Abstract
The p33ING1 protein is a regulator of cell cycle, senescence, and apoptosis. Three alternatively spliced transcripts of p33ING1 encode p47ING1a, p33ING1b, and p24ING1c. We cloned an additional ING family member, p33ING2/ING1L. Unlike p33ING1b, p33ING2 is induced by the DNA-damaging agents etoposide and neocarzinostatin. p33ING1b and p33ING2 negatively regulate cell growth and survival in a p53-dependent manner through induction of G1-phase cell-cycle arrest and apoptosis. p33ING2 strongly enhances the transcriptional-transactivation activity of p53. Furthermore, p33ING2 expression increases the acetylation of p53 at Lys-382. Taken together, p33ING2 is a DNA damage-inducible gene that negatively regulates cell proliferation through activation of p53 by enhancing its acetylation.
Keywords: p33ING1, PHD-finger, apoptosis, cell cycle
The p33ING1 gene was identified by using a strategy for tumor-suppressor gene isolation (1). p33ING1 encodes a nuclear protein and is located on chromosome 13q33-34 (2). The properties of p33ING1 suggest its involvement in the regulation of cell proliferation, senescence, and apoptosis (3, 4). Recently, three alternatively spliced transcripts of p33ING1 were reported (5–7). The three transcripts encode p47ING1a, p33ING1b, and p24ING1c, which share the C-terminal region. The sequence of p33ING1 originally reported is incorrect because of a cloning error; it is a partial sequence that is frame-shifted and truncated at the 5′ end of p47ING1a (5).
The tumor-suppressor gene product p53 maintains genomic integrity (8). p53 can negatively regulate cell growth through induction of cell-cycle arrest, senescence, and apoptosis after cellular stresses, including DNA damage, hypoxia, and nucleotide deprivation (9–13). p53 also plays a critical role in other cellular functions such as nucleotide-excision repair and base-excision repair (14–18). p53 is a transcriptional activator and repressor (19–22). Wild-type p53 transactivates genes (e.g., p21/waf1 and bax) through its binding to specific DNA sequences in the promoter regions of the genes (23–26). p53 down-regulates another set of genes (e.g., Map4 and stathmin) during apoptosis and growth arrest (27, 28). Activation of sequence-specific DNA-binding by p53 in response to these stresses is mediated through posttranslational modifications, such as phosphorylation, dephosphorylation, acetylation, and sumolation (27, 29–34).
p33ING1 has been reported to interact physically with p53, and both act to exert growth-inhibitory effects (35). Interpretation of the data in reports on p33ING1 is quite complicated because of the inadvertent use of an incorrect construct (1, 3, 4, 35). Thus, the true functions of p33ING1b are unclear. Both others and we have recently cloned an additional ING family member, p33ING2/ING1L, by homology search (36). Here, we report the differing biological mechanisms of authentic p33ING1b and p33ING2 mediated through their physical and functional interactions with p53.
Materials and Methods
cDNA Cloning of p33ING2 Gene.
We cloned the p33ING2 gene by a homology search with the p33ING1b cDNA sequence and the blast program (National Center for Biotechnology Information). We isolated the coding regions of the p33ING1b and p33ING2 genes from human placenta Marathon-Ready cDNA (CLONTECH) by reverse transcription–PCR and ligated them into the pcDNA3.1 expression vector (Invitrogen), producing pcDNA3.1-ING1b, pcDNA3.1-ING2, and pcDNA3.1-Anti-ING2, respectively.
Generation of p33ING1b and p33ING2 Antibodies.
Rabbit polyclonal antibodies for p33ING1b (Ping1) and p33ING2 (Ping2) were raised against chemically synthesized, keyhole limpet hemocyanin-conjugated peptides: p33ING1b sequence MLSPANGEQLHLVNYVE (amino acids 1–17) and p33ING2 sequence QQLYSSAALLTGERSRLLTC (amino acid 8–27). The antisera from immunized rabbits were affinity-purified with the respective peptide coupled to SulfoLink (Pierce). The specificities of Ping1 and Ping2 were determined by Western blot analysis with p33ING1b and p33ING2 proteins that were produced by an in vitro transcription/translation system (Promega).
Preparation of Cell Lysates and Western Blot Analysis.
