Abstract
Cytokines often deliver simultaneous, yet distinct, cell growth and cell survival signals. The 70-kDa ribosomal protein S6 kinase (p70S6K) is known to regulate cell growth by inducing protein synthesis components. We purified membrane-based p70S6K as a kinase responsible for site-specific phosphorylation of BAD, which inactivates this proapoptotic molecule. Rapamycin inhibited mitochondrial-based p70S6K, which prevented phosphorylation of Ser-136 on BAD and blocked cell survival induced by insulin-like growth factor 1 (IGF-1). Moreover, IGF-1-induced phosphorylation of BAD Ser-136 was abolished in p70S6K-deficient cells. Thus, p70S6K is itself a dual pathway kinase, signaling cell survival as well as growth through differential substrates which include mitochondrial BAD and the ribosomal subunit S6, respectively.
The maintenance of cellular homeostasis within organs is mediated in part by a critical interdependence between cells of different types. This includes a cellular dependence on a series of factors, such as IGF-1, nerve growth factor, or interleukin-3 (IL-3), that transduce signals through surface receptors to repress apoptosis and stimulate growth of target cells (1–5). The BCL-2 family of proteins that regulate cell death is frequently a target of posttranslational modification downstream of both survival and death signal transduction cascades (6, 7). Such modifications to BCL-2 members often dictate their active-versus-inactive conformation, subcellular localization, and partner proteins. BAD is one such target, a “BH3 domain-only” proapoptotic member sharing sequence homology only within the BH3 amphipathic α-helical domain (8–11). In the presence of requisite survival factors, cells phosphorylate BAD on two serine residues (S112 and S136) embedded within 14-3-3 consensus binding sites. Phosphorylated BAD appears to be the inactive moiety sequestered in the cytosol bound to 14–3-3, freeing BCL-XL or BCL-2 to promote survival. Only the active, nonphosphorylated BAD heterodimerizes with BCL-XL or BCL-2 at membrane sites to promote cell death (12). It has been recently reported that S155 in the BH3 domain is also phosphorylated to disrupt the binding of BAD to BCL-XL or BCL-2 (13–17).
Several kinases have been noted to phosphorylate BAD (18–25). For example, 90-kDa Ribosomal S6 Kinase in the mitogen-activated protein kinase pathway phosphorylates S112 of BAD (19, 20). We noted that IL-3-induced BAD kinase activity resides predominantly at membrane sites. In this context, we purified protein kinase A tethered to mitochondria by an A kinase anchoring protein as the kinase responsible for phosphorylating S112 of BAD at that target organelle (21). In multiple systems, activation of the phosphatidylinositol 3-kinase pathway appears responsible for the phosphorylation of BAD on S136. For example, the downstream kinase AKT (protein kinase B) is capable of phosphorylating S136 and inactivating BAD (22–24). Yet when AKT was immunodepleted from an hematopoietic cell line, considerable activity remained, which makes the argument that AKT may not be the principal kinase directly responsible for phosphorylating BAD S136 in this system (21).
Here, we purified membrane-based 70-kDa ribosomal protein S6 kinase (p70S6K) as a BAD S136-specific kinase. Rapamycin, which blocks mitochondrial-based p70S6K activity, inhibits IGF-1-induced BAD S136 phosphorylation and cell survival. The p70S6K-deficient cells support the importance of IGF-1-induced BAD S136 phosphorylation by this kinase.
Materials and Methods
Purification of S136-Specific BAD Kinase.
