ABSTRACT
Tau protein is involved in numerous human neurodegenerative diseases, and Tau hyper-phosphorylation has been linked to Tau aggregation and toxicity. Previous studies have addressed toxicity and phospho-biology of human Tau (hTau) in Drosophila melanogaster. However, hTau transgenes have most often been randomly inserted in the genome, thus making it difficult to compare between different hTau isoforms and phospho-mutants. In addition, many studies have expressed hTau also in mitotic cells, causing non-physiological toxic effects. Here, we overcome these confounds by integrating UAS-hTau isoform transgenes into specific genomic loci, and express hTau post-mitotically in the Drosophila nervous system. Lifespan and locomotor analyses show that all six of the hTau isoforms elicit similar toxicity in flies, although hTau2N3R showed somewhat elevated toxicity. To determine if Tau phosphorylation is responsible for toxicity, we analyzed the effects of co-expressing hTau isoforms together with Tau-kinases, focusing on TTBK1, TTBK2 and MARK1. We observed toxicity when expressing each of the three kinases alone, or in combination. Kinase toxicity was enhanced by hTau co-expression, with strongest co-toxicity for TTBK1. Mutagenesis and phosphorylation analysis indicates that hTau-MARK1 combinatorial toxicity may be due to direct phosphorylation of hTau, while hTau-TTBK1/2 combinatorial toxicity may result from independent toxicity mechanisms.
KEY WORDS: Tauopathy, Neuronal toxicity, Drosophila, Tau-kinases
Summary: Tau hyper-phosphorylation has been linked to toxicity, but the Tau isoforms, kinases and residues remain unclear. Using the Drosophila model, we find evidence for involvement of TTBK and MARK kinases.
INTRODUCTION
Tau is a neuronal, microtubule-associated protein that plays a key role during microtubule assembly and stabilization. Tau is also heavily implicated in human disease, in particular in Alzheimer's disease (AD) where intracellular neurofibrillary tangles (NFTs) of hyper-phosphorylated human Tau (hTau) is one of the two major histopathological hallmarks found in patients with the disease (Braak and Braak, 1991). Additional tauopathies have been identified, characterized by intracellular aggregates of hyper-phosphorylated hTau as well as mutations in hTau, e.g. fronto-temporal dementia with parkinsonism-17 (FTDP-17), Pick's disease and corticobasal degeneration (CBD) (Coppola et al., 2012; Kouri et al., 2014). Although the pathophysiology of hTau in these neurodegenerative diseases has been intensively studied over recent years, its contribution to disease initiation and progression remains unclear.
In the adult human brain, there are six main hTau isoforms, ranging from 352 to 441 amino acids, which are differentially expressed during development (Goedert et al., 1989; Takemura et al., 1991; Takuma et al., 2003). These isoforms derive from alternative mRNA splicing of the hTau gene (van Slegtenhorst et al., 2000), and differ from each other in the absence or presence of two N-terminal domains (N1, N2), and three or four carboxy-terminal microtubule binding domains (R1-R4) (Fig. 1A). These isoforms may have specific roles during development, for example with regards to axonal generation and their role in microtubule cytoskeleton stability (van Slegtenhorst et al., 2000). However, it has also become clear that specific isoforms are either more abundant or absent from brains of patients with various forms of dementia (Andreadis, 2012; Niblock and Gallo, 2012). Tau has an unusually large proportion of phosphorylation sites; the 2N4R isoform has a total of 441 amino acids with 85 serines, threonines and tyrosines, 45 of which have been observed to be phosphorylated (Hanger et al., 2009). Tau phosphorylation is also regulated developmentally, and during early fetal development Tau is highly phosphorylated and modified at many sites, broadly similar to the phosphorylation status of Tau in AD (van Slegtenhorst et al., 2000). Interestingly, although highly phosphorylated, Tau is functional in the fetus and does not form NFTs as it does in AD.
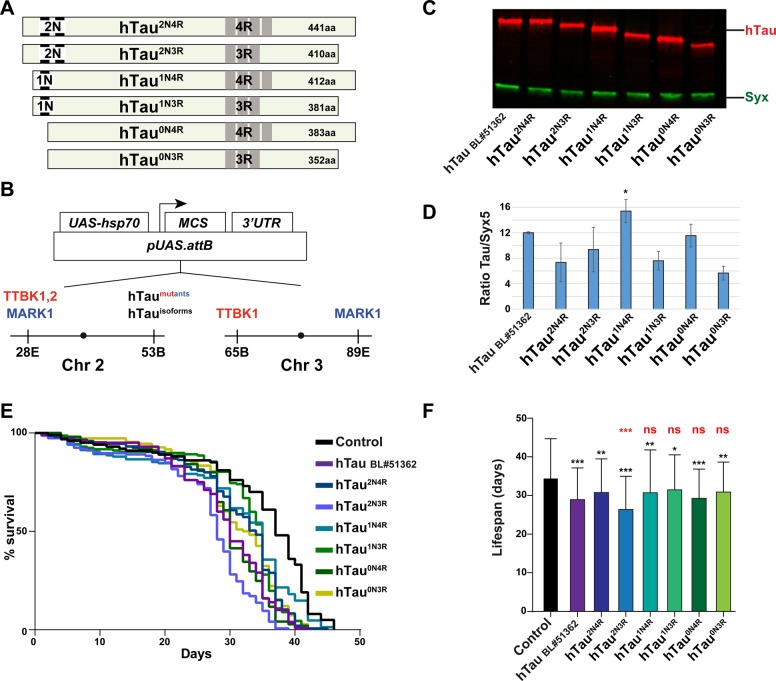
Fig. 1.
Expression of human Tau isoforms in post-mitotic neurons results in reduced lifespan in Drosophila. (A) Schematic diagram of the six main hTau isoforms, which vary in the N-terminal domains and the R-repeats. (B) Constructs were inserted in the pUAS.attB vector, and landed at the indicated attP landing sites. (C) Western blot analysis of hTau protein levels in fly head extracts, using Odyssey CLX Licor system. Syntaxin (Syx) was used as a loading control. (D) Quantification of protein expression using Licor Image Studio software, showing average signal ratio of hTau/Syx (three independent experiments, 20 adult heads each; asterisks depict pair-wise comparison with hTau2N4R; * P≤0.05; Student's two-tailed t-test; mean±s.d.). (E,F) Lifespan assay measuring the toxicity of the six common hTau isoforms, landed in the same genomic location, driven by the post-mitotic, pan-neuronal n-Syb-Gal4 driver. Results are depicted as Kaplan–Meier survival curves (E) (Kaplan and Meier, 1958) and bar graph (F). Control flies (n-Syb-Gal4 crossed to the attP landing site strain, BL#9750) were compared to a previously published human Tau2N4R construct (UAS-hTau BL#51362), and all six isoforms of UAS-hTau landed on chromosome 2 (53B). Three independent experiments were performed, which gave similar results, and hence the graphs show the average lifespan of the combination of the three experiments (≥100 flies). Error bars in (F) indicate mean±s.d.; black asterisks depict pair-wise comparison with control, and red asterisks depict pair-wise comparison with hTau2N4R (*P≤0.05, **P≤0.01, ***P≤0.001; Student's two-tailed t-test); ns, not significant.
Several protein kinases and phosphatases regulate Tau phosphorylation state (Wang and Mandelkow, 2016). Genetic screens in Drosophila and Caenorhabditis elegans have identified several kinases as suppressors/enhancers of Tau toxicity (Shulman and Feany, 2003; Kraemer et al., 2006; Ambegaokar and Jackson, 2011). Among these are the microtubule affinity regulating kinase (MARK) and Tau tubulin binding kinase1 and 2 (TTBK1, TTBK2).
Drosophila has been extensively used to study Tau biology due to its sophisticated genetic tools, short lifespan, relative low cost, and the powerful Gal4/UAS transgenic expression system (Gistelinck et al., 2012; Fernandez-Funez et al., 2015; Sun and Chen, 2015). However, previous studies mostly used P element UAS transgenes, which randomly insert into the genome and hence are susceptible to position effects and copy number complications. This problem makes it difficult to compare numbers of Tau isoforms and conduct controlled structure-function studies. Moreover, many studies have relied on expressing hTau both in progenitor and post-mitotic cells, using the eye Gal4 driver GMR-Gal4, expressed throughout eye disc development, or the nervous system driver elav-Gal4, which also expresses in actively dividing neural progenitor cells (Freeman, 1996; Berger et al., 2007). Because Tau is a microtubule-binding protein, it will likely interfere with mitosis in cycling cells, a notion that was recently confirmed by detailed studies in the wing disc (Bouge and Parmentier, 2016).