Human cell lines (n = 13) were grown in the recommended media. Cell lines used were as follows: one normal lymphoblastoid (C3ABR), one ataxia-telangiectasia mutated (ATM)-null lymphoblastoid (L3), one hepatoblastoma (Hep G2), two osteosarcomas (OsA-CL, Saos-2), three colorectal carcinomas (HCT 116, RKO, RKO E6), one hepatocellular carcinoma (Hep 3B), one prostate cancer (PC-3), two non-small cell lung cancers (Calu-6, NCI-H157), and one pancreatic cancer (AsPC-1). Cell lines (n = 12) were used to screen protein expression of p33ING1b or p33ING2. C3ABR cells were treated with γ-irradiation (10 Gy), doxorubicin (0.2 μg/ml), etoposide (30 μg/ml), bleomycin (0.03 units/ml), neocarzinostatin (0.3 μg/ml), or cis-platinum (2 μg/ml) to analyze the response of p53, p33ING1b, p33ING2, or p53 acetylated at Lys-382. L3 or Calu-6 cells were treated with etoposide (30 μg/ml) to analyze the response of p33ING2. Subconfluent cultures of cells were harvested and lysed in buffer of 10 mM Tris⋅HCl, pH 7.5/1 mM EDTA/400 mM NaCl/10% (vol/vol) glycerol/0.5% Nonidet P-40/5 mM NaF/0.5 mM sodium orthovanadate/1 mM DTT and protease inhibitor mixture (Roche Molecular Biochemicals). Equal amounts of cell lysates (20 μg) were resuspended in 2× Tris-glycine SDS sample buffer, electrophoresed on SDS/10% polyacrylamide gels, and electrophoretically transferred to Immobilon-P membrane (Millipore). Detection of proteins was done with anti-p53 Ab-6 antibody (1:1,000, Calbiochem), anti-p33ING1b Ping1 antibody (1:200), anti-p33ING2 Ping2 antibody (1:200), acetylated p53 Ab-1 (1:100, Oncogene Research Products), or anti-actin Ab-1 antibody (1:5,000, Oncogene Research Products), followed by enhanced chemiluminescence (Amersham Pharmacia).
Colony Formation Assay.
Cells were plated into 6-cm dishes at 2 × 104 cells per cm2 and cultured for 24 hr at 37°C. Cells were transfected with 2 μg of pcDNA3.1-ING1b, pcDNA3.1-ING2, or pcDNA3.1 (empty vector control) with Lipofectamine Plus reagent (Life Technologies, Rockville, MD), as recommended by the manufacturer. After 2 weeks of G418 selection (1 mg/ml), colonies were fixed, stained with crystal violet, and counted.
Detection of Apoptosis.
Cells were plated into eight-well chamber slides at 1 × 104 cells per cm2 and cultured for 24 hr at 37°C. Cells were transfected with 50 ng of pcDNA3.1-ING1b, pcDNA3.1-ING2, or pcDNA3.1 (empty vector control) with Lipofectamine Plus reagent. Cells were cotransfected with 5 ng of pEGFP-F Amp as a transfection marker. Cells were fixed with 4% (vol/vol) paraformaldehyde in Dulbecco's PBS 24 hr after transfection. Fragmented DNA was labeled by terminal transferase (Oncogene Research Products) with tetramethylrhodamine 5–2′-deoxy-uridine 5′-triphosphate (Roche Molecular Biochemicals) by terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assay. Nuclear DNA was stained with 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories). Apoptosis was analyzed with fluorescence microscopy. Cells with a green fluorescent signal were considered to be transfection-positive. Apoptotic cells were scored by both TUNEL and DAPI staining.
Cell-Cycle Analysis.
Cells were plated into 10-cm dishes at 2 × 104 cells per cm2 and cultured for 24 hr at 37°C. Cells were transfected with 4 μg of pcDNA3.1-ING1b, pcDNA3.1-ING2, or pcDNA3.1 (empty vector control) with Lipofectamine Plus. Cells were cotransfected with 0.4 μg of pEGFP-F Amp as a transfection marker. At 48 hr after transfection, cells were harvested, fixed with 70% (vol/vol) ethanol, and then suspended in Dulbecco's PBS containing 20 μg/ml propidium iodide and 100 μg/ml RNaseA. The propidium iodide signal was used as a measure for DNA content to determine cell-cycle profiles on a FACSCalibur flow cytometer (Becton-Dickinson). The percentages of the cells in each cell-cycle phase (G0/G1, S, and G2/M) were calculated by the modfit lt program (Verity Software House, Topsham, ME). Cells with a green fluorescent signal at least two times stronger than that in the negative cells were considered to be transfection-positive cells. The cell-cycle profiles of green fluorescent protein (GFP)-positive populations were compared. Each analysis contains data from at least 10,000 GFP-positive cells.
Reporter Gene Assay.
Cells were plated into six-well plates at 2 × 104 cells per cm2 and cultured for 24 hr at 37°C. Cells were transfected with 500 ng of pcDNA3.1-ING1b, pcDNA3.1-ING2, or pcDNA3.1 (empty vector control) with Lipofectamine Plus reagent. The transcriptional-transactivation activities of p53 were examined with the Luciferase Reporter system (Promega) by cotransfection with 100 ng of p53-responsive reporter constructs WWP-Luc-p21 or PGL3-Luc-Bax. The cells were lysed, and the lysates were collected to measure the promoter activities 24 hr after transfection. The activities of luciferase were quantified with a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego).
