Abstract
Background: DNA methylation in sputum has been an attractive candidate biomarker for the non-invasive screening and detection of lung cancer. Materials and Methods: Databases including PubMed, Ovid, Cochrane library, Web of Science databases, Chinese Biological Medicine (CBM), Chinese National Knowledge Infrastructure (CNKI), Wanfang, Vip Databases and Google Scholar were searched to collect the diagnostic trials on aberrant DNA methylation in the screening and detection of lung cancer published until 1 December 2016. Indirect comparison meta-analysis was used to evaluate the diagnostic value of the included candidate genes. Results: The systematic literature search yielded a total of 33 studies including a total of 4801 subjects (2238 patients with lung cancer and 2563 controls) and covering 32 genes. We identified that methylated genes in sputum samples for the early screening and auxiliary detection of lung cancer yielded an overall sensitivity of 0.46 (0.41–0.50) and specificity of 0.83 (0.80–0.86). Combined indirect comparisons identified the superior gene of SOX17 (sensitivity: 0.84, specificity: 0.88), CDO1 (sensitivity: 0.78, specificity: 0.67), ZFP42 (sensitivity: 0.87, specificity: 0.63) and TAC1 (sensitivity: 0.86, specificity: 0.75). Conclusions: The present meta-analysis demonstrates that methylated SOX17, CDO1, ZFP42, TAC1, FAM19A4, FHIT, MGMT, p16, and RASSF1A are potential superior biomarkers for the screening and auxiliary detection of lung cancer.
Keywords: DNA methylation, lung cancer, sputum, detection, meta-analysis, indirect efficacy comparison
1. Introduction
Lung cancer is the leading cause of malignant tumor death, with the morbidity and mortality of lung cancer gradually increasing over the past decades [1]. Only 13% of lung cancer patients survive more than 5 years, and the mortality is close to the morbidity (the ratio of mortality to morbidity is 0.87) [1,2,3]. Despite research on the diagnosis of lung cancer and the use of increasingly advanced technology in its treatment, the prognosis remains poor because of the predominant diagnosis of III, IV-stage disease [3]. Therefore, early diagnosis, auxiliary detection, and treatment have become a major focus to reduce the mortality caused by lung cancer.
With the emergence and development of molecular epidemiology, which opens the “black box” of the disease process, molecular biomarkers have much potential to improve the understanding of the occurrence, development, and prognosis of disease for the early detection of these lesions at the pre-invasive stage, even predicting the progress of the disease [4,5]. Nowadays, it is clearly acknowledged that genetic alterations are accompanied by equally important epigenetic modifications in the pathogenesis of lung cancer [6,7]. In particular, DNA methylation is one of the earliest epigenetic modifications; it is closely associated with the occurrence and development of lung cancer, and it appears earlier than obvious malignant phenotype [7]. Abundant evidence manifests that a variety of well-known DNA methylations can be used as a promising biomarker for the early diagnosis of lung cancer, such as p16 [8], RASSF1A [9], APC [10], MGMT [11], DAPK [12] and RARβ [13]. Many studies have demonstrated that the aberrant methylation of some genes in sputum samples could be a novel “remote medium” for the early detection of lung cancer, which avoids the necessity of invasive procedures [5,14]. The development of next-generation sequencing technology and the maturation of methylation detection technology [15,16] have made methylation detection much more stable and cheaper, and could provide the necessary conditions for clinical utility. Taken together, findings suggest that methylation could serve as efficient diagnostic biomarkers for lung cancer.
However, the DNA methylation of specific genes in sputum samples do not yet provide a convincing and superior gene panel with high sensitivity and specificity for clinical utility in lung cancer. In addition, there was still no comparative evaluation on the diagnostic accuracy of methylation in sputum samples for the screening and detection of lung cancer. Recently, network meta-analysis has been developed to assess the comparative effectiveness of several interventions and to synthesize evidence across a network of studies [17,18]. This method makes it possible to estimate the comparative diagnostic accuracy of multiple biomarkers which have not been directly compared with each other in one study but have been reported by multiple studies under a common comparator. Therefore, we performed a network meta-analysis to indirectly compare the efficacy of the DNA methylation of multiple genes in the diagnosis of lung cancer.
2. Materials and Methods
We followed the reporting guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analysis (PRISMA) statement when conducting this review. The PRISMA statement has guidelines that include a four-phase flow diagram to systematically guide the inclusion and exclusion of research papers [19]. In addition, the guidelines provide a 27-item checklist that describes the requirements per review section (e.g., title, abstract, introduction, methods, results, discussions, and funding) to ensure that systematic reviews are properly conducted and reported [19].
2.1. Search Strategy
We conducted a comprehensive literature search in PubMed, Ovid, Cochrane library, Web of Science databases, Chinese Biological Medicine (CBM), Chinese National Knowledge Infrastructure (CNKI), Wanfang, Vip Databases, and Google Scholar. The main search terms included: lung cancer or lung carcinoma or non-small cell lung cancer or “NSCLC”, “sputum or flema”, “diagnostic”, “sensitivity and specificity” and “methylation or hypermethylation or hypomethylation or demethylation”. All articles published until December 2016 were considered. In addition, the reference lists of all identified studies were manually searched to identify any additional studies.
2.2. Inclusion Criteria and Exclusion Criteria
The articles, which could not be excluded based on the title and abstract, were retrieved for full-text review. Studies were included in this meta-analysis if they met the following criteria: (1) the diagnostic potential of sputum DNA methylation for lung cancer; (2) study design being case-control; (3) the patients being diagnosed with lung cancer by pathology; (4) provided data on the numbers of true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN); (5) the methods of detecting methylation based on methylation-specific polymerase chain reaction (MSP) or quantitative methylation-specific PCR (qMSP).
Studies were excluded from the meta-analysis for the following reasons: (1) abstracts, letters, reviews, expert opinions, case reports, or nonclinical studies; (2) studies had duplicate or overlapping data; (3) study was based on tissue, blood, or animals.
2.3. Data Extraction and Quality Assessment
Data extraction was conducted in duplicate by two investigators (Di Liu and Hongli Peng) based on title, abstract, author, year of publication, country of origin, sample size, assay methods, and diagnostic performance (sensitivity (SEN), specificity (SPE), TP, FP, FN, TN), target gene(s), and the score of the quality assessment of studies of diagnostic accuracy (QUADAS) [20] and the standards for reporting of diagnostic accuracy (STARD) [21]. Any disagreements in data extraction were resolved by consensus.
STARD and QUADAS guidelines were utilized to assess the methodological quality of each study. There are 25 items in the STARD initiative checklist, and a score of 1 was given when the item was yielded [21]. Fourteen items were included in the QUADAS tool, whereby a score of 1 was given when a specific item was fulfilled, 0 if this item was unclear, and −1 for the item not achieved [20]. All of these studies were evaluated independently and discussed by the reviewers until a consensus was reached.
2.4. Statistical Analysis
We used standard methods recommended for the direct meta-analysis which estimated the diagnostic test evaluation of DNA methylation compared with the gold standard [22]. The number of TP, TN, FP, and FN were retrieved from each article. The SEN, SPE, positive likelihood ratio (PLR), negative likelihood ratio (NLR), diagnostic odds ratio (DOR) estimates with 95% confidence interval (CI) from each study were analyzed using a random-effect model and the bivariate summary receiver operating characteristic (SROC) curve was generated. The area under the curve (AUC) represents an analytical summary of the test performance and illustrates the trade-off between sensitivity and specificity [23]. The heterogeneity among studies was assessed on the basis of the Chi Square test using the Cochran Q statistic. The I2 statistic, which measures the extent of inconsistency between studies, was also assessed [24]. Spearman’s correlation coefficient of logarithm sensitivity and 1-specificity for each gene was assessed to determine the threshold effect [25]. Analyses were performed using two statistical software programs (Meta-Disc 1.4 for Windows and Stata version 12.0, Stata Corp, College Station, TX, USA).
For indirect comparisons, the comparative diagnostic accuracy of all biomarkers was estimated according to common comparator (the gold standard). We did not assume consistency (which was evaluated by comparing the direct estimates with the indirect estimates for each comparison) of two biomarkers without direct analyses. We took the step-wise approach, which was suitable for the simple star-network meta-analysis to obtain an indirect analysis. The Deeks’ test and Egger’s test were utilized to estimate the funnel plot asymmetry and the publication bias [26]. The indirect meta-analysis was conducted using indirect treatment comparison (ITC) software and Stata 12.0 (Stata Corp, College Station, TX, USA) [27]. A two-sided p-value of less than 0.05 was considered significant.