To circumvent these problems we have (1) used attP UAS ‘landing sites’ to avoid position and copy number confounds (Bischof et al., 2007), and (2) used post-mitotic neuronal Gal4 drivers, either expressed throughout life or specifically induced in adults. Combined, this enables precise comparative structure-function analysis of hTau isoforms and phospho-sites, and addresses the effects of various kinases on toxicity. Using these improvements, we addressed hTau isoform toxicity, and found similar toxicity for all of the six main hTau isoforms, although hTau2N3R showed somewhat elevated toxicity. We furthermore analyzed the toxic effects of co-expressing hTau with three Tau-directed kinases: TTBK1, TTBK2, and MARK1. We observed combinatorial hTau-kinase toxicity, as judged by reduced lifespan and locomotor activity, in particular when co-expressing the TTBK1 kinase along with hTau. Focusing of TTBK1 and hTau, we find similar combinatorial toxicity for all six hTau isoforms. Phospho-residue mutagenesis of hTau, as well as phosphorylation analysis, indicates that in the case of MARK1, combinatorial toxicity may be due to kinase acting directly upon hTau. In contrast, the enhanced toxicity with TTBK1, -2, does not appear to be related to altered hTau phosphorylation.
RESULTS
Expression of the six main human Tau isoforms in post-mitotic neurons in Drosophila results in moderate toxicity
We generated DNA constructs for the six main hTau isoforms (Fig. 1A) by gene synthesis. Open reading frames were codon-optimized for Drosophila expression, and a ‘Cavener start-ATG’ was added to the 5′ (Cavener and Ray, 1991) (see Supplementary Materials). Constructs were inserted into the pUAS.attB landing site transgenic vector (Bischof et al., 2007) and all six isoform transgenes were landed at the same site on chromosome 2 (53B) (Fig. 1B). To enable robust, post-mitotic expression in most neurons in the nervous system, we used the previously described n-Syb-Gal4 driver; a driver where Gal4 expression is under control of the neuronal Synaptobrevin promoter (Jonson et al., 2015). Combined with landing site transgenesis, this enables quantitative comparison of toxicity of the six different hTau isoforms in post-mitotic neurons.
Immunoblot analysis indicated robust expression of all six isoforms of hTau, with similar expression levels, although hTau1N4R did show a significantly increased protein expression when compared to the hTau2N4R (Fig. 1C,D). Lifespan experiments revealed that all six hTau isoforms showed significant reduction of lifespan when compared to control flies (n-Syb-Gal4 crossed to the attP landing site strain BL#9750), with evidence for a somewhat elevated toxicity for hTau2N3R when compared to the hTau2N4R (Fig. 1E,F). We also analyzed a previously published human UAS-Tau transgene for comparison (Chatterjee et al., 2009). This transgene displayed expression levels and a reduction in lifespan that was generally comparable to our novel transgenes (Fig. 1C-F).
Expression of the TTBK1, TTBK2 and MARK1 hTau-directed kinases reveals strong toxicity for TTBK1 and TTBK2
The low toxicity observed upon post-mitotic expression of hTau in the Drosophila CNS facilitated the systematic testing of combinatorial toxicity of hTau when co-expressed with hTau-directed kinases; TTBK1, TTBK2 and MARK1 kinases were identified in genetic screens (Shulman and Feany, 2003; Kraemer et al., 2006; Ambegaokar and Jackson, 2011). We generated UAS attB transgenes for these kinases (Fig. 2A), codon-optimized for expression in Drosophila, and tagged at N- and C-terminals with Myc and HA epitope tags, respectively (Fig. 2A; see Supplementary Materials). All three kinase transgenes were integrated into the same site on chromosome 2 (28E) (Fig. 1B). To assess the combinatorial contribution of multiple kinases to hTau-related toxicity, TTBK1 and MARK1 kinase transgenes were also integrated at site 65B or 89E, respectively, on chromosome 3 (Fig. 1B).
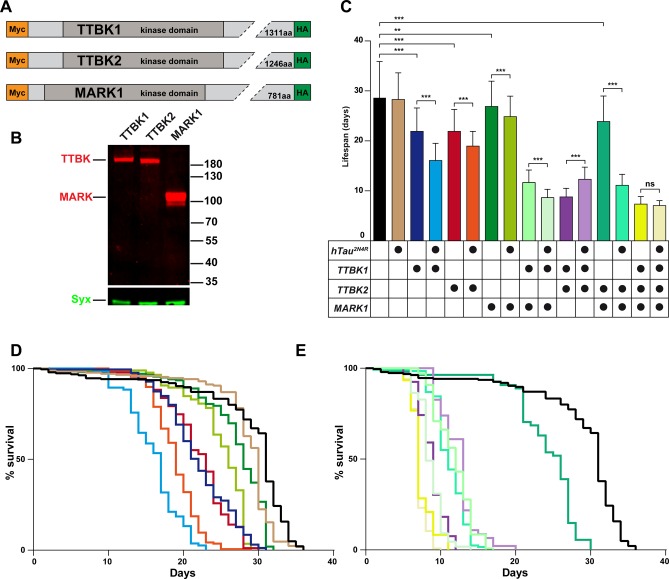
Fig. 2.
MARK1 and TTBK1, TTBK2 kinases reduce lifespan, and the effects are increased by co-expression with hTau. (A) Human MARK and TTBK kinase constructs; a c-Myc and HA antibody epitope tag was fused to the N- or C-terminus, for detection by western blot. (B) Western blot analysis of fly head extracts expressing human TTBK or MARK kinases. Kinases were detected with anti-HA antibody, and Syx was used as loading control. There is robust protein expression of all three kinases. TTBK1 and TTBK2 migrated substantially slower than their molecular weight. (C-E) Lifespan analysis of flies expressing kinases with or without co-expression of hTau2N4R, using the inducible n-Syb-Gal4;Gal80[ts] driver. Results are depicted as Kaplan–Meier survival curves (D,E) (Kaplan and Meier, 1958) and bar graph (C). Flies were crossed and reared at 20°C until they hatched, then transferred to 29°C. Using this inducible driver, there was no significant reduction in lifespan of hTau2N4R when compared to control flies (n-Syb-Gal4;Gal80[ts] crossed to the attP landing site strain BL#9750). However, all other flies showed significantly reduced lifespan when compared to control flies, including flies expressing each of the three kinases alone. There was combinatorial toxicity when co-expressing hTau2N4R with hTau-directed kinases TTBK1, TTBK2 or MARK1. The graphs show the average lifespan from a combination of 3-5 independent experiments (≥46 flies). Error bars in (C) indicate mean±s.d. (*P≤0.05, **P≤0.01, ***P≤0.001; ANOVA multiple comparison Tukey's test).
First, we analyzed protein expression using western blot of fly head protein extracts, followed by detection using the epitope antibodies, and noted that all three kinases were readily detectable (Fig. 2B). Next, we addressed the toxicity of each kinase when expressed alone, without hTau, by crossing them to the n-Syb-Gal4 driver. Expression of TTBK1 kinase was larval lethal, with no adult flies emerging, and expression of TTBK2 kinase also elicited severe toxic effects with very few adults emerging. These results indicate that broad post-mitotic neuronal expression of TTBK1 and TTBK2 kinases is toxic to the fly, even without co-expression of hTau.
Inducible expression of TTBK1, TTBK2 and MARK1 kinases along with hTau2N4R results in combinatorial toxicity
Because of complete lethality of TTBK1 and partial lethality of TTBK2 using the n-Syb-Gal4 driver, we generated an inducible driver to enable development into adulthood before inducing expression. To achieve this, we used Gal80[ts], a temperature-sensitive Gal80 allele that represses Gal4 protein activity at 20°C, but allows Gal4 activity at 29°C (McGuire et al., 2003). We generated flies carrying two copies of n-Syb-Gal4 and two copies tubP-Gal80[ts] transgenes, for robust Gal4 and Gal80[ts] activities. These flies were crossed to UAS-eGFP, and the resulting progeny did not express eGFP if maintained at 18°C, but eGFP was strongly induced in all neurons after shifting flies to 29°C as adults (Fig. S1). Crossing this driver to the UAS-TTBK1 and UAS-TTBK2 kinase transgenes allowed adults to emerge, enabling comparison of toxicity with UAS-hTau alone. Because kinases are known to activate each other by priming phosphorylation, we generated flies expressing one, two or three kinases, with or without hTau to analyze combinatorial toxic effects on hTau.
Focusing first on the kinases, using the inducible n-Syb-Gal4;Gal80[ts] driver we noted reduced lifespan as a result of expressing the TTBK1, TTBK2 or MARK1 kinases, when compared to control (n-Syb-Gal4;Gal80[ts] crossed to the attP landing site strain BL#9750) (Fig. 2C-E; Table S1). Because kinases have been found to prime each other by phosphorylating neighboring residues in proteins (Cesaro and Pinna, 2015), we generated flies carrying multiple UAS kinase transgenes. Flies expressing multiple kinases showed enhanced toxicity compared to fly strains expressing a single kinase. This was true both for TTBK1-TTBK2 co-expression, as well as for TTBK1-MARK1 co-expression. Surprisingly, TTBK2-MARK1 co-expression did however not show enhanced toxicity (Fig. 2C-E; Table S1).