Interaction and Posttranslational Modification.
RKO cells were plated into 10-cm dishes at 2 × 104 cells per cm2 and cultured for 24 hr at 37°C. Cells were transfected with 4 μg of pcDNA3.1-ING1b, pcDNA3.1-ING2, or pcDNA3.1 (empty vector control) with Lipofectamine Plus. Cells were treated with 5 μM trichostatin A (Wako Biochemicals, Osaka) for 3 hr to aid in the detection of p53 acetylation. Cells were then harvested and lysed in a buffer of 20 mM Tris⋅HCl, pH 7.5/150 mM NaCl/2 mM EDTA/0.2% Triton X-100 with protease inhibitor mixture. Two mg of each cell lysate was immunoprecipitated with agarose-linked anti-p53 Ab-6 (Calbiochem) and Pab 240 (Santa Cruz Biotechnology) or anti-p33ING1b Ping1 with Protein G Plus/Protein A Agarose (Calbiochem). Agarose-linked normal mouse IgG or agarose-linked rabbit IgG (both from Santa Cruz Biotechnology) was used as a negative control. Each immunoprecipitation was washed four times in the same lysis buffer and was analyzed on SDS/10% polyacrylamide gel. p33ING1b, p33ING2, or acetylation of p53 at Lys-382 in the p53-immunoprecipitated complex was detected by Ping1, Ping2, or acetylated p53 Ab-1 antibody, respectively. p53 in the p33ING1b-immunoprecipitated complex was detected by anti-p53 Ab-6. Phosphorylation of p53 at Ser-15 or Ser-392 was analyzed by Phospho-p53 or Phospho-p53 antibody (Cell Signaling Technology, Beverly, MA), respectively. C3ABR cells treated with doxorubicin (0.2 μg/ml) were used as positive controls for posttranslational modifications of p53. OsA-CL cells were transfected with pcDNA3.1-ING2, pcDNA3.1-Anti-ING2, or pcDNA3.1 (empty vector control). Cells were treated with etoposide (30 μg/ml) for 8 hr and trichostatin A for 3 hr to analyze p53 acetylation at Lys-382.
Results
cDNA Cloning of the p33ING2 Gene.
We cloned an additional human ING family member, p33ING2, by conducting a blast search with the human p33ING1 cDNA sequence. The entire cDNA sequence of p33ING2 and the coding region of p33ING1/p33ING1b were determined from human placenta cDNA. The sequence data were reported to GenBank (accession nos. AF078835 and AF053537).
Protein Expression of p33ING1b and p33ING2.
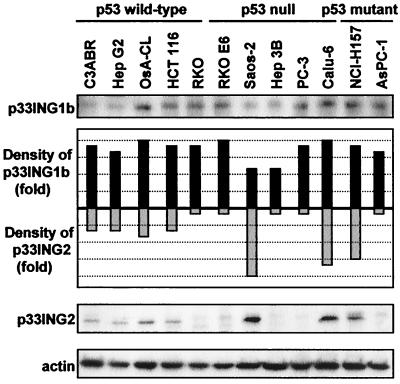
Specific rabbit polyclonal antibodies against p33ING1b and p33ING2 were produced—Ping1 and Ping2, respectively. Cell lines (n = 12) were screened for expression of p33ING1b or p33ING2 by Western blot analysis. All cell lines examined showed some level of p33ING1b expression. In contrast, the level of p33ING2 was highly variable between the cell lines with no visible expression in 5 of the 12. Although the expression profile of p33ING2 did not correlate with the mutational status of the p53 protein as reported for p33ING1, the three cell lines with the most abundant p33ING2 contained either null or mutant p53 (Fig. 1; ref. 37).
Figure 1.
p33ING1b and p33ING2 protein expression. Expression of p33ING1b, p33ING2, and actin was analyzed in 12 human cell lines by Western blotting. The steady-state levels of p33ING1b and p33ING2 were quantified by densitometry and normalized with actin.
DNA Damage and Protein Accumulation.
We then determined whether p53, p33ING1b, p33ING2, or p53 acetylation is induced by DNA damage in the normal lymphoblastoid cell line C3ABR, the ATM-null lymphoblastoid cell line L3, and the p53-null lung cancer cell line Calu-6. In C3ABR cells, p53 accumulated after exposure to each of the DNA-damaging agents examined, but p33ING1b levels did not change. However, p33ING2 protein was specifically induced by etoposide or neocarzinostatin, but not by γ-irradiation, doxorubicin, bleomycin, or cis-platinum. Etoposide treatment of L3 cells or Calu-6 cells also led to induction of p33ING2. p53 was acetylated at Lys-382 after exposure to the DNA-damaging agents etoposide, neocarzinostatin, or doxorubicin (Fig. 2).