3. Results
3.1. Subsection
3.1.1. Study Characteristics and Quality of Included Studies
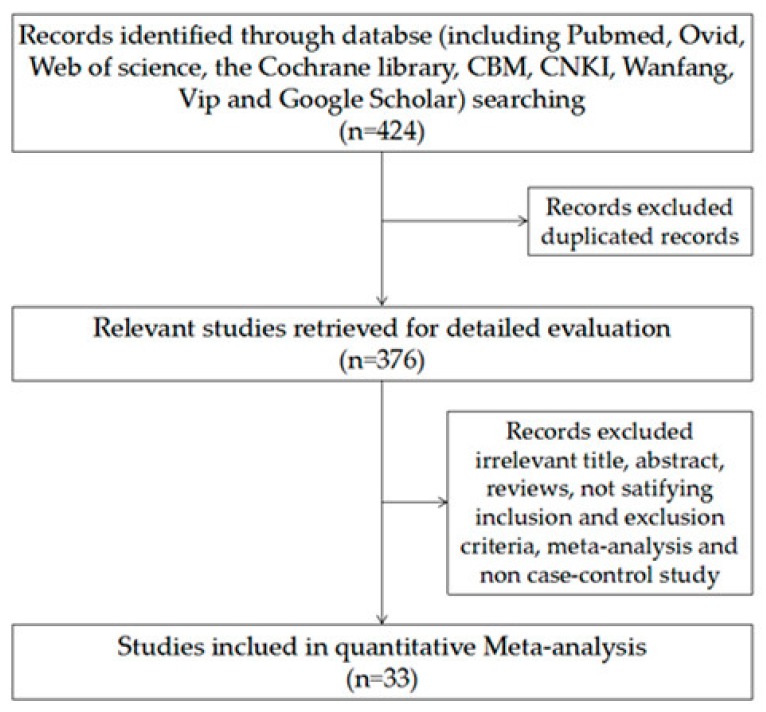
The flowchart of included studies was presented in Figure 1. A total of 424 studies were preliminarily reviewed, of which 33 were available for the indirect meta-analysis [5,14,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58]. The characteristics of each study are shown in Table 1, including name of the study, number of patients and controls, biomarkers, and quality assessment based on STARD and QUADAS. The systematic literature search yielded a total of 33 studies including a total of 4801 subjects (2238 patients with lung cancer and 2563 controls) and covering 32 genes (P16, RASSF1A, APC, MGMT, PAX5, CGB, GATA, DAPK, RARβ, MAGE, HOXA, 3OST2, PRDM14, FAM19A4, PHACTR3, PCDH20, CXCL, Dal-1, Dab2, Dcr2, SULF2, Kifla, Jph3, SOX17, CDO1, ZFP42, TAC1, CDH1, H-cadherin, FHIT, RASSF2, TCF2l).
Figure 1.
Flow diagram of studies through the review process.
Table 1.
Baseline characteristics of all included studies.
| Study/Year | Country | Cases/Controls | Biomarkers | STARD | QUADAS |
|---|---|---|---|---|---|
| Destro/2004 [41] | Italy | 24/100 | p16 | 21 | 12 |
| Zhang/2004 [48] | China | 44/20 | p16 | 18 | 8 |
| Wang/2004 [57] | China | 34/21 | p16 | 17 | 7 |
| Konno/2004 [31] | Japan | 78/94 | p16, APC, RARβ | 20 | 11 |
| Belinsky/2005 [39] | USA | 53/118 | p16, MGMT, RASSF1A, DAP, H-cadherin, PAX5 | 20 | 9 |
| Olaussen/2005 [32] | France | 20/17 | HOXA9, p16, MAGE | 17 | 8 |
| Cirincione/2006 [44] | Italy | 18/112 | RARβ, p16, RASSF1A | 15 | 7 |
| Wang/2006 [45] | China | 79/22 | FHIT, p16, RARβ | 20 | 10 |
| Liu/2006 [54] | China | 77/30 | MGMT | 18 | 8 |
| Belinsky/2006 [47] | USA | 98/92 | p16, PAX5, MGMT, DAPK, GATA, RASSF1ASFRP1, HLHPBETA3, IGFBP3HCAD, LAMC2 | 20 | 10 |
| Georgiou/2007 [30] | Greece | 80/40 | p16 | 16 | 9 |
| Hsu/2007 [5] | China | 82/37 | p16, RARβ | 16 | 7 |
| Liu/2008 [29] | China | 58/107 | p16 | 18 | 10 |
| Guo/2008 [53] | China | 100/50 | p16 | 16 | 7 |
| Van der Drift/2008 [33] | Netherlands | 28/68 | RASSF1A | 19 | 11 |
| Hu/2009 [51] | China | 42/25 | p16 | 16 | 9 |
| Ye/2010 [49] | China | 30/27 | RASSF1A | 16 | 7 |
| Zhang/2010 [49] | China | 82/25 | RASSF1A, p16, DAPK | 18 | 8 |
| Hwang/2011 [36] | Korea | 76/109 | HOXA | 20 | 9 |
| Song/2011 [28] | China | 42/9 | p16, MGMT | 17 | 8 |
| Zhang/2011 [52] | China | 41/15 | p16 | 17 | 9 |
| Hang/2011 [38] | China | 47/24 | FHIT | 20 | 8 |
| Sun/2012 [55] | China | 120/120 | p16, RASSF1A | 19 | 7 |
| Hubers/2012 [46] | The Netherlands | 53/47 | RASSF1A, APC, CYGB | 18 | 10 |
| Guzmán/2012 [14] | Chile | 26/33 | p16, CDH1, MGMT | 18 | 11 |
| Shin/2012 [42] | Korea | 65/30 | MAGE, p16 | 17 | 9 |
| Leng/2012 [35] | USA | 64/64 | p16, MGMT, DAPK, PAX5, GATA, Dal-1, PCDH20, Jph3, Kifla, SULF2, RASSFlA, GATA, Dab2, Dcr2, RASSF2, TCF2l | 20 | 11 |
| Leng/2012 [35] | USA | 40/90 | p16, MGMT, DAPK, PAX5, GATA, Dal-1, PCDH20, Jph3, Kifla, SULF2, CXCL, RASSFlA, Dab2, Dcr2, RASSF2, TCF2l | 20 | 11 |
| Pan/2013 [56] | China | 20/13 | p16 | 19 | 8 |
| Hubers/2014 [34] | The Netherlands | 20/31 | RASSF1A, APC, CYGB, 3OST, PRDM14, FAM19A4, PHACTR3 | 19 | 8 |
| Hubers/2014 [43] | The Netherlands | 98/90 | RASSF1A, APC, CYGB | 20 | 10 |
| Hubers/2014 [43] | The Netherlands | 60/445 | RASSF1A, APC, CYGB | 20 | 10 |
| Hubers/2015 [58] | The Netherlands | 73/86 | RASSF1A, APC, CYGB, 3OST2, PRDM14, FAM19A4, PHACTR3 | 21 | 11 |
| Hubers/2015 [58] | The Netherlands | 159/154 | RASSF1A, APC, CYGB, 3OST2, PRDM14, FAM19A4, PHACTR3 | 21 | 11 |
| Su/2016 [40] | China | 117/174 | RASSF1A, 3OST2, PRDM14 | 18 | 7 |
| Hulbert/2016 [37] | USA | 90/24 | SOX17, TAC1, CDO1, HOXA, ZFP42 | 16 | 9 |
STARD: Standards for reporting of diagnostic accuracy; QUADAS: Quality assessment of studies of diagnostic accuracy
3.1.2. Summary Performance of Diagnostic Estimates
As shown in Table 2, of the 32 genes, the SEN (sensitivity) ranged from 0.03 to 0.87 (pooled 0.46; 95% CI: 0.41–0.50), whereas SPE ranged from 0.25 to 0.99 (pooled 0.83; 95% CI: 0.80–0.86).The PLR (positive likelihood ratio) ranged from 1.03 to 6.76 (pooled 2.72; 95% CI: 2.32–3.22),NLR (negative likelihood ratio) ranged from 0.18 to 0.98 (pooled 0.64; 95% CI: 0.60–0.68), and DOR (diagnostic odds ratio) ranged from 1.05 to 38.00 (pooled 4.28; 95% CI: 3.50–5.20). Of the 32 genes, the higher of SEN were ZFP42: 0.87 (0.78–0.93), TAC1: 0.86 (0.77–0.92), SOX17: 0.84 (0.75–0.91), FAM19A4: 0.80 (0.74–0.85), and HOXA: 0.79 (0.63–0.89); whereas the higher of PLR were SOX17: 6.76 (2.34–19.54), FHIT: 5.93 (2.29–15.36), RASSF1A: 5.61 (3.73–8.43), MGMT: 4.78 (1.47–15.55), and p16: 4.71 (2.53–8.78).
