Abstract
The HIV-1 capsid protein (CA) forms the capsid shell that encloses RNA within a mature HIV-1 virion. Previous studies by electron microscopy have shown that the capsid shell is primarily a triangular lattice of CA hexamers, with variable curvature that destroys the ideal symmetry of a planar lattice. The mature CA lattice depends on CA dimerization, which occurs through interactions between helix 9 segments of the C-terminal domain (CTD) of CA. Several high-resolution structures of the CTD-CTD dimerization interface have been reported, based on x-ray crystallography and multidimensional solution nuclear magnetic resonance (NMR), with significant differences in amino acid sidechain conformations and helix 9-helix 9 orientations. In a structural model for tubular CA assemblies based on cryogenic electron microscopy (cryoEM) [Zhao et al. (2013) Nature 497:643–646], the dimerization interface is substantially disordered. The dimerization interface structure in noncrystalline CA assemblies and the extent to which this interface is structurally ordered within a curved lattice have therefore been unclear. Here we describe solid state NMR measurements on the dimerization interface in tubular CA assemblies, which contain the curved triangular lattice of a mature virion, including quantitative measurements of intermolecular and intramolecular distances using dipolar recoupling techniques, solid state NMR chemical shifts, and long-range sidechain-sidechain contacts. When combined with restraints on the distance and orientation between helix 9 segments from the cryoEM study, the solid state NMR data lead to a unique high-resolution structure for the dimerization interface in the noncrystalline lattice of CA tubes. These results demonstrate that CA lattice curvature is not dependent on disorder or variability in the dimerization interface. This work also demonstrates the feasibility of local structure determination within large noncrystalline assemblies formed by high-molecular-weight proteins, using modern solid state NMR methods.
Graphical Abstract
INTRODUCTION
The mature state of the type 1 human immunodeficiency virus (HIV-1) is characterized by a capsid core that encloses the viral RNA and readies the virion for infection of a new host cell.1 The capsid shell is composed of about 1500 copies of the 231-residue capsid protein (CA)2, which contains independently folding N-terminal and C-terminal domains (NTD and CTD), separated by a short linker. In solution, CA dimerizes via interactions between CTD subunits.3 In supramolecular assemblies, CA forms hexamers that are stabilized by intermolecular NTD-NTD and NTD-CTD interactions and linked to one another by CTD-CTD dimerization.4
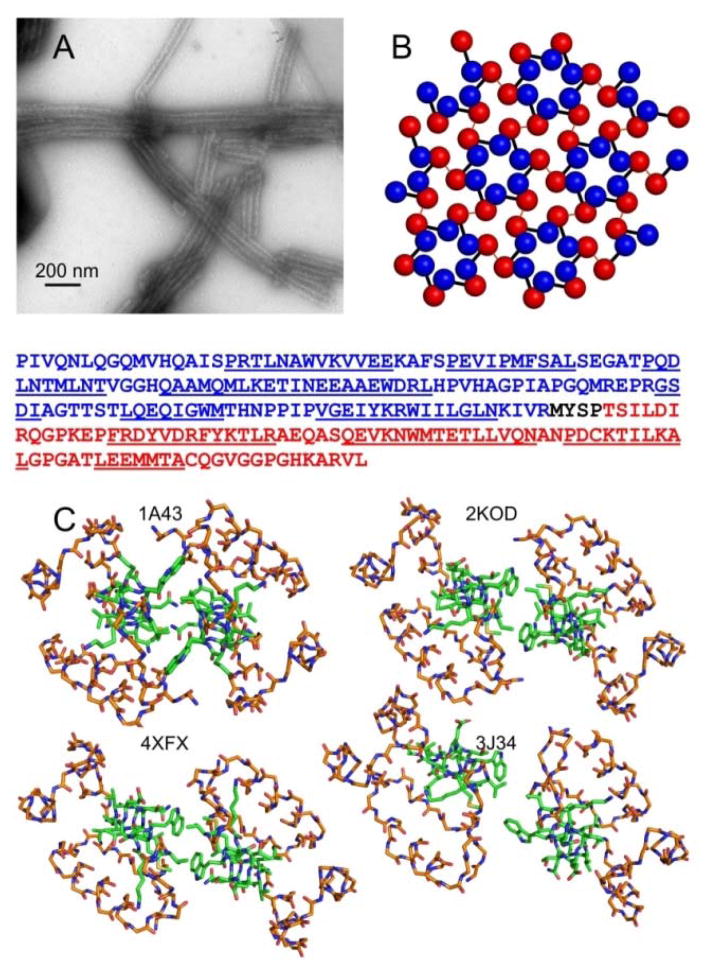
Much of our understanding of the supramolecular architecture of the mature capsid shell comes from studies of CA assemblies that form in vitro.2,3c,4a,4b,4d,5 Tubular CA assemblies (Fig. 1A) are particularly valuable models for mature capsids, as they exhibit the variable surface curvature and imperfect symmetry of mature capsids. CA tubes contain the characteristic triangular lattice of CA hexamers3c,4a,4d (Fig. 1B). Studies of native virions by cryogenic electron microscopy (cryoEM) confirm that the same lattice exists in the mature capsid core 6, although with the likely addition of CA pentamers to permit closure of the lattice.4d,7
Figure 1.
Diverse dimerization interface structures from crystallography, solution NMR, and cryoEM. (A) TEM image of typical tubular HIV-1 CA assemblies studied by solid state NMR. (B) Ideal CA lattice, with NTD and CTD represented by blue and red spheres, respectively. Black bars connect NTD and CTD of the same CA molecule. Red lines represent intermolecular dimerization interfaces. The CA sequence is shown below panels A and B, with the NTD in blue, the CTD in red, and helical segments underlined. (C) CTD dimer structures from the indicated PDB files, viewed down the two-fold (or approximate two-fold) symmetry axes. Backbone atoms of residues 148–180 and 195–218 are shown, using orange to represent carbon atoms. Backbone and sidechain atoms of residues 181–194 are shown, using green to represent carbon atoms. For PDB 2KOD, the first of 30 NMR-based models is depicted. For PDB 3J34, one of the 12 cryoEM-based dimerization interfaces is depicted.