The entire purification was done at 4°C or on ice. The crude membrane fraction from 20 liters of FL5.12 BCL-XL/BAD cells (about 5 × 1010 cells) was applied to a HiTrap Q column (Amersham Pharmacia) equilibrated with buffer A [20 mM Tris (pH 7.5)/100 mM NaCl/25 mM NaF/1 mM Na3VO4/10 mM β-glycerophosphate/1 mM DTT/0.15 unit per ml aprotinin/20 mM leupeptin/1 mM phenylmethylsulfonyl fluoride]. After washing with buffer A, bound proteins were eluted with a linear gradient of 0.1–0.5 M NaCl in buffer A. Fractions were assayed for BAD S136-specific kinase activity by using glutathione S-transferase (GST)-BAD S112A as a substrate. The active fractions were pooled and buffer was exchanged into buffer B [50 mM sodium phosphate (pH 7.0)/1 mM EDTA/1 mM DTT/0.05% Brij-35] by using a Sephadex G-25 column (Amersham Pharmacia). Appropriate fractions were loaded onto a HiTrap SP column (Amersham Pharmacia) equilibrated with buffer B. The active fractions were pooled, made 1 M in ammonium sulfate, and loaded onto a phenyl-Sepharose column (Amersham Pharmacia) equilibrated with buffer B containing 1 M ammonium sulfate. The column was eluted with a linear gradient of 1.0–0 M ammonium sulfate in buffer B. The active fractions were pooled and buffer was exchanged into buffer C [10 mM sodium phosphate (pH 6.8)/1 mM EDTA/1 mM DTT/0.05% Brij-35] by using a Sephadex G-25 column. Appropriate fractions were loaded onto a hydroxylapatite column (Bio-Rad) equilibrated with buffer C. The column was eluted with a linear gradient of 10–400 mM sodium phosphate in buffer C. The active fractions were pooled, the buffer was exchanged into buffer B, and the preparation was loaded onto a Mono S 5/5 column (Amersham Pharmacia). The column was eluted with a linear gradient of 0–0.5 M NaCl in buffer B. Then the active fractions were pooled, buffer was exchanged into buffer A, and the preparation was loaded onto a Mono Q 5/5 column (Amersham Pharmacia). The column was eluted with a linear gradient of 0–0.5 M NaCl in buffer A.
In-Gel Kinase Assay.
In-gel kinase assay was carried out essentially as described (26). GST-BAD S112A (0.3 mg/ml) was incorporated in a polyacrylamide gel as a substrate.
Transfection and Luciferase Assay.
Transient transfections were performed in Rat-1a cells with pcDNA3-derived constructs. The luciferase reporter plasmid (100 ng) was mixed with 50 ng of BAD expression vectors and 350 ng of p70S6K constructs. The plasmids were mixed with 3 μl of Fugene 6 (Roche) in a volume of 0.5 ml added to Rat-1a (1 × 105 cells). Twenty-four hours after transfection, serum was withdrawn for 16 h. Then cells were lysed and luciferase assays were performed by using a standard substrate (PharMingen). The reduction of the luciferase activity is because of cell death, as previously defined (9, 27).
Western Blot Analyses.
The analyses were carried out as described (12). The antibodies were obtained and diluted for use as follows: anti-p70S6K, Santa Cruz Biotechnology, 1:1000; anti-BAD, Santa Cruz Biotechnology, 1:250; anti-phospho-specific BAD, from M. Greenberg (Children's Hospital, Boston), 1:1000; anti-AKT, from P. Tsichlis (Kimmel Cancer Center, Philadelphia), 1:1000; anti-phospho-AKT, New England Biolabs, 1:1000.
In Vitro Kinase Assays.
The BAD kinase assays were carried out as described (21).
Results
Purification of p70S6K as the Membrane-Based BAD S136 Kinase.
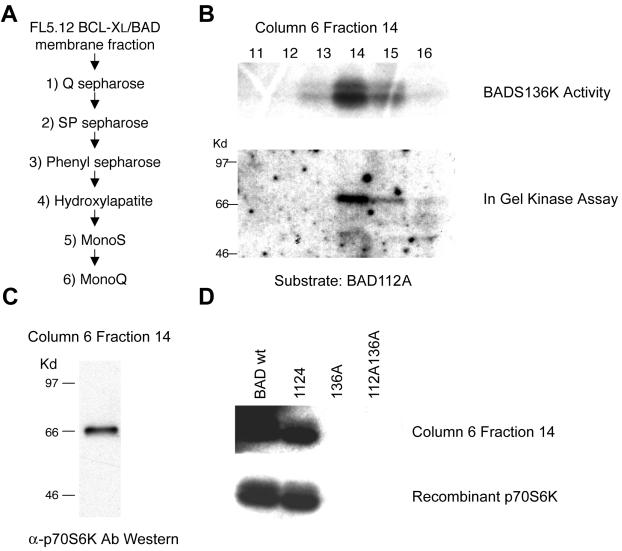
We turned to the membrane fraction of factor-dependent hematopoietic cells in which BAD kinase activity is predominantly localized to purify BAD S136-specific kinase(s). Kinase activity was enriched by fractionating this membrane preparation through a series of chromatographic purification steps, selecting for activity that phosphorylated a BAD S112A peptide, but not a BAD S112AS136A peptide (neither S112 nor S136 available) (Fig. 1A) (21). The peak kinase activity specific for the BAD S136 site was present principally in fraction 14 of the final Mono Q column (Fig. 1B). An in-gel kinase assay was performed with BAD S112A as a substrate, and it revealed a single band of ≈70 kDa in fraction 14 and weakly in fraction 15, consistent with the kinase activity of the corresponding fractions (Fig. 1B). Nano-electrospray mass spectroscopy of tryptic peptides derived from the corresponding sized band from a silver-stained gel (28) identified the protein as 70-kDa ribosomal protein S6 kinase (p70S6K). This identification included detection of a tryptic fragment unique to p70S6K that is not present in the alternative p60S6K2 enzyme (29). Subsequent Western blot analysis of fraction 14 confirmed the 70-kDa protein present as p70S6K (Fig. 1C). Finally, recombinant p70S6K displayed the same S136 specificity as the purified BAD kinase within fraction 14 (Fig. 1D).