Table 2.
The summary performance of diagnostic estimates.
| Genes | Study-Case/Control | SEN (95% CI) | SPE (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) |
|---|---|---|---|---|---|---|
| CDH1 | 1–26/33 | 0.35 (0.17–0.56) | 0.70 (0.51–0.84) | 1.14 (0.55–2.39) | 0.94 (0.66–1.00) | 1.22 (0.41–3.65) |
| SOX17 | 1–90/24 | 0.84 (0.75–0.91 | 0.88 (0.68–0.97) | 6.76 (2.34–19.54) | 0.18 (0.11–0.29) | 38.00 (9.98–144.73) |
| CDO1 | 1–90/24 | 0.78 (0.68–0.86) | 0.67 (0.45–0.84) | 2.32 (1.31–4.15) | 0.33 (0.21–0.54) | 7.00 (2.62–18.72) |
| ZFP42 | 1–90/24 | 0.87 (0.78–0.93) | 0.63 (0.41–0.81) | 2.31 (1.37–3.90) | 0.21 (0.12–0.39) | 10.83 (3.88–30.22 |
| TAC1 | 1–90/24 | 0.86 (0.77–0.92) | 0.75 (0.53–0.90) | 3.42 (1.70–6.88) | 0.19 (0.11–0.33) | 17.77 (5.94–53.12) |
| H-cadherin | 1–53/118 | 0.50 (0.23–0.77) | 0.57 (0.46–0.68) | 1.18 (0.66–2.11) | 0.87 (0.50–1.00) | 1.35 (0.43–4.22) |
| FHIT | 2–126/46 | 0.52 (0.43–0.61) | 0.91 (0.79–0.98) | 5.93 (2.29–15.36) | 0.53 (0.43–0.65) | 11.19 (3.79–33.06) |
| PCDH20 | 2–104/154 | 0.58 (0.48–0.67) | 0.49 (0.41–0.58) | 1.14 (0.91–1.43) | 0.86 (0.65–1.00) | 1.33 (0.80–2.19) |
| Dab2 | 2–104/154 | 0.03 (0.01–0.08) | 0.99 (0.95–1.00) | 2.22 (0.38–13.06) | 0.98 (0.95–1.00) | 2.26 (0.37–13.75) |
| Dcr2 | 2–104/154 | 0.41 (0.32–0.51) | 0.60 (0.52–0.68) | 1.03 (0.76–1.38) | 0.98 (0.80–1.00) | 1.05 (0.63–1.73) |
| SULF2 | 2–104/154 | 0.51 (0.41–0.61) | 0.57 (0.49–0.65) | 1.19 (0.91–1.55) | 0.86 (0.68–1.00) | 1.39 (0.84–2.28) |
| Kifla | 2–104/154 | 0.44 (0.34–0.54 | 0.62 (0.54–0.70 | 1.17 (0.87–1.58) | 0.89 (0.72–1.00) | 1.31 (0.79–2.18) |
| Dal-1 | 2–104/154 | 0.30 (0.21–0.39 | 0.86 (0.80–0.91 | 2.17 (1.32–3.55) | 0.82 (0.71–0.94) | 2.65 (1.42–4.94) |
| Jph3 | 2–104/154 | 0.31 (0.23–0.41 | 0.79 (0.721–0.85 | 1.47 (0.97–2.22) | 0.87 (0.75–1.00) | 1.68 (0.96–2.95) |
| RASSF2 | 2–104/154 | 0.08 (0.03–0.15) | 0.95 (0.91–0.98) | 1.69 (0.63–4.52) | 0.97 (0.91–1.00) | 1.75 (0.61–4.98) |
| TCF2l | 2–104/154 | 0.29 (0.20–0.39) | 0.71 (0.63–0.78) | 0.99 (0.67–1.46) | 1.01 (0.86–1.00) | 0.98 (0.57–1.70) |
| CXCL | 2–80/180 | 0.36 (0.26–0.48 | 0.79 (0.72–0.85 | 1.72 (1.15–2.58) | 0.81 (0.67–0.97) | 2.12 (1.19–23.79) |
| MAGE | 4–202/118 | 0.45 (0.34–0.55 | 0.82 (0.56–0.94 | 2.44 (0.75–7.96 | 0.68 (0.47–0.99 | 3.60 (0.76–16.98 |
| HOXA | 4–276/174 | 0.79 (0.63–0.89) | 0.50 (0.16–0.84) | 1.56 (0.80–3.07) | 0.43 (0.28–0.66) | 3.63 (1.28–10.26) |
| RARβ | 4–257/223 | 0.44 (0.29–0.60) | 0.79 (0.58–0.91) | 2.09 (0.93–4.70) | 0.71 (0.52–0.98) | 2.93 (0.99–8.69) |
| FAM19A4 | 3–252/271 | 0.80 (0.74–0.85 | 0.25 (0.20–0.30) | 1.06 (0.97–1.16) | 0.82 (0.59–1.00) | 1.29 (0.86–1.96) |
| PHACTR3 | 3–252/271 | 0.60 (0.53–0.66 | 0.68 (0.62–0.73) | 1.85 (1.52–2.27) | 0.60 (0.50–0.71) | 3.11 (2.17–4.45) |
| DAPK | 5–337/389 | 0.45 (0.40–0.51) | 0.79 (0.64–0.89) | 2.16 (1.13–4.14) | 0.69 (0.58–0.86) | 3.12 (1.32–7.36) |
| 3OST2 | 4–369/445 | 0.50 (0.45–0.55) | 0.85 (0.82–0.88) | 3.36 (2.63–4.30) | 0.59 (0.53–0.66) | 5.71 (4.10–7.96) |
| PRDM14 | 4–369/445 | 0.62 (0.57–0.67) | 0.76 (0.72–0.80) | 2.63 (2.19–3.17) | 0.50 (0.43–0.57) | 5.30 (3.91–7.17) |
| GATA | 6–404/492 | 0.66 (0.31–0.90) | 0.53 (0.33–0.71) | 1.40 (1.16–1.69) | 0.64 (0.34–1.21) | 2.20 (1.01–4.83) |
| MGMT | 8–447/460 | 0.42 (0.32–0.52) | 0.91 (0.77–0.97) | 4.78 (1.47–15.55) | 0.64 (0.50–0.81) | 7.48 (1.87–29.91) |
| PAX5 | 8–510/728 | 0.37 (0.29–0.45) | 0.78 (0.70–0.84) | 1.65 (1.28–2.12) | 0.81 (0.74–0.90) | 2.02 (1.45–2.83) |
| CYGB | 6–453/853 | 0.51 (0.45–0.57) | 0.79 (0.69–0.88) | 2.39 (1.61–3.56) | 0.62 (0.54–0.72) | 3.83 (2.28–6.44) |
| APC | 8–588/928 | 0.43 (0.34–0.53) | 0.87 (0.71–0.95) | 3.30 (1.67–6.51) | 0.65 (0.59–0.72) | 5.06 (2.55–10.04) |
| p16 | 24–1357/1249 | 0.48 (0.40–0.56) | 0.90 (0.82–0.95) | 4.71 (2.53–8.78) | 0.58 (0.50–0.68) | 8.11 (3.94–16.70) |
| RASSF1A | 17–1160/1767 | 0.28 (0.20–0.38) | 0.95 (0.93–0.97) | 5.61 (3.73–8.43) | 0.76 (0.67–0.85) | 7.40 (4.54–12.06) |
| Summary | 33–2238/2563 | 0.46 (0.41–0.50) | 0.83 (0.80–0.86) | 2.72 (2.32–3.22) | 0.64 (0.60–0.68) | 4.28 (3.50–5.20) |
DOR: Diagnostic odds ratio; NLR: Negative likelihood ratio; PLR: Positive likelihood ratio; SEN: Sensitivity; SPE: Specificity. Bold text: The top five values.
