Abstract
Objective
Cerebral Edema (CE) in TBI is the consequence of multiple underlying mechanisms, and is associated with unfavorable outcomes. Genetic variability in these pathways likely explains some of the clinical heterogeneity observed in edema development. A role for Sulfonylurea-receptor-1 (Sur1) in CE is supported. However, there are no prior studies examining the effect of genetic variability in the Sur1 gene (ABCC8) on the development of CE. We hypothesize that ABCC8 single nucleotide polymorphisms (SNP) are predictive of CE.
Methods
DNA was extracted from 385 patients. SNPs in ABCC8 were genotyped using the Human Core Exome v1.2 (Illumina). CE measurements included acute CT edema, mean and peak intracranial pressure (ICP), and need for decompressive craniotomy.
Results
14 SNPs with minor-allele frequency>0.2 were identified. 4 SNPS rs2283261, rs3819521, rs2283258 and rs1799857 were associated with CE measures. In multiple regression models, homozygote-variant genotypes in rs2283261, rs3819521, and rs2283258 had increased odds of CT edema (OR=2.45, p=0.007; OR=2.95, p=0.025; OR=3.00, p=0.013), had higher mean (β=3.13,p=0.000; β=2.95,p=0.005; β=3.20,p=0.008) and peak (β=8.00,p=0.001; β=7.64,p=0.007; β=6.89,p=0.034) ICP. The homozygote wild-type genotype of rs1799857 had decreased odds of decompressive craniotomy (OR=0.47, p=0.004).
Conclusions
This is the first report assessing the impact of ABCC8 genetic variability on CE development in TBI. Minor allele ABCC8 SNP genotypes had increased risk of CE, while major SNP alleles were protective—potentially suggesting an evolutionary advantage. These findings could guide risk stratification, treatment responders, and the development of novel targeted or gene-based therapies against CE in TBI and other neurological disorders.
Keywords: ABCC8, cerebral edema, traumatic brain injury, single nucleotide polymorphism (SNP), sulfonylurea receptor-1 (Sur1)
INTRODUCTION
Traumatic brain injury (TBI) is a heterogeneous disease. Clinical variability develops immediately from initial impact through recovery, making this population challenging to study. Non-modifiable differences include demographic factors (age, sex, race), initial impact mechanism (blunt vs penetrating, velocity, force), and type of primary injury (diffuse axonal injury, subdural vs epidural vs intraparenchymal hemorrhages). However, these factors only partially explain ensuing clinical heterogeneity with respect to extent of primary injury, secondary injury development (e.g. cerebral edema(CE), tissue hypoxia, seizures), treatment response, and outcomes. Genetic variability has been increasingly implicated in contributing to observed clinical variability after TBI[1,2].
Genetic differences in TBI have primarily been examined with regards to outcome associations including genes like ApoE, p53, IL[2]. However, there has been limited evaluation of pathways contributing to CE /intracranial hypertension-both pivotal pathophysiologic factors associated with mortality and unfavorable prognosis in TBI. CE occurs in > 60% of severe TBI (sTBI) patients with mass lesions and 15% of those with normal computed tomography(CT) scans on presentation[3]. The bulk of critical care in sTBI is currently dedicated towards management of intracranial hypertension and CE.
Multiple underlying pathways have been implicated in CE formation after sTBI and are potential candidates for exploration of genetic variability. These include aquaporin-4 (AQP-4), high-mobility-group-box protein-1 with toll-like-receptor 4, Na+-K(+)-2Cl(-) cotransporter and sulfonylurea receptor (Sur1)-transient receptor potential cation channel member-4 (Trpm4) [4–11]. Single nucleotide polymorphisms (SNP) in the AQP-4 gene are associated with functional outcome, however have not been evaluated with regards to CE generation[1]. Although current therapies against CE are reactionary and non-specific (hyperosmolar treatment, neuromuscular blockade, cerebrospinal fluid (CSF) drainage, decompressive craniotomy), ongoing identification of causative pathways involved in CE has the potential to lead to the development of targeted treatment[7,10,12,13]. Genetic polymorphisms in these pathways could alter gene expression and regulation, as well as modify protein structure and function. This may affect degree and timing of CE development as well as individual responses to specific therapies. A priori identification of a patient’s risk for CE, dominance of a certain pathway, and likelihood of response to targeted therapy could have practical and important theranostic implications.
Of the known mechanisms involved in CE development, the Sur1 pathway is particularly unique. Sur1 is a sulfonylurea-receptor and transmembrane protein, encoded by ABCC8. Other members of the ABC-transporter family have been recognized as important mediators of solute transport at blood-brain and blood-CSF barriers, and genetic variations have been associated with outcomes after TBI[14]. Sur1 is an ideal target for investigation in central nervous system (CNS) diseases because it is not normally expressed in the brain, but is upregulated in injury[7,15]. Additionally, an existing FDA approved anti-diabetes medication (glyburide) inhibits Sur1 at non-hypoglycemic doses and has shown promise against cerebral edema in Phase 1 and 2 clinical trials[16–18]. Upregulated Sur1 by itself performs no recognized function; rather, it is a regulatory protein. It undergoes obligate association with a non-selective cation channel (Trpm4) allowing for channel opening, depolarization, water influx and oncotic edema, eventually followed by cell death.
Given the combination of Sur1’s regulatory function and specific upregulation during CNS injury, genetic polymorphisms in ABCC8 have the potential to significantly influence CNS-specific regulation and expression, CE development, and response to targeted therapy. To our knowledge, this association remains currently undefined. We undertook an exploratory candidate gene approach to determine potential associations between ABCC8 SNPs and measures of CE. We hypothesized that coding and non-coding SNP variations in ABCC8 would be associated with measures of CE in sTBI.
METHODS
Study Design
Subjects were prospectively enrolled through the Brain-Trauma Research Center at the University of Pittsburgh. Eligibility was determined by the presence of sTBI defined as Glasgow Coma Scale (GCS) score <9, placement of an external ventricular drain (EVD) for therapeutic CSF drainage per standard care, and age 16–80 years. Exclusion criteria were penetrating brain injury, brain death and pregnancy. 385 consecutively enrolled patients who consented to the study primarily of North-American ancestry (CEU population) between 2006–2013 were included. All subjects/health-care proxies provided informed consent including the collection of genetic material. The University of Pittsburgh Institutional Review Board approved the study.
Genotyping and SNP identification
DNA samples were obtained on admission-while the timing of DNA collection is not relevant to a research study involving SNPs (since these polymorphisms are constant per individual)[19], obtaining SNP analyses prior to the development of cerebral edema may eventually be important in clinical practice in order to serve as a useful genetic biomarker. Blood was centrifuged and DNA extracted from white blood cells by the simple salting out method[20]. If blood was unavailable, DNA was extracted from excess CSF in the ventriculostomy bag that would have otherwise been discarded[21]. All extracted DNA was stored at 4°C in 1XTE buffer.