Figure 1.
Purification of the BAD S136 specific kinase. (A) The purification scheme used for a BAD S136-specific kinase. (B Upper) Fractions 11–16 from the Mono Q column were assayed for BAD kinase activity with GST-BAD S112A as a substrate. (Lower) In-gel kinase assay was performed by using the same fractions as above with GST-BAD S112A as a substrate. (C) A Western blot of fraction 14 from the Mono Q column using an anti-p70S6K antibody. (D) Purified BAD kinase (Mono Q, fraction 14) was tested for its ability to phosphorylate GST-BAD wild-type (BAD wt), and GST-BAD whose S112, S136, or both were changed to Ala (112A, 136A, and 112A136A). Recombinant p70S6K (Upstate Biotechnology, Lake Placid, NY) was used to show S136 specificity of BAD phosphorylation in vitro.
Rapamycin, Which Blocks p70S6K, Inhibits IGF-1- but Not IL-3-Mediated Survival.
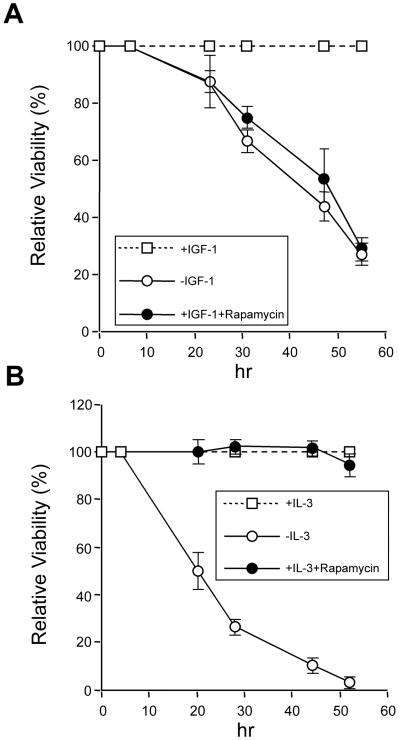
We next determined whether the candidate kinase p70S6K effected phosphorylation of BAD S136 in vivo and influenced cell survival. Rapamycin blocks p70S6K activity in cells by binding FKBP12, which complexes and inhibits a critical upstream kinase, termed “mTOR/FRAP” (30, 31). To examine whether p70S6K could mediate survival-factor signaling through phosphorylation of BAD S136, we assessed the effects of rapamycin on a BAD-dependent system. FL5.12 BCL-XL/BAD S112A cells provide this opportunity, as they possess an intact BAD S136 site. In this paradigm, cells were deprived of survival factor for 4 h to initiate a death signal. After that incubation, either IGF-1 or IL-3 was added back in the presence or absence of rapamycin. Rapamycin effectively inhibited IGF-1-dependent survival of FL5.12 BCL-XL/BAD S112A cells (Fig. 2A). Consistent with the specificity of p70S6K for BAD S136, rapamycin had no effect on IGF-1-mediated survival of FL5.12 BCL-XL/BAD S136A cells, which possess only the BAD S112 site (data not shown). Of note, rapamycin had no effect on IL-3-mediated survival in the very same cell, revealing that IL-3 induces kinase(s), in addition to p70S6K, that phosphorylate BAD on S136 and promote cell survival. The data suggest that rapamycin, which has been noted to interfere with IGF-1-mediated survival in additional cell types (32), mediates its effects in this system through p70S6K inactivation of BAD on S136.