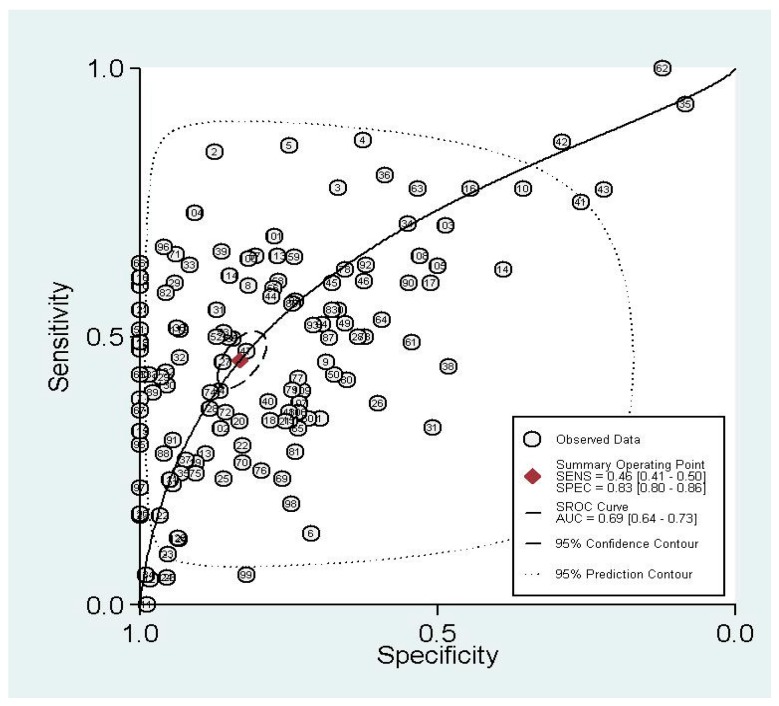
The graph of the SROC (summary receiver operating characteristic) cure is shown in Figure 2, which demonstrates the trade-off between SEN and SPE values in multiple studies. The SROC curve results showed that AUC (area under the curve) of 32 different methylated genes was 0.69 (0.64–0.73), indicating the ability of 32 pooled gene methylations to differentiate lung cancer patients from non-lung-cancer patients with a mid-level accuracy.
Figure 2.
Summary receiver operating characteristic (SROC) curve with pooled estimates of sensitivity, specificity, and area under the curve (AUC) on the diagnostic value of gene methylation in lung cancer.
3.1.3. Indirect Comparisons of Diagnostic Analysis
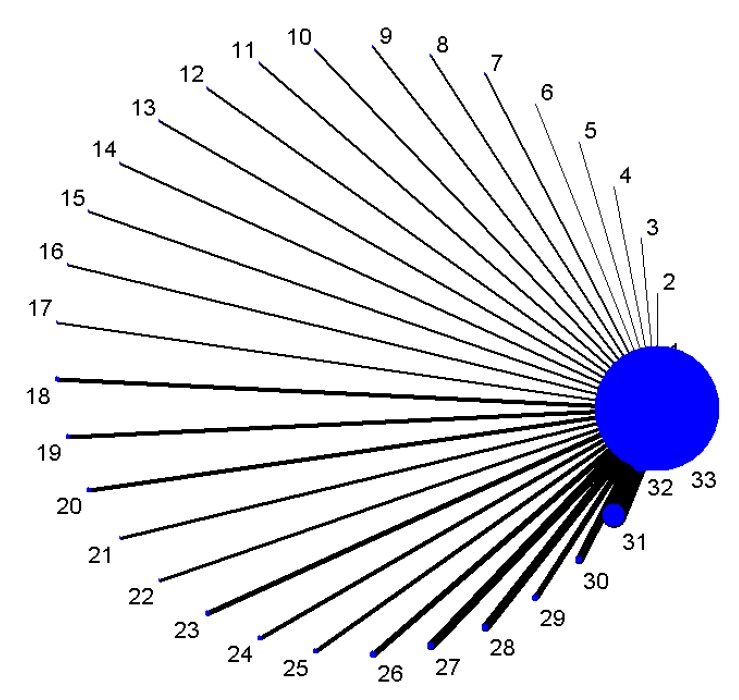
Figure 3 shows the star-network of comparisons for methylated genes (1–32) with the gold standard of being diagnosed with lung cancer (33). We established a network to compare the diagnostic accuracy of 32 methylated genes, with the results of indirect comparisons presented in Table S1. The OR and 95% CI of SOX17, TAC1, ZFP42, CDO1 differed significantly from the other 28 methylated genes and were higher than 1, indicating that SOX17, TAC1, ZFP42, and CDO1 have a higher diagnostic accuracy. More information is shown in the supplemental content (Figures S1–S32). We combined indirect comparisons to evaluate the comparative efficacy of different methylated genes; the superior genes were performed by SOX17 (sensitivity: 0.84, specificity: 0.88), CDO1 (sensitivity: 0.78, specificity: 0.67), ZFP42 (sensitivity: 0.87, specificity: 0.63), TAC1 (sensitivity: 0.86, specificity: 0.75) (Table 2).
Figure 3.
The star-network plot of different methylated genes for 1–32 (1/CDH1, 2/SOX17, 3/CDO1, 4/ZFP42, 5/TAC1, 6/H-cadherin, 7/FHIT, 8/PCDH20, 9/Dab2, 10/Dcr2, 11/SULF2, 12/Kifla, 13/Dal-1, 14/Jph3, 15/RASSF2, 16/TCF2l, 17/CXCL, 18/MAGE, 19/HOXA, 20/RARβ, 21/FAM19A4, 22/PHACTR3, 23/DAPK, 24/3OST2, 25/PRDM14, 26/GATA, 27/MGMT, 28/PAX5, 29/CYGB, 30/APC, 31/p16, 32/RASSF1A) and 33/the gold standard of being diagnosed with lung cancer.
3.1.4. Test of Heterogeneity and Meta-Regression
In the meta-analysis, computation of the Spearman’s rank correlation coefficient between the logit of sensitivity and that of 1-specificity of sputum DNA testing was 0.465 (p < 0.001), indicating the heterogeneity of threshold effect. We also investigated the non-threshold effects; the results indicated the existence of significant heterogeneity in the overall sensitivity (I2 = 91.2%, p < 0.001), specificity (I2 = 93.5%, p < 0.001), PLR (I2 = 85.4%, p < 0.001), NLR (I2 = 88.8%, p < 0.001), and DOR (I2 = 72.6%, p < 0.001). Therefore, a bivariate binomial mixed model was applied to summarize the pooled estimates in this study. To determine the sources of heterogeneity, we performed meta-regression to test the effect of ethnicity (Asian/others), sample size (n = 0–100/101–200/201–), and the quality of study (low/medium/high). Multivariable regression showed that ethnicity (coefficient = −0.785, p = 0.001) and the sample size (coefficient = −0.324, p = 0.036) had statistically significant differences, while the quality of study (coefficient = −0.074, p = 0.552) showed no significant difference. Then, we conducted subgroup analysis based on ethnicity and the sample size, as shown in Table 3.
Table 3.
Detailed information of subgroup analysis.
| Analysis | SEN (95% CI) | SPE (95% CI) | PLR (95% CI) | NLR (95% CI) | DOR (95% CI) |
|---|---|---|---|---|---|
| Ethnicity | |||||
| Asian | 0.46 (0.44–0.48) | 0.84 (0.93–0.86) | 4.04 (2.91–5.62) | 0.61 (0.55–0.66) | 6.50 (4.73–8.92) |
| Others | 0.47 (0.46–0.48) | 0.75 (0.74–0.75) | 1.88 (1.68–2.10) | 0.72 (0.68–0.77) | 2.92 (2.46–3.47) |
| Sample size | |||||
| 0–100 | 0.48 (0.46–0.51) | 0.85 (0.83–0.88) | 4.13 (2.80–6.08) | 0.59 (0.54–0.65) | 7.80 (5.22–11.65) |
| 101–200 | 0.45 (0.44–0.46) | 0.75 (0.74–0.76) | 1.78 (1.58–2.00) | 0.73 (0.69–0.78) | 2.75 (2.29–3.31) |
| 201– | 0.51 (0.48–0.53) | 0.76 (0.74–0.77) | 2.52 (1.85–3.44) | 0.65 (0.59–0.73) | 3.94 (2.68–5.42) |
In addition, we performed a meta-regression to test the effect of ethnicity, sample size, and the quality of study with different genes. The results showed that ethnicity (coefficient = −1.117, p = 0.048) and the sample size (coefficient = −1.177, p = 0.026) were of statistically significant bias for p16, while not significant bias for other candidate genes (p > 0.050).