Genotype data was generated using Human Core Exome v1.2 from Illumina in the University of Pittsburgh Genomics Core laboratory. For quality control, blind technical duplicates were included and any discrepancies were rectified using raw data or re-genotyped. For additional quality control, samples or SNPs that did not have a minimum 95% call rate were excluded. Data for the ABCC8 gene was abstracted using its position in genome build 37/hg19, to correspond with the build for Human Core Exome v1.2. Research assistants involved in genotyping SNPs were blinded to CE development outcomes. Pairwise linkage disequilibrium (LD) between SNPs (r2 and D’) was determined using JLIN software[22].
Data Collection
Demographic and outcome data were collected by a research assistant masked to ABCC8 SNPs. Three outcome measures assessed development of CE.
Edema on initial head CT (size of ventricles, basilar cisterns; transfalcine herniation; loss of gray/white matter differentiation) is associated with raised ICP after TBI [23,24], and was included as an outcome. Determinations were made by official radiologist reports.
Hourly ICP - subcategorized into mean and peak ICP during the course of neuromonitoring (at our institution this is typically for 5 days).
Decompressive craniotomy for intracranial hypertension. These surgeries were not performed for hematoma evacuations, but were operations involving skull removal or lobectomies due to intraoperative concerns for CE. Based on our institutional protocol, decompressive craniotomy is considered when the patient has elevated ICP despite adequate sedation/analgesia, continuous CSF drainage by EVD, hyperosmolar therapy, propofol infusion to a maximum of 60 mcg/kg/minute, and neuromuscular blockade. The need for decompressive craniotomy was determined by the attending neurosurgeon responsible for the patient due to concerns for swelling. This information was gained objectively from the medical charts in the operative report: concern for cerebral edema as an indication for surgery, the decision to leave the skull flap off due to the subjective intraoperative edematous nature of the brain, or craniotomies where a lobectomy was performed due to swelling concerns.
Statistical Analysis
Descriptive statistics of baseline variables were reported as means±standard deviations. Linear regression models evaluated the independent relationship between ABCC8 SNPs and continuous outcomes (ICP). Logistic regression models evaluated the independent relationship between ABCC8 SNPs and categorical outcomes (CT edema, decompressive craniotomy). Odds ratios, β-coefficients, and 95% confidence intervals (CI) were calculated based on modes of inheritance: dominant (AA vs Ab+bb), recessive (bb vs AA+Ab), heterogzygous (Ab vs AA+bb), as well as by an allele difference model (A vs b)[1,25]. Multiple variable logistic and linear regression models were developed with clinically relevant variables (age, gender, initial GCS score) to control for confounders in evaluating the independent relationship between ABCC8 SNPs and CE development. Tests were two-tailed. Although exploratory, multiple comparisons were adjusted using the established Benjamin-Yekutieli (B-Y) method-a more conservative modification of the standard False Discovery Rate (FDR) correction used in genetics studies, since the Bonferroni method has been previously cited as inappropriately conservative for SNP evaluation[26,27]. In the B-Y method, the critical significance level is determined by: where α = pre-correction significance level (0.05), k= number of hypotheses, and i= ith comparison. Analyses were performed using Stata 14.0(StataCorp, TX).
RESULTS
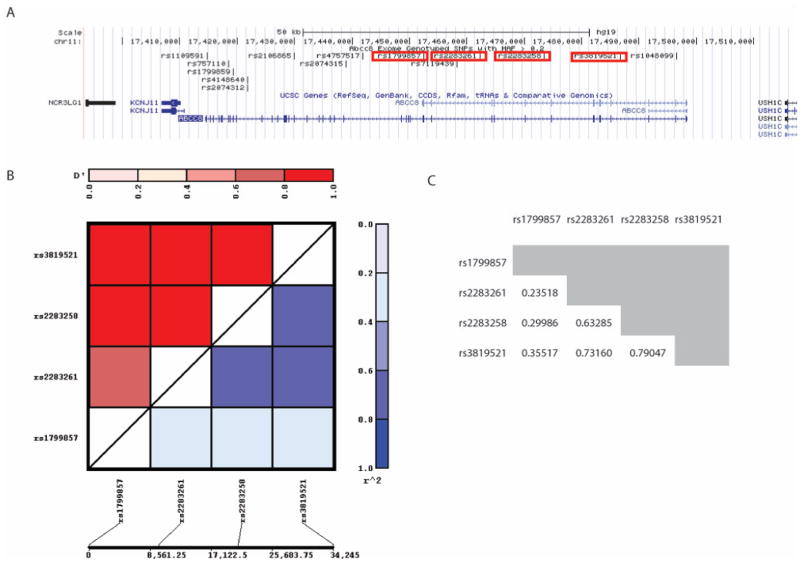
Demographic and clinical characteristics are summarized in Table 1. 44% of patients had edema noted on acute CT, and 33% underwent decompressive craniotomy. Of those who underwent this surgery, 68% were performed on the day of admission, 14% occurred on day 1, and 11% occurred between days 2–5. Fourteen ABCC8 SNPs with MAF>0.2 were identified. The p-value after adjusting for multiple hypotheses testing (14 SNPS, 4 outcomes) was 0.0108. Four SNPs (rs2283261, rs3819521, rs2283258 and rs1799857) were associated with measures of CE in both univariate and multivariate single locus analyses. The four SNPs were in close physical proximity (Figure 1A), and there was likely some, but not complete LD with each other (all r2 < 0.8, Figure 1B–C).
TABLE 1.
PATIENT CHARACTERISTICS
| Variable (n=385) | Mean (SD, Range) |
|---|---|
| Age | 37.9 (16.8, 16–77) |
| Initial GCS | 5.83 (1.51, 3–8) |
| Frequency n (%) | |
| Gender (M) | 304 (79.0%) |
| Race | |
| • White | 380 (98.7%) |
| • Asian/Pacific Islander | 2 (0.52%) |
| • Other | 3 (0.78%) |
| Edema noted on acute CT (Y) | 146 (0.44) |
| Decompressive Craniotomy (Y) | 128/385 (0.33) |
| • Day 0 | 87/128 (0.68) |
| • Day 1 | 18/128 (0.14) |
| • Days 2, 3, 4 or 5 | 14/128 (0.11) |
Table 1 summarizes the patient characteristics of the severe TBI population overall (n=385). All patients had severe TBI (GCS 3–8. Mean 5.83), and were predominantly male (79%).
Figure 1.