High-resolution structural models from solution nuclear magnetic resonance (NMR)3c,8 and x-ray crystallography3a,3b,4c,4e,7b,9 have brought a molecular understanding to the extensive biochemical and mutagenic studies on the function of CA and its mechanism of assembly.2,3c,10 The dimerization interface, formed by hydrophobic residues in helix 9 segments of CTD (residues 179–193 of full-length CA), is especially critical. Amino acid substitutions in helix 9 result in non-infectious virions in vivo10, due to disruption of the HIV-1 maturation process, and preclude CA lattice assembly and tube formation in vitro.2,8c,8d,11
According to earlier studies, intermolecular interactions involving W184 and M185 are the main contributors to the stability of the dimerization interface.3,4e A variety of conformations have been observed in the various constructs utilized in these studies. As shown in Fig. 1C, the orientation of W184 sidechains in a crystal structure of CTD (residues 146–231, PDB 1A43)3b is clearly different from the orientation in a solution NMR structure of CTD (PDB 2KOD).3c Although the dimerization interface in a recent crystal structure of full-length CA (PDB 4XFX)4e is similar to that in the solution NMR structure of CTD, sidechain conformations and helix 9-helix 9 orientations are somewhat different (e.g., intra-residue M185 Cα-Cε and intermolecular L189 Cα-L189 Cα distances of 5.3 Å and 11.4 Å in PDB 4XFX, compared to 4.4–5.2 Å and 14.2 Å in PDB 2KOD). Moreover, the relevance of structural results from solution NMR and crystallography to noncrystalline assemblies of full-length CA has been unclear. In a recent model for CA tubes developed from cryoEM (EMDB-5582 and PDB 3J34)4d, sidechain conformations and helix 9-helix 9 distances in the dimerization interface are significantly different and highly disordered, with large variations from one CTD-CTD interface to another (see below). These differences from structural studies in different contexts suggest that the dimerization interface may have inherent structural plasticity, which may contribute to the variable curvature of the CA lattice in mature capsids.3c,4d,12 On the other hand, solid state NMR spectra of CA tubes show a single set of sharp signals for residues in the dimerization interface, indicating a unique and well-defined molecular structure even in non-planar, noncrystalline CA assemblies13
Here we describe solid state NMR experiments on HIV-1 CA tubes aimed at elucidating the structure of the dimerization interface in noncrystalline assemblies at atomic resolution. By combining a variety of residue-specific isotopic labeling patterns (Table 1) with a variety of solid state NMR techniques, we obtain a set of quantitative restraints on the dimerization interface structure. When supplemented with restraints on the distance and relative orientation between helix 9 segments from the reported cryoEM density of CA tubes4d, the solid state NMR data are sufficient to determine a unique dimerization interface structure, which represents the first high-resolution characterization of an intermolecular interface in noncrystalline HIV-1 CA assemblies that mimic the mature HIV-1 capsid.
Table 1.
HIV-1 CA samples for solid state NMR
| HIV-1 CA sample | labeling pattern | experimental purpose |
|---|---|---|
| I | U-15N,13C-Met and U-15N | Met chemical shift assignments, Met BroBaRR |
| II | 2-13C-glycerol and U-15N | Trp chemical shift assignments, long-range aromatic/aliphatic correlations |
| III | 2-13C-glycerol, U-15N, unlabeled Tyr, and unlabeled Phe | Trp chemical shift assignments, long-range aromatic/aliphatic correlations, Trp NCCN |
| IV | 13Cε-Met or 15N-indole (1:1 mixture) | LG-CP, intermolecular REDOR |
| V | 13Cε-Meta, 2-13C-indole, and U-15N | intramolecular REDOR |
Although the precursor methionine was not 15N-labeled, aminotransferase-catalyzed amino exchange results in incorporation of 15N at amide sites of Met residues.
MATERIALS AND METHODS
Sample preparation
HIV-1 CA protein (plasmid pNL4–3) was expressed and purified as described previously.13 Purified protein solutions were concentrated to 30 mg/ml in Tris buffer (50 mM, pH 8.0) prior to addition of NaCl to 1.0 M concentration to promote self-assembly. The tubular morphology and sample homogeneity were verified with TEM images, using an FEI Morgagni microscope operating at 80 keV. All CA samples were grown in media containing unlabeled glucose, except samples II and III, which used 2-13C-glycerol as the carbon source. Both samples mixed to make sample IV were produced using unlabeled NH4Cl as the nitrogen source. For amino-acid-specific labeling, the appropriate precursors were added to the cell culture one hour before induction, including U-13C,15N-L-methionine, 13Cε-L-methionine, 2-13C-indole (for labeling of Trp Cδ1), or 15N-indole (120 mg/l each). For sample III, aromatic 13C signals from Tyr and Phe residues were suppressed by supplying unlabeled L-tyrosine and L-phenylalanine (150 mg/l of each amino acid). Absence of Tyr and Phe signals from solid state NMR spectra of sample III confirmed their reverse labeling.
For sample V, mass spectrometry indicates 98% incorporation of 15N at amide sites of the 11 Met residues, through aminotransferase-catalyzed exchange with 15N-labeled amino acids. Specifically, the molecular mass of sample V was determined experimentally to be 25924 Da, compared with an experimental value of 25913 Da for CA with only 15N labeling. Uncertainties in molecular masses were 0.2 Da. 13C labeling patterns for samples II and III were deduced from 2D 13C-13C spectra, and were found to be in general agreement with expectations based on earlier publications.14 However, in addition to the expected Cγ, Cδ2, and Cζ3 sites of Trp residues, signals from Cδ1 and Cε3 were also observed.
Solid state NMR spectroscopy
2D NMR spectra, rotational echo double resonance15 (REDOR) data, and NCCN16 data were obtained with a Varian InfinityPlus spectrometer operating at a 1H NMR frequency of 599.2 MHz, equipped with a Varian BioMAS probe with a 3.2-mm MAS module. Lee-Goldburg cross-polarization17 (LG-CP) and broadband rotational resonance18 (BroBaRR) data were obtained with a Varian Infinity spectrometer operating at a 1H NMR frequency of 746.4 MHz, equipped with a Bruker Efree probe with a 3.2-mm MAS module. Sample temperatures were maintained near 20° C with cooled nitrogen gas. 2D 13C-13C and 15N-13C-13C correlation spectra were recorded with magic-angel spinning (MAS) at 11.00 kHz (sample I) or 16.00 kHz (samples II and III), using spin diffusion mixing periods τSD = 200 ms (sample I) or τSD = 700 ms (samples II and III) for 13C-13C longitudinal exchange, 3.0 ms periods for 15N-13C polarization transfer by cross-polarization, and 80 kHz proton decoupling levels, with two-pulse phase modulation(ref) (TPPM). 1H-13C LG-CP measurements used 11.00 kHz MAS, a 50 kHz 13C rf field amplitude, and a 61 kHz effective 1H field. 1H-15N LG-CP measurements used 11.00 kHz MAS, a 30 kHz 15N rf field amplitude, and a 41 kHz effective 1H field. REDOR data were recorded with 8.93 kHz MAS, using 85 kHz proton decoupling with TPPM and 12.5 μs 15N π pulses during the REDOR recoupling period. BroBaRR experiments were performed with 8.00 kHz, to match approximately the chemical shift difference between M185 Cα and Cε resonances. During the BroBaRR recoupling period, the 13C rf carrier frequency was set to 38.5 ppm, the effective 13C π pulse lengths were 500 μs with MLEV-32 phases,19 and the 1H decoupling level was 85 kHz. NCCN vector angle measurements were performed on sample III with REDOR recoupling conditions as described above and with τSD = 200 ms for 13C-13C longitudinal exchange between REDOR periods. Additional details of NMR measurements are given in Table S1.