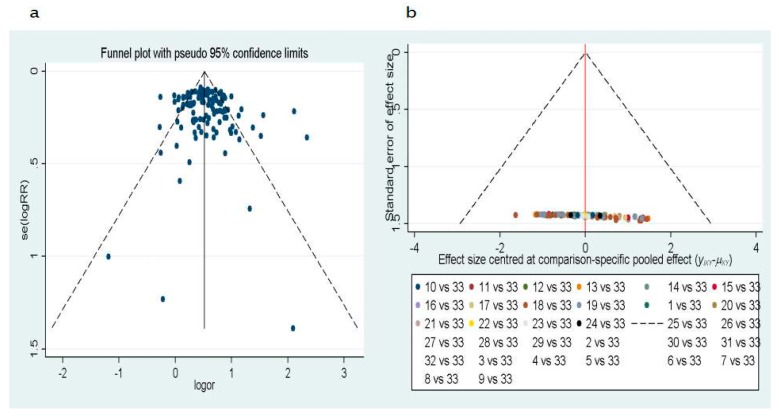
Publication bias was evaluated by Deeks’ test and Egger’s test. The funnel plots for publication bias showed no asymmetry (Figure 4). The result of Deeks’ test showed that p = 0.008, indicating that publication bias could exist in the meta-analysis.
Figure 4.
Funnel plot of the publication bias (a) from Deeks’ test, the data is based on the article of cases and controls. (b) From Egger’s test, the data is based on the article indirect comparisons of methylated gene for 1–32 (1/CDH1, 2/SOX17, 3/CDO1, 4/ZFP42, 5/TAC1, 6/H-cadherin, 7/FHIT, 8/PCDH20, 9/Dab2, 10/Dcr2, 11/SULF2, 12/Kifla, 13/Dal-1, 14/Jph3, 15/RASSF2, 16/TCF2l, 17/CXCL, 18/MAGE, 19/HOXA, 20/RARβ, 21/FAM19A4, 22/PHACTR3, 23/DAPK, 24/3OST2, 25/PRDM14, 26/GATA, 27/MGMT, 28/PAX5, 29/CYGB, 30/APC, 31/p16, 32/RASSF1A) and 33/the gold standard of being diagnosed with lung cancer.
4. Discussion
Lung cancer has become a global burden, further substantiating the need for early screening and auxiliary detection [1,3]. The key to accomplishing both these goals is the better understanding of the genes or pathways disrupted in causing lung cancer [6,45,59]. The fact that silencing genes through hypermethylation or activating genes through hypomethylation play an important role in the initiation and progression of lung cancer has stimulated the development of screening approaches to identify additional genes and pathways that are disrupted within the epigenome [59]. In addition, DNA methylation in sputum samples has the potential to serve as a non-invasive screening method for the identification of specific biomarkers, enabling the early detection of lung cancer [31,39].
In the direct meta-analysis, we identified that methylated genes in sputum samples for the early screening and auxiliary detection of lung cancer yielded an overall sensitivity of 0.46 at the same specificity of 0.83. Furthermore, the PLR (positive likelihood ratio) was 2.72, NLR (negative likelihood ratio) was 0.64, and DOR (diagnostic odds ratio) value was 4.28, and the AUC (area under the curve) was 0.69, indicating a mid-level accuracy. Therefore, we should pick the superior genes for clinical utility as diagnostic biomarkers for lung cancer. Combined indirect comparisons identified the superior genes as SOX17 (sensitivity: 0.84, specificity: 0.88), CDO1 (sensitivity: 0.78, specificity: 0.67), ZFP42 (sensitivity: 0.87, specificity: 0.63), and TAC1 (sensitivity: 0.86, specificity: 0.75). A single DNA methylation biomarker cannot be expected to detect all cases of lung cancer. Some studies demonstrated that combined multiple methylated genes could improve the diagnostic value of cancers [37,60]. We identified that the sensitivity value of methylated FAM19A4 and PLR value of methylated RASSF1A, FHIT, MGMT, and p16 are relatively high, suggesting that they are comprehensive parameters for the screening test [61]. In addition, methylated RASSF1A and p16 genes are reported to be promising driving molecules in many cancers under the concept of precision medicine [9,62,63,64,65,66,67]. In addition, the methylation of FAM19A4, FHIT, and MGMT were reported to play important roles in the occurrence and deterioration of lung cancer [68,69,70,71]. Therefore, we advocate that methylated SOX17, CDO1, ZFP42, TAC1, FAM19A4, FHIT, MGMT, p16, and RASSF1A are useful in the screening and auxiliary detection of lung cancer.
To our knowledge, this study is the first systemic review and indirect meta-analysis to assess the comparative diagnostic effectiveness of the methylation profile of multiple candidate gens in sputum samples for the early screening and detection of lung cancer. According to the method of network meta-analysis [18,27], we used indirect comparison to estimate the comparative diagnostic accuracy of two methylated genes based on a common comparator (the gold standard of being diagnosed with lung cancer). Therefore, the inconsistency between the direct and indirect comparison is not available to address.
However, we observed a large degree of heterogeneity among studies investigating methylation profile in sputum samples used for lung cancer. Threshold effect is one of the primary causes of heterogeneity among diagnostic accuracy studies [24]. In the present meta-analysis, we found obvious heterogeneity as a result of threshold effect, which may be caused by different genes. There is a clear and unified cut-off value for methylation/unmethylation regardless of whether it is based on qualitative analysis or quantitative analysis for each gene tested by the method of MSP [16]. Moreover, we performed meta-regression to test the heterogeneity caused by ethnicity, sample size, and the quality of study. The results suggested that study region (p = 0.001) and the sample size (p = 0.036) might be a source of heterogeneity for this meta-analysis. The results of subgroup analyses showed that large-sample studies had higher sensitivity than the small- and moderate-sample studies, while studies in Asia had lower sensitivity than other regions.
In addition, publication bias could exist in the meta-analysis. This meta-analysis was only based on published studies, therefore inducing the possibility of publication bias. The Deeks’ test and Egger’s test not only detect publication bias, but also indicate the heterogeneity due to the effect of ethnicity, sample size, the quality of study, etc. [72,73]. Therefore, we proposed that the heterogeneity was potentially due to different genes, study region, and sample size. However, this speculation needs to be investigated in the future study.
The present network meta-analysis included 33 articles and 32 candidate genes, with the majority of genes only included in one article. These make it difficult to directly compare the diagnostic efficacy among multiple genes; thus, only indirect comparisons were evaluated in this study. However, the absence of direct comparisons may lead to bias. Pairwise meta-analysis and network meta-analysis were carried out sequentially for direct and indirect comparisons of migraine headache days among three interventions compared with those treated by three placebos, and the results showed that there was no significant inconsistency between the direct and indirect evidence for the majority of comparisons [74]. Another network meta-analysis was performed to directly and indirectly compare the effectiveness of several oral antidiabetic drugs in the prevention of cardiovascular mortality and morbidity, and the results indicated that the inconsistency between direct and indirect estimates of all-cause mortality, cardiovascular-related mortality, acute coronary syndrome, and myocardial infraction were significant low [75]. In summary, the present results from indirect comparisons should be reliable and acceptable.
Based on the focus of diagnostic accuracy studies, we identified other common limitations and insufficiency. Firstly, all the publications included in this analysis were reported on case-control studies, indicating that the selection bias could possibly lead to over-estimations of diagnostic accuracy compared with the cross-sectional study and cohort study [76,77]. In addition, the effects of language selection bias and publication bias cannot be ignored in any meta-analysis [19]. Finally, the detection utilizing sputum DNA testing was not good enough. We think two methods might have the potential to screen valid and good biomarkers for the advancement of the field. Firstly, a panel with multiple methylated genes may be of high performance in diagnostic models. Secondly, instead of qualitative methods (MSP), quantitative methods for the determination of the methylation patterns in candidate genes may increase the diagnostic performance.
5. Conclusions
In conclusion, despite these limitations, our meta-analysis advocates that methylated SOX17, CDO1, ZFP42, TAC1, FAM19A4, FHIT, MGMT, p16, and RASSF1A are useful biomarkers in the screening and auxiliary detection of lung cancer. Our findings provide new avenues for assessing the comparative diagnostic effectiveness of several methylations in lung cancer based on the method of network meta-analysis. Further high-quality and large-scale studies are needed to confirm our analysis.