(A) The chromosomal locations of all 14 SNPs identified in ABCC8 with MAF > 0.2 based on the UCSC genome browser (GRCh38/hg38). The SNPS significantly associated with CE measures (rs2283261, rs3819521, rs2283258 and rs1799857) are boxed in red. LD plot (B) and r2 values (C) between rs2283261, rs3819521, rs2283258 and rs1799857 were generated using the JLIN program. The results show that none of the r2 values between any of the 4 SNPs are > 0.8.
rs2283261 is associated with acute CT edema, mean and peak ICP
There were 50.4% heterozygotes (AC) for rs2283261; 14% of patients were homozygous for the minor-allele (CC, Table 2). Homozygous-minor patients had higher mean and peak ICPs, and frequency of acute CT edema (Table 2). In univariate analyses (Table 3), rs2283261 homozygote-variant genotype increased odds of acute CT edema (OR 2.46, p=0.006), mean ICP (β=3.60, p <0.001) and peak ICP (β=8.29, p=0.001). Heterozygotes were protected against CT edema (OR 0.54, p=0.006), had lower peak ICP (β=−5.87, p=0.002) and tended to have lower mean ICP (β= −1.57, p=0.029-however this did not survive the B-Y correction). Concordantly, in the allele difference model, the wild-type allele(A) was protective for CT edema (OR 0.41, p=0.006), and these patients had lower mean (β= −3.70, p=0.000) and peak (β= −8.29, p=0.001) ICPs. These results retained significance in multivariate analyses (Table 4) where homozygote-variants had increased odds of CT edema (OR=2.45, p=0.007), higher mean ICP by 3.13 mmHg (p=0.000) and higher peak ICP by 8 mmHg (p=0.001). The wild-type allele remained protective against acute CT edema (p=0.007) and ICP (mean p=0.000; peak p=0.001).
TABLE 2.
SNP ALLELE AND GENOTYPE FREQUENCIES IN THE SEVERE TBI POPULATION
| SNP | Homozygous Major | Heterozygous | Homozygous Minor | Major Allele | Minor Allele | |
|---|---|---|---|---|---|---|
|
| ||||||
| rs2283261 | Total Frequency (%) | 137 (35.6%) | 194 (50.4%) | 54 (14%) | 331 (86%) | 248 (64.4%) |
| A=major C= minor |
• Average ICP, Mean (SD) | 10.0 (3.1) | 9.6 (3.6) | 13.5 (9.2) | 9.8 (3.4) | 24.5 (11.0) |
| • Peak ICP, Mean (SD) | 27.0 (13.0) | 23.1 (9.4) | 32.8 (18.8) | 10.5 (5.7) | 25.4 (12.9) | |
| • Acute CT Edema* Frequency Y (%) | 53 (47.3%) | 64 (36.8%) | 29 (63%) | 117 (40.9) | 93 (42.3) | |
| • Decompressive Surgery Frequency Y (%) | 45 (32.9%) | 65 (33.5) | 18 (33.5%) | 110 (33.2) | 83 (33.5) | |
|
| ||||||
| rs3819521 | Total Frequency (%) | 164 (42.6) | 181 (47.0) | 40 (10.4) | 345 (89.6) | 221 (57.4) |
| C= major T= minor |
• Average ICP, Mean (SD) | 10.3 (3.2) | 9.7 (5.1) | 13.4 (7.8) | 9.9 (4.32) | 10.5 (5.9) |
| • Peak ICP, Mean (SD) | 27.2 (12.4) | 22.9 (10.7) | 33.1 (18.5) | 24.9 (11.7) | 25.0 (13.3) | |
| • Acute CT Edema* Frequency Y (%) | 67 (48.6) | 58 (36.2) | 21 (63.6) | 125 (41.8) | 79 (40.7) | |
| • Decompressive Surgery Frequency Y (%) | 58 (35.4) | 58 (35.4) | 12 (30.0) | 116 (33.6) | 70 (31.7) | |
|
| ||||||
| rs2283258 | Total Frequency (%) | 184 (47.8) | 171 (44.4) | 20 (7.8) | 355 (92.2) | 201 (52.2) |
| G=major A=minor |
• Average ICP, Mean (SD) | 10.2 (3.3) | 9.9 (5.5) | 13.9 (7.5) | 10.0 (4.5) | 10.6 |
| • Peak ICP, Mean (SD) | 26.6 (12.3) | 23.7 (12.2) | 33.0 (16.9) | 25.2 (12.3) | 25.3 (13.4) | |
| • Acute CT Edema*v Frequency Y (%) | 72 (46.8) | 56 (36.8) | 18 (69.2) | 128 (41.8) | 74 (41.6) | |
| • Decompressive Surgery Frequency Y (%) | 61 (33.2) | 57 (33.3) | 10 (33.3) | 118 (33.2) | 67 (33.3) | |
|
| ||||||
| rs1799857 | Total Frequency (%) | 124 (32.2) | 189 (49.1) | 72 (18.7) | 313 (81.3) | 261 (67.8) |
| G=major A=minor |
• Average ICP, Mean (SD) | 10.9 (5.6) | 10.2 (5.0) | 9.7 (3.2) | 10.5 (5.3) | 10.1 (4.6) |
| • Peak ICP, Mean (SD) | 27.2 (14.0) | 25.1 (13.1) | 25.4 (10.0) | 26.0 (10) | 25.1 (12.3) | |
| • Acute CT Edema* Frequency Y (%) | 40 (37.4) | 76 (46.9) | 30 (47.6) | 116 (43.1) | 106 (47.1) | |
| • Decompressive Surgery Frequency Y (%) | 28 (22.6) | 72 (38.1) | 28 (38.9) | 100 (32.0) | 100 (38.3) | |
Table 2 summarizes the relative frequencies of the SNP genotypes and alleles in the TBI population, as well as for the various measures of cerebral edema. For example, row 1 evaluates rs2283261. 35.6% of the TBI population were wild-type (homozygous-major), 50.4% were heterozygotes, and 14% were homozygous for the variant SNP. Focusing on the homozygous major genotype (AA) for rs2283261, in patients with this genotype, acute CT edema occured in 47.3%, the mean ICP was 10.0 ± 3.1 mmHg, peak ICP was 27.0 ±13.0, and decompressive craniotomies were performed 32.9% of the patients. In contrast, the TBI sample contained 14% of patients with homozygous minor genotype of the rs2283261 SNP (CC) in. In these patients (CC), acute CT edema occured in a higher percentage (63%) of patients, mean and peak ICP was also higher 13.5 ±9.2 and 32.8 ±g18.8.
CT edema data were available for 332 patients of the total 385.
TABLE 3.