Turning next to hTau-kinase co-expression, we first noted that hTau2N4R expression alone, using adult-induced Gal4 activity, caused no significant reduction in lifespan. However, adult-induced co-expression of the TTBK1, TTBK2 or MARK1 kinase with hTau2N4R showed a significant combinatorial toxicity (Fig. 2C-E; Table S1). This combinatorial effect was strongest for TTBK1. Co-expression of both TTBK1 and MARK1 kinases with hTau2N4R resulted in further increased toxicity. Similarly, TTBK2-MARK1 co-expression, which was weakly toxic on its own, showed severe toxicity together with hTau2N4R (Fig. 2C-E; Table S1). In contrast, the strong toxicity of TTBK1, TTBK2 co-expression was somewhat alleviated by co-expression of hTau2N4R. Expression of the three kinases together was strongly toxic, and was not further enhanced by co-expression of hTau2N4R. These results reveal that co-expression of kinases results in combinatorial toxicity, and that co-expression of kinases together with hTau2N4R results in enhanced combinatorial toxicity.
All six hTau isoforms are combinatorially toxic with TTBK1
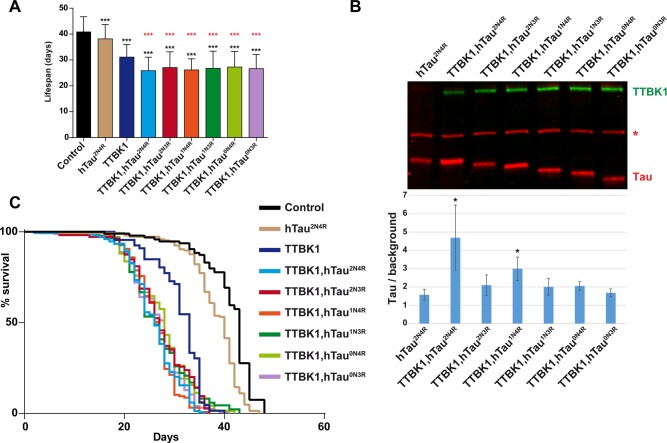
The above experiments were conducted using the 2N4R isoform of hTau. Next, we tested whether the combinatorial hTau-kinase toxicity was similar for all six different hTau isoforms, focusing on the TTBK1 kinase, due to the strong effects of TTBK1 on lifespan. To this end, the other five isoforms, landed in the same genomic location as 2N4R, were also combined with TTBK1. We noted similar combinatorial effects on lifespan between TTBK1 and all six hTau isoforms when compared to control (n-Syb-Gal4;Gal80[ts] crossed to the attP landing site strain BL#9750) (Fig. 3A,C). Analyzing hTau protein levels we noted significantly elevated hTau2N4R and hTau1N4R protein levels when co-expressed with TTBK1 (Fig. 3B).
Fig. 3.
TTBK1 toxicity is enhanced by all six hTau isoforms. (A,B) Lifespan and western blot analysis of flies expressing TTBK1 with hTau isoforms, using the inducible n-Syb-Gal4;Gal80[ts] driver. Results are depicted as Kaplan–Meier survival curves (C) (Kaplan and Meier, 1958) and bar graph (A). (A,C) All six hTau isoforms showed significantly shortened lifespan when co-expressed with TTBK1, when compared to control flies (n-Syb-Gal4;Gal80[ts] crossed to the attP landing site strain BL9750). Error bars in (A) indicate mean±s.d.; black asterisks depict pair-wise comparison with control, and red asterisks depict pair-wise comparison with TTBK1 (***P≤0.001; Student's two-tailed t-test; n=3 independent experiments; ≥70 flies). (B) Western blot, using Licor, of fly head extracts after 5 days expression of the transgenes at 29°C. TTBK1 kinase was detected using anti-HA antibody, hTau was detected using a rabbit anti-hTau antibody (asterisk denotes antibody background reactivity to an endogenous Drosophila protein). We observed increased hTau levels in the hTau2N4R and hTau1N4R isoforms when co-expressed with TTBK1. Bar graph in B represents the average value of three independent gels, showing the value of the ratio of Tau/ background. Error bars indicate mean±s.d. and black asterisks depict pair-wise comparison with hTau2N4R.
Co-expression of TTBK1, TTBK2 or MARK1 kinase with hTau2N4R in motor neurons resulted in combinatorial locomotor defects
To further analyze combinatorial hTau-kinase toxicity, we complemented the lifespan assays with a motor output behavioral assay. For these studies we used OK371-Gal4, which drives expression in most/all fly motor neurons (Mahr and Aberle, 2006). We used a negative geotaxis assay, where the flies' ability to climb up the side of a vial is assessed, at days 1, 7 and 14 after emerging as adults. Briefly, the percentage of flies climbing up to a 5 cm mark on the vial within 30 s was quantified (see Materials and Methods). Flies expressing hTau2N4R alone did not show any significant effect upon climbing ability when compared to control at Day 1 and 7, but a minor effect at Day 14 (Fig. 4). In contrast, flies expressing TTBK1 alone displayed significantly reduced climbing ability when compared to control (OK371-Gal4 crossed to the attP landing site strain BL#9750), apparent already at Day 1 and more pronounced at Day 7. By Day 14, all TTBK1 flies had died (Fig. 4). Expression of TTBK2 alone showed milder but significant effects, apparent only at Day 14, while MARK1 alone showed no effects (Fig. 4).
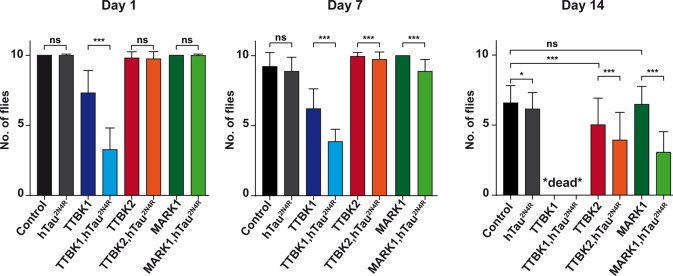
Fig. 4.
Combinatorial geotaxis effects between hTau and TTBK/MARK kinases. Negative geotaxis assay shows combinatorial effects from co-expressing hTau2N4R with TTBK1, TTBK2 or MARK1 kinase, expressed using the OK371-Gal4 driver. Flies were assessed for climbing ability at Day 1, 7 and 14. Ten empty vials with ten flies per vial were counted ten times each, and the average number of flies per vial that climbed up to the 5 cm mark within 30 s is plotted in the graphs (error bars indicate mean±s.d.; *P≤0.05, **P≤0.01, ***P≤0.001; Student's two-tailed t-test; n=3 independent experiments; ≥100 flies). Expression of each kinase alone revealed effects of TTBK1 at Day 1 and Day 7, and for TTBK2 at Day 14, when compared to control flies (OK371-Gal4 crossed to the attP landing site strain BL#9750). At Day 14, TTBK1 flies had died. MARK1 expression alone showed no toxicity. Co-expression revealed that on Day 1 there was significant difference between flies expressing TTBK1 when compared to TTBK1-hTau2N4R flies, and this effect persisted into Day 7; by Day 14 these genotypes had died. On Day 7 there was significant difference between flies expressing MARK1 when compared to MARK1-hTau2N4R, as well as between TTBK2 and TTBK2-hTau2N4R. These effects persisted into Day 14. Expression of hTau2N4R alone showed significant only at Day 14. ns, not significant.
Next, we turned to addressing possible combinatorial effects. We found that the climbing ability of flies expressing TTBK1, TTBK2 or MARK1 kinases were all significantly reduced by the co-expression of hTau2N4R (Fig. 4). These effects were apparent already at Day 1 for TTBK1-hTau2N4R, and at Days 7 and 14 for TTBK2-hTau2N4R and MARK1-hTau2N4R (Fig. 4). We attempted to test additional combinations of these three kinases with or without hTau2N4R in the climbing assay, but all such UAS combinations were larval lethal using the OK371-Gal4 driver, and adult flies never emerged.
We conclude that expression of hTau2N4R in motor neurons, using the OK371-Gal4 driver, has no effect upon locomotor activity, i.e. climbing ability. Single expression of TTBK1 or TTBK2 kinases affected geotaxis, while MARK1 had no effect. The TTBK1, TTBK2, effects were worsened by co-expression of hTau2N4R. MARK1 also had effects when co-expressed with hTau. Combinations of two or three kinases, with and without hTau, were larval lethal using the OK371-Gal4 driver.