Acknowledgments
This work was supported by grants from National Natural Science Foundation of China (81370083, 81673247), the National Natural Sciences Foundation of China (NSFC)-Australian National Health and Medical Research Council (NHMRC) Joint Research Project (NSFC 81561128020-NHMRC APP1112767), and Beijing Nova Program (Z141107001814058).
Supplementary Materials
The following are available online at www.mdpi.com/1660-4601/14/7/679/s1, Figures S1–S32: Indirect comparisons between one methylated gene and 31 other genes; Table S1: Indirect comparisons of 32 genes.
Author Contributions
Youxin Wang designed the study, wrote and revised the manuscript. Di Liu performed the analysis and interpretation of data, and drafted the manuscript. Hongli Peng, Qi Sun and Zhongyao Zhao performed the analysis and interpretation of data. Xinwei Yu, Siqi Ge, Hao Wang, Honghong Fang, Qing Gao, Jiaonan Liu, Lijuan Wu, Manshu Song provided technical support for the analysis and critical revision of the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., He J. Cancer statistics in China, 2015. Cancer J. Clin. 2016;66:115. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Sun X., Tong L.P., Wang Y.T., Wu Y.X., Sheng H.S., Lu L.J., Wang W. Can global variation of nasopharynx cancer be retrieved from the combined analyses of IARC Cancer Information (CIN) databases? PLoS ONE. 2011;6:e22039. doi: 10.1371/journal.pone.0022039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teng A.M., Atkinson J., Disney G., Wilson N., Blakely T. Changing socioeconomic inequalities in cancer incidence and mortality: Cohort study with 54 million person-years follow-up 1981–2011. Int. J. Cancer. 2016;140:1306–1316. doi: 10.1002/ijc.30555. [DOI] [PubMed] [Google Scholar]
- 4.Ghasemi M., Nabipour I., Omrani A., Alipour Z., Assadi M. Precision medicine and molecular imaging: New targeted approaches toward cancer therapeutic and diagnosis. Am. J. Nucl. Med. Mol. Imaging. 2016;6:310–327. [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu H.-S., Chen T.P., Wen C.K., Hung C.H., Chen C.Y., Chen J.T., Wang Y.C. Multiple genetic and epigenetic biomarkers for lung cancer detection in cytologically negative sputum and a nested case–control study for risk assessment. J. Pathol. 2007;213:412–419. doi: 10.1002/path.2246. [DOI] [PubMed] [Google Scholar]
- 6.Dietel M. Molecular Pathology: A Requirement for Precision Medicine in Cancer. Oncol. Res. Treat. 2016;39:804–810. doi: 10.1159/000453085. [DOI] [PubMed] [Google Scholar]
- 7.Dong N., Shi L., Wang D.C., Chen C., Wang X. Role of epigenetics in lung cancer heterogeneity and clinical implication. Semin. Cell Dev. Biol. 2016;64:18–25. doi: 10.1016/j.semcdb.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 8.Yan R., Chi L., Zheng X., Sun R., You J., Ye X. A meta-analysis of serum p16 gene promoter methylation for diagnosis of nonsmall cell lung cancer. Indian J. Cancer. 2015;52(Suppl. S2):e116. doi: 10.4103/0019-509X.172507. [DOI] [PubMed] [Google Scholar]
- 9.Huang Y.Z., Wu W., Wu K., Xu X.N., Tang W.R. Association of RASSF1A Promoter Methylation with Lung Cancer Risk: A Meta-analysis. Asian Pac. J. Cancer Prev. 2014;15:10325–10328. doi: 10.7314/APJCP.2014.15.23.10325. [DOI] [PubMed] [Google Scholar]
- 10.Guo S., Tan L., Pu W., Wu J., Xu K., Wu J., Li Q., Ma Y., Xu J., Jin L. Quantitative assessment of the diagnostic role of APC promoter methylation in non-small cell lung cancer. Clin. Epigenet. 2014;6:5. doi: 10.1186/1868-7083-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen C., Hua H., Han C., Cheng Y., Cheng Y., Wang Z., Bao J. Prognosis value of MGMT promoter methylation for patients with lung cancer: A meta-analysis. Int. J. Clin. Exp. Pathol. 2015;8:11560–11564. [PMC free article] [PubMed] [Google Scholar]
- 12.Li F.F., Yang Y., Wang X.L., Hong Y.Y., Wang N.F., Chen Z.D. Promoter methylation of DAPK gene may contribute to the pathogenesis of nonsmall cell lung cancer: A meta-analysis. Tumor Biol. 2014;35:6011–6020. doi: 10.1007/s13277-014-1796-1. [DOI] [PubMed] [Google Scholar]
- 13.Song X., Shi K., Zhou S.J., Yu D.P., Liu Z., Han Y. Clinicopathological significance and a potential drugtarget of RARβ in non-small-cell lung carcinoma: A meta-analysis and a systematic review. Drug Des. Dev. Ther. 2016;10:1345–1354. doi: 10.2147/DDDT.S96766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guzmán L., Depix M.S., Salinas A.M., Roldán R., Aguayo F., Silva A., Vinet R. Analysis of aberrant methylation on promoter sequences of tumor suppressor genes and total DNA in sputum samples: A promising tool for early detection of COPD and lung cancer in smokers. Diagn. Pathol. 2012;7:87. doi: 10.1186/1746-1596-7-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fariashesson E., Erikson J., Atkins A., Shen P., Davis R.W., Scharfe C., Pourmand N. Semi-Automated Library Preparation for High-Throughput DNA Sequencing Platforms. Biomed. Res. Int. 2010;2010:617469. doi: 10.1155/2010/617469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herman J.G., Graff J.R., Myöhänen S., Nelkin B.D., Baylin S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cipriani A., Higgins J.P., Geddes J.R., Salanti G. Conceptual and technical challenges in network meta-analysis. Ann. Intern. Med. 2013;159:130–137. doi: 10.7326/0003-4819-159-2-201307160-00008. [DOI] [PubMed] [Google Scholar]
- 18.Mills E.J., Ioannidis J.P., Thorlund K., Schünemann H.J., Puhan M.A., Guyatt G.H. How to use an article reporting a multiple treatment comparison meta-analysis. JAMA. 2012;308:1246–1253. doi: 10.1001/2012.jama.11228. [DOI] [PubMed] [Google Scholar]
- 19.Knobloch K., Yoon U., Vogt P.M. Preferred reporting items for systematic reviews and meta-analyses (PRISMA) statement and publication bias. J. Cranio-Maxillo Fac. Surg. 2010;39:91–92. doi: 10.1016/j.jcms.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Whiting P., Rutjes A.W., Dinnes J., Reitsma J., Bossuyt P.M., Kleijnen J. Development and validation of methods for assessing the quality of diagnostic accuracy studies. Health Technol. Assess. 2004;8:1–234. doi: 10.3310/hta8250. [DOI] [PubMed] [Google Scholar]
- 21.Bossuyt P.M., Reitsma J.B., Bruns D.E., Gatsonis C.A., Glasziou P.P., Irwig L.M., Lijmer J.G., Moher D., Rennie D., de Vet H.C. Toward complete and accurate reporting of studies of diagnostic accuracy. The STARD initiative. Am. J. Clin. Pathol. 2003;119:18–22. doi: 10.1309/8EXCCM6YR1THUBAF. [DOI] [PubMed] [Google Scholar]
- 22.Devillé W.L., Buntinx F., Bouter L.M., Montori V.M., de Vet H.C., van der Windt D.A., Bezemer P.D. Conducting systematic reviews of diagnostic studies: Didactic guidelines. BMC Med. Res. Methodol. 2002;2:9. doi: 10.1186/1471-2288-2-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K.W., Lee J., Sang H.C., Huh J., Park S.H. Systematic Review and Meta-Analysis of Studies Evaluating Diagnostic Test Accuracy: A Practical Review for Clinical Researchers—Part I. General Guidance and Tips. Korean J. Radiol. 2015;16:1175–1187. doi: 10.3348/kjr.2015.16.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dinnes J., Deeks J., Kirby J., Roderick P. A methodological review of how heterogeneity has been examined in systematic reviews of diagnostic test accuracy. Health Technol. Assess. 2005;9:1–113. doi: 10.3310/hta9120. [DOI] [PubMed] [Google Scholar]