Figure 2.
p53, p33ING1b, and p33ING2 protein expression, and p53 acetylation after DNA damage (A–E). After cells were treated with 10 Gy of γ-irradiation (IR), 0.2 μg/ml doxorubicin (DOX), 30 μg/ml etoposide (ETO), 0.3 μg/ml neocarzinostatin (NCS), 0.03 units/ml bleomycin (BLEO), or 2 μg/ml cis-platinum (CDDP), protein expression of p53, p33ING1b, p33ING2, and actin and p53 acetylation at Lys-382 at each time point were analyzed by Western blotting. (F) The steady-state level of each protein was quantified by densitometry and normalized with actin. p53, p33ING1b, p33ING2, and p53 acetylation at Lys-382 after exposure to ETO.
Regulation of Cell Proliferation.
The effects of p33ING1b and p33ING2 on cell growth, apoptosis, and cell cycle were investigated in two isogenic cell lines: RKO (with wild-type p53) and RKO E6 (with p53 inactivated by the HPV E6 protein; refs. 38 and 39). Transfection of p33ING1b or p33ING2 strongly inhibited colony formation in RKO cells but not as completely as in RKO E6 cells (Fig. 3A). Expression of p33ING1b or p33ING2 also induced apoptosis within 24 hr of transfection in RKO cells, but apoptosis was minimal in RKO E6 cells (Fig. 3B). In addition, expression of p33ING1b or p33ING2 induced G1-phase cell-cycle arrest in RKO cells within 48 hr of transfection but not in RKO E6 cells (Fig. 3C).
Figure 3.
Negative regulation of cell proliferation by p33ING1b or p33ING2. RKO or RKO E6 cells were transfected with empty control, p33ING1b, or p33ING2 expression vector. (A) Colony-formation assay. After 2 weeks of G418 selection, colonies were fixed, stained with crystal violet, and counted. Data represent the means ± SD. Statistical analysis was carried out with the Student's t test. (B) Induction of apoptosis by p33ING1b or p33ING2 was analyzed. Data represent the means ± SD of three independent experiments. Statistical analysis was carried out with the Student's t test. (C) Cell-cycle regulation by p33ING1b or p33ING2 was analyzed by flow cytometry. Cell-cycle profiles (G0/G1, S, and G2/M) were calculated with the modfit program.
Effects on Transcriptional-Transactivation Activity of p53.
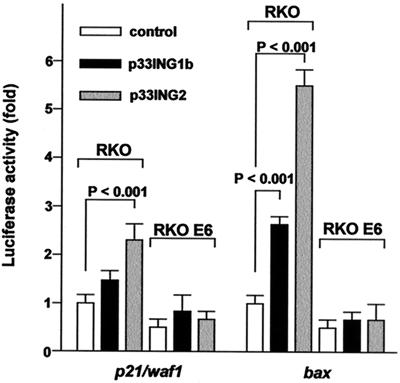
To test the effect of p33ING1b and p33ING2 on the transcriptional-transactivation activity of p53, the p53-regulated p21/waf1 and bax promoter activities were examined. The activities of the p21/waf1 and bax promoters were enhanced by p33ING1b or p33ING2 in RKO cells. p33ING2 was significantly more effective in enhancing p53 transcriptional-transactivation activity than was p33ING1b. p33ING1b or p33ING2 did not have any enhancing effect on the promoter activities in RKO E6 cells (Fig. 4).
Figure 4.
Sequence-specific transcriptional-transactivation activity of p53. Promoter activities of p21/waf1 and bax were detected after cotransfection of p53-responsive reporter constructs with empty control, p33ING1b, or p33ING2 expression vectors in RKO or RKO E6 cells. The luciferase activities from the reporter constructs were measured with a luminometer. Relative luciferase activities (RLAs) were calculated by dividing the luciferase activity by the protein concentration. Results represent the means ± SD of six independent experiments. Differences in the relative luciferase activities were analyzed by using the Student's t test.
Interaction Between p53 and ING Proteins and Posttranslational Modification of p53.