Simulations of solid state NMR data
Numerical simulations of LG-CP data were performed with SPINEVOLUTION software.20 REDOR simulations were performed with SIMPSON.21 NCCN and BroBaRR measurements were simulated with custom programs (available upon request) that calculated spin density matrix evolution under the relevant rf pulse sequences, with time-dependent dipole-dipole couplings under MAS. In NCCN simulations, 13C-13C polarization transfer was assumed to be orientation-independent. BroBaRR simulations included transverse (T2) relaxation as damping of appropriate density matrix elements, using T2 values measured for 13Cα and 13Cε under the BroBaRR pulse sequence (7 ms and 25 ms, respectively).
Structure calculations
Simulated annealing calculations were carried out with Xplor-NIH22, using experimental restraints from solid state NMR measurements and the cryoEM density of CA tubes in the Electron Microscopy Data Bank (EMD-5582). CryoEM densities for interacting helix 9 segments were extracted within the Chimera program (https://www.cgl.ucsf.edu/chimera/), by fitting pairs of helix 9 segments (residues 175–193) from PDB 3J34 to the density and then using the “Zone” function to select the density within 3.0 Å of the atomic coordinates. Xplor-NIH calculations included two copies of residues 175–193, with initial atomic coordinates taken from PDB files 4XFX, 2KOC, or 1A43. Prior to simulated annealing, a preliminary rigid-body fit of the initial atomic coordinates to the cryoEM density (Fig. S7) was performed, without allowing changes in interhelical distance or orientation, using the probDistPot potential energy term and the randomizeDomainPos function of Xplor-NIH. Simulated annealing was then performed from 3500 K to 25 K in 12,500 steps, with 0.2 ps of torsion angle dynamics at each temperature step. Distance restraints from REDOR and BroBaRR measurements (Figs. 3C–3F) were represented by NOE potentials with distance ranges equal to the experimental error ranges stated in the main text. Distance restraints from long-range correlation experiments (Fig. 2C) were assigned an upper limit of 7.0 Å. The force constant for NOE potential terms increased geometrically from 50 to 200 kcal/mol-Å2 during annealing. Vector angle restraints from NCCN data (Fig. 3G) were represented by VEAN potential in Xplor-NIH and a force constant increasing from 1 to 10 kcal/mol-rad2. The cryoEM density was applied as a restraint on backbone heavy atoms only, using the probDistPot potential23 with a scale factor of 40 kcal/mol-Å4, and allowing the two copies of residues 179–193 to move independently of each other. The observation of a single set of chemical shifts for each residue in solid state NMR spectra of CA tubes implies a single protein conformation. Therefore, a noncrystallographic symmetry potential (NCS) was applied, with a scale factor of 10 kcal/mol-Å2. In order to preserve the integrity of the α-helical conformation, hydrogen bond restraints between residues i and i+4 were included in the NOE potential, for i = 178–189. In addition, backbone ϕ and ψ torsion angle restraints from our previous work13, which are consistent with an α-helical conformation, were applied with the CDIH potential, using a scale factor of 400 kcal/mol-rad2. The scale factor for van der Waals-like repulsions increased from 0.004 to 4 kcal/mol-Å4, with a radius scale factor decreasing from 0.9 to 0.8. Standard bond length, bond angle, and improper angle potentials were also used. After annealing, energy minimization was performed. In each set of structure calculations, the 20 lowest-energy structures out of 200 independent calculations were selected for each structure ensemble.
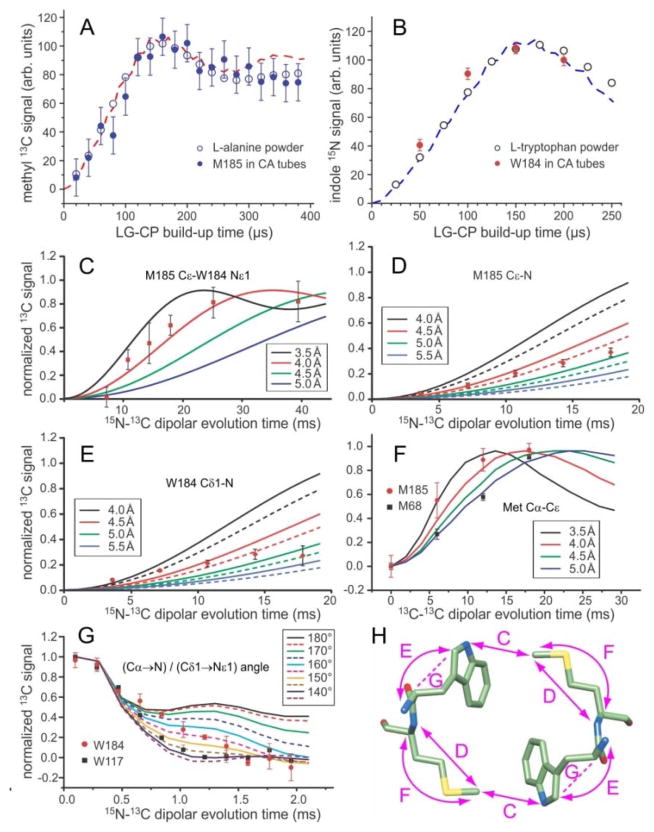
Figure 3.
Quantitative restraints on the dimerization interface structure from solid state NMR. (A,B) Comparison of 1H-13C and 1H-15N LG-CP build-up data for M185 Cε and W184 Nε1 in HIV-1 CA tubes with data for the methyl site in L-alanine powder and the Nε1 site in L-tryptophan powder. The similarity of these data for CA tubes and amino acid powders indicates the absence of large-amplitude motions of M185 and W184 sidechains on sub-millisecond timescales. Dashed lines are ideal LG-CP simulations for four-spin (panel A) and two-spin (panel B) systems, assuming methyl C-H bond lengths of 1.08 Å, with rapid methyl rotation, and indole N-H bond lengths of 0.98 Å. (C,D,E) 15N-13C REDOR data that restrain the intermolecular M185 Cε-W184 Nε1 distance, the intra-residue M185 Cε-N distance, and the intra-residue W184 Cε1-N distance in HIV-1 CA tubes. Solid curves in panel C are ideal two-spin simulations for isolated 15N-13C pairs with the indicated distances. Solid and dashed curves in panels D and E are three-spin simulations, including an additional 15N at the farthest and closest possible distance (see text). (F) BroBaRR data that restrain the intra-residue Cα-Cε distance in M185. Data for M68 are also shown for comparison. Solid lines are two-spin simulations for the indicated 13C-13C distances. (G) NCCN data that restrain the angle between the Cα-N and Cδ1-Nε1 bond vectors in W184. Data for W184 are also shown for comparison. Solid and dashed lines are ideal simulations for the indicated angles. (H) Dimer of W184-M185 units, showing the distances (double-headed arrows) and vector angles (dashed lines) restrained by data in panels C-G. Error bars in panels A–G represent uncertainties calculated from the root-mean-squared noise in the corresponding solid state NMR spectra.