Co-expression of hTau with kinases increases phosphorylation
The combinatorial toxicity observed when we co-expressed hTau with kinases in this system may be caused by increased phosphorylation of hTau by the kinases. To investigate this we used pan-hTau and phospho-specific hTau primary antibodies, and fluorescent secondary antibodies to determine relative levels of specific phospho-hTau epitopes, when compared to pan-hTau levels, using adult fly head extracts. Due to the lethality of TTBK1, and partial lethality for TTBK2, we again used the inducible n-Syb-Gal4;Gal80[ts] driver to obtain adult heads.
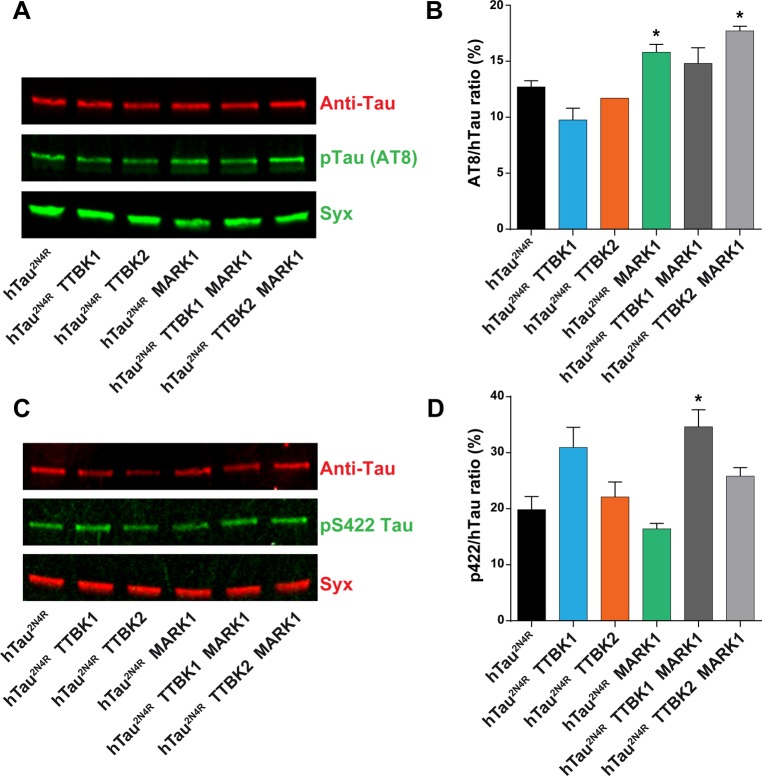
The AT8 monoclonal antibody detects phosphorylated S202 and T205, epitopes known to be phosphorylated by TTBK1 and TTBK2. We observed that hTau2N4R extracts stained for AT8, even without kinase co-expression, and hence hTau2N4R is already phosphorylated at these sites by endogenous Drosophila kinases (Fig. 5A). Surprisingly, we found that the levels of AT8 in hTau2N4R-TTBK1 or hTau2N4R-TTBK2 co-expression flies were comparable to those in flies expressing hTau2N4R alone (Fig. 5A,B). However, we observed an increase in the AT8 signal in flies co-expressing hTau2N4R with MARK1, with or without TTBK2 (Fig. 5A,B).
Fig. 5.
Phospho-analysis of flies expressing hTau2N4R and Tau kinases. (A,C) Western blot, using Licor, of fly head extracts after 5 days expression of the transgenes at 29°C (two independent experiments, 20 adult heads each). Transgene expression was induced by incubating the flies at 29°C for five days using the n-Syb-Gal4;Gal80[ts] driver. (A) Total hTau2N4R was detected using a rabbit hTau antibody and phospho-S202 and S205 were detected using mouse monoclonal AT8 antibody. Syx was used as a loading control. (B) Quantification of AT8/total hTau from two similar experiments in A, using Licor software Studio Image. We observed an increase in the AT8/total hTau ratio in flies expressing MARK1-hTau2N4R and TTBK2-hTau2N4R-MARK1, when compared to hTau2N4R alone (error bars indicate mean±s.d.; *P≤0.05; Student's two-tailed t-test). (C) Total hTau was detected using a mouse hTau5 antibody and phospho-S422 was detected using rabbit polyclonal pS422 antibody. Syx was used as loading control. (D) Quantifications of pS422/total hTau from two similar experiments in C, using Licor software Studio Image. We observed an increase in the AT8/total hTau ratio in flies expressing TTBK1-hTau2N4R-MARK1, when compared to hTau2N4R alone (error bars indicate mean±s.d.; *P≤0.05; Student's two-tailed t-test).
Next, we used a rabbit polyclonal antibody which detects phosphorylated S422, also known to be phosphorylated by TTBK1 and TTBK2 kinases (Fig. 6A). Again, we detected phosphorylation of hTau2N4R at S422 in extracts from flies expressing hTau2N4R alone, showing that hTau2N4R is phosphorylated also at this site by endogenous Drosophila kinases (Fig. 5C). The phosphorylation of hTau at p422 by endogenous kinases has been previously observed (Grammenoudi et al., 2006). Looking at co-expressing flies, we observed an increase in the pS422 signal over total hTau2N4R in flies co-expressing hTau2N4R with TTBK1-MARK1 (Fig. 5C,D). In contrast, all other co-expression of hTau2N4R with kinases did not result in elevated p422 level (Fig. 5C,D).
Fig. 6.
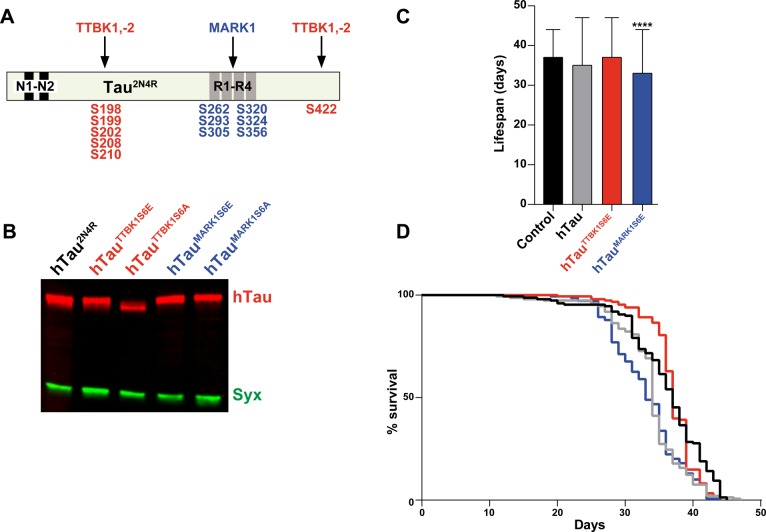
Mutational analysis of phospho-residues in hTau. (A) hTau2N4R with the different phospho-serines that are known to be targeted by TTBK1, TTBK2 and MARK1. (B) Western blot analysis of mutant UAS-hTau2N4R transgenes driven by the n-Syb-Gal4;Gal80[ts] driver. hTau2N4R was detected using a polyclonal hTau antibody; Syx was used as loading control (two independent experiments, 20 adult heads each). hTauTTBK1S6A displayed faster migration than the other hTau2N4R variants. (C,D) Lifespan analysis of mutant hTau2N4R strains driven by the n-Syb-Gal4;Gal80[ts] driver. Results are depicted as Kaplan–Meier survival curves (D) (Kaplan and Meier, 1958) and bar graph (C). hTau2N4R and hTauTTBK1S6E did not show any toxicity while hTauMARK1S6E toxicity was significant when compared to control flies (n-Syb-Gal4;Gal80[ts] crossed to the attP landing site strain BL#9750; error bars indicate mean±s.d.; ***P≤0.001; Dunnett's multiple comparison; n=3 independent experiments; ≥96 flies).
Together, these results indicate that while hTau2N4R is already phosphorylated at several sites by endogenous kinases, this can be elevated by the co-expression of human TTBK1, TTBK2 or MARK1. Surprisingly, in spite of the co-toxicity of TTBK2 with hTau2N4R, as evidenced by lifespan and geotaxis, we did not observe any apparent effects on hTau2N4R phosphorylation with TTBK2- hTau2N4R co-misexpression.