Figure 2.
Inhibition of p70S6K activation induced by IGF-1 enhances cell death. FL5.12 BCL-XL/BAD S112A cells were deprived of IL-3 with or without rapamycin (20 ng/ml, Calbiochem) to initiate a death signal, whereas IGF-1 (50 ng/ml, Life Technologies) or IL-3 (10 ng/ml, Genzyme) was added back at 4 h before irreversible damage occurred. At each time point, viability was assessed by trypan blue exclusion. The IGF-1- or IL-3-dependent survival is compared with and without rapamycin. Data shown are representative of three independent experiments.
IGF-1 Induces Mitochondrial-Based p70S6K, Which Phosphorylates S136 on BAD.
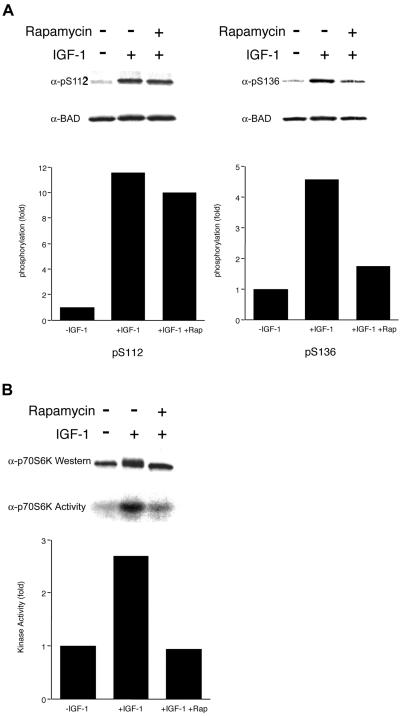
To examine whether IGF-1 induces the phosphorylation of BAD and whether rapamycin selectively prevents phosphorylation of S136, we directly assessed phosphorylation in cells in which only S136 or S112 was available. IGF-1 induced BAD phosphorylation at S112 as well as S136 sites, as detected by pS112 and pS136 phosphospecific antibodies. However, rapamycin strongly inhibited the pS136, but had no substantial effect on pS112 phosphorylation (Fig. 3A). These in vivo data are consistent with the in vitro observation that p70S6K phosphorylates only the S136 site (Fig. 1D). Consistent with cell survival studies, rapamycin did not substantially block IL-3-induced BAD phosphorylation on either S112 or S136 (data not shown).
Figure 3.
Rapamycin specifically inhibits IGF-1-induced BAD S136 phosphorylation and p70S6K activity in the mitochondrial fraction. (A) Survival factors were withdrawn from FL5.12 BCL-XL/BAD S112A or FL5.12 BCL-XL/BAD S136A cells for 4 h (−IGF-1), followed by addition of IGF-1 (50 ng/ml) for 15 min (+IGF-1). Rapamycin (20 ng/ml) was added when factors were withdrawn. Western blot analyses were done with antibodies that recognize phosphorylated forms of BAD at S112 or S136 (19, 22). The filters were reprobed with an antibody that recognizes BAD regardless of its phosphorylation state. Data shown are representative of two independent experiments. Histograms present the fold increase in intensity of the phosphorylated band relative to total BAD, in which − IGF-1 is set at 1.0. (B) FL5.12 BCL-XL/BAD S112A cells were treated as in A. The mitochondrial heavy membrane fractions were analyzed by Western blot with an anti-p70S6K Ab (Upper), or immunoprecipitated with an anti-p70S6K Ab, followed by an in vitro kinase assay using GST-S6 as substrate. Data shown are representative of two independent experiments. Histogram presents the fold increase in intensity of the phosphorylated S6.
We next examined whether IGF-1 would regulate the p70S6K activity associated with mitochondria. IGF-1 induced the activity of p70S6K in the mitochondrial fraction. The p70S6K activation was diminished to nearly basal levels by treating cells with rapamycin (Fig. 3B). In contrast, AKT, a kinase found in the cytosol that is also activated by IGF-1 treatment, was not affected by rapamycin (data not shown). In total, these results argue that mitochondria-associated p70S6K directly phosphorylates BAD S136 to promote cell survival.
p70S6K Deficiency Eliminates IGF-1-Induced Phosphorylation of BAD S136.