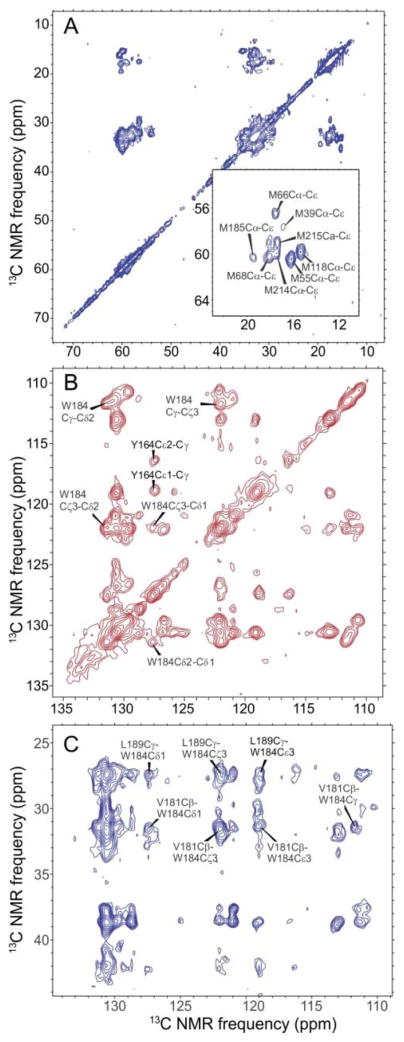
Figure 2.
2D solid state NMR spectra of HIV-1 CA tubes. (A) Aliphatic region of a 2D 13C-13C spectrum of sample I, obtained with a spin diffusion mixing period τSD = 200 ms. The inset shows assignments of Met Cα-Cε crosspeaks. (B) Aromatic region of a 2D 13C-13C spectrum of sample II, obtained with τSD = 700 ms. (C) Aromatic/aliphatic region of a 2D 13C-13C correlation spectrum of sample II, obtained with τSD = 700 ms. Crosspeaks indicating long-range contacts from W184 to V181 and L189 are labeled. Intraresidue crosspeaks from the four other Trp residues (W23, W80, W117, and W133) are also resolved13, but for clarity are not labeled. Contour levels increase by factors of 1.2 in panels A–C.
RESULTS
Assignment of solid state NMR signals in the dimerization interface
In a previous study13, we obtained 15N and 13C chemical shift assignments for most backbone N, CO, Cα, and Cβ sites in CA tubes, as well as some sidechain assignments. Additional sidechain assignments, especially for W184 and M185, were required for the structural measurements described below. To obtain 13C chemical shift assignments for Met sidechains, we prepared a CA tube sample that was uniformly 15N-labeled at all residues and uniformly 13C-labeled only at Met residues (sample I). From 2D NCACX and NCOCX spectra (Fig. S1) and 2D 13C-13C spectra (Fig. 2A, Fig. S2), we obtained full assignments for eight of the 11 Met residues in CA, including M185. Importantly, the 13Cε signal of M185 is resolved from those of all other Met residues.
Chemical shift assignments for aromatic sidechains were obtained from 2D 13C-13C spectra of CA tubes that were partially 13C-labeled (sample II), using 2-13C-glycerol as the carbon source for protein expression (Fig. 2B). Most carbon sites in Trp sidechains were partially 13C-labeled in this sample, allowing us to trace connectivities among sidechain signals for W184, as well as for the other Trp residues. Additional spectra of a sample labeled with 2-13C glycerol and reverse-labeled with natural-abundance Tyr and Phe amino acids (sample III) helped us identify partially overlapping aromatic signals.
Rigidity of the dimerization interface
Before attempting quantitative structural measurements, it was important to establish that sidechains in the dimerization interface are not highly dynamic. Large-amplitude motions of sidechains on sub-millisecond time scales attenuate one-bond 1H-13C and 1H-15N dipole-dipole couplings in solid state NMR measurements on proteins.24 Therefore, we measured the one-bond 1H-13Cε coupling for M185 and the one-bond 1H-15Nε1 coupling for W184 in CA tubes, using the LG-CP technique.17 As shown in Figs. 3A and 3B, build-up curves for M185 13Cε and W184 15Nε1 signals in CA tubes under LG-CP are nearly identical to build-up curves for amino acid powders, in which large-amplitude sidechain motions are not present, and are in good agreement with numerical simulations for rigid sites (but with rapid uniaxial methyl rotation). Together with the observation of sharp crosspeak signals for M185 and W184 (Figs. 2A and 2B), the LG-CP data imply a well-defined, rigid molecular structure in the dimerization interface. In contrast, the M185 13Cε signal is absent from solution NMR spectra of soluble CA dimers, presumably due to conformational exchange (Fig. S3).
In addition, one-dimensional 13C and 15N solid state NMR spectra showed no significant changes in signal intensities from M185 and W184 sidechain over the 0–30° C temperature range. Previously reported 2D 13C-13C spectra of CA tubes, acquired with conditions similar to those in solution NMR (i.e., low-power proton decoupling and spin polarization transfers driven by scalar couplings), did not show signals attributable to M185 or W184, indicating an absence of rapid, isotropic reorientation.
Quantitative structural restraints from solid state NMR
Quantitative restraints on specific interatomic distances in protein assemblies can be obtained from quantitative measurements of nuclear magnetic dipole-dipole couplings, using a class of solid state NMR techniques with magic-angle spinning (MAS) called “dipolar recoupling” techniques.25 These techniques require that signals from the sites of interest be sufficiently well resolved that their intensities can be measured accurately. For this purpose, we used samples I, IV, and V, which allowed us to measure specific intermolecular and intra-residue distances involving W184 and M185.
The intermolecular M185 Cε-W184 Nε1 distance across the dimerization interface was measured with REDOR.15a Sample IV was used for these measurements to ensure that only intermolecular 15N-13C couplings were measured and that the detected M185 13Cε signals were resolved in 1D. REDOR data are plotted in Fig. 3C as the normalized difference (S0–S1)/S0, where S1 and S0 are 13C NMR signal areas with and without a central 15N π pulse that determines whether a net recoupling of 15N-13C interactions occurs (Fig. S4A). The data are additionally scaled by a factor of 2.0 to account for the 50% 15N labeling in sample IV. As shown in Fig. 3C, the REDOR data can be fit with simulations for a 15N-13C distance of 3.5–4.0 Å. This distance agrees well with the 4.1 Å intermolecular M185 Cε-W184 Nε1 distance in PDB 4XFX (2.43 Å crystallographic resolution), but is shorter than distances in PDB 2KOD (NMR structure with 0.9 Å heavy atom root-mean-squared deviation) (4.3–5.2 Å) and PDB 1A43 (2.60 Å crystallographic resolution) (7.5 Å).