Phospho-residue mutagenesis supports a link between kinases and hTau toxicity
To further explore the nature of the hTau-kinase combinatorial toxicity, we next performed mutational analysis of hTau2N4R. To this end, we created mutant forms of hTau2N4R that either prevents (S-to-A) or mimics (S-to-E) phosphorylation of the residues known to be phosphorylated by either TTBK1 (S198, S199, S202, S208, S210, S422) or MARK1 (S262, S293, S305, S320, S324, S356) (Fig. 6A) (Drewes et al., 1997; Sato et al., 2006; Timm et al., 2006; Chatterjee et al., 2009; Hanger et al., 2009; Lund et al., 2013; Ikezu and Ikezu, 2014). First, we analyzed the protein levels and mobility of these mutants by western blot (Fig. 6B). While the protein levels were comparable, we noticed that the hTau2N4R mutant with S-to-A mutation in the TTBK1 sites (hTauTTBK1S6A) migrated faster than wild-type hTau2N4R or hTauMARK1S6A (Fig. 6B). This is in line with the positive signal for AT8 and pS422 already in flies expressing hTau2N4R alone (Fig. 5). In contrast, we did not observe a clear mobility shift for the hTauMARK1S6A mutant (Fig. 6B). Both of the phospho-mimic mutants (hTauTTBK1S6E and hTauMARK1S6E) migrated with similar mobility as wild-type hTau2N4R (Fig. 6B). To confirm the molecular behavior of these phospho-mutants, we analyzed western blots using phospho-antibodies. This revealed the anticipated loss of signal for the corresponding mutant-antibody pairs (Fig. S2A-C). In most cases the S6E mutants also resulted in loss of staining, presumably due to the difference in the structure of the p-Ser residue when compared to Glu (Fig. S2B,C). Noteworthy is that all phospho-antibodies tested reacted robustly with hTau2N4R without co-expression of human kinases, demonstrating substantial phosphorylation of hTau2N4R by endogenous Drosophila kinases (Fig. S2B,C).
Lifespan analysis, using the n-Syb-Gal4;Gal80[ts] driver, revealed that the hTauTTBK1S6E phospho-mimetic mutant did not show toxicity when compared to wild-type hTau2N4R or to control (n-Syb-Gal4;Gal80[ts] crossed to the attP landing site strain BL#9750) (Fig. 6C). However, in line with the fact that the MARK sites may not be highly phosphorylated by endogenous kinases, the hTauMARK1S6E phospho-mimetic mutant showed more enhanced toxicity (Fig. 6C,D). To further address combinatorial toxicity between TTBK1 and hTau, we combined TTBK1 with an hTau mutant where the TTBK1 phospo-residues were mutated to alanine (hTauTTBK1S6A). This revealed that TTBK1 was still combinatorially toxic with hTauTTBK1S6A, when compared to TTBK1 alone (Fig. S3A,B).
DISCUSSION
Post-mitotic expression of hTau results in weak toxicity, and may more closely model the slow progression of tauopathies in post-mitotic neurons
A number of previous studies have relied on the GMR-Gal4 and elav-Gal4 drivers, both of which express in dividing cells: the developing eye disc and neural precursors, respectively (Freeman, 1996; Berger et al., 2007). Because of the microtubule-binding properties of Tau, its expression in mitotic cells may have toxic effects due to interference with mitotic spindles in dividing cells, and this notion is supported by recent studies (Bouge and Parmentier, 2016). In line with the potential of Tau toxicity in mitotic cells, expression of Tau, although widespread in the developing nervous system, is generally not observed in neural progenitors (Takemura et al., 1991). To circumvent the potentially non-physiological effects of Tau in dividing cells we have performed our toxicity studies of hTau in Drosophila using a post-mitotic pan-nervous system driver, the n-Syb-Gal4 driver, driving throughout development and adulthood, or in an inducible version combined with Gal80[ts] to achieve selective expression only in adulthood. We also use the post-mitotic OK371-Gal4 driver – a Gal4 P element enhancer trap insertion in the Vesicular glutamate transporter gene – expressed in cholinergic motor neurons, to address locomotor effects in a geotaxis assay. We would argue that the post-mitotic expression described here, achieved by using n-Syb-Gal4 or OK371-Gal4, is more in line with the expression of Tau in post-mitotic cells, its function and dysfunction in neurons, as well as the generally slow progress of tauopathies in humans. Moreover, given the normally robust expression of Tau in most/all post-mitotic neurons throughout life, it is perhaps not surprising that we observe comparatively mild toxicity for hTau using post-mitotic drivers. Similar observations regarding the comparatively mild effects of hTau when expressed post-mitotically as opposed to mitotically has also been noted by others (Williams et al., 2000; Mudher et al., 2004; Folwell et al., 2010; Sinadinos et al., 2013).
The six main isoforms of hTau show similar toxicity
Integrase-based site-specific transgenesis allowed us to address hTau isoform toxicity without the complicating issue of transgenic position effects upon expression levels. Other studies have provided support for the notion that certain isoforms are more toxic (van Slegtenhorst et al., 2000; Grammenoudi et al., 2008; Kosmidis et al., 2010). We did not find striking differences between isoforms when integrated at the same genomic position and driving them from the n- Syb-Gal4 driver, although the hTau2N3R isoform did show significantly higher toxicity when compared to hTau2N4R. This apparent lack of difference in toxicity for five of the isoforms and elevated toxicity for hTau2N3R was not reflected by the protein analysis, where hTau1N4R was the only isoform that showed significantly elevated protein levels, when compared to hTau2N4R. Combining the six hTau isoforms with TTBK1 we noted combinatorial toxicity, but we did not observe any striking differences in co-toxicity when comparing the six isoforms. We conclude that the six isoforms may well have differential toxicity in different settings, but would argue that these are rather minor differences.
Combinatorial hTau-kinase expression results in increased toxicity
We find that post-mitotic expression of human TTBK1, TTBK2 and MARK1 kinases is toxic to the fly, even without co-expression of hTau. Comparing TTBK1 with TTBK2, we noted stronger effects on lifespan and geotaxis for TTBK1 than TTBK2, apparent in all three expression paradigms – direct n-Syb-Gal4, inducible n-Syb-Gal4 (n-Syb-Gal4;Gal80[ts]) and OK371-Gal4 (with or without hTau expression). Because both UAS transgenes were synthetic and contain identical 5′ and 3′ UTRs, are landed in the identical genomic location and the western blot show similar expression levels (Fig. 2B), we would speculate that these differences in toxicity truly reveal differences in the protein function of TTBK1 when compared to TTBK2. Intriguingly, TTBK1 and TTBK2 contain highly homologous kinase domains, but most of the sequences outside are not highly homologous (reviewed in Liao et al., 2015).
However, in spite of the toxicity observed when expressing these human kinases alone, we do observe combinatorial toxicity when they are co-expressed individually with hTau. This is apparent both in the lifespan and geotaxis assays. In addition, site-specific transgenesis allowed us to readily combine hTau with 2-3 kinases. This resulted in several interesting findings. First, regarding combinatorial kinase toxicity without hTau2N4R, we find that the combination of TTBK1 with MARK1 is highly combinatorially toxic. Specifically, while MARK1 alone is weakly toxic and TTBK1 reduces lifespan with 23%, their co-expression reduces lifespan by a non-additive 60% (Fig. 2C). Such combinatorial kinase toxicity may be due to action of common targets, on interacting targets, or on each other. Second, in contrast to combinatorial toxicity of TTBK1-MARK1, we did not find any enhancement of TTBK2 toxicity by co-expression of MARK1. Third, the TTBK1-TTBK2 double kinase flies show enhanced toxicity when compared to either kinase alone. Fourth, triple kinase flies are even more toxic, reduced down to 26% lifespan of control. These results point to a perhaps surprising selectivity in combinatorial kinase toxicity.
Next, focusing on the hTau-kinase combinations, we find that all hTau plus single kinase combinations were combinatorially more toxic than hTau or kinase alone. For double-kinase co-expression with hTau2N4R, the strongest effect was in the TTBK2-MARK1 combination, which was strongly enhanced by hTau2N4R co-expression. In contrast, triple kinase expression, which again gave only a 26% lifespan when compared to control, was not significantly enhanced by hTau2N4R co-expression (although there was a trend down). Surprisingly, TTBK1-TTBK2 co-expression, which is quite toxic, showed decreased toxicity when co-expressed with hTau2N4R.
We conclude that all three human kinases are toxic in flies, in a TTBK1>TTBK2>MARK1 order. Furthermore, all three are combinatorially toxic with hTau, and two of three double-kinase flies were also enhanced by hTau2N4R. However, triple kinase expression is highly toxic, and we can no longer observe any combinatorial toxic effect when co-expressing hTau2N4R. These results demonstrate that there is considerable toxicity of TTBK1, TTBK2 and MARK1 kinase expression even without hTau co-expression.
Combinatorial hTau-kinase toxicity: direct or indirect action?
We observe combinatorial effects on toxicity when co-expressing TTBK1, TTBK2 or MARK1 kinase with hTau. A central question is whether or not this reflects a direct action of the kinases upon hTau, and hence increased hTau toxicity, or alternatively, totally independent toxicity mechanisms, which only combine with respect to shortening the lifespan or affecting geotaxis.