UNIVARIATE ANALYSIS OF ABCC8 SNP ASSOCIATION WITH CEREBRAL EDEMA IN TBI
| SNP | Acute CT Edema | Average ICP | Peak ICP | Decompressive Craniotomy | |||||
|---|---|---|---|---|---|---|---|---|---|
| Odds Ratio (95% CI) | p | β-coefficient (95% CI) | p | β-coefficient (95% CI) | p | Odds ratio (95% CI) | P | ||
| rs2283261 Intron 10 |
Homozygous major (AA) | 1.23 (0.78–1.94) | 0.38 | −0.51 (−2.04–1.01) | 0.51 | 1.56 (−2.4–5.5) | 0.44 | 0.97 (0.62–1.51) | 0.91 |
| Homozygous minor (CC) | 2.46 (1.29–4.69) | 0.006 | 3.70 (1.85–5.56) | 0.000 | 8.29 (3.42–13.17) | 0.001 | 1.005 (0.55–1.85) | 0.99 | |
| Heterozygous (AC) | 0.54 (0.35–0.84) | 0.006 | −1.57 (−2.98–−0.16) | 0.029 | −5.87 (−9.48–−2.27) | 0.002 | 1.023 (0.67–1.56) | 0.91 | |
| Allele A | 0.41 (0.21–0.77) | 0.006 | −3.70 (−5.56–−1.84) | 0.000 | −8.29 (−13.2–−3.42) | 0.001 | 1.00 (0.54–1.83) | 0.99 | |
| Allele C | 0.82 (0.52–1.29) | 0.38 | 0.52 (−1.01–2.04) | 0.51 | −1.56 (−5.52–2.41) | 0.44 | 1.03 (0.66–1.60) | 0.90 | |
|
| |||||||||
| rs3819521 Intron 3 |
Homozygous major (CC) | 1.37 (0.88–2.13) | 0.16 | −0.17 (−1.62–1.28) | 0.82 | 2.24 (−1.51–6.00) | 0.24 | 1.18 (0.77–1.81) | 0.45 |
| Homozygous minor (TT) | 2.43 (1.16–5.13) | 0.019 | 3.42 (1.29–5.55) | 0.002 | 8.23 (2.67–13.80) | 0.004 | 0.85 (0.42–1.72) | 0.65 | |
| Heterozygous (CT) | 0.53 (0.34–0.82) | 0.005 | −1.29 (−2.7–0.12) | 0.07 | −5.67 (−9.29–−2.06) | 0.002 | 0.90 (0.59–1.38) | 0.64 | |
| Allele C | 0.41 (0.19–0.87) | 0.019 | −3.42 (−5.55–−1.29) | 0.002 | −8.23 (−13.8–−2.67) | 0.004 | 1.18 (0.58–2.41) | 0.65 | |
| Allele T | 0.73 (0.47–1.13) | 0.16 | 0.17 (−1.28–1.62) | 0.82 | −2.24 (−6.00–1.51) | 0.24 | 0.85 (0.55–1.30) | 0.45 | |
|
| |||||||||
| rs2283258 Intron 7 |
Homozygous major (GG) | 1.23 (0.80–1.91) | 0.72 | −0.41 (−1.84–1.01) | 0.57 | 1.36 (−2.35–5.07) | 0.47 | 0.99 (0.65–1.52) | 0.97 |
| Homozygous minor (AA) | 3.13 (1.32–7.42) | 0.01 | 3.84 (1.49–6.28) | 0.002 | 7.80 (1.40–14.21) | 0.017 | 1.00 (0.46–2.21) | 0.99 | |
| Heterozygous (GA) | 0.58 (0.38–0.91) | 0.016 | −0.84 (−2.27–0.58) | 0.25 | −3.92 (−7.60–−0.25) | 0.037 | 1.01 (0.66–1.54) | 0.97 | |
| Allele G | 0.32 (0.18–0.76) | 0.010 | −3.84 (−6.28–−1.40) | 0.002 | −7.80 (−14.21–−1.4) | 0.017 | 1.00 (0.45–2.20) | 0.99 | |
| Allele A | 0.81 (0.52–1.25) | 0.34 | -.41 (−1.01–1.84) | 0.57 | −1.36 (−5.07–2.35) | 0.47 | 1.01 (0.66–1.54) | 0.97 | |
|
| |||||||||
| rs1799857 Exon 12 |
Homozygous major (GG) | 0.67 (0.42–1.07) | 0.096 | 0.85 (−0.64–2.32) | 0.26 | 2.02 (−1.83–5.87) | 0.3 | 0.47 (0.29–0.77) | 0.002 |
| Homozygous minor (AA) | 1.20 (0.69–2.08) | 0.52 | −0.83 (−2.71–1.05) | 0.39 | −0.60 (−5.50–4.30) | 0.81 | 1.36 (0.80–2.30) | 0.26 | |
| Heterozygous (GA) | 1.26 (0.82–1.95) | 0.29 | −0.303 (−1.73–1.12) | 0.68 | −1.52 (−5.22–2.19) | 0.42 | 1.54 (1.004–2.36) | 0.048 | |
| Allele G | 0.83 (0.48–1.45) | 0.52 | 0.83 (−1.05–2.71) | 0.39 | 0.60 (−4.30–5.50) | 0.81 | 0.74 (0.43–1.25) | 0.26 | |
| Allele A | 1.49 (0.93–2.39) | 0.096 | −0.85 (−2.32–0.64) | 0.26 | −2.02 (−5.88–1.84) | 0.30 | 2.13 (1.31–3.47) | 0.002 | |
Table 3: Univariate analyses demonstrating the association of each of the 4 SNPs with measures of cerebral edema including mean and peak ICP, acute CT edema, and need for decompressive craniotomy. Odds ratios (OR) are provided for binary outcomes (CT edema, decompressive craniotomy), and βcoefficients for continuous outcomes (mean and peak ICP).
TABLE 4.
MULTIVARIATE ANALYSIS OF ABCC8 SNP ASSOCIATION WITH MEASURES OF CEREBRAL EDEMA IN TBI
| SNP | Acute CT Edema | Average ICP | Peak ICP | Decompressive Craniotomy | |||||
|---|---|---|---|---|---|---|---|---|---|
| Odds Ratio (95% CI) | p | β-coefficient (95% CI) | p | β-coefficient (95% CI) | p | Odds ratio (95% CI) | P | ||
| rs2283261 | Homozygous minor (CC) | 2.45 (1.28–4.70) | 0.007 | 3.13 (1.50–5.13) | 0.000 | 8.00 (3.13–12.86) | 0.001 | 0.99 (0.53–1.87) | 0.99 |
| Heterozygous (AC) | 0.53 (0.34–0.83) | 0.005 | −1.20 (−2.56–−0.16) | 0.084 | −5.17 (−8.74–−1.60) | 0.005 | 0.96 (0.61–1.48) | 0.84 | |
| Allele A | 0.41 (0.21–0.78) | 0.007 | −3.31 (−5.13–−1.50) | 0.000 | −8.00 (−12.86–−3.13) | 0.001 | 1.01 (0.53–1.89) | 0.99 | |
|
| |||||||||
| rs3819521 | Homozygous minor (TT) | 2.36 (1.11–5.01) | 0.025 | 2.95 (0.88–5.02) | 0.005 | 7.64 (2.11–13.16) | 0.007 | 0.83 (0.40–1.74) | 0.63 |
| Heterozygous (CT) | 0.52 (0.33–0.82) | 0.004 | −0.70 (−2.08–0.69) | 0.32 | −4.61 (−8.24–−0.96) | 0.013 | 0.80 (0.51–1.25) | 0.33 | |
| Allele C | 0.42 (0.20–0.90) | 0.025 | −2.95 (−5.02–−0.88) | 0.005 | −7.64 (−13.2–−2.11) | 0.007 | 1.20 (0.58–2.50) | 0.63 | |
|
| |||||||||
| rs2283258 | Homozygous minor (AA) | 3.00 (1.26–7.18) | 0.013 | 3.20 (0.83–5.58) | 0.008 | 6.89 (0.52–13.25) | 0.034 | 1.03 (0.46–2.34) | 0.94 |
| Allele G | 0.33 (0.14–0.80) | 0.013 | −3.20 (−5.58–−0.83) | 0.008 | −6.89 (−13.25–−0.52) | 0.034 | 0.97 (0.43–2.19) | 0.94 | |
|
| |||||||||
| rs1799857 | Homozygous major (GG) | 0.67 (0.42–1.08) | 0.102 | 0.82 (−0.61–2.23) | 0.26 | 1.90 (−1.88–5.68) | 0.32 | 0.47 (0.28–0.78) | 0.004 |
| Allele A | 1.49 (0.92–2.39) | 0.102 | −0.81 (−2.23–0.61) | 0.26 | −90 (−5.68–1.88) | 0.32 | 2.12 (1.28–3.51) | 0.004 | |
Table 4: Multivariate analyses demonstrating the association of each of the 4 SNPs with measures of cerebral edema including mean and peak ICP, acute CT edema, and need for decompressive craniotomy after controlling for age, gender, and initial GCS score. Adjusted Odds ratios (OR) are provided for binary outcomes (CT edema, decompressive craniotomy), and β-coefficients for continuous outcomes (mean and peak ICP).
