Disease: Genetics, Epigenetics, and Pathogenesis. Front Immunol. 2015;6:551. doi: 10.3389/fimmu.2015.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scarpa M, Stylianou E. Epigenetics: Concepts and relevance to IBD pathogenesis. Inflamm Bowel Dis. 2012;18:1982–1996. doi: 10.1002/ibd.22934. [DOI] [PubMed] [Google Scholar]
- 14.Ranjha R, Paul J. Micro-RNAs in inflammatory diseases and as a link between inflammation and cancer. Inflamm Res. 2013;62:343–355. doi: 10.1007/s00011-013-0600-9. [DOI] [PubMed] [Google Scholar]
- 15.Fasseu M, Tréton X, Guichard C, Pedruzzi E, Cazals-Hatem D, Richard C, Aparicio T, Daniel F, Soulé JC, Moreau R, et al. Identification of restricted subsets of mature microRNA abnormally expressed in inactive colonic mucosa of patients with inflammatory bowel disease. PLoS One. 2010;5 doi: 10.1371/journal.pone.0013160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dignass A, Eliakim R, Magro F, Maaser C, Chowers Y, Geboes K, Mantzaris G, Reinisch W, Colombel JF, Vermeire S, et al. Second European evidence-based consensus on the diagnosis and management of ulcerative colitis part 1: definitions and diagnosis. J Crohns Colitis. 2012;6:965–990. doi: 10.1016/j.crohns.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR, et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. doi: 10.1093/nar/gni178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Dalal SR, Kwon JH. The Role of MicroRNA in Inflammatory Bowel Disease. Gastroenterol Hepatol (N Y) 2010;6:714–722. [PMC free article] [PubMed] [Google Scholar]
- 20.Schaefer JS, Attumi T, Opekun AR, Abraham B, Hou J, Shelby H, Graham DY, Streckfus C, Klein JR. MicroRNA signatures differentiate Crohn’s disease from ulcerative colitis. BMC Immunol. 2015;16:5. doi: 10.1186/s12865-015-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bai J, Li Y, Shao T, Zhao Z, Wang Y, Wu A, Chen H, Li S, Jiang C, Xu J, et al. Integrating analysis reveals microRNA-mediated pathway crosstalk among Crohn’s disease, ulcerative colitis and colorectal cancer. Mol Biosyst. 2014;10:2317–2328. doi: 10.1039/c4mb00169a. [DOI] [PubMed] [Google Scholar]
- 22.Van der Goten J, Vanhove W, Lemaire K, Van Lommel L, Machiels K, Wollants WJ, De Preter V, De Hertogh G, Ferrante M, Van Assche G, et al. Integrated miRNA and mRNA expression profiling in inflamed colon of patients with ulcerative colitis. PLoS One. 2014;9:e116117. doi: 10.1371/journal.pone.0116117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Zhang S, Yu Q, Yang G, Guo J, Li M, Zeng Z, He Y, Chen B, Chen M. Circulating MicroRNA223 is a New Biomarker for Inflammatory Bowel Disease. Medicine (Baltimore) 2016;95:e2703. doi: 10.1097/MD.0000000000002703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu XM, Zhang HJ. miRNAs as new molecular insights into inflammatory bowel disease: Crucial regulators in autoimmunity and inflammation. World J Gastroenterol. 2016;22:2206–2218. doi: 10.3748/wjg.v22.i7.2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim SW, Ramasamy K, Bouamar H, Lin AP, Jiang D, Aguiar RC. MicroRNAs miR-125a and miR-125b constitutively activate the NF-κB pathway by targeting the tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20) Proc Natl Acad Sci USA. 2012;109:7865–7870. doi: 10.1073/pnas.1200081109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo X, Mao XH, Zou QM, Yu PW, Zuo QF, et al. Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One. 2012;7:e41629. doi: 10.1371/journal.pone.0041629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, Sun L, Ji G, Shi Y, Han Z, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9:824–833. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 28.Wang H, Chao K, Ng SC, Bai AH, Yu Q, Yu J, Li M, Cui Y, Chen M, Hu JF, et al. Pro-inflammatory miR-223 mediates the cross-talk between the IL23 pathway and the intestinal barrier in inflammatory bowel disease. Genome Biol. 2016;17:58. doi: 10.1186/s13059-016-0901-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mattiske S, Suetani RJ, Neilsen PM, Callen DF. The oncogenic role of miR-155 in breast cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:1236–1243. doi: 10.1158/1055-9965.EPI-12-0173. [DOI] [PubMed] [Google Scholar]
- 30.Jiang S, Zhang HW, Lu MH, He XH, Li Y, Gu H, Liu MF, Wang ED. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70:3119–3127. doi: 10.1158/0008-5472.CAN-09-4250. [DOI] [PubMed] [Google Scholar]
- 31.Kong W, He L, Richards EJ, Challa S, Xu CX, Permuth-Wey J, Lancaster JM, Coppola D, Sellers TA, Djeu JY, et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–689. doi: 10.1038/onc.2012.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fisher K, Lin J. MicroRNA in inflammatory bowel disease: Translational research and clinical implication. World J Gastroenterol. 2015;21:12274–12282. doi: 10.3748/wjg.v21.i43.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K, Wu M, Liang Y, Liu P, Tang J, et al. MiR-138 suppressed nasopharyngeal carcinoma growth and tumorigenesis by targeting the CCND1 oncogene. Cell Cycle. 2012;11:2495–2506. doi: 10.4161/cc.20898. [DOI] [PubMed] [Google Scholar]
- 34.Liu X, Jiang L, Wang A, Yu J, Shi F, Zhou X. MicroRNA-138 suppresses invasion and promotes apoptosis in head and neck squamous cell carcinoma cell lines. Cancer Lett. 2009;286:217–222. doi: 10.1016/j.canlet.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature. 2011;474:307–317. doi: 10.1038/nature10209. [DOI] [PMC free article] [PubMed] [Google Scholar]