A number of studies have addressed hTau phospho-biology using Drosophila (Gistelinck et al., 2012; Fernandez-Funez et al., 2015; Sun and Chen, 2015). Several of these studies have provided support for the well-established notion that phosphorylation of specific hTau residues enhances toxicity. In addition, genetic screens in Drosophila and C. elegans have identified numerous kinases and phosphatases as suppressors/enhancers of Tau toxicity (reviewed in Hannan et al., 2016). Among the kinases identified were TTBK and MARK kinases (Shulman and Feany, 2003; Kraemer et al., 2006; Ambegaokar and Jackson, 2011).
Focusing on the MARK kinases, denoted Par-1 in Drosophila, previous studies have found that overexpression of Par-1 in the eye triggers a mild rough eye phenotype, which could be partly suppressed by removing one gene copy of dTau (Nishimura et al., 2004). Studies furthermore supported a role for the S262 and S356 residues, which are targeted by Par-1, and hTau toxicity (Nishimura et al., 2004; Chatterjee et al., 2009; Folwell et al., 2010; Iijima-Ando et al., 2010). We mutated the six main MARK kinase target sites in hTau, S-to-A or S-to-E, but did not observe any obvious mobility shifts in western blots for either mutant. However, we observed increased toxicity of the S6E mutant, when compared to hTau2N4R. Based upon these findings, we would postulate that hTau2N4R is not extensively phosphorylated on the MARK sites by endogenous kinases, in spite of the presence of Drosophila Par-1. We further propose that MARK1 kinase over-expression increases toxicity, at least in part, via these sites. In relation to these observations it is noteworthy that the MARK1 sites on hTau are known to be involved in microtubule stabilization, i.e. when they are un-phosphorylated the microtubules are stabilized (Schneider et al., 1999; Matenia et al., 2005). Thus, this might be part of the explanation for MARK1-hTau combinatorial toxicity, especially in neurons given their reliance on fast axonal transport.
The TTBK kinases have been less studied with regard to hTau toxicity in Drosophila. However, studies have found enhanced toxicity linked to TTBK target sites in hTau. Single or double S-to-A mutations in two of the TTBK target sites (S199/S202) did not reduce toxicity (Steinhilb et al., 2007b). However, a 14 amino acid S/T-to-A mutant (including three TTBK sites: S199/S202/S422) did reduce hTau toxicity, and the converse S-to-E phospho-mimetic mutant increased toxicity (Steinhilb et al., 2007a). We find combinatorial toxicity between TTBK1 or TTBK2, when expressed with hTau. However, this combinatorial toxicity is still present in TTBK1 hTauTTBK1S6A flies, and we did not observe enhanced toxicity in the phosho-mimetic hTauTTBK1S6E mutant when expressed alone. Moreover, when co-expressing TTBK1, TTBK2 with hTau2N4R, we do not find a clear increase in pTau using AT8 (Fig. 5A,B). In contrast, TTBK1, but not TTBK2, resulted in an increase in pS422 (Fig. 5C,D). We observe faster migration of the S-to-A mutant on the western blots (Fig. 6B). Combined with the lack of enhanced toxicity in the S-to-E mutant, we propose that although pS422 can be enhanced, the TTBK target serines have a high degree of phosphorylation even without co-expression of TTBK1 or TTBK2 kinases, potentially by the Drosophila TTBK homolog Asator. Hence, in contrast to MARK1-hTau combinatorial toxicity, we propose that TTBK-hTau combinatorial toxicity may largely result from independent toxic effects of each protein.
One surprising finding was that while neither TTBK1 nor TTBK2 co-expression with hTau increased the AT8 signal, MARK1 triggered a minor increase (Fig. 5A,B). Because MARK1 is not known to target AT8 this is likely an indirect effect, either from triggering AT8 phosphorylation by acting on other ‘priming’ phospho-sites in hTau, or by triggering the activity of other kinases, e.g. TTBK1. It is tempting to speculate that such kinase cross-talk may help explain the strong co-toxicity observed when co-expressing MARK1 with TTBK1.
Recent studies have identified another kinase, Nuak1, which also phosphorylates Tau on S356 and hence shares a phospho-target site with MARK kinases (Lasagna-Reeves et al., 2016). However, unlike MARK kinases, which target several phospho-residues, Nuak1 exclusively targets S356. Nuak1 phosphorylation stabilizes Tau, and Nuak1 enhances Tau toxicity, both in flies and mice. It would be interesting to address combinatorial toxicity of MARK with Nuak1.
Finally, it should be noted that a growing number of studies are indicating that Tau phosphorylation may not be the only, or even primary, toxicity driver. For example, in a screen for modifiers of hTau toxicity, there was an apparent disconnect between modifier action and the extent of Tau phosphorylation, as well as the interaction of modifiers and wild type versus S11A phosphorylation-resistant mutant hTau (Ambegaokar and Jackson, 2011). In addition, zinc has been found to enhance Tau aggregation (Mo et al., 2009), and recent studies indicate that this operates independently of Tau phosphorylation status (Huang et al., 2014). Finally, recent studies of Tau toxicity have revealed a role for Tau toxicity and the loss of heterochromatin, indicating a nuclear role for Tau (Frost et al., 2014). Given the role of Tau in a number of human tauopathies, uncovering the function and dysfunction of Tau, its relationship to the growing list of Tau-kinases, and possible new roles for Tau, will continue to be important for future development of therapeutics.
MATERIALS AND METHODS
Fly stocks
From Bloomington Drosophila Stock Center: UAS-hTau (BL#51362) and OK371-Gal4 (BL#26160). For this study, a double transgenic line, n-Syb-Gal4#2-1, 1M was used (Jonson et al., 2015). To generate Gal80[ts]/n-Syb-Gal4, because of the strong expression from the n-Syb-Gal4 driver, two tubP-Gal80[ts] transgenes on chromosome 2 (BL#7019 and BL#7108) (McGuire et al., 2003) were recombined, and then combined with n-Syb-Gal4#2-1, 1M. This resulted in a driver line that showed no apparent eGFP expression in the adult fly brain when crossed to UAS-eGFP at 18°C, but robust expression when flies were moved to 29°C (Fig. S1).
Generation of UAS Transgenic flies
Both hTau and kinases were codon optimized for expression in Drosophila (http://www.kazusa.or.jp/codon/). Sequences were added to the 5′: a consensus start codon (Cavener and Ray, 1991) and an EcoRI site, as well as to the 3′; three different stop codons (amb, och, opa) and an XbaI site (see Supplementary Materials for all DNA sequences). DNAs were generated by gene-synthesis (Genscript, New Jersey, USA), and cloned into pUAS.attB (Bischof et al., 2007), as EcoRI/XbaI fragments. DNAs were control-sequenced on both strands in the pUAS.attB vector (GATC BioTech, Germany), and injected into landing site strains BL#9723 (28E); BL#9736 (53B); BL#9750 (65B) and BL#9744 (89E) (BestGene, CA, USA).
Lifespan assay
Lifespan assay using n-Syb-Gal4 was performed as previously described (Jonson et al., 2015). Briefly, flies were reared at 25°C under a 12 h light:12 h dark cycle until eclosion and at 29°C post eclosion. Flies were maintained at 29°C in 50 ml vials (20 flies per vial) containing Drosophila food [water, potato mash powder, corn flour, yeast, agar, syrup, propionic acid (diluted: 48.5 ml propionic acid+∼950 ml H2O)]. Every 2-3 days the flies were transferred to fresh vials and the number of surviving flies was recorded throughout the lifetime of all flies. The assay was repeated three times and the data was pooled and analyzed together. For n-Syb-Gal4;Gal80[ts], flies were kept at 20°C until eclosion, and flies were then reared at 29°C. As control, the different Gal4 drivers used were crossed to the attP landing site strain BL#9750. GraphPad Prism 6.0a software (GrapPad Software Inc., San Diego, CA, USA) was used to generate Kaplan–Meier survival curves (Kaplan and Meier, 1958) and to perform all statistical analysis.
Negative geotaxis assay
Transgenic UAS flies were crossed with OK371-Gal4 line and reared at 26°C until eclosion. The adult flies were sorted and placed in 10 vials of 10 flies per vial and placed at 29°C. Only males were used in order to avoid differences in mobility due to amount of eggs carried by the fly. Flies were examined on Day 1, 7 and 14. Flies were always allowed to recover from CO2 for at least 3 h until assayed. Flies were transferred into new, empty vials and allowed to acclimatize for 30 s before the assay was started. The flies were gently shaken to the bottom of the vial and the percentage of flies that climbed up to a 5 cm mark on the vial within 30 s was counted, and the procedure was repeated 10 times for each vial. The mean with standard deviation is plotted.