- 25.Zamora J., Abraira V., Muriel A., Khan K., Coomarasamy A. Meta-DiSc: A software for meta-analysis of test accuracy data. BMC Med. Res. Methodol. 2006;6:31. doi: 10.1186/1471-2288-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deeks J.J., Macaskill P., Irwig L. The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J. Clin. Epidemiol. 2005;58:882–893. doi: 10.1016/j.jclinepi.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 27.Song F., Clark A.C., Bachmann M.O., Maas J. Simulation evaluation of statistical properties of methods for indirect and mixed treatment comparisons. BMC Med. Res. Methodol. 2012;12:138. doi: 10.1186/1471-2288-12-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Q., Chen L., Tang Y., Jing W. The promoter hypermethylation of DAPK gene and p16 gene in sera from Chinese non-small cell lung cancer patients. Chin.-Ger. J. Clin. Oncol. 2006;5:184–188. doi: 10.1007/s10330-005-0430-5. [DOI] [Google Scholar]
- 29.Liu Y., Lan Q., Shen M., Mumford J., Keohavong P. Aberrant gene promoter methylation in sputum from individuals exposed to smoky coal emissions. Anticancer Res. 2008;28:2061–2066. [PMC free article] [PubMed] [Google Scholar]
- 30.Georgiou E., Valeri R., Tzimagiorgis G., Anzel J., Krikelis D., Tsilikas C., Sarikos G., Destouni C., Dimitriadou A., Kouidou S. Aberrant p16 promoter methylation among Greek lung cancer patients and smokers: Correlation with smoking. Eur. J. Cancer Prev. 2007;16:396–402. doi: 10.1097/01.cej.0000236260.26265.d6. [DOI] [PubMed] [Google Scholar]
- 31.Sato K.C.T., Yukio M.M.D., Masakatsu F.C.T., Yujian S.M.D., Daye W.M.D., Ryota T.M.D., Yuko M.M.D., Iijima T., Noguchi M. Anthracotic index and DNA methylation status of sputum contents can be used for identifying the population at risk of lung carcinoma. Cancer Cytopathol. 2004;102:348–354. doi: 10.1002/cncr.20643. [DOI] [PubMed] [Google Scholar]
- 32.Olaussen K.A., Soria J.C., Park Y.W., Kim H.J., Kim S.H., Ro J.Y., André F., Jang S.J. Assessing abnormal gene promoter methylation in paraffin-embedded sputum from patients with NSCLC. Eur. J. Cancer. 2005;41:2112–2119. doi: 10.1016/j.ejca.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Ma V.D.D., Prinsen C.F., Hol B.E., Bolijn A.S., Jeunink M.A., Dekhuijzen P.N., Thunnissen F.B. Can free DNA be detected in sputum of lung cancer patients? Lung Cancer. 2008;61:385–390. doi: 10.1016/j.lungcan.2008.01.007. [DOI] [PubMed] [Google Scholar]
- 34.Hubers A.J., Brinkman P., Boksem R.J., Rhodius R.J., Witte B.I., Zwinderman A.H., Heideman D.A.M., Duin S., Koning R., Steenbergen R.D.M. Combined sputum hypermethylation and eNose analysis for lung cancer diagnosis. J. Clin. Pathol. 2014;67:707–711. doi: 10.1136/jclinpath-2014-202414. [DOI] [PubMed] [Google Scholar]
- 35.Leng S., Do K., Yingling C.M., Picchi M.A., Wolf H.J., Kennedy T.C., Feser W.J., Baron A.E., Franklin W.A., Brock M.V. Defining a Gene Promoter Methylation Signature in Sputum for Lung Cancer Risk Assessment. Clin. Cancer Res. 2012;18:3387–3395. doi: 10.1158/1078-0432.CCR-11-3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang S.H., Kim K.U., Kim J.E., Kim H.H., Lee M.K., Lee C.H., Lee S.Y., Oh T., An S. Detection of HOXA9 gene methylation in tumor tissues and induced sputum samples from primary lung cancer patients. Clin. Chem. Lab. Med. 2011;49:699–704. doi: 10.1515/CCLM.2011.108. [DOI] [PubMed] [Google Scholar]
- 37.Hulbert A., Jusue-Torres I., Stark A., Chen C., Rodgers K., Lee B., Griffin C., Yang A., Huang P., Wrangle J. Early Detection of Lung Cancer Using DNA Promoter Hypermethylation in Plasma and Sputum. Clin. Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kang C.Y., Wang D.D., Tang S.P., Xiao H. Effect of promoter hypermethylation of FHIT, P16, MGMT, RASSF1A and APC genes in sputum specimens on diagnosis of lung cancer. Chin. J. Clin. Exp. Pathol. 2011;27:869–873. [Google Scholar]
- 39.Belinsky S.A., Klinge D.M., Dekker J.D., Smith M.W., Bocklage T.J., Gilliland F.D., Crowell R.E., Karp D.D., Stidley C.A., Picchi M.A. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin. Cancer Res. 2005;11:6505–6511. doi: 10.1158/1078-0432.CCR-05-0625. [DOI] [PubMed] [Google Scholar]
- 40.Yun S., Fang H.B., Feng J. Integrating DNA methylation and microRNA biomarkers in sputum for lung cancer detection. Clin. Epigenet. 2016;8:109. doi: 10.1186/s13148-016-0275-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Destro A., Bianchi P., Alloisio M., Laghi L., Di G.S., Malesci A., Cariboni U., Gribaudi G., Bulfamante G., Marchetti A. K-ras and p16(INK4A)alterations in sputum of NSCLC patients and in heavy asymptomatic chronic smokers. Lung Cancer. 2004;44:23–32. doi: 10.1016/j.lungcan.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 42.Shin K.C., Lee K.H., Lee C.H., Shin I.H., Suh H.S., Jeon C.H. MAGE A1–A6 RT-PCR and MAGE A3 and p16 methylation analysis in induced sputum from patients with lung cancer and non-malignant lung diseases. Oncol. Rep. 2012;27:911–916. doi: 10.3892/or.2011.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hubers A.J., Ma V.D.D., Prinsen C.F., Witte B.I., Wang Y., Shivapurkar N., Stastny V., Bolijn A.S., Hol B.E., Feng Z. Methylation analysis in spontaneous sputum for lung cancer diagnosis. Lung Cancer. 2014;84:127–133. doi: 10.1016/j.lungcan.2014.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Cirincione R., Lintas C., Conte D., Mariani L., Roz L., Vignola A.M., Pastorino U., Sozzi G. Methylation profile in tumor and sputum samples of lung cancer patients detected by spiral computed tomography: A nested case–control study. Int. J. Cancer. 2006;118:1248. doi: 10.1002/ijc.21473. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y.C., Hsu H.S., Chen T.P., Chen J.T. Molecular Diagnostic Markers for Lung Cancer in Sputum and Plasma. Ann. N. Y. Acad. Sci. 2006;1075:179–184. doi: 10.1196/annals.1368.024. [DOI] [PubMed] [Google Scholar]
- 46.Hubers A.J., Heideman D.A., Herder G.J., Burgers S.A., Sterk P.J., Kunst P.W., Smit H.J., Postmus P.E., Witte B.I., Duin S. Prolonged sampling of spontaneous sputum improves sensitivity of hypermethylation analysis for lung cancer. J. Clin. Pathol. 2012;65:541–545. doi: 10.1136/jclinpath-2012-200712. [DOI] [PubMed] [Google Scholar]
- 47.Belinsky S.A., Liechty K.C., Gentry F.D., Wolf H.J., Rogers J., Vu K., Haney J., Kennedy T.C., Hirsch F.R., Miller Y. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W., Sun Y., Lu G. The diagnostic value of determination of p16 methylation of sputum exfoliated cells for peripheral lung cancer. China J. Mod. Med. 2004;7:46–49. doi: 10.3779/j.issn.1009-3419.2004.01.12. [DOI] [PubMed] [Google Scholar]
- 49.Peng Z., Shan C., Wang H. Value of promoter methylation of RASSF1A, p16, and DAPK genes in induced sputum in diagnosing lung cancers. J. Cent. South Univ. Med. Sci. 2010;35:247–253. doi: 10.3969/j.issn.1672-7347.2010.03.010. [DOI] [PubMed] [Google Scholar]