rs3819521 is associated with acute CT edema, mean and peak ICP
There were 47 % heterozygotes (CT) for rs3819521. 10.4% of patients were homozygous for the minor-allele (TT, Table 2). Homozygous-minor patients had higher mean and peak ICPs, and frequency of acute CT edema (Table 2). Similar to rs2283261, wild-type rs3819521 allele (C) was protective against ICP and acute CT edema in both univariate (Table 3) and multivariate (Table 4) analyses. Homozygote-variant genotypes had higher mean ICP (β=3.42, p=0.002), higher peak ICP (β=8.23, p=0.004), and tended to have higher odds of CT edema (OR=2.43, p=0.019). Maintaining internal consistency, the wild-type allele protected against higher mean (p=0.002) and peak ICP (p=0.004). Heterozygotes were protected against the measures of CE including acute CT edema (OR=0.53, p=0.005) and peak ICP (β=−5.67, p=0.002). These findings remained robust in multivariate analyses where the homozygous-variant genotype remained associated with increased risk of edema, and the wild-type allele was protective (Table 4).
rs2283258 is associated with acute CT edema, mean and peak ICP
There were 44.4 % heterozygotes (AG) for rs2283258, and 7.8% of patients were homozygous for the minor allele (AA, Table 2). Homozygous minor patients had higher mean and peak ICPs, and frequency of acute CT edema (Table 2). In univariate analyses, patients with homozygous-variant genotype had increased odds of acute CT edema (OR=3.13, p=0.01), and had higher mean ICP (β=3.84, p =0.002). Peak ICP was also higher by 6.89 mmHg in patients with the homozygous-variant genotype (p=0.034) but this did not maintain significance after correcting for multiple comparisons. The presence of the wild-type allele (G) was protective against acute CT edema (OR=0.32, p=0.01) and mean ICP (β=−3.84, p =0.002). Heterozygotes had a trend towards decreased CT edema and lower peak ICP like the other SNPs but this was not statistically significant. These findings were confirmed in multivariate analyses (Table 4) where the homozygous-variant genotype was associated with increased odds of acute CT edema (OR=3.0, p=0.013) and mean ICP(β=3.2, p=0.008), and the wild-type allele was protective.
rs1799857 is associated with Decompressive Craniotomy
There were 49.1% heterozygotes (GA) for rs1799857, and 18.7% of patients were homozygous for the minor-allele (AA, Table 2). This SNP was associated with odds of decompressive craniotomy. In univariate analyses (Table 3), the homozygous wild-type genotype (GG) decreased odds of decompressive craniotomy (OR=0.47, p=0.002), and presence of the variant-allele (A) increased odds of the surgery (OR=2.13, p=0.002). This retained robustness in the multivariate model (Table 4).
DISCUSSION
Much of neurocritical care in sTBI focuses on monitoring and treating CE and intracranial hypertension. Sur1 is a key regulatory protein involved in CE generation in multiple CNS disorders including TBI and ischemic stroke[7–9,15]. Sur1 expression has been demonstrated in human contusional tissue[28]. We have exciting preliminary data suggesting human CSF Sur1 levels are undetectable in non-injured controls, elevated in patients with sTBI, and trajectories may correlate with CE and outcomes (In Press, Critical Care Medicine, abstract presented at American Academy of Neurology, 2016[29]). The efficacy of inhibiting this pathway with Glyburide has shown promise, and is being evaluated by ongoing clinical trials[16,17]. Particularly in light of its regulated expression and regulatory function, genetic variability in the Sur1 gene, ABCC8, could provide important information regarding patient risk-stratification, monitoring, efficacy of targeted therapies, and prognosis - not only in TBI but also in other neurologic diseases affected by CE like ischemic stroke.
To our knowledge, this is the first analysis evaluating the relationship between genetic variability in ABCC8 and development of CE in sTBI. This study identifies 3 ABCC8 SNPs (rs2283261, rs3819521, and rs2283258) significantly associated with multiple measures of CE including radiographic edema on initial CT, and clinical measurements of ICP. A fourth SNP (rs1799857) was associated with odds of having a decompressive craniotomy.
CE associations with rs2283261, rs3819521, rs2283258 and rs1799857
For rs2283261, rs3819521, and rs2283258, homozygous-variant genotypes appeared to confer increased risk of CE and presence of wild-type alleles was protective-potentially reflecting an evolutionary advantage. In rs2283261 and rs3819521, the increased risk of CE measures was eliminated in heterozygotes with 1 SNP/minor-allele. Interestingly, heterozygotes may potentially be more protected than homozygous wild-type-one potential explanation is that the Sur1-Trpm4 cation channel is a hetero-octameric structure comprised of four Sur1 subunits, and four Trpm4 subunits[15]. Any heterogeneity in Sur1 splicing/protein structure (vs. a homogeneous structure in homozygotes) may possibly reduce optimal protein-protein interactions and therefore channel efficiency and subsequent edema development.