Western blotting of adult fly heads and antibodies
20 fly heads were homogenized in ice-cold RIPA buffer (10 mM Tris pH 8, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl), 1 × protease inhibitors, 1 × PhosSTOP (both from Roche, Basel, Switzerland), and silica beads in a Fastprep24 homogenizer (MP Biomedical, Santa Ana, CA, USA) at 6.5 m/s for 30 s, and quickly placed on ice. 2 × Laemmli protein sample buffer (BioRad) with 5% β-mercaptoethanol was added and the samples were then boiled for 5 min. The samples were then centrifuged at 12,000 g, the supernatant transferred to a new tube and centrifuged again. SDS-PAGE, western blotting, blocking and antibody incubations were all performed according to standard procedures. Antibodies used were: mouse AT8 at 1:1000 dilution (Thermo Scientific), mouse Tau5 at 1:1000 dilution (Thermo Scientific), rabbit TauH-150 at 1:1000 dilution (Santa Cruz Biotechnology), mouse Syntaxin at 1:4000 dilution (mAb 8C3-s; Developmental Studies Hybridoma Bank), and mouse PHF1-Tau at 1:1000 dilution (kind gift from Peter Davies, Albert Einstein College of Medicine, Bronx, NY, USA). Rabbit monoclonal antibodies to pSTau198, pSTau199, pSTau202, pSTau262 and pSTau356, and rabbit polyclonal pSTau422, were all used at 1:1000 (Abcam). Secondary antibodies were all IRDye from Li-COR diluted 1:15,000: goat anti mouse 680RD, goat anti rabbit 680RD, goat anti rat 680RD, goat anti mouse 800CW, goat anti rabbit 800CW and goat anti rat 800CW. The nitrocellulose (Amersham Protran, GE Healthcare, Uppsala, Sweden) was blocked in 5% milk in PBS with 0.1% Tween for 30 min. Antibodies were diluted in 5% milk PBS with 0.1% tween.
Acknowledgements
We are grateful to Johannes Bischof (University of Zurich, Switzerland), Konrad Basler (University of Zurich, Switzerland), Peter Davies (Albert Einstein College of Medicine, Bronx, NY, USA), the Developmental Studies Hybridoma Bank at the University of Iowa, and the Bloomington Stock Center for sharing antibodies, fly lines and DNAs. We thank Per Hammarstrom and Catherine M. Cowan for critically reading the manuscript. Helen Ekman and Carolin Jonsson provided excellent technical assistance.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: S.T., J.F.; Methodology: S.T., J.F., M.P., A.S.; Formal analysis: J.F., A.S.; Data curation: M.P.; Writing - original draft: S.T., J.F.; Writing - review & editing: S.T., J.F., M.P., A.S.; Supervision: S.T.; Project administration: S.T.; Funding acquisition: S.T.
Funding
This work was supported by Konung Gustaf V:s och Drottning Victorias Frimurarestiftelse to S.T. (grant # 700-0557). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Supplementary information
Supplementary information available online at http://bio.biologists.org/lookup/doi/10.1242/bio.022749.supplemental
References
- Ambegaokar S. S. and Jackson G. R. (2011). Functional genomic screen and network analysis reveal novel modifiers of tauopathy dissociated from tau phosphorylation. Hum. Mol. Genet. 20, 4947-4977. 10.1093/hmg/ddr432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreadis A. (2012). Tau splicing and the intricacies of dementia. J. Cell. Physiol. 227, 1220-1225. 10.1002/jcp.22842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger C., Renner S., Lüer K. and Technau G. M. (2007). The commonly used marker ELAV is transiently expressed in neuroblasts and glial cells in the Drosophila embryonic CNS. Dev. Dyn. 236, 3562-3568. 10.1002/dvdy.21372 [DOI] [PubMed] [Google Scholar]
- Bischof J., Maeda R. K., Hediger M., Karch F. and Basler K. (2007). An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc. Natl. Acad. Sci. USA 104, 3312-3317. 10.1073/pnas.0611511104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bougé A. L. and Parmentier M. L. (2016). Tau excess impairs mitosis and kinesin-5 function, leading to aneuploidy and cell death. Dis. Model Mech. 9, 307-319. 10.1242/dmm.022558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H. and Braak E. (1991). Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239-259. 10.1007/BF00308809 [DOI] [PubMed] [Google Scholar]
- Cavener D. R. and Ray S. C. (1991). Eukaryotic start and stop translation sites. Nucleic Acids Res. 19, 3185-3192. 10.1093/nar/19.12.3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesaro L. and Pinna L. A. (2015). The generation of phosphoserine stretches in phosphoproteins: mechanism and significance. Mol. Biosyst. 11, 2666-2679. 10.1039/C5MB00337G [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Sang T.-K., Lawless G. M. and Jackson G. R. (2009). Dissociation of tau toxicity and phosphorylation: role of GSK-3beta, MARK and Cdk5 in a Drosophila model. Hum. Mol. Genet. 18, 164-177. 10.1093/hmg/ddn326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola G., Chinnathambi S., Lee J. J., Dombroski B. A., Baker M. C., Soto-Ortolaza A. I., Lee S. E., Klein E., Huang A. Y., Sears R. et al. (2012). Evidence for a role of the rare p.A152T variant in MAPT in increasing the risk for FTD-spectrum and Alzheimer's diseases. Hum. Mol. Genet. 21, 3500-3512. 10.1093/hmg/dds161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewes G., Ebneth A., Preuss U., Mandelkow E.-M. and Mandelkow E. (1997). MARK, a novel family of protein kinases that phosphorylate microtubule-associated proteins and trigger microtubule disruption. Cell 89, 297-308. 10.1016/S0092-8674(00)80208-1 [DOI] [PubMed] [Google Scholar]
- Fernandez-Funez P., de Mena L. and Rincon-Limas D. E. (2015). Modeling the complex pathology of Alzheimer's disease in Drosophila. Exp. Neurol. 274, 58-71. 10.1016/j.expneurol.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folwell J., Cowan C. M., Ubhi K. K., Shiabh H., Newman T. A., Shepherd D. and Mudher A. (2010). Aβ exacerbates the neuronal dysfunction caused by human tau expression in a Drosophila model of Alzheimer's disease. Exp. Neurol. 223, 401-409. 10.1016/j.expneurol.2009.09.014 [DOI] [PubMed] [Google Scholar]
- Freeman M. (1996). Reiterative use of the EGF receptor triggers differentiation of all cell types in the Drosophila eye. Cell 87, 651-660. 10.1016/S0092-8674(00)81385-9 [DOI] [PubMed] [Google Scholar]
- Frost B., Hemberg M., Lewis J. and Feany M. B. (2014). Tau promotes neurodegeneration through global chromatin relaxation. Nat. Neurosci. 17, 357-366. 10.1038/nn.3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gistelinck M., Lambert J. C., Callaerts P., Dermaut B. and Dourlen P. (2012). Drosophila models of tauopathies: what have we learned? Int J Alzheimers Dis 2012, 970980 10.1155/2012/970980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M., Spillantini M. G., Potier M. C., Ulrich J. and Crowther R. A. (1989). Cloning and sequencing of the cDNA encoding an isoform of microtubule-associated protein tau containing four tandem repeats: differential expression of tau protein mRNAs in human brain. EMBO J. 8, 393-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grammenoudi S., Kosmidis S. and Skoulakis E. M. (2006). Cell type-specific processing of human Tau proteins in Drosophila. FEBS Lett. 580, 4602-4606. 10.1016/j.febslet.2006.07.045 [DOI] [PubMed] [Google Scholar]
- Grammenoudi S., Anezaki M., Kosmidis S. and Skoulakis E. M. (2008). Modelling cell and isoform type specificity of tauopathies in Drosophila. SEB Exp. Biol. Ser. 60, 39-56. [PubMed] [Google Scholar]
- Hanger D. P., Anderton B. H. and Noble W. (2009). Tau phosphorylation: the therapeutic challenge for neurodegenerative disease. Trends Mol. Med. 15, 112-119. 10.1016/j.molmed.2009.01.003 [DOI] [PubMed] [Google Scholar]
- Hannan S. B., Drager N. M., Rasse T. M., Voigt A. and Jahn T. R. (2016). Cellular and molecular modifier pathways in tauopathies: the big picture from screening invertebrate models. J. Neurochem. 137, 12-25. 10.1111/jnc.13532 [DOI] [PubMed] [Google Scholar]
