Abstract
The environmental spread of antibiotic-resistant bacteria has been recognised as a growing public health threat for which hospitals play a significant role. The aims of this study were to investigate the prevalence of antibiotic resistance and antibiotic resistance genes (ARGs) in Escherichia coli isolates from hospital wastewater in Vietnam. Wastewater samples before and after treatment were collected using continuous sampling every month over a year. Standard disk diffusion and E-test were used for antibiotic susceptibility testing. Extended-spectrum beta-lactamase (ESBL) production was tested using combined disk diffusion. ARGs were detected by polymerase chain reactions. Resistance to at least one antibiotic was detected in 83% of isolates; multidrug resistance was found in 32%. The highest resistance prevalence was found for co-trimoxazole (70%) and the lowest for imipenem (1%). Forty-three percent of isolates were ESBL-producing, with the blaTEM gene being more common than blaCTX-M. Co-harbouring of the blaCTX-M, blaTEM and qepA genes was found in 46% of isolates resistant to ciprofloxacin. The large presence of antibiotic-resistant E. coli isolates combined with ARGs in hospital wastewater, even post-treatment, poses a threat to public health. It highlights the need to develop effective processes for hospital wastewater treatment plants to eliminate antibiotic resistant bacteria and ARGs.
Keywords: antibiotic resistance, antibiotic resistance genes, blaCTX-M, blaTEM, qepA, hospital wastewater
1. Introduction
The environmental spread of antibiotic resistant bacteria has been recognized as a growing public health threat [1,2]. Hospitals are “hotspots” for antibiotic use and not only play an important role in antibiotic dissemination but also in the release of antibiotic resistant bacteria into the environment. Hospital wastewater treatment plants containing antibiotic residues can favour the development of antibiotic resistance due to the selective pressure placed on bacteria [3,4]. Moreover, antibiotic resistance genes (ARGs) carried by bacterial contaminants can be transferred to other bacterial populations including pathogenic bacteria found in hospital wastewater [1]. Hospital effluents can reach water bodies used in agriculture or for domestic purposes. From there, antibiotic resistant bacteria and/or ARGs can be transferred to humans.
In recent years, the presence of antibiotic resistant Escherichia coli (E. coli), particularly extended-spectrum beta-lactamase (ESBL)-producing isolates, in surface water has attracted attention [5]. A direct relationship between clinical E. coli isolates and the quantity of ESBL-producing E. coli strains found in hospital wastewater has been demonstrated [6]. Consequently, the existence of ESBL-producing E. coli carriers in hospitals may lead to their environmental spread [6].
Antibiotic resistance causes prolonged illness, excess mortality, and higher costs for patients and health systems [7,8,9]. Despite increased warnings and numerous efforts to contain it, antibiotic resistance has been increasing [10,11,12,13]. At the recent United Nations general assembly, it was highlighted that antibiotic resistance is among the greatest global health risks, requiring urgent attention [14].
The risks are potentially more serious in low- and middle-income countries where many hospitals either do not have wastewater treatment plants or they are ineffective. To make matters worse, in many places, but particularly rural areas, surface water is used for agriculture and domestic purposes or even consumed untreated. Most research on antibiotic resistant bacteria in hospital wastewater originates from high-income countries [15].
Therefore, this study sought to investigate the prevalence of resistant E. coli isolates to commonly used antibiotics, ESBL-producing isolates along with genes coding for cephalosporin resistance blaCTX-M and blaTEM, and a gene coding for ciprofloxacin resistance qepA, in hospital wastewater in a rural and an urban hospital in Vietnam.
2. Materials and Methods
This is a repeated cross-sectional study with monthly data collection in one rural and one urban hospital in Vietnam, a lower middle-income country. The rural hospital has 220 beds and is situated 60 km northwest of central Hanoi. The 520-bedded urban hospital is located in central Hanoi. Both hospitals’ wastewater is routed to wastewater treatment plants (WWTPs) where it is treated using filtering, microbiological, and biochemical mechanisms. After treatment, hospital effluents are discharged into sewer systems, which lead to nearby rivers.
2.1. Collection of Water Samples
The collection of water samples is described in detail elsewhere [4]. Briefly, samples of wastewater before treatment (WBT) as well as wastewater after treatment (WAT) were collected using 24 h continuous sampling on a weekday during the last week of every month in 2013. The water samples were stored in closed containers surrounded by ice and transferred to the microbiological laboratory in Bach Mai Hospital in central Hanoi within 6 h of testing. The urban hospital and its wastewater treatment plant was under reconstruction from June to August 2013, therefore sampling was ceased during this period.