- 50.Wu X.M., Chen Y., Shao Y., Zhou X.L., Tang W.R. Association between cigarette smoking and RASSF1A gene promoter hypermethylation in lung cancer patients: A meta- analysis. Asian Pac. J. Cancer Prev. 2014;15:8451–8454. doi: 10.7314/APJCP.2014.15.19.8451. [DOI] [PubMed] [Google Scholar]
- 51.Hu Z.J., Liu D.Y., Hu H.B., Chen Y.P., Chen J.P., Tan H.C., Zhou J.G., Liu H.L., Zhong Y. Clinical significance at diagnosis of lung cancer by detecting aberrant p16 promoter methylation in suspicious lung cancer patients. Chin. J. Pract. Intern. Med. 2009;72:302–308. [Google Scholar]
- 52.Zhuo-Jun H.U., Hong-Bo H.U., Liu D.Y., Chen Y.P. Clinical significance of detection of p16 gene methylation in early diagnosis of lung cancer. Chin. J. Pathophysiol. 2009;25:1941–1945. [Google Scholar]
- 53.Guo X.J. Diagnostic value of combinative detection of hypermethylation of p16 gene and cytokeratin 19 fragment and CA15-3 antigen for non-small cell lung cancer patients. Clin. Focus. 2008;23:1067–1070. [Google Scholar]
- 54.Ming L., Liu J., Bing L., Zhai F., Wang A., Yuanqiu X. Diagnostic value of hypermethylation of p16 and MGMT genes in sputum specimens for lung cancer. Tumor. 2006;26:1020–1023. [Google Scholar]
- 55.Sun N., Zhang L., Liu Y.Y., Zheng S.Y., Zhao X. Methylation of P16 and RASSF1A Genes in Sputum Samples Associated with Peripheral Non-Small Cell Lung Cancer. Prog. Mod. Biomed. 2012;13:045. [Google Scholar]
- 56.Pan J., Fan H.J., Jiang Y., Dan M.A., Wang J., Xia C. P16~(INK4a) gene methylation in sputum, blood and bronchoscopic biopsies and lung cancer. J. Clin. Pulm. Med. 2013;6:058. [Google Scholar]
- 57.Wang X., Cao A., Peng M., Chunfeng H.U., Liu D., Gu T., Liu H. The value of chest CT scan and tumor markers detection in sputum for early diagnosis of peripheral lung cancer. Chin. J. Lung Cancer. 2004;7:58–63. doi: 10.3779/j.issn.1009-3419.2004.01.15. [DOI] [PubMed] [Google Scholar]
- 58.Hubers A.J., Heideman D.A.M., Burgers S.A., Herder G.J.M., Sterk P.J., Rhodius R.J., Smit H.J., Krouwels F., Welling A., Witte B.I. DNA hypermethylation analysis in sputum for the diagnosis of lung cancer: Training validation set approach. Br. J. Cancer. 2015;112:1105–1113. doi: 10.1038/bjc.2014.636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sinčić N., Herceg Z. DNA methylation and cancer: Ghosts and angels above the genes. Curr. Opin. Oncol. 2011;23:69–76. doi: 10.1097/CCO.0b013e3283412eb4. [DOI] [PubMed] [Google Scholar]
- 60.Wang X., Ling L., Su H., Cheng J., Jin L. Aberrant methylation of genes in sputum samples as diagnostic biomarkers for non-small cell lung cancer: A meta-analysis. Asian Pac. J. Cancer Prev. 2014;15:4467–4474. doi: 10.7314/APJCP.2014.15.11.4467. [DOI] [PubMed] [Google Scholar]
- 61.Shiga T., Wajima Z., Inoue T., Sakamoto A. Predicting difficult intubation in apparently normal patients: A meta-analysis of bedside screening test performance. Anesthesiology. 2005;103:429–437. doi: 10.1097/00000542-200508000-00027. [DOI] [PubMed] [Google Scholar]
- 62.Jiang Y., Cui L., Chen W.D., Shen S.H., Ding L.D. The prognostic role of RASSF1A promoter methylation in breast cancer: A meta-analysis of published data. PLoS ONE. 2012;7:e36780. doi: 10.1371/journal.pone.0036780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shou F., Feng X., Gang L., Zhao Z., Ying M., Yang F., Wang H., Guo H. RASSF1A promoter methylation is associated with increased risk of thyroid cancer: A meta-analysis. Oncotargets Ther. 2017;10:247–257. doi: 10.2147/OTT.S124417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Si J.G., Su Y.Y., Han Y.H., Chen R.H. Role of RASSF1A promoter methylation in the pathogenesis of ovarian cancer: A meta-analysis. Genet. Test. Mol. Biomark. 2014;18:394–402. doi: 10.1089/gtmb.2014.0022. [DOI] [PubMed] [Google Scholar]
- 65.Wang H.L., Zhang Y., Liu P., Zhou P.Y. Aberrant promoter methylation of RASSF1A gene may be correlated with colorectal carcinogenesis: A meta-analysis. Mol. Biol. Rep. 2014;41:3991–3999. doi: 10.1007/s11033-014-3267-6. [DOI] [PubMed] [Google Scholar]
- 66.Qi D., Li J., Jiang M., Liu C., Hu Y., Li M.X., Su J.L., Que B., Ji W.D. The relationship between promoter methylation of p16 gene and bladder cancer risk: A meta-analysis. Int. J. Clin. Exp. Med. 2015;8:20701–20711. [PMC free article] [PubMed] [Google Scholar]
- 67.Feng W., Han Z., Zhu R., Liu P., Liu S. Association of p16 gene methylation with prostate cancer risk: A meta-analysis. J. BUON. 2015;20:1074–1080. [PubMed] [Google Scholar]
- 68.Wang W., Li T., Wang X.L., Yuan W.X., Cheng Y.Y., Zhang H.Y., Xu E.Q., Zhang Y.M., Shi S., Ma D.L., et al. FAM19A4 is a novel cytokine ligand of formyl peptide receptor 1 (FPR1) and is able to promote the migration and phagocytosis of macrophages. Cell. Mol. Immunol. 2015;12:615–624. doi: 10.1038/cmi.2014.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan W., Xu N., Han X., Zhou X.M., He B. The clinicopathological significance of FHIT hypermethylation in non-small cell lung cancer, a meta-analysis and literature review. Sci. Rep. 2016;6:19303. doi: 10.1038/srep19303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fong K.M., Biesterveld E.J., Virmani A., Wistuba I., Sekido Y., Bader S.A., Ahmadian M., Rassool F.V., Zimmerman P.V., Giaccone G., et al. FHIT and FRA3B 3p14.2 allele loss are common in lung cancer and preneoplastic bronchial lesions and are associated with cancer-related FHIT cDNA splicing aberrations. Cancer Res. 1997;57:2256–2267. [PubMed] [Google Scholar]
- 71.Gu C.M., Lu J.C., Cui T.P., Lu C., Shi H., Xu W.M., Yuan X.L., Yang X.B., Huang Y.X., Lu M.X. Association between MGMT Promoter Methylation and Non-Small Cell Lung Cancer: A Meta-Analysis. PLoS ONE. 2013;8:e72633. doi: 10.1371/journal.pone.0072633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Atakpo P., Vassar M. Publication bias in dermatology systematic reviews and meta-analyses. J. Dermatol. Sci. 2016;82:69–74. doi: 10.1016/j.jdermsci.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 73.Lorenc T., Felix L., Petticrew M., Melendez-Torres G.J., Thomas J., Thomas S., O’Mara-Eves A., Richardson M. Meta-analysis, complexity, and heterogeneity: A qualitative interview study of researchers’ methodological values and practices. Syst. Rev. 2016;5:192. doi: 10.1186/s13643-016-0366-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.He A., Song D., Zhang L., Li C. Unveiling the relative efficacy, safety and tolerability of prophylactic medications for migraine: Pairwise and network-meta analysis. J. Headache Pain. 2017;18:26. doi: 10.1186/s10194-017-0720-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee G., Oh S.W., Hwang S.S., Yoon J.W., Kang S., Joh H.K. Comparative effectiveness of oral antidiabetic drugs in preventing cardiovascular mortality and morbidity: A network meta-analysis. PLoS ONE. 2017;12:e0177646. doi: 10.1371/journal.pone.0177646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jupiter D.C. Made to Measure: Designs Tailored to Your Study Needs. J. Foot Ankle Surg. 2015;54:1001–1002. doi: 10.1053/j.jfas.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 77.Setia M.S. Methodology Series Module 2: Case-control Studies. Indian J. Dermatol. 2016;61:146–151. doi: 10.4103/0019-5154.177773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.