- Huang Y., Wu Z., Cao Y., Lang M., Lu B. and Zhou B. (2014). Zinc binding directly regulates tau toxicity independent of tau hyperphosphorylation. Cell Rep. 8, 831-842. 10.1016/j.celrep.2014.06.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima-Ando K., Zhao L. J., Gatt A., Shenton C. and Iijima K. (2010). A DNA damage-activated checkpoint kinase phosphorylates tau and enhances tau-induced neurodegeneration. Hum. Mol. Genet. 19, 1930-1938. 10.1093/hmg/ddq068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezu S. and Ikezu T. (2014). Tau-tubulin kinase. Front. Mol. Neurosci. 7, 33 10.3389/fnmol.2014.00033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonson M., Pokrzywa M., Starkenberg A., Hammarstrom P. and Thor S. (2015). Systematic abeta analysis in drosophila reveals high toxicity for the 1-42, 3-42 and 11-42 peptides, and emphasizes N- and C-terminal residues. PLoS ONE 10, e0133272 10.1371/journal.pone.0133272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan E. and Meier P. (1958). Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 53, 457-481. 10.1080/01621459.1958.10501452 [DOI] [Google Scholar]
- Kosmidis S., Grammenoudi S., Papanikolopoulou K. and Skoulakis E. M. (2010). Differential effects of Tau on the integrity and function of neurons essential for learning in Drosophila. J. Neurosci. 30, 464-477. 10.1523/JNEUROSCI.1490-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouri N., Carlomagno Y., Baker M., Liesinger A. M., Caselli R. J., Wszolek Z. K., Petrucelli L., Boeve B. F., Parisi J. E., Josephs K. A. et al. (2014). Novel mutation in MAPT exon 13 (p.N410H) causes corticobasal degeneration. Acta Neuropathol. 127, 271-282. 10.1007/s00401-013-1193-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraemer B. C., Burgess J. K., Chen J. H., Thomas J. H. and Schellenberg G. D. (2006). Molecular pathways that influence human tau-induced pathology in Caenorhabditis elegans. Hum. Mol. Genet. 15, 1483-1496. 10.1093/hmg/ddl067 [DOI] [PubMed] [Google Scholar]
- Lasagna-Reeves C. A., de Haro M., Hao S., Park J., Rousseaux M. W. C., Al-Ramahi I., Jafar-Nejad P., Vilanova-Velez L., See L., De Maio A. et al. (2016). Reduction of Nuak1 Decreases Tau and Reverses Phenotypes in a Tauopathy Mouse Model. Neuron 92: 407-418. 10.1016/j.neuron.2016.09.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J. C., Yang T. T., Weng R. R., Kuo C. T. and Chang C. W. (2015). TTBK2: a tau protein kinase beyond tau phosphorylation. Biomed. Res. Int. 2015, 575170 10.1155/2015/575170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund H., Cowburn R. F., Gustafsson E., Stromberg K., Svensson A., Dahllund L., Malinowsky D. and Sunnemark D. (2013). Tau-tubulin kinase 1 expression, phosphorylation and co-localization with phospho-Ser422 tau in the Alzheimer's disease brain. Brain Pathol. 23, 378-389. 10.1111/bpa.12001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahr A. and Aberle H. (2006). The expression pattern of the Drosophila vesicular glutamate transporter: a marker protein for motoneurons and glutamatergic centers in the brain. Gene Expr. Patterns 6, 299-309. 10.1016/j.modgep.2005.07.006 [DOI] [PubMed] [Google Scholar]
- Matenia D., Griesshaber B., Li X. Y., Thiessen A., Johne C., Jiao J., Mandelkow E. and Mandelkow E. M. (2005). PAK5 kinase is an inhibitor of MARK/Par-1, which leads to stable microtubules and dynamic actin. Mol. Biol. Cell 16, 4410-4422. 10.1091/mbc.E05-01-0081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire S. E., Le P. T., Osborn A. J., Matsumoto K. and Davis R. L. (2003). Spatiotemporal rescue of memory dysfunction in Drosophila. Science 302, 1765-1768. 10.1126/science.1089035 [DOI] [PubMed] [Google Scholar]
- Mo Z.-Y., Zhu Y. Z., Zhu H.-L., Fan J.-B., Chen J. and Liang Y. (2009). Low micromolar zinc accelerates the fibrillization of human tau via bridging of Cys-291 and Cys-322. J. Biol. Chem. 284, 34648-34657. 10.1074/jbc.M109.058883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudher A., Shepherd D., Newman T. A., Mildren P., Jukes J. P., Squire A., Mears A., Berg S., MacKay D., Asuni A. A. et al. (2004). GSK-3beta inhibition reverses axonal transport defects and behavioural phenotypes in Drosophila. Mol. Psychiatry 9, 522-530. 10.1038/sj.mp.4001483 [DOI] [PubMed] [Google Scholar]
- Niblock M. and Gallo J.-M. (2012). Tau alternative splicing in familial and sporadic tauopathies. Biochem. Soc. Trans. 40, 677-680. 10.1042/BST20120091 [DOI] [PubMed] [Google Scholar]
- Nishimura I., Yang Y. and Lu B. (2004). PAR-1 kinase plays an initiator role in a temporally ordered phosphorylation process that confers tau toxicity in Drosophila. Cell 116, 671-682. 10.1016/S0092-8674(04)00170-9 [DOI] [PubMed] [Google Scholar]
- Sato S., Cerny R. L., Buescher J. L. and Ikezu T. (2006). Tau-tubulin kinase 1 (TTBK1), a neuron-specific tau kinase candidate, is involved in tau phosphorylation and aggregation. J. Neurochem. 98, 1573-1584. 10.1111/j.1471-4159.2006.04059.x [DOI] [PubMed] [Google Scholar]
- Schneider A., Biernat J., von Bergen M., Mandelkow E. and Mandelkow E.-M. (1999). Phosphorylation that detaches tau protein from microtubules (Ser262, Ser214) also protects it against aggregation into Alzheimer paired helical filaments. Biochemistry 38, 3549-3558. 10.1021/bi981874p [DOI] [PubMed] [Google Scholar]
- Shulman J. M. and Feany M. B. (2003). Genetic modifiers of tauopathy in Drosophila. Genetics 165, 1233-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinadinos C., Quraishe S., Sealey M., Samson P. B., Mudher A. and Wyttenbach A. (2013). Low endogenous and chemical induced heat shock protein induction in a 0N3Rtau-expressing Drosophila larval model of Alzheimer's disease. J. Alzheimers Dis. 33, 1117-1133. 10.3233/JAD-2012-121534. [DOI] [PubMed] [Google Scholar]
- Steinhilb M. L., Dias-Santagata D., Fulga T. A., Felch D. L. and Feany M. B. (2007a). Tau phosphorylation sites work in concert to promote neurotoxicity in vivo. Mol. Biol. Cell 18, 5060-5068. 10.1091/mbc.E07-04-0327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhilb M. L., Dias-Santagata D., Mulkearns E. E., Shulman J. M., Biernat J., Mandelkow E.-M. and Feany M. B. (2007b). S/P and T/P phosphorylation is critical for tau neurotoxicity in Drosophila. J. Neurosci. Res. 85, 1271-1278. 10.1002/jnr.21232 [DOI] [PubMed] [Google Scholar]
- Sun M. K. and Chen L. (2015). Studying tauopathies in Drosophila: A fruitful model. Exp. Neurol. 274, 52-57. 10.1016/j.expneurol.2015.03.029 [DOI] [PubMed] [Google Scholar]
- Takemura R., Kanai Y. and Hirokawa N. (1991). In situ localization of tau mRNA in developing rat brain. Neuroscience 44, 393-407. 10.1016/0306-4522(91)90064-U [DOI] [PubMed] [Google Scholar]
- Takuma H., Arawaka S. and Mori H. (2003). Isoforms changes of tau protein during development in various species. Brain Res. Dev. Brain Res. 142, 121-127. 10.1016/S0165-3806(03)00056-7 [DOI] [PubMed] [Google Scholar]
- Timm T., Matenia D., Li X.-Y., Griesshaber B. and Mandelkow E. M. (2006). Signaling from MARK to tau: regulation, cytoskeletal crosstalk, and pathological phosphorylation. Neurodegener. Dis. 3, 207-217. 10.1159/000095258 [DOI] [PubMed] [Google Scholar]
- van Slegtenhorst M., Lewis J. and Hutton M. (2000). The molecular genetics of the tauopathies. Exp. Gerontol. 35, 461-471. 10.1016/S0531-5565(00)00114-5 [DOI] [PubMed] [Google Scholar]
- Wang Y. and Mandelkow E. (2016). Tau in physiology and pathology. Nat. Rev. Neurosci. 17, 22-35. 10.1038/nrn.2015.1 [DOI] [PubMed] [Google Scholar]
- Williams D. W., Tyrer M. and Shepherd D. (2000). Tau and tau reporters disrupt central projections of sensory neurons in Drosophila. J. Comp. Neurol. 428, 630-640. [DOI] [PubMed] [Google Scholar]