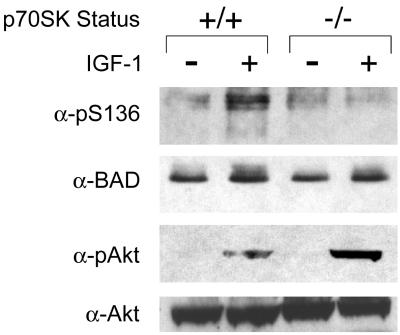
We turned to p70S6K-deficient cells to directly test the importance of this particular kinase in IGF-1-induced phosphorylation of BAD (29, 33). In wild-type embryonic stem (ES) cells, IGF-1 induced the phosphorylation of endogenous BAD on S136 (Fig. 4). In contrast, IGF-1 induced essentially no BAD S136 phosphorylation in p70S6K−/− ES cells. However, IGF-1-induced activation of AKT as assessed by an anti-phospho-AKT Ab was even stronger in p70S6K−/− cells. This in vivo loss of function study provides further support for p70S6K, rather than AKT in this system, as a kinase responsible for BAD S136 phosphorylation downstream of IGF-1.
Figure 4.
IGF-1-induced phosphorylation of BAD S136 is abolished in p70S6K-deficient cells. Serum was withdrawn from p70S6K+/+ or p70S6K−/− ES cells for 4 h (−IGF-1), followed by addition of IGF-1 (50 ng/ml) for 15 min (+IGF-1). Whole-cell extracts were immunoprecipitated with an anti-BAD antibody, followed by Western blot analyses with a phospho-S136-specific anti-BAD antibody or an antibody that recognizes BAD regardless of its phosphorylation state. Western blot analyses were also done by using an antibody that recognizes the phosphorylated form of AKT or an antibody that recognizes AKT regardless of its phosphorylation state. Data shown are representative of two independent experiments.
p70S6K Blocks BAD-Induced Apoptosis by Phosphorylation of S136.
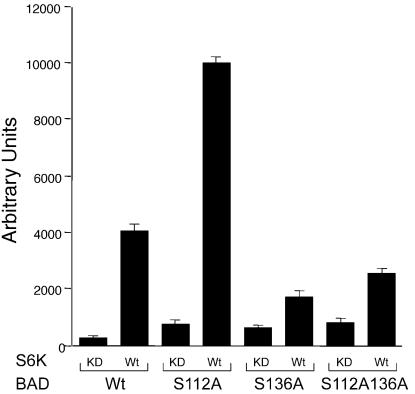
To assess whether p70S6K is capable of suppressing BAD-induced cell death in a S136 site-dependent manner, we compared the capacity of wild-type versus kinase-dead p70S6K expression to counter BAD in Rat-1a cells. BAD wild-type and serine-replaced mutants induced cell death in 80–90% of transfected cells. Wild-type, but not kinase dead p70S6K effectively suppressed the death induced by BAD wild-type or BAD S112A (Fig. 5). In contrast, death induced by BAD S136A or BAD S112AS136A was not substantially rescued by cotransfection with wild-type p70S6K. These results indicate that p70S6K can potently block BAD-induced apoptosis by phosphorylation of the S136 site. In total, these results indicate that mitochondria-associated p70S6K is a prominent kinase in a survival signaling pathway downstream of IGF-1, where it is responsible for inactivating BAD.
Figure 5.
p70S6K suppresses BAD-mediated death in a S136-dependent manner. Rat-1a cells were transiently cotransfected with a luciferase reporter together with constructs expressing either BAD wild-type (Wt), S112A, S136A, or S112AS136A together with constructs expressing either p70S6K Wt or a kinase-dead (KD) mutant (Lys-100 → Gln) that does not bind ATP. Twenty-four hours after transfection, serum was withdrawn for 16 h. Luciferase activity (ordinate, arbitrary units) has been shown to represent the viability of the cells defined (9, 27), and this assay is representative of three independent experiments.
Discussion
It has been shown that “BH3 domain-only” pro-apoptotic members connect proximal death and survival signals to the core apoptotic pathway at the level of the “multidomain” BCL-2 family members (34). For example, BID proved important in hepatocytes for cell death after Fas activation in vivo (35). BIM is important in neuronal as well as hematopoietic cells for cell death after survival factor withdrawal (36, 37). These results, coupled with the observations on BAD, indicate that each BH3 domain-only member plays a distinct role in selected cell types after specified death and survival signals.