Intra-residue M185 Cε-N and W184 Cδ1-N distances, which restrain the M185 and W184 sidechain conformations, were also measured with REDOR. Sample V, in which only Met Cε and Trp Cδ1 sites were 13C-labeled and all backbone nitrogens (but not Trp sidechain nitrogens) were 15N-labeled, was used. Experimental (S0–S1)/S0 data are shown in Figs. 3D and 3E, without additional scaling. In these measurements, depending on the sidechain conformations, inter-residue 15N-13C couplings involving backbone nitrogens of neighboring residues are not necessarily negligible. Therefore, three-spin simulations were performed, including the intra-residue 15N-13C pair and one additional 15N spin at a variable position chosen to be consistent with α-helical structure in residues 183–186. Solid and dashed lines in Figs. 3D and 3E represent simulations with the additional 15N spin at positions that produce the most and least rapid build-up of REDOR difference signals, respectively. Comparisons of simulations with experimental data then indicated an intra-residue M185 Cε-N distance of 4.3–5.0 Å and an intra-residue W184 Cε1-N distance of 4.0–5.0 Å. For comparison, M185 Cε-N distances in PDB 4XFX, 2KOD, and 1A43 are 5.7 Å, 4.8–5.7 Å, and 5.1 Å, respectively. W184 Cδ1-N distances in PDB 4XFX, 2KOD, and 1A43 are 4.7 Å, 4.5–4.6 Å, and 3.6 Å.
Experimental REDOR data in Figs. 3D and 3D clearly deviate from the simulated curves at larger values of the evolution time. This effect is attributable to insufficiently large proton decoupling fields26, which lead to an accumulating imperfection in 15N spin inversion by the train of MAS-synchronized 15N π pulses in the REDOR sequence (Fig. S4A). The uncertainties in 15N-13C distances stated above take this effect into account.
The intra-residue M185 Cα-Cε distance was measured with BroBaRR18, which allows the dipole-dipole coupling between two specific 13C-labeled sites to be measured in a uniformly 13C-labeled residue, provided that at least one of the two 13C chemical shifts is resolved. Selectivity in the BroBaRR technique is achieved by proper choice of the 13C radio-frequency (rf) carrier frequency, the MAS frequency, and the amplitude of a train of weak rf pulses that is applied during the BroBaRR evolution period (Fig. S4B). Experimental BroBaRR data, obtained with sample I, are compared with simulations in Fig. 3F. For M185, the Cα-Cε distance was found to be 3.5–4.3 Å, shorter than the 5.3 Å and 4.4–5.2 Å distances in PDB 4XFX and 2KOD, and similar to the 4.1 Å distance in PDB 1A43. BroBaRR data for M68 were obtained simultaneously, indicating a Cα-Cε distance of 4.9 +/− 0.2 Å that is in good agreement with the corresponding distances of 4.5 Å, 4.8 Å, and 4.9 Å in crystal structures of full-length CA (PDB 4XFX, 3MGE, and 3P05).4c,4e,7b Representative spectra from REDOR and BroBaRR experiments are shown in Fig. S5. In addition to distance measurements with dipolar recoupling techniques, structural restraints can be obtained with “tensor correlation” techniques in solid state NMR, which measure the orientations of two different chemical bond vectors or functional groups relative to one another.27 As a further restraint on the W184 sidechain conformation in CA tubes, we recorded NCCN tensor correlation data16 to probe the angle between the backbone Cα-N bond vector and the sidechain Cδ1-Nε1 bond vector. Sample III, which was uniformly 15N-labeled and partially 13C-labeled, but not 13C-labeled at Tyr and Phe residues, was chosen for these measurements because the 13Cδ1 signal of W184 was fully resolved. In our version of the NCCN pulse sequence (Fig. S4C), 13C polarization on Cα was prepared by 1H-15N cross-polarization followed by band-selective 15N-13Cα cross-polarization. After a REDOR period for evolution under the 15N-13Cα dipole-dipole coupling, the polarization was transferred to Cδ1 during a 200 ms 13C-13C spin diffusion period (Fig. S6). After a second REDOR period for evolution under the 15Nε1-13Cδ1 dipole-dipole coupling, 13Cδ1 signals were measured. Experimental NCCN data are compared with simulations in Fig. 3F. For W184, the data indicate an angle in the 150°–170° range (absolute value). Values of the same angle are 160°, 164–174°, and 113° in PDB 4XFX, 2KOD, and 1A43, respectively. NCCN data were also obtained for W117, implying an angle between Cα-N and Cδ1-Nε1 bond vectors that is less than 150° (absolute value), in a range where the NCCN signals decay rapidly and are not sensitive to the precise angle. For comparison, values of the same angle for W117 in PDB 4XFX, 3MGE, and 3P05 are 78°, 80°, and 77°.
Fig. 3H summarizes the aspects of the dimerization interface that are restrained by the quantitative structural measurements in Figs. 3C–3G. As discussed above, these measurements for CA tubes are not fully consistent with previous high-resolution structures, showing that the dimerization interface structure in noncrystalline CA tubes is not the same as in the soluble or crystalline constructs examined previously. The good agreement between solid state NMR data for M68 and W117 (Figs. 3F and 3G) and results from crystallography is reassuring, since both M68 and W117 are in NTD and are not involved in dimerization or other potentially variable intermolecular interfaces.
Sidechain-sidechain contacts
While W184 and M185 constitute the core of the dimerization interface, other hydrophobic residues of helix 9 segments also participate in intermolecular interactions in previously described structures.3,4e Since various interactions are possible, depending on the sidechain conformations, we recorded additional 2D 13C-13C spectra with long spin diffusion mixing periods (τSD = 700 ms) to reveal sites close in space to W184. Fig. 2C shows the aromatic-aliphatic section of a 2D spectrum of sample III, under conditions where crosspeaks are expected between 13C pairs with internuclear distances up to roughly 7 Å. Multiple crosspeaks are observed between aromatic 13C sites of W184 and aliphatic sidechain sites of V181 and L189, with signal-to-noise values in the 9–15 range. These crosspeaks help to define the conformation of W184 in the context of interactions with surrounding hydrophobic residues.