Unlike the 3 intronic SNPs where homozygous-variant genotypes conferred increased risk of CE measures, in rs1799857 (exon 12), the homozygous wild-type genotype was protective against odds of decompressive craniotomy. Presence of the variant-allele eliminated the protective effect and increased odds of surgery. Although in our population rs1799857 was not significantly associated with acute CT edema, the same trend as decompressive craniotomy was observed; the homozygous wild-type genotype decreased odds of acute radiographic edema and the minor allele increased those odds (p=0.096). It is not unexpected that rs1799857 was associated with decompressive craniotomy, but not ICP. As evident from Table 1, >80% of the surgeries were performed on D0-D1 after injury, thereby likely obviating risks of subsequent intracranial hypertension.
Potential significance of rs2283261, rs3819521, rs2283258 and rs1799857
Much of the existing research on genetic variation in ABCC8 is reported in the diabetes literature. The role of Sur1 was initially appreciated in pancreatic β cells, where Sur1 associates with the potassium channel (Kir6.2) [15,30–32]. In this literature, ABCC8 SNPs are associated with disorders of glucose metabolism like congenital hyperinsulinism and neonatal diabetes[30,32,33]. Reported SNPs for these diseases are located throughout coding (exons 1–16, 18–39) and non-coding (promoter, introns 3, 8, 10–11, 14–16, 18–19, 22, 24–25, 29, 32, 36, 38) regions of the ABCC8 gene[30].
Our study identifies 3 novel SNPs (rs2283261, rs3819521, and rs2283258) associated with acute CT edema, mean and peak ICP in a sTBI population. These SNPs are in different non-coding portions of the ABCC8 gene (Figure 1); rs3819521 is in intron 3, rs2283258 is in intron 7, and rs2283261 is in intron 10. As with most human genomic SNPs, these non-coding portions may be significant in protein regulation, expression and modifications (only 60,000 of the originally reported 1.42 million SNPs in the human genome were within exons)[19]. Previously reported polymorphisms in introns 7 and 10 have been associated with aberrant splicing and hyperinsulinism[30,34–38]. Aberrant splicing due to an intron 10 SNP has also been shown to influence a transmembrane domain of Sur1 protein[36]. Although SNPs in intron 3 have been reported in connection with insulin disorders, these remain unclassified and of unclear functional significance[30,36].
The fourth significant SNP identified in our population, rs1799857, is located in exon 12 that encodes a Sur1 transmembrane domain. Substitution of variant-allele A in place of G, results in a synonymous variant at position 562 (of 1581); although there is no change in the amino acid Histidine, even in synonymous variants the effect of the polymorphism may not be silent, could impact mRNA stability, and can be further evaluated at the mRNA level[39].
While there is a strong pathophysiologic basis for genetic variability in ABCC8 influencing the presence and degree of brain edema development, the precise mechanisms remain to be elucidated. The specific impact of rs2283261, rs3819521, rs2283258 and rs1799857 on Sur1 protein structure, function, expression, splicing or regulation requires further investigation beyond the scope of this study. Although any potential causality of the relationships is presently unknown, the identified SNPs could nonetheless provide valuable insights as risk-stratification biomarkers. This study lays the groundwork for additional research to assess the functional impact of these SNPs on the Sur1 pathway and CE.
Limitations
This was a single-center population thereby introducing the possibility of selection bias. SNP identification was limited based on the existing coverage of the ABCC8 gene. Since treatment with hyperosmolar therapy could affect measures used to assess the development of cerebral edema such as ICP or CT findings, incorporating this information using a validated measure such as ‘Therapeutic Intensity Level’ would have been useful, but was unavailable. Fortunately, since these maneuvers reduce measures of cerebral edema, this limitation while important reduces the likelihood of a falsely positive relationship. Incorporation of the use of hyperosmolar therapy using measures such as the TIL is nonetheless warranted in further confirmatory studies. Additionally, our sample size was relatively small for genetic studies and requires a validation cohort. However, notwithstanding the limited sample, our findings demonstrated relatively large effect sizes, were robust to adjusting for potential confounders in multivariate regression models, and survived statistical correction for multiple comparisons. Given our limited sample size, we used a candidate gene approach and did not evaluate the impact of genetic variability on other related proteins in the pathway, such as the Sur1 regulated channel Trpm4. Moreover, although we are limited in terms of sample size due to the incidence of severe TBI, this is one of the largest polymorphism studies reported in this disease[1,14,21,25,40]. We eagerly anticipate validation in future cohorts facilitated by emerging multi-center studies such as Transforming Research and Clinical Knowledge in TBI (TRACK-TBI). We did not include outcome measures in our study–this was intentional; given the underlying pathophysiology of the Sur1 pathway, the focus of our study was limited to genetic variability related to CE development which is a unique target relevant to neurointensivists. Finally, as with most genetic association studies, our study is limited in that the relationships between identified SNPs and CE measures are currently associative in nature.
CONCLUSIONS
As the molecular understanding of underlying pathways contributing to CE becomes increasingly sophisticated and preclinical findings are translated to humans, recognition of potential genetic influences becomes progressively more relevant. The Sur1 pathway is distinctive. Since Sur1 is normally absent in the CNS, not only is it a potentially specific therapeutic target against CE, but the degree of upregulation and expression of this regulatory protein (and resultant CE) may vary based on genetic differences. This study demonstrates the involvement of genetic variations in ABCC8 (rs2283261, rs3819521, rs2283258 and rs1799857) in CE development in sTBI. Our data provide the foundation for further research into understanding the functional implications of these and other ABCC8 polymorphisms on CE. Our findings need to be validated in additional populations. If validated, this could also inform the use of certain ABCC8 polymorphisms as biomarkers to a priori identify sTBI patients with altered Sur1 expression, structure or function and an increased risk of CE. In the rapidly evolving world of individualized medicine, understanding the impact of genetic variability in the Sur1 protein and pathway could eventually guide CE evaluation including categorization of treatment responders, and advance the development of targeted or gene-based therapies.
Acknowledgments
Study Funding:
We are grateful to NIH Grants #: KL2 TR000146, R00 NR013176, and R01NR013342 for their generous support.
Footnotes
Author Contributions:
Ruchira M. Jha–study concept, design, data analysis and interpretation, manuscript generation.
Ava Puccio - acquisition of data.
David O. Okonkwo - acquisition of data.
Benjamin E. Zusman-acquisition of data
Seo-Young Park–statistical analysis review
Jessica Wallisch - content expertise, critical revision of manuscript.
Philip E. Empey - content expertise, critical revision of manuscript.
Lori A. Shutter–content expertise, critical revision of manuscript.
Robert S.B. Clark–content expertise, critical revision of manuscript.
Patrick M. Kochanek–study concept, content expertise, critical revision of manuscript.
Yvette P. Conley–study concept, data acquisition and interpretation, supervision, critical revision of manuscript.
Financial Disclosure Statements
Ruchira M. Jha reports no disclosures.
Ava Puccio reports no disclosures.
David O. Okonkwo reports no disclosures.
Benjamin E. Zusman reports no disclosures.
Seo-Young Park reports no disclosures.
Jessica Wallisch reports no disclosures.
Philip E. Empey reports no disclosures.