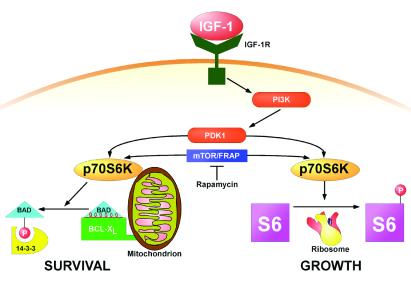
A model factor, IGF-1, promotes cell growth, including volume control, protein synthesis and metabolic rate (1, 38), as well as represses apoptosis (2, 3) (Fig. 6). IGF-1 activates downstream p70S6K by multisite phosphorylation, which includes phosphoinositide-dependent protein kinase 1 phosphorylating the activation loop of p70S6K as well as rapamycin-sensitive mTOR/FRAP, which appears to regulate T389 phosphorylation (30, 31, 39–41). Although an immunosuppressive agent, rapamycin, has also been reported to induce apoptosis in multiple cell types (32, 42–46), the elimination of a p70S6K survival pathway by rapamycin as defined here provides a mechanistic explanation for the noted apoptosis. This expands the role of p70S6K beyond its documented affects by means of S6 to increase ribosomal biosynthesis, to now include cell survival. These observations should promote a detailed search for the reciprocal effects of BAD and p70S6K on selected cells in whole animals. Mitochondrial-based BAD represents one bona fide substrate for p70S6K that resides in the apoptotic pathway. Additional mitochondrial substrates in the apoptotic or perhaps metabolic growth pathways may well exist. Of note, evolving evidence indicates that a unique 55–60S subset of ribosomes is present in mammalian mitochondria, perhaps associated with the inner membrane, which may represent a mitochondrial protein biosynthetic system (47). Although p70S6K appears to perform a focused kinase/substrate interaction at this organelle (48, 49), it does not become an integral mitochondrial membrane protein (data not shown), suggesting it is not localized to any internal ribosome and may prove more analogous to protein kinase A, which is part of a mitochondrial-tethered complex that phosphorylates the BAD S112 site (21). Other evidence indicates that ribosomes synthesizing nuclear-encoded mitochondrial proteins may bind specifically to the surface of mitochondria, providing an alternative mechanism whereby p70S6K could have mitochondrial as well as ribosomal substrates (50). p70S6K, which has been proposed to serve as a key coordinator between ribosome biogenesis and cell-cycle progression (1, 38, 51), may also help to integrate apoptosis with these processes. We propose that the dual pathway capability of p70S6K evolved as a tightly coupled mechanism to ensure the inhibition of apoptosis during periods of increased metabolic stress that accompany cell growth.
Figure 6.
p70S6K as a dual pathway kinase. A schematic diagram of survival and growth substrates of S6K.IGF-1R, IGF-1 receptor; PI3K, phosphatidylinositol 3-kinase; PDK-1, 3-phosphoinositide-dependent protein kinase 1.
Acknowledgments
We are grateful to M. Greenberg and P. Tsichlis for providing the phosphospecific BAD antibodies and anti-AKT antibody, respectively. We thank E. D. Smith for preparing the manuscript. H.H. is a Special Fellow of the Leukemia and Lymphoma Society. This work was supported by National Institutes of Health Grant CA50239-13. Work in M.M.'s laboratory is funded by a generous grant from the Danish National Research Foundation to the Center for Experimental Bioinformatics.