Structure calculations with combined cryoEM density and solid state NMR restraints
Structure calculations were performed by restrained simulated annealing in Xplor-NIH.22 Restraints from solid state NMR, described above, are sufficient to define the structural arrangement of hydrophobic sidechains within the dimerization interface, but do not determine the distance and orientation between the two helix 9 segments. Therefore, we made use of the cryoEM density map for CA tubes reported by Zhao et al.4d, deposited in the Electron Microscopy Data Bank as file EMD-5582. Densities for helix 9 pairs (Fig. S7) were extracted manually, using the Chimera program (https://www.cgl.ucsf.edu/chimera/). These densities were applied as restraints in simulated annealing structure calculations, using the probDistPot potential energy function of Xplor-NIH. Restraints based on solid state NMR data included quantitative interatomic distance restraints from REDOR and BroBaRR data, vector angle restraints from the NCCN data (using the VEAN potential term of Xplor-NIH), upper-bound distance restraints from long-range crosspeaks in 2D 13C-13C spectra with τSD = 700 ms, and backbone torsion angle restraints obtained previously from 15N and 13C chemical shifts.13 Starting structures were pairs of interacting helix 9 segments (residues 175–193) from PDB files 4XFX, 2KOD, and 1A43 (Figs. 1C and S7A–D). Thus, these calculations can be considered to be structure refinements, rather than de novo structure determinations, in which we have used structures from soluble or crystalline samples as the starting point for high-resolution characterization of the dimerization interface structure in the noncrystalline, curved lattice of CA tubes.
Fig. 4A shows a superposition of the three starting structures, highlighting the differences in helix orientations and sidechain conformations. Fig. 4B shows a superposition of the corresponding lowest-energy final structures. Pairwise root-mean-squared deviations (rmsd) among these structures, including backbone heavy atoms of residues 179–193 and sidechain heavy atoms of V181, W184, M185, and L189, are reduced from 1.35–3.50 Å to 0.85–1.16Å. The convergence of refined structures from three different starting points demonstrates that the combination of solid state NMR and cryoEM-based restraints results in a unique result, which represents the dimerization interface structure in CA tubes.
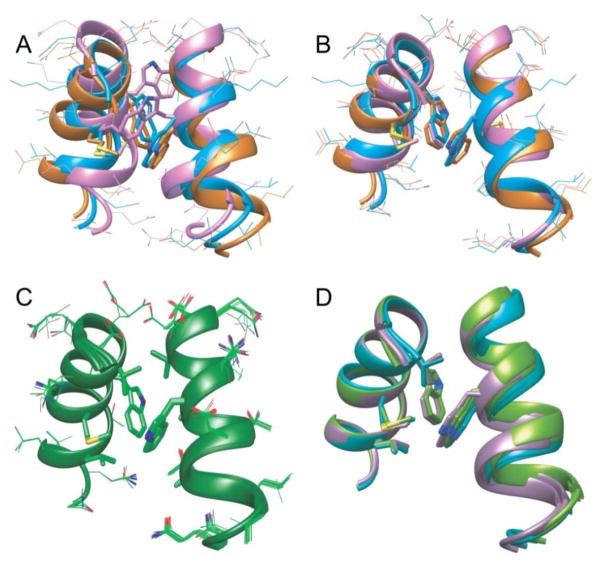
Figure 4.
Structure refinement based on solid state NMR restraints and cryoEM densities. (A) Superposition of residues 178–193 in three different starting structures, taken from studies of crystalline full-length CA (PDB 4XFX, orange), CTD dimers in solution (PDB 2KOD, cyan), and crystalline CTD dimers (PDB 1A43, pink). Alignment of the three structures highlights differences in the relative helix orientations and the positions of W184 and M185 sidechains. Other amino acid sidechains are represented by thinner sticks. (B) Final structures obtained from the three starting structures in panel A, with the same color scheme. Lowest-energy structures are shown from three sets of simulated annealing calculations, using the structural restraints from solid state NMR data in Figs. 2 and 3 and cryoEM densities from one of the dimer interfaces in EMD-5582. (C) Superposition of 20 low-energy structures from 100 independent calculations, using the dimer interface from PDB 4XFX as the starting structure. Sidechains of W184, M185, V181, and L189 are represented by thick sticks. (D) Superposition of six lowest-energy final structures from calculations that used cryoEM densities from six different dimer interfaces in EMD-5582. Starting structures were taken from PDB 4XFX. Based on two-fold symmetry in the cryoEM reconstruction, the dimer interface densities are grouped into three pairs, and the resulting refined structures are colored purple, cyan, and green. The six final structures are optimally aligned for one helix, on which sidechains of W184, M185, V181, and L189 are shown. Only W184 is shown on the other helix.
Rmsd values (as defined above) between starting structures and lowest-energy final structures (Figs. 4A and 4B) are 0.92 Å, 1.40 Å, and 3.48 Å for starting structures taken from PDB 4XFX, 2KOD, and 1A43, respectively. In all cases, restrained simulated annealing results in structural changes, indicating that the dimerization interface structure in CA tubes is not identical to structures in soluble or crystalline states. In calculations starting with PDB 4XFX, the most significant changes are in the values of sidechain torsion angles for W184 (from χ1= 176°, χ2 = 85° to χ1= 165°, χ2 = 92°), which result in displacements of sidechain indole atoms by as much as 1.1 Å. Similar displacements of other hydrophobic sidechains also occur in the dimerization interface. In calculations starting with PDB 2KOD, displacements of indole atoms are somewhat larger (up to 1.6 Å), reducing the nearest-neighbor carbon-carbon distance between W184 sidechains by 1.7 Å. The intermolecular angle between α-helices also increases by approximately 8°. In calculations starting with PDB 1A43, sidechain conformations in the dimerization interface change dramatically, with displacements of sidechain indole atoms up to 7.0 Å. The intermolecular angle between α-helices increases by approximately 28°.
Fig. 4C shows a bundle of 20 final structures from a set of 100 independent calculations, all starting with coordinates from PDB 4XFX (see also Fig. S8E). The rmsd for all heavy atom coordinates in this bundle is 1.04 Å. The largest rmsd contributions come from sidechains that are not involved in intermolecular contacts, which were unrestrained. For backbone heavy atoms of all residues and sidechain heavy atoms of V181, W184, M185, and L189, the rmsd is 0.41 Å. Thus, the experimental data are sufficient to define the dimerization interface structure with high precision. Atomic coordinates from Fig. 4C have been deposited in the Protein Data Bank as PDB file 5IRT. Structural restraints and corresponding values in PDB 5IRT are summarized in Table S2.
Finally, Fig. 4D shows a superposition of six lowest-energy final structures from calculations in which six different cryoEM densities were used (Fig. S7), with the same starting structure (from PDB 4XFX) in each case. The rmsd value among these six final structures, including backbone heavy atoms of all residues and sidechain heavy atoms of V181, W184, M185, and L189, is 0.50 Å. Thus, the specific choice of helix 9 pair from the cryoEM density map is unimportant. This observation is consistent with our finding that the lowering of symmetry induced by lattice curvature and by the variations in CA tube diameters in solid state NMR samples does not produce significant inhomogeneous broadening of solid state NMR signals from the dimerization interface, relative to signals from other regions of CA.13 In contrast, dimerization interface structures from molecular dynamics flexible fitting (MDFF) calculations (PDB 3J34)4d exhibit substantially greater variability and disorder (Fig. S7), apparently reflecting the lack of restraints with sufficiently high resolution in the MDFF calculations.