Lori A. Shutter reports DOD grant W81XWH-08-2-0159 and NINDS grant 1U10NS069498. This funding was not related to this study and there are no conflicts of interest.
Robert S.B. Clark reports no disclosures.
- Dr. Kochanek has Grants from NIH and the U.S. DoD
- Dr. Kochanek is editor in chief of the journal Pediatric Critical Care Medicine and receives an editor Stipend
- Dr. Kochanek is one of the editor of the Textbook of Critical Care and receives royalties for that work
- Dr. Kochanek is a co-author of several US patents or provisional patents (below):
-
United States Patent: US 8,628,512 B2
- Title: Method of Inducing EPR Following Cardiopulmonary Arrest
- Filing Date: 6/22/05
- Inventors: PM Kochanek, SA Tisherman, X Wu, SW Stezoski, LJ Yaffe
-
United States Provisional Patent:
- Title: Compositions and Methods for Identifying Subjects at Risk for Traumatic Brain Injury
- Serial No: 62/113,292
- Filing Date: February 6, 2015
- Inventors: RP Berger, PM Kochanek, BJ Pak, PT Smith, MD Kolesnikova
-
United States Invention Disclosure:
- Title: Small Molecule Inhibitors of RNA Binding MOTIF (RBM) Proteins for the Treatment of Acute Cellular Injury
- University of Pittsburgh
- Filing Date: November 13, 2014
- Inventors: TC Jackson, J Verrier, PM Kochanek
-
United States Invention Disclosure:
- Title: Method to improve neurologic outcomes in temperature managed patients
- Application No.: 62/164,205
- Country: United States
- Innovators: Travis C. Jackson (University of Pittsburgh); Patrick M. Kochanek
Yvette P. Conley reports no disclosures.
References
- 1.Dardiotis E, Paterakis K, Tsivgoulis G, Tsintou M, Hadjigeorgiou GF, Dardioti M, et al. AQP4 Tag Single Nucleotide Polymorphisms in Patients with Traumatic Brain Injury. J Neurotrauma. 2014;31:1920–6. doi: 10.1089/neu.2014.3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dardiotis E, Fountas KN, Dardioti M, Xiromerisiou G, Kapsalaki E, Tasiou A, et al. Genetic association studies in patients with traumatic brain injury. Neurosurg Focus. 2010;28:E9. doi: 10.3171/2009.10.FOCUS09215. [DOI] [PubMed] [Google Scholar]
- 3.Narayan RK, Kishore PR, Becker DP, Ward JD, Enas GG, Greenberg RP, et al. Intracranial pressure: to monitor or not to monitor? A review of our experience with severe head injury. J Neurosurg Journal of Neurosurgery Publishing Group. 1982;56:650–9. doi: 10.3171/jns.1982.56.5.0650. [DOI] [PubMed] [Google Scholar]
- 4.Liang F, Luo C, Xu G, Su F, He X, Long S, et al. Deletion of aquaporin-4 is neuroprotective during the acute stage of micro traumatic brain injury in mice. Neuroscience Letters. 2015;598:29–35. doi: 10.1016/j.neulet.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 5.Yao X, Uchida K, Papadopoulos MC, Zador Z, Manley GT, Verkman AS. Mildly Reduced Brain Swelling and Improved Neurological Outcome in Aquaporin-4 Knockout Mice Following Controlled Cortical Impact Brain Injury. J Neurotrauma. 2015 doi: 10.1089/neu.2014.3675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laird MD, Shields JS, Sukumari-Ramesh S, Kimbler DE, Fessler RD, Shakir B, et al. High mobility group box protein-1 promotes cerebral edema after traumatic brain injury via activation of toll-like receptor 4. Glia. 2014;62:26–38. doi: 10.1002/glia.22581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Simard JM, Chen M, Tarasov KV, Bhatta S, Ivanova S, Melnitchenko L, et al. Newly expressed SUR1-regulated NC(Ca-ATP) channel mediates cerebral edema after ischemic stroke. Nat Med. 2006;12:433–40. doi: 10.1038/nm1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patel AD, Meriggioli MN, Sanders DB, Gerzanich V, Geng Z, Simard JM. Glibenclamide reduces hippocampal injury and preserves rapid spatial learning in a model of traumatic brain injury. J Neuropathol Exp Neurol. 2010;69:1177–90. doi: 10.1097/NEN.0b013e3181fbf6d6. [DOI] [PubMed] [Google Scholar]
- 9.Simard JM, Kilbourne M, Tsymbalyuk O, Tosun C, Caridi J, Ivanova S, et al. Key role of sulfonylurea receptor 1 in progressive secondary hemorrhage after brain contusion. J Neurotrauma. 2009;26:2257–67. doi: 10.1089/neu.2009.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walcott BP, Kahle KT, Simard JM. Novel Treatment Targets for Cerebral Edema. Neurotherapeutics. 2011;9:65–72. doi: 10.1007/s13311-011-0087-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang L-Q, Zhu G-F, Deng Y-Y, Jiang W-Q, Fang M, Chen C-B, et al. Hypertonic saline alleviates cerebral edema by inhibiting microglia-derived TNF-α and IL-1β-induced Na-K-Cl Cotransporter up-regulation. J Neuroinflammation. 2014;11:102. doi: 10.1186/1742-2094-11-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marmarou CR, Liang X, Abidi NH, Parveen S, Taya K, Henderson SC, et al. Selective vasopressin-1a receptor antagonist prevents brain edema, reduces astrocytic cell swelling and GFAP, V1aR and AQP4 expression after focal traumatic brain injury. Brain Res. 2014;1581:89–102. doi: 10.1016/j.brainres.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J-H, Yang L-K, Chen L, Wang Y-H, Wu Y, Jiang B-J, et al. Atorvastatin ameliorates early brain injury after subarachnoid hemorrhage via inhibition of AQP4 expression in rabbits. International Journal of Molecular Medicine Spandidos Publications. 2016;37:1059–66. doi: 10.3892/ijmm.2016.2506. [DOI] [PubMed] [Google Scholar]
- 14.Cousar JL, Conley YP, Willyerd FA, Sarnaik AA, Puccio AM, Empey PE, et al. Influence of ATP-binding cassette polymorphisms on neurological outcome after traumatic brain injury. Neurocrit Care. 2013;19:192–8. doi: 10.1007/s12028-013-9881-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simard JM, Woo SK, Schwartzbauer GT, Gerzanich V. Sulfonylurea receptor 1 in central nervous system injury: a focused review. J Cereb Blood Flow Metab. 2012;32:1699–717. doi: 10.1038/jcbfm.2012.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Remedy Pharmaceuticals, Inc. A Phase I Randomized, Double-Blind, Placebo-Controlled Study to Assess the Safety, Tolerability, and Pharmacokinetics of Escalating Doses of RP-1127 (Glyburide for Injection) in Healthy Male and Female Volunteers. 2016:1–3. Available from: https://clinicaltrials.gov/ct2/show/NCT01132703?term=glibenclamide+traumatic+brain+injury&rank=2.