Abbreviations
- IGF-1
insulin-like growth factor 1
- p70S6K
70-kDa ribosomal protein S6 kinase
- ES cells
embryonic stem cells
References
- 1.Conlon I, Raff M. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 2.Harrington E A, Bennett M R, Fanidi A, Evan G I. EMBO J. 1994;13:3286–3295. doi: 10.1002/j.1460-2075.1994.tb06630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peruzzi F, Prisco M, Dews M, Salomoni P, Grassilli E, Romano G, Calabretta B, Baserga R. Mol Cell Biol. 1999;19:7203–7215. doi: 10.1128/mcb.19.10.7203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yao R, Cooper G M. Science. 1995;267:2003–2006. doi: 10.1126/science.7701324. [DOI] [PubMed] [Google Scholar]
- 5.Kinoshita T, Yokota T, Arai K, Miyajima A. EMBO J. 1995;14:266–275. doi: 10.1002/j.1460-2075.1995.tb07000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams J M, Cory S. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 7.Gross A, McDonnell J M, Korsmeyer S J. Genes Dev. 1999;13:1899–1911. doi: 10.1101/gad.13.15.1899. [DOI] [PubMed] [Google Scholar]
- 8.Yang E, Zha J, Jockel J, Boise L H, Thompson C B, Korsmeyer S J. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 9.Zha J, Harada H, Osipov K, Jockel J, Waksman G, Korsmeyer S J. J Biol Chem. 1997;272:24101–24104. doi: 10.1074/jbc.272.39.24101. [DOI] [PubMed] [Google Scholar]
- 10.Kelekar A, Chang B S, Harlan J E, Fesik S W, Thompson C B. Mol Cell Biol. 1997;17:7040–7046. doi: 10.1128/mcb.17.12.7040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ottilie S, Diaz J L, Horne W, Chang J, Wang Y, Wilson G, Chang S, Weeks S, Fritz L C, Oltersdorf T. J Biol Chem. 1997;272:30866–30872. doi: 10.1074/jbc.272.49.30866. [DOI] [PubMed] [Google Scholar]
- 12.Zha J, Harada H, Yang E, Jockel J, Korsmeyer S J. Cell. 1996;87:619–628. doi: 10.1016/s0092-8674(00)81382-3. [DOI] [PubMed] [Google Scholar]
- 13.Datta S R, Katsov A, Hu L, Petros A, Fesik S W, Yaffe M B, Greenberg M E. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- 14.Zhou X M, Liu Y, Payne G, Lutz R J, Chittenden T. J Biol Chem. 2000;275:25046–25051. doi: 10.1074/jbc.M002526200. [DOI] [PubMed] [Google Scholar]
- 15.Tan Y, Demeter M R, Ruan H, Comb M J. J Biol Chem. 2000;275:25865–25869. doi: 10.1074/jbc.M004199200. [DOI] [PubMed] [Google Scholar]
- 16.Lizcano J M, Morrice N, Cohen P. Biochem J. 2000;349:547–557. doi: 10.1042/0264-6021:3490547. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Virdee K, Parone P A, Tolkovsky A M. Curr Biol. 2000;10:1151–1154. doi: 10.1016/s0960-9822(00)00702-8. [DOI] [PubMed] [Google Scholar]
- 18.Downward J. Nat Cell Biol. 1999;1:E33–E35. doi: 10.1038/10026. [DOI] [PubMed] [Google Scholar]
- 19.Bonni A, Brunet A, West A E, Datta S R, Takasu M A, Greenberg M E. Science. 1999;286:1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 20.Shimamura A, Ballif B A, Richards S A, Blenis J. Curr Biol. 2000;10:127–135. doi: 10.1016/s0960-9822(00)00310-9. [DOI] [PubMed] [Google Scholar]
- 21.Harada H, Becknell B, Wilm M, Mann M, Huang L J, Taylor S S, Scott J D, Korsmeyer S J. Mol Cell. 1999;3:413–422. doi: 10.1016/s1097-2765(00)80469-4. [DOI] [PubMed] [Google Scholar]
- 22.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 23.del Peso L, González-García M, Page C, Herrera R, Nuñez G. Science. 1997;278:687–689. doi: 10.1126/science.278.5338.687. [DOI] [PubMed] [Google Scholar]
- 24.Blume-Jensen P, Janknecht R, Hunter T. Curr Biol. 1998;8:779–782. doi: 10.1016/s0960-9822(98)70302-1. [DOI] [PubMed] [Google Scholar]
- 25.Schurmann A, Mooney A F, Sanders L C, Sells M A, Wang H G, Reed J C, Bokoch G M. Mol Cell Biol. 2000;20:453–461. doi: 10.1128/mcb.20.2.453-461.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kameshita I, Fujisawa H. Anal Biochem. 1989;183:139–143. doi: 10.1016/0003-2697(89)90181-4. [DOI] [PubMed] [Google Scholar]