DISCUSSION
Results described above demonstrate a unique role for solid state NMR in applications to noncrystalline supramolecular assemblies formed by proteins of relatively high molecular weight. For assemblies such as HIV-1 CA tubes, full structure determination based entirely on solid state NMR data would be extremely challenging, primarily due to limited resolution in the multidimensional spectra from which the necessary long-range structural restraints would be obtained. However, full structure determination is not required, since a wealth of structural information already exists from solution NMR studies of soluble NTD, CTD, and full-length CA3c,8, from crystallographic studies of NTD, CTD, and full-length CA3a,3b,4c,4e,7b,9, and electron microscopy of CA tubes and sheets.3c,4a,4b,4d What has been missing is high-resolution structural characterization of intermolecular interfaces within the curved, noncrystalline CA lattice that exists in mature HIV-1 capsids, for which CA tubes are an appropriate model. As demonstrated above, this missing information can be provided by solid state NMR data, including quantitative measurements of interatomic distances and bond vector orientations, conformation-dependent chemical shifts, and long-range inter-residue contacts. With appropriate isotopic labeling patterns, specific interfaces can be targeted.
It is notable that the solid state NMR data lead to a unique structure for the dimerization interface, despite the variable curvature of the CA lattice and the heterogeneity of tube diameters and helicities in solid state NMR samples, and even when inequivalent cryoEM densities are used (Figs. 4D and S6). Structural plasticity in intermolecular interfaces, including the dimerization and trimerization interfaces formed by CTD domains3c,4d,12, has been suggested to play a role in CA lattice curvature. Previous solid state NMR studies of CA tubes by our laboratory13,28 and by Polenova and coworkers12,29 provided evidence for dynamic disorder in the NTD-CTD linker and surrounding segments, which is consistent with variability in the intramolecular orientation between NTD and CTD.7b,30 The observation of a well-structured dimerization interface is further evidence that plasticity of intermolecular CTD-CTD interactions does not make a major contribution to the variable curvature of the mature capsid surface.
A recent crystal structure of full-length CA from Gres et al. (PDB 4XFX)4e provides a high-resolution picture of a planar lattice of CA hexamers, building on earlier electron diffraction studies of planar CA assemblies by Ganser-Pornillos et al.4b The dimerization interface in the curved lattice derived from our solid state NMR data, as described above, is very similar to the interface in the planar lattice, with differences in the precise conformations of W184 and M185 sidechains. The dimerization interface in the curved lattice is also similar to the interface in soluble CTD dimers determined by Byeon et al. (PDB 2KOD)3c (which is also consistent with solution NMR data for full-length CA dimers8e), but with somewhat larger differences in sidechain conformations and helix 9-helix 9 orientations. The dimerization interface in crystalline CTD reported by Worthylake et al. (PDB 1A43)3b is quite different from the interface structure in CA tubes. Dimerization interfaces in an atomic model for CA tubes developed by MDFF4d, using the cryoEM density as a restraint, are substantially disordered (Fig. S7), in strong disagreement with our solid state NMR data.
The similarity of dimerization interface structures in noncrystalline CA tubes, determined from our solid state NMR data, and in the crystalline, planar 2D lattice of Gres et al. (PDB 4XFX) supports the idea that lattice curvature is not dependent on plasticity of the dimerization interface. Dissimilarity with the interface structure in the CTD dimer crystal structure of Worthylake et al. (PDB 1A43) may be attributable to the fact that their CTD construct included only residues 146–231, with an additional N-terminal Met residue. Similarity with the interface structure in the CTD dimer solution NMR structure of Byeong et al. (PDB 2KOD), which included residues 145–231, supports their proposal that Y145 has a significant impact on the dimerization interface.
CONCLUSIONS
We have shown that the dimerization interface, one of the key sites of intermolecular interactions within the HIV-1 CA lattice, is rigid and structurally ordered in CA tubes. From solid state NMR restraints on interatomic distances and conformations for sidechains of hydrophobic residues within the dimerization interface, supplemented by densities for helix 9 pairs from cryoEM, we have developed an atomic resolution structural model for the dimerization interface in CA tubes. Goals for future work include the acquisition of quantitative structural restraints from solid state NMR for the trimerization interface, involving intermolecular interactions of helix 10 and helix 11 segments, and for intermolecular NTD-CTD interactions that stabilize CA hexamers. Such data will lead to improved models for the complete CA lattice in noncrystalline assemblies and to a more comprehensive understanding of variable lattice curvature. The same approach can also be applied to the spherical Gag lattice of immature HIV-1, which has been proposed to involve two separate intermolecular dimerization interfaces, formed by helix 9 segments of CTD and by helix 1 segments of NTD.31
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases of the National Institutes of Health, and by the NIH Intramural AIDS Targeted Antiviral Program. We thank Dr. Eric Moore for assistance with simulations of NMR data and Drs. Charles Schwieters and Guillermo Bermejo for assistance with Xplor-NIH calculations.