- 17.Sheth KN, Elm JJ, Beslow LA, Sze GK, Kimberly WT. Glyburide Advantage in Malignant Edema and Stroke (GAMES-RP) Trial: Rationale and Design. Neurocrit Care. 2016;24:132–9. doi: 10.1007/s12028-015-0189-7. [DOI] [PubMed] [Google Scholar]
- 18.Kimberly WT, Battey TWK, Pham L, Wu O, Wu O, Yoo AJ, et al. Glyburide is Associated with Attenuated Vasogenic Edema in Stroke Patients. Neurocrit Care. 2013 doi: 10.1007/s12028-013-9917-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409:928–33. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 20.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conley YP, Okonkwo DO, Deslouches S, Alexander S, Puccio AM, Beers SR, et al. Mitochondrial polymorphisms impact outcomes after severe traumatic brain injury. J Neurotrauma. 2014;31:34–41. doi: 10.1089/neu.2013.2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carter KW, McCaskie PA, Palmer LJ. JLIN: a java based linkage disequilibrium plotter. BMC Bioinformatics. 2006;7:60. doi: 10.1186/1471-2105-7-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eisenberg HM, Gary HE, Aldrich EF, Saydjari C, Turner B, Foulkes MA, et al. Initial CT findings in 753 patients with severe head injury. A report from the NIH Traumatic Coma Data Bank. J Neurosurg. 1990;73:688–98. doi: 10.3171/jns.1990.73.5.0688. [DOI] [PubMed] [Google Scholar]
- 24.Miller MT, Pasquale M, Kurek S, White J, Martin P, Bannon K, et al. Initial head computed tomographic scan characteristics have a linear relationship with initial intracranial pressure after trauma. Journal of Trauma and Acute Care Surgery. 2004;56:967–73. doi: 10.1097/01.ta.0000123699.16465.8b. [DOI] [PubMed] [Google Scholar]
- 25.Dardiotis E, Jagiella J, Xiromerisiou G, Dardioti M, Vogiatzi C, Urbanik A, et al. Angiotensin-converting enzyme tag single nucleotide polymorphisms in patients with intracerebral hemorrhage. Pharmacogenet Genomics. 2011;21:136–41. doi: 10.1097/FPC.0b013e328343ab15. [DOI] [PubMed] [Google Scholar]
- 26.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–91. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 27.Narum SR. Beyond Bonferroni: Less conservative analyses for conservation genetics. Conserv Genet. 2006;7:783–7. [Google Scholar]
- 28.Martínez-Valverde T, Vidal-Jorge M, Martinez-Saez E, Castro L, Arikan F, Cordero E, et al. Sulfonylurea Receptor 1 in Humans with Post-Traumatic Brain Contusions. J Neurotrauma. 2015;32:1478–87. doi: 10.1089/neu.2014.3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jha R, Puccio A, Chou S, Chang C-C, Wallisch J, Molyneaux B, et al. Neurology. Vol. 86. Lippincott Williams & Wilkins; 2016. Sulfonylurea Receptor-1 as a Novel Biomarker for Cerebral Edema in Patients with Severe Traumatic Brain Injury (S46.001) p. S46.001. [Google Scholar]
- 30.Flanagan SE, Clauin S, Bellanné-Chantelot C, de Lonlay P, Harries LW, Gloyn AL, et al. Update of mutations in the genes encoding the pancreatic beta-cell K ATPchannel subunits Kir6.2 ( KCNJ11) and sulfonylurea receptor 1 ( ABCC8) in diabetes mellitus and hyperinsulinism. Hum Mutat. 2008;30:170–80. doi: 10.1002/humu.20838. [DOI] [PubMed] [Google Scholar]
- 31.Proverbio MC, Mangano E, Gessi A, Bordoni R, Spinelli R, Asselta R, et al. Whole Genome SNP Genotyping and Exome Sequencing Reveal Novel Genetic Variants and Putative Causative Genes in Congenital Hyperinsulinism. In: Zhi D, editor. PLoS ONE. Vol. 8. 2013. pp. e68740–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aittoniemi J, Fotinou C, Craig TJ, de Wet H, Proks P, Ashcroft FM. SUR1: a unique ATP-binding cassette protein that functions as an ion channel regulator. Philosophical Transactions of the Royal Society B: Biological Sciences. 2009;364:257–67. doi: 10.1098/rstb.2008.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Denton JS, Jacobson DA. Channeling dysglycemia: ion-channel variations perturbing glucose homeostasis. Trends in Endocrinology & Metabolism Elsevier Ltd. 2012;23:41–8. doi: 10.1016/j.tem.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aguilar-Bryan L, Bryan J. Molecular biology of adenosine triphosphate-sensitive potassium channels. Endocr Rev. 1999;20:101–35. doi: 10.1210/edrv.20.2.0361. [DOI] [PubMed] [Google Scholar]
- 35.Hardy OT, Hernandez-Pampaloni M, Saffer JR, Suchi M, Ruchelli E, Zhuang H, et al. Diagnosis and localization of focal congenital hyperinsulinism by 18F-fluorodopa PET scan. J Pediatr. 2007;150:140–5. doi: 10.1016/j.jpeds.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 36.Nestorowicz A, Glaser B, Wilson BA, Shyng S-L, Nichols CG, Stanley CA, et al. Genetic heterogeneity in familial hyperinsulinism. Human molecular Genetics. 1998;7:1119–28. doi: 10.1093/hmg/7.7.1119. [DOI] [PubMed] [Google Scholar]
- 37.Glaser B, Ryan F, Donath M, Landau H, Stanley CA. Hyperinsulinism caused by paternal-specific inheritance of a recessive mutation in the sulfonylurea-receptor gene. Diabetes. 1999;48:1652–7. doi: 10.2337/diabetes.48.8.1652. [DOI] [PubMed] [Google Scholar]
- 38.Stanley CA, Thornton PS, Ganguly A, MacMullen C, Underwood P, Bhatia P, et al. Preoperative evaluation of infants with focal or diffuse congenital hyperinsulinism by intravenous acute insulin response tests and selective pancreatic arterial calcium stimulation. The Journal of Clinical Endocrinology & Metabolism. 2004;89:288–96. doi: 10.1210/jc.2003-030965. [DOI] [PubMed] [Google Scholar]
- 39.Knight JC. Functional implications of genetic variation in non-coding DNA for disease susceptibility and gene regulation. Clin Sci Portland Press Limited. 2003;104:493–501. doi: 10.1042/CS20020304. [DOI] [PubMed] [Google Scholar]
- 40.Ritter AC, Kammerer CM, Brooks MM, Conley YP, Wagner AK. Genetic variation in neuronal glutamate transport genes and associations with posttraumatic seizure. Epilepsia. 2016;57:984–93. doi: 10.1111/epi.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]