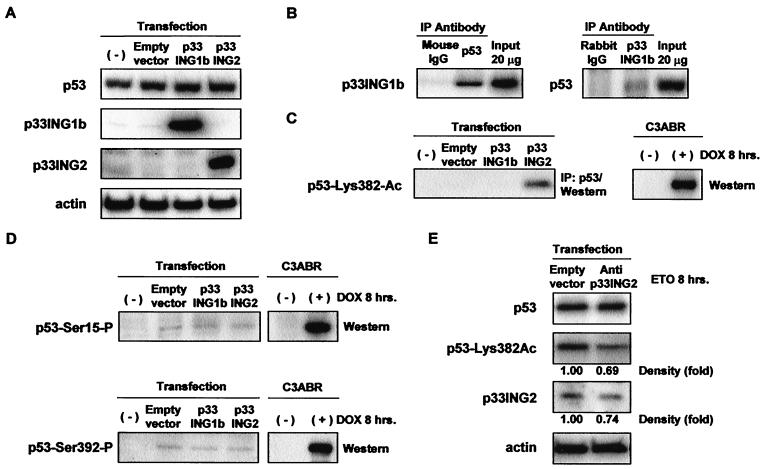
We next analyzed the physical interactions between p53 and the ING proteins by coimmunoprecipitation in RKO cells after transfection of p33ING1b or p33ING2. Expression of p53, p33ING1b, and p33ING2 was detected by Western blot analysis (Fig. 5A). p33ING1b was detected in p53 immunoprecipitates (Fig. 5B Left), but p33ING2 was not detected (data not shown). p53 was also detected in p33ING1b immunoprecipitates in RKO cells after transfection of p33ING1b (Fig. 5B Right). To help elucidate the mechanism by which the ING proteins might activate p53, we investigated the posttranslational modifications of p53 induced by p33ING1b or p33ING2 expression. Although p33ING1b did not increase the acetylation of p53, p33ING2 expression led to acetylation of p53 at Lys-382 (Fig. 5C). The expression of p33ING1b or p33ING2 did not alter the phosphorylation of p53 at either Ser-15 or Ser-392 (Fig. 5D). Expression of antisense-p33ING2 led to a proportional decrease in both p33ING2 and p53 at Lys-382 in OsA-CL cells after exposure to etoposide (Fig. 5E).
Figure 5.
Interaction of p53 with p33ING1b- and p33ING2-mediated acetylation of p53. Immunoprecipitation (IP)-Western analyses were performed with RKO or OsA-CL cell extracts after transfection with empty control, p33ING1b, p33ING2, or anti-p33ING2 expression vector. (A) Expression of p53, p33ING1b, p33ING2, and actin in RKO cells after transfection was detected by Western blot analysis. (B) Anti-p53 (Ab-6 and Pab 240), anti-p33ING1b (Ping1), normal mouse IgG (negative control), or normal rabbit IgG (negative control) were used for immunoprecipitation to analyze the interaction of p53 with p33ING1b in RKO cells. (C) Acetylation of p53 at Lys-382 was detected in anti-p53 (Ab-6 and Pab 240) immunoprecipitates from RKO cells. (D) Phosphorylation of p53 at either Ser-15 or Ser-392 was investigated in RKO cells by Western blot analysis. C3ABR cells were treated with doxorubicin for 8 hr and were used as positive controls for acetylated and phosphorylated p53. (E) Acetylation of p53 at Lys-382 in OsA-CL cells treated for 8 hr with ETO 24 hr after transfection with empty control or anti-p33ING2 expression vector.
Discussion
The human p33ING2 gene was identified by a homology search with human p33ING1. Sequences of the p33ING1 gene originally reported (accession nos. AF001954 and AF044076) were incorrect because of cloning errors and the presence of multiple-splicing isoforms. The amino acid sequence of p33ING2 has 70% homology with the authentic p33ING1/p33ING1b sequence (36). The chromosomal location of ING1 is 13q33-34, and p33ING2 is located on chromosome 4q35. Loss of heterozygosity (LOH) at chromosome 13q33-34 was reported in head and neck squamous cell carcinoma, and LOH at chromosome 4q35 was reported in hepatocellular carcinoma (6, 40–43). A low rate of p33ING1b mutation in head and neck squamous cell carcinoma has been shown (refs. 6 and 44 and K.R., unpublished data). Further mutational analysis of p33ING1b and p33ING2 in these cancer types will be of interest.
Northern blot analysis has revealed ubiquitous expression of p33ING1b and p33ING2 mRNAs in all normal tissues examined (36). Although p33ING1b protein was detected in all cell lines examined by Western blot analysis, the steady-state expression levels of p33ING2 protein in the cancer cell lines were quite variable; expression was very low or absent in several of the cell lines. Analysis of the mechanism of inactivation of p33ING2 expression in cancer cells is warranted.
p33ING2 was specifically induced by the DNA double-strand break-inducing agents etoposide and neocarzinostatin (45, 46). This induction was shown in the ATM-null lymphoblastoid cell line L3 and the p53-null lung cancer cell line Calu-6 (47, 48). Thus, p33ING2 accumulation is independent of either ATM or p53 function. Interestingly, some of the other agents that are expected to induce double-strand breaks did not induce p33ING2. It is possible that structural differences in the breaks caused by these agents can differentially affect downstream pathways.