Footnotes
- Summary of solid state NMR measurement conditions (Table S1)
- Summary of structural restraints in Xplor-NIH calculations (Table S2)
- Solid state NMR pulse sequences (Figure S4)
- Additional solid state NMR spectra (Figures S1, S2, S3, S5, and S6)
- Additional structure calculation results (Figures S7 and S8)
References
- 1.Ganser-Pornillos BK, Yeager M, Sundquist WI. Curr Opin Struct Biol. 2008;18:203. doi: 10.1016/j.sbi.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganser-Pornillos BK, von Schwedler UK, Stray KM, Aiken C, Sundquist WI. J Virol. 2004;78:2545. doi: 10.1128/JVI.78.5.2545-2552.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Gamble TR, Yoo SH, Vajdos FF, vonSchwedler UK, Worthylake DK, Wang H, McCutcheon JP, Sundquist WI, Hill CP. Science. 1997;278:849. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]; (b) Worthylake DK, Wang H, Yoo SH, Sundquist WI, Hill CP. Acta Crystallogr Sect D-Biol Crystallogr. 1999;55:85. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]; (c) Byeon IJL, Meng X, Jung JW, Zhao GP, Yang RF, Ahn JW, Shi J, Concel J, Aiken C, Zhang PJ, Gronenborn AM. Cell. 2009;139:780. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Li S, Hill CP, Sundquist WI, Finch JT. Nature. 2000;407:409. doi: 10.1038/35030177. [DOI] [PubMed] [Google Scholar]; (b) Ganser-Pornillos BK, Cheng A, Yeager M. Cell. 2007;131:70. doi: 10.1016/j.cell.2007.08.018. [DOI] [PubMed] [Google Scholar]; (c) Pornillos O, Ganser-Pornillos BK, Kelly BN, Hua YZ, Whitby FG, Stout CD, Sundquist WI, Hill CP, Yeager M. Cell. 2009;137:1282. doi: 10.1016/j.cell.2009.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Zhao GP, Perilla JR, Yufenyuy EL, Meng X, Chen B, Ning JY, Ahn J, Gronenborn AM, Schulten K, Aiken C, Zhang PJ. Nature. 2013;497:643. doi: 10.1038/nature12162. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Gres AT, Kirby KA, KewalRamani VN, Tanner JJ, Pornillos O, Sarafianos SG. Science. 2015;349:99. doi: 10.1126/science.aaa5936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrlich LS, Liu TB, Scarlata S, Chu B, Carter CA. Biophys J. 2001;81:586. doi: 10.1016/S0006-3495(01)75725-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briggs JAG, Wilk T, Welker R, Krausslich HG, Fuller SD. Embo J. 2003;22:1707. doi: 10.1093/emboj/cdg143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.(a) Ganser BK, Li S, Klishko VY, Finch JT, Sundquist WI. Science. 1999;283:80. doi: 10.1126/science.283.5398.80. [DOI] [PubMed] [Google Scholar]; (b) Pornillos O, Ganser-Pornillos BK, Yeager M. Nature. 2011;469:424. doi: 10.1038/nature09640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Gitti RK, Lee BM, Walker J, Summers MF, Yoo S, Sundquist WI. Science. 1996;273:231. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]; (b) Tang C, Ndassa Y, Summers MF. Nat Struct Biol. 2002;9:537. doi: 10.1038/nsb806. [DOI] [PubMed] [Google Scholar]; (c) Wong HC, Shin R, Krishna NR. Biochemistry. 2008;47:2289. doi: 10.1021/bi7022128. [DOI] [PubMed] [Google Scholar]; (d) Shin R, Tzou YM, Krishna NR. Biochemistry. 2011;50:9457. doi: 10.1021/bi2011493. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Deshmukh L, Schwieters CD, Grishaev A, Ghirlando R, Baber JL, Clore GM. J Am Chem Soc. 2013;135:16133. doi: 10.1021/ja406246z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.(a) Momany C, Kovari LC, Prongay AJ, Keller W, Gitti RK, Lee BM, Gorbalenya AE, Tong L, McClure J, Ehrlich LS, Summers MF, Carter C, Rossmann MG. Nat Struct Biol. 1996;3:763. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]; (b) Gamble TR, Vajdos FF, Yoo SH, Worthylake DK, Houseweart M, Sundquist WI, Hill CP. Cell. 1996;87:1285. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 10.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. J Virol. 2003;77:5439. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pornillos O, Ganser-Pornillos BK, Banumathi S, Hua YZ, Yeager M. J Mol Biol. 2010;401:985. doi: 10.1016/j.jmb.2010.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byeon IJL, Hou GJ, Han Y, Suiter CL, Ahn J, Jung J, Byeon CH, Gronenborn AM, Polenova T. J Am Chem Soc. 2012;134:6455. doi: 10.1021/ja300937v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bayro MJ, Chen B, Yau WM, Tycko R. J Mol Biol. 2014;426:1109. doi: 10.1016/j.jmb.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higman VA, Flinders J, Hiller M, Jehle S, Markovic S, Fiedler S, van Rossum BJ, Oschkinat H. J Biomol NMR. 2009;44:245. doi: 10.1007/s10858-009-9338-7. [DOI] [PubMed] [Google Scholar]
- 15.(a) Gullion T, Schaefer J. J Magn Reson. 1989;81:196. [Google Scholar]; (b) Gullion T, Baker DB, Conradi MSJ. Magn Reson. 1990;89:479. [Google Scholar]
- 16.Costa PR, Gross JD, Hong M, Griffin RG. Chem Phys Lett. 1997;280:95. [Google Scholar]
- 17.van Rossum BJ, de Groot CP, Ladizhansky V, Vega S, de Groot HJM. J Am Chem Soc. 2000;122:3465. [Google Scholar]
- 18.Chan JCC, Tycko R. J Chem Phys. 2004;120:8349. doi: 10.1063/1.1737369. [DOI] [PubMed] [Google Scholar]
- 19.Levitt MH, Freeman R, Frenkiel T. J Magn Reson. 1982;47:328. [Google Scholar]
- 20.Veshtort M, Griffin RG. J Magn Reson. 2006;178:248. doi: 10.1016/j.jmr.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 21.Bak M, Rasmussen JT, Nielsen NC. J Magn Reson. 2000;147:296. doi: 10.1006/jmre.2000.2179. [DOI] [PubMed] [Google Scholar]
- 22.Schwieters CD, Kuszewski JJ, Clore GM. Prog Nucl Magn Reson Spectrosc. 2006;48:47. doi: 10.1016/j.pnmrs.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong Z, Schwieters CD, Tang C. PLoS One. 2015;10:e0120445. doi: 10.1371/journal.pone.0120445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.(a) Lorieau JL, McDermott AE. J Am Chem Soc. 2006;128:11505. doi: 10.1021/ja062443u. [DOI] [PubMed] [Google Scholar]; (b) Schanda P, Meier BH, Ernst M. J Am Chem Soc. 2010;132:15957. doi: 10.1021/ja100726a. [DOI] [PubMed] [Google Scholar]
- 25.De Paepe G. In: Annual Review of Physical Chemistry. Johnson MA, Martinez TJ, editors. Vol. 63. Vol. 63. 2012. p. 661. [Google Scholar]
- 26.(a) Sharpe S, Kessler N, Anglister JA, Yau WM, Tycko R. J Am Chem Soc. 2004;126:4979. doi: 10.1021/ja0392162. [DOI] [PubMed] [Google Scholar]; (b) Yang J, Weliky DP. Biochemistry. 2003;42:11879. doi: 10.1021/bi0348157. [DOI] [PubMed] [Google Scholar]
- 27.Dabbagh G, Weliky DP, Tycko R. Macromolecules. 1994;27:6183. [Google Scholar]
- 28.Lu JX, Bayro MJ, Tycko R. J Biol Chem. 2016 doi: 10.1074/jbc.M116.720557. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Y, Hou GJ, Suiter CL, Ahn J, Byeon IJL, Lipton AS, Burton S, Hung I, Gor’kov PL, Gan ZH, Brey W, Rice D, Gronenborn AM, Polenova T. J Am Chem Soc. 2013;135:17793. doi: 10.1021/ja406907h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey GD, Hyun JK, Mitra AK, Kingston RL. J Mol Biol. 2012;417:212. doi: 10.1016/j.jmb.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Schur FKM, Hagen WJH, Rumlova M, Ruml T, Muller B, Krausslich HG, Briggs JAG. Nature. 2015;517:505. doi: 10.1038/nature13838. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.