2.2. Antibacterial Susceptibility Testing and Detection of ARGs
Coliforms were detected with the most probable number procedure [16]. A presumptive test involved three subsets of tubes containing different amounts of lactose or lauryl tryptose broth. Each subset contained five tubes with inverted Durham tubes to collect gas produced by fermentation. The three subsets were inoculated with water samples of 10, 1.0, and 0.1 mL, respectively. The tubes were then incubated for 24 h at 35–37 °C. A positive test for gas formation was presumptive evidence of coliforms. A confirmatory test for coliforms was made by inoculating another broth from one of the positive tubes. The test was completed by final isolation of the coliforms on selective and differential media, Gram staining the isolates, and reconfirming gas production. Coliform isolates were then sub-cultured on BrillianceTM UTI agar to collect presumptive E. coli isolates. Following biochemical confirmation using standard tests, identified E. coli isolates were tested for antibiotic susceptibility using the standard Kirby Bauer disc diffusion method for: (i) amoxicillin/clavulanic acid; (ii) ceftazidime; (iii) ceftriaxone; (iv) ciprofloxacin; (v) co-trimoxazole (trimethoprim/sulfamethoxazole); (vi) fosfomycin; (vii) gentamicin; and (viii) imipenem. The selected antibiotics were commonly used in the hospitals and routinely tested in clinical laboratories. Antibiotic susceptibility test results were interpreted as resistant, intermediate, and susceptible using Clinical and Laboratory Standard Institute guidelines (CLSI M100-2013) [17]. Minimum inhibitory concentrations (MICs) were determined for ciprofloxacin and ceftazidime using E-test. Disc diffusion zone diameters were also compared with epidemiological cut-off values (ECOFF) [18].
For E. coli isolates resistant to third generation cephalosporins, extended-spectrum beta-lactamase (ESBL) production was tested using combined disc diffusion. Genotypic confirmation was done through polymerase chain reactions. Genes coding for beta-lactam resistance, blaCTX-M and blaTEM, were tested in ESBL-producing isolates and qepA gene was tested for in ciprofloxacin-resistant isolates [19,20].
2.3. Data Analysis
Prevalence of resistance to at least one of the studied antibiotics, to each studied antibiotic, and multidrug resistance (MDR) in E. coli isolates were analysed. The definition of MDR reported by Magiorakos et al. (2012) from ECDC Joint Expert Meetings was applied; according to that, bacterial isolates were considered multidrug-resistant if they were non-susceptible to at least one agent in three or more antibiotic categories [21]. Fisher’s exact test was applied to test the difference between the prevalence of antibiotic resistant E. coli isolates before and after wastewater treatment in each hospital using Stata 12 (StataCorp LP, College Station, TX, USA).
3. Results
In total, 265 E. coli isolates were collected from both the hospitals during the study period; 158 from the rural hospital (WBT = 84; WAT = 74) and 107 isolates from the urban hospital (WBT = 60; WAT = 47).
3.1. Resistance to Studied Antibiotics
In the rural hospital, 85% of E. coli isolates were resistant to at least one of the tested antibiotics (WBT = 94%; WAT = 74%). Resistance was most common towards co-trimoxazole, with 70% of isolates being resistant to it (WBT = 86%; WAT = 53%). Resistance to ceftriaxone was found in 49% of isolates (WBT = 55%; WAT = 42%), and resistance to ceftazidime, gentamicin, and amoxicillin/clavulanic acid was around 40%, respectively. Thirty percent of the isolates were resistant to ciprofloxacin (WBT = 25%; WAT = 35%), and 2% were resistant to fosfomycin (WBT = 1%; WAT = 3%). Resistance to imipenem was only detected in one isolate (1%). MDR was found in 35% of isolates (WBT = 44%; WAT = 26%). Prevalence of resistance to the studied antibiotics in E. coli isolates from WAT was less common than WBT, with the exception of ciprofloxacin and fosfomycin, for which enrichment of resistant E. coli isolates after wastewater treatment was found. The differences are statistically significant for amoxicillin/clavulanic acid, co-trimoxazole, gentamicin, resistance to at least one studied antibiotic, and MDR (Table 1).
Table 1.