The PHD-finger motif present in both p33ING1b and p33ING2 is found in nuclear proteins thought to be involved in chromatin-mediated transcriptional regulation, e.g., RBP2 (Rb-binding protein), human IFN-induced nuclear phosphoprotein, and human TIF1 (a putative transcriptional mediator for nuclear receptors; refs. 49 and 50). Analogous to the LIM domain, the PHD-finger motif could be involved in protein–protein interactions for the assembly or activity of multicomponent complexes involved in transcriptional activation or repression.
We have demonstrated that expression of p33ING1b or p33ING2 negatively regulates cell growth through induction of apoptosis and G1-phase cell-cycle arrest in a p53-dependent manner. Confirming previous results (35), p33ING1b slightly enhances p53-dependent transcriptional activities, but it does not enhance acetylation or alter phosphorylation of p53. p33ING1b is reported to be functionally associated with histone deacetylase complex (HDAC)1-dependent transcriptional repression (51). p53 not only can activate gene transcription but also can repress the expression of specific genes (19–22, 27). p53 physically associates with HDACs through the corepressor mSin3a (28). The p53–mSin3a–HDAC1 complex has a role in transcriptional repression of gene expression in a non-sequence-specific manner. Thus, it is possible that the modest p33ING1b-mediated enhancement of p53 sequence-specific target promoters reported both elsewhere (35) and in our report is an artifact of transient overexpression, and that the growth inhibition is the result of p53-dependent transcriptional repression (28).
In contrast with p33ING1b (35), p33ING2 was not detected in a direct association with p53, but it led to an increased acetylation of p53 at Lys-382 in RKO cells. Expression of antisense-p33ING2 reduced the level of acetylated p53 in OsA-CL cells in which p33ING2 was endogenously expressed. Compared with p33ING1b, p33ING2 was a stronger enhancer of p53-dependent transcriptional activities at the p21/waf1 and bax promoters. Yeast ING1-related proteins and human ING1 proteins associate with histone acetyltransferase, and acetylation of lysine residues in histones has been implicated in the regulation of transcriptional activities (ref. 52 and K.R., unpublished data). In addition to a general role in acetylating histones, cAMP response element binding protein (CBP)/p300 and p33/CBP associated factor (PCAF) have been shown to acetylate lysine residues within the C terminus of p53 (53, 54). Sequence-specific transcriptional activation by p53 correlates with its biological function to suppress cell proliferation. Posttranslational modifications such as phosphorylation and acetylation within the C-terminal region of p53 stimulate p53 sequence-specific DNA-binding activity as a result of converting p53 from an inert to an active form (31, 32, 55). We propose that p33ING1b functions in a transcriptional-repression pathway mediated by HDAC activity (28, 51), and that p33ING2 acts in a p53-dependent transcriptional-activation pathway mediated by histone acetyltransferase activity.
DNA damage-induced acetylation of p53 strongly enhances its site-specific DNA-binding and transcriptional-activation functions (31, 32, 55). Inhibition of acetylation can counteract p53-mediated G1-phase cell-cycle arrest and induction of apoptosis (56, 57). Our demonstrations that p33ING2 is induced by DNA damage, that it can enhance p53 acetylation, and that it is absent in a number of tumor cell lines suggest that p33ING2 plays a significant role in these crucial tumor-suppressor pathways. As such, it may prove to be an important marker and/or therapeutic target.
Acknowledgments
We thank Dr. K. K. Khanna for providing the C3ABR and L3 cell lines, Drs. B. Vogelstein and W. El-Deiry for providing the WWP-Luc-p21 and PGL3-Luc-Bax vectors, Drs. Y. Pommier and L. E. Huang for comments on the manuscripts, Drs. Y. H. Higashimoto and E. Appella for advice on preparing the Ping 1 and Ping 2 antibodies, and D. Dudek for editorial and graphic assistance.