- 27.Wang K, Yin X M, Chao D T, Milliman C L, Korsmeyer S J. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 28.Shevchenko A, Wilm M, Vorm O, Mann M. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 29.Shima H, Pende M, Chen Y, Fumagalli S, Thomas G, Kozma S C. EMBO J. 1998;17:6649–6659. doi: 10.1093/emboj/17.22.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peterson R T, Schreiber S L. Curr Biol. 1998;8:R248–R250. doi: 10.1016/s0960-9822(98)70152-6. [DOI] [PubMed] [Google Scholar]
- 31.Dufner A, Thomas G. Exp Cell Res. 1999;253:100–109. doi: 10.1006/excr.1999.4683. [DOI] [PubMed] [Google Scholar]
- 32.Hosoi H, Dilling M B, Shikata T, Liu L N, Shu L, Ashmun R A, Germain G S, Abraham R T, Houghton P J. Cancer Res. 1999;59:886–894. [PubMed] [Google Scholar]
- 33.Kawasome H, Papst P, Webb S, Keller G M, Johnson G L, Gelfand E W, Terada N. Proc Natl Acad Sci USA. 1998;95:5033–5038. doi: 10.1073/pnas.95.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei M C, Zong W X, Cheng E H, Lindsten T, Panoutsakopoulou V, Ross A J, Roth K A, MacGregor G R, Thompson C B, Korsmeyer S J. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yin X M, Wang K, Gross A, Zhao Y, Zinkel S, Klocke B, Roth K A, Korsmeyer S J. Nature (London) 1999;400:886–891. doi: 10.1038/23730. [DOI] [PubMed] [Google Scholar]
- 36.Putcha G V, Moulder K L, Golden J P, Bouillet P, Adams J A, Strasser A, Johnson E M. Neuron. 2001;29:615–628. doi: 10.1016/s0896-6273(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 37.Bouillet P, Metcalf D, Huang D C, Tarlinton D M, Kay T W, Kontgen F, Adams J M, Strasser A. Science. 1999;286:1735–1738. doi: 10.1126/science.286.5445.1735. [DOI] [PubMed] [Google Scholar]
- 38.Neufeld T P, de la Cruz A F, Johnston L A, Edgar B A. Cell. 1998;93:1183–1193. doi: 10.1016/s0092-8674(00)81462-2. [DOI] [PubMed] [Google Scholar]
- 39.Alessi D R, Kozlowski M T, Weng Q P, Morrice N, Avruch J. Curr Biol. 1998;8:69–81. doi: 10.1016/s0960-9822(98)70037-5. [DOI] [PubMed] [Google Scholar]
- 40.Pullen N, Dennis P B, Andjelkovic M, Dufner A, Kozma S C, Hemmings B A, Thomas G. Science. 1998;279:707–710. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 41.Williams M R, Arthur J S, Balendran A, van der Kaay J, Poli V, Cohen P, Alessi D R. Curr Biol. 2000;10:439–448. doi: 10.1016/s0960-9822(00)00441-3. [DOI] [PubMed] [Google Scholar]
- 42.Gottschalk A R, Boise L H, Thompson C B, Quintans J. Proc Natl Acad Sci USA. 1994;91:7350–7354. doi: 10.1073/pnas.91.15.7350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi Y, Frankel A, Radvanyi L G, Penn L Z, Miller R G, Mills G B. Cancer Res. 1995;55:1982–1988. [PubMed] [Google Scholar]
- 44.Muthukkumar S, Ramesh T M, Bondada S. Transplantation. 1995;60:264–270. doi: 10.1097/00007890-199508000-00010. [DOI] [PubMed] [Google Scholar]
- 45.Ishizuka T, Sakata N, Johnson G L, Gelfand E W, Terada N. Biochem Biophys Res Commun. 1997;230:386–391. doi: 10.1006/bbrc.1996.5967. [DOI] [PubMed] [Google Scholar]
- 46.Koh J S, Lieberthal W, Heydrick S, Levine J S. J Clin Invest. 1998;102:716–727. doi: 10.1172/JCI1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu M, Spremulli L. J Biol Chem. 2000;275:29400–29406. doi: 10.1074/jbc.M002173200. [DOI] [PubMed] [Google Scholar]
- 48.Hubbard M J, Cohen P. Trends Biochem Sci. 1993;18:172–177. doi: 10.1016/0968-0004(93)90109-z. [DOI] [PubMed] [Google Scholar]
- 49.Pawson T, Scott J D. Science. 1997;278:2075–2080. doi: 10.1126/science.278.5346.2075. [DOI] [PubMed] [Google Scholar]
- 50.Crowley K S, Payne R M. J Biol Chem. 1998;273:17278–17285. doi: 10.1074/jbc.273.27.17278. [DOI] [PubMed] [Google Scholar]
- 51.Thomas G. Nat Cell Biol. 2000;2:E71–E72. doi: 10.1038/35010581. [DOI] [PubMed] [Google Scholar]