Abbreviations
- Pingn
rabbit polyclonal antibodies for p33INGn
- HDAC
histone deacetylase complex
- ATM
ataxia-telangiectasia mutated
Footnotes
References
- 1.Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Nat Genet. 1996;14:415–420. doi: 10.1038/ng1296-415. [DOI] [PubMed] [Google Scholar]
- 2.Garkavtsev I, Demetrick D, Riabowol K. Cytogenet Cell Genet. 1997;76:176–178. doi: 10.1159/000134539. [DOI] [PubMed] [Google Scholar]
- 3.Helbing C C, Veillette C, Riabowol K, Johnston R N, Garkavtsev I. Cancer Res. 1997;57:1255–1258. [PubMed] [Google Scholar]
- 4.Garkavtsev I, Riabowol K. Mol Cell Biol. 1997;17:2014–2019. doi: 10.1128/mcb.17.4.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garkavtsev I. Nat Genet. 1999;23:373. doi: 10.1038/15566. [DOI] [PubMed] [Google Scholar]
- 6.Gunduz M, Ouchida M, Fukushima K, Hanafusa H, Etani T, Nishioka S, Nishizaki K, Shimizu K. Cancer Res. 2000;60:3143–3146. [PubMed] [Google Scholar]
- 7.Saito A, Furukawa T, Fukushige S, Koyama S, Hoshi M, Hayashi Y, Horii A. J Hum Genet. 2000;45:177–181. doi: 10.1007/s100380050206. [DOI] [PubMed] [Google Scholar]
- 8.Lane D P. Nature (London) 1992;358:15–16. doi: 10.1038/358015a0. [DOI] [PubMed] [Google Scholar]
- 9.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 10.Dulic V, Kaufmann W K, Wilson S J, Tlsty T D, Lees E, Harper J W, Elledge S J, Reed S I. Cell. 1994;76:1013–1023. doi: 10.1016/0092-8674(94)90379-4. [DOI] [PubMed] [Google Scholar]
- 11.Di Leonardo A, Linke S P, Clarkin K, Wahl G M. Genes Dev. 1994;8:2540–2551. doi: 10.1101/gad.8.21.2540. [DOI] [PubMed] [Google Scholar]
- 12.Canman C E, Kastan M B. Adv Pharmacol. 1997;41:429–460. doi: 10.1016/s1054-3589(08)61068-6. [DOI] [PubMed] [Google Scholar]
- 13.Serrano M, Lin A W, McCurrach M E, Beach D, Lowe S W. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 14.Wang X W, Yeh H, Schaeffer L, Roy R, Moncollin V, Egly J M, Wang Z, Freidberg E C, Evans M K, Taffe B G, et al. Nat Genet. 1995;10:188–195. doi: 10.1038/ng0695-188. [DOI] [PubMed] [Google Scholar]
- 15.Smith M L, Chen I T, Zhan Q, O'Connor P M, Fornace A J., Jr Oncogene. 1995;10:1053–1059. [PubMed] [Google Scholar]
- 16.Ford J M, Hanawalt P C. Proc Natl Acad Sci USA. 1995;92:8876–8880. doi: 10.1073/pnas.92.19.8876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Offer H, Wolkowicz R, Matas D, Blumenstein S, Livneh Z, Rotter V. FEBS Lett. 1999;450:197–204. doi: 10.1016/s0014-5793(99)00505-0. [DOI] [PubMed] [Google Scholar]
- 18.Zhou J, Ahn J, Wilson S H, Prives C. EMBO J. 2001;20:914–923. doi: 10.1093/emboj/20.4.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine A J, Perry M E, Chang A, Silver A, Dittmer D, Wu M, Welsh D. Br J Cancer. 1994;69:409–416. doi: 10.1038/bjc.1994.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zambetti G P, Bargonetti J, Walker K, Prives C, Levine A J. Genes Dev. 1992;6:1143–1152. doi: 10.1101/gad.6.7.1143. [DOI] [PubMed] [Google Scholar]
- 21.Farmer G, Bargonetti J, Zhu H, Friedman P, Prywes R, Prives C. Nature (London) 1992;358:83–86. doi: 10.1038/358083a0. [DOI] [PubMed] [Google Scholar]
- 22.Ginsberg D, Mechta F, Yaniv M, Oren M. Proc Natl Acad Sci USA. 1991;88:9979–9983. doi: 10.1073/pnas.88.22.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Deiry W S, Tokino T, Velculescu V E, Levy D B, Parsons R, Trent J M, Lin D, Mercer W E, Kinzler K W, Vogelstein B. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 24.Miyashita T, Reed J C. Cell. 1995;80:293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 25.Rajah R, Valentinis B, Cohen P. J Biol Chem. 1997;272:12181–12188. doi: 10.1074/jbc.272.18.12181. [DOI] [PubMed] [Google Scholar]
- 26.Zhou X, Wang X W, Xu L, Hagiwara K, Nagashima M, Wolkowicz R, Zurer I, Rotter V, Harris C C. Cancer Res. 1999;59:843–848. [PubMed] [Google Scholar]
- 27.Ko L J, Prives C. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 28.Murphy M, Ahn J, Walker K K, Hoffman W H, Evans R M, Levine A J, George D L. Genes Dev. 1999;13:2490–2501. doi: 10.1101/gad.13.19.2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shieh S Y, Ikeda M, Taya Y, Prives C. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- 30.Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, et al. Cell. 2000;102:849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 31.Gu W, Roeder R G. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 32.Sakaguchi K, Herrera J E, Saito S, Miki T, Bustin M, Vassilev A, Anderson C W, Appella E. Genes Dev. 1998;12:2831–2841. doi: 10.1101/gad.12.18.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez M S, Desterro J M, Lain S, Midgley C A, Lane D P, Hay R T. EMBO J. 1999;18:6455–6461. doi: 10.1093/emboj/18.22.6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Giaccia A J, Kastan M B. Genes Dev. 1998;12:2973–2983. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 35.Garkavtsev I, Grigorian I A, Ossovskaya V S, Chernov M V, Chumakov P M, Gudkov A V. Nature (London) 1998;391:295–298. doi: 10.1038/34675. [DOI] [PubMed] [Google Scholar]
- 36.Shimada Y, Saito A, Suzuki M, Takahashi E, Horie M. Cytogenet Cell Genet. 1998;83:232–235. doi: 10.1159/000015188. [DOI] [PubMed] [Google Scholar]
- 37.Cheung K J, Bush J A, Jia W, Li G. Br J Cancer. 2000;83:1468–1472. doi: 10.1054/bjoc.2000.1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kessis T D, Slebos R J, Nelson W G, Kastan M B, Plunkett B S, Han S M, Lorincz A T, Hedrick L, Cho K R. Proc Natl Acad Sci USA. 1993;90:3988–3992. doi: 10.1073/pnas.90.9.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Havre P A, Yuan J, Hedrick L, Cho K R, Glazer P M. Cancer Res. 1995;55:4420–4424. [PubMed] [Google Scholar]
- 40.Maestro R, Piccinin S, Doglioni C, Gasparotto D, Vukosavljevic T, Sulfaro S, Barzan L, Boiocchi M. Cancer Res. 1996;56:1146–1150. [PubMed] [Google Scholar]
- 41.Kuroki T, Fujiwara Y, Tsuchiya E, Nakamori S, Imaoka S, Kanematsu T, Nakamura Y. Genes Chromosomes Cancer. 1995;13:163–167. doi: 10.1002/gcc.2870130305. [DOI] [PubMed] [Google Scholar]
- 42.Rashid A, Wang J S, Qian G S, Lu B X, Hamilton S R, Groopman J D. Br J Cancer. 1999;80:59–66. doi: 10.1038/sj.bjc.6690321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bando K, Nagai H, Matsumoto S, Koyama M, Kawamura N, Onda M, Emi M. Genes Chromosomes Cancer. 1999;25:284–289. [PubMed] [Google Scholar]
- 44.Sanchez-Cespedes M, Okami K, Cairns P, Sidransky D. Genes Chromosomes Cancer. 2000;27:319–322. doi: 10.1002/(sici)1098-2264(200003)27:3<319::aid-gcc13>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 45.Maser R S, Monsen K J, Nelms B E, Petrini J H. Mol Cell Biol. 1997;17:6087–6096. doi: 10.1128/mcb.17.10.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shao R G, Cao C X, Zhang H, Kohn K W, Wold M S, Pommier Y. EMBO J. 1999;18:1397–1406. doi: 10.1093/emboj/18.5.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Banin S, Moyal L, Shieh S, Taya Y, Anderson C W, Chessa L, Smorodinsky N I, Prives C, Reiss Y, Shiloh Y, et al. Science. 1998;281:1674–1677. doi: 10.1126/science.281.5383.1674. [DOI] [PubMed] [Google Scholar]
- 48.Canman C E, Lim D S, Cimprich K A, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan M B, Siliciano J D. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 49.Aasland R, Gibson T J, Stewart A F. Trends Biochem Sci. 1995;20:56–59. doi: 10.1016/s0968-0004(00)88957-4. [DOI] [PubMed] [Google Scholar]
- 50.Moosmann P, Georgiev O, Le Douarin B, Bourquin J P, Schaffner W. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skowyra D, Zeremski M, Neznanov N, Li M, Choi Y, Uesugi M, Hauser C A, Gu W, Gudkov A V, Qin J. J Biol Chem. 2001;276:8734–8739. doi: 10.1074/jbc.M007664200. [DOI] [PubMed] [Google Scholar]
- 52.Loewith R, Meijer M, Lees-Miller S P, Riabowol K, Young D. Mol Cell Biol. 2000;20:3807–3816. doi: 10.1128/mcb.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grunstein M. Nature (London) 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 54.Kouzarides T. Curr Opin Genet Dev. 1999;9:40–48. doi: 10.1016/s0959-437x(99)80006-9. [DOI] [PubMed] [Google Scholar]
- 55.Liu L, Scolnick D M, Trievel R C, Zhang H B, Marmorstein R, Halazonetis T D, Berger S L. Mol Cell Biol. 1999;19:1202–1209. doi: 10.1128/mcb.19.2.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lill N L, Grossman S R, Ginsberg D, DeCaprio J, Livingston D M. Nature (London) 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 57.Avantaggiati M L, Ogryzko V, Gardner K, Giordano A, Levine A S, Kelly K. Cell. 1997;89:1175–1184. doi: 10.1016/s0092-8674(00)80304-9. [DOI] [PubMed] [Google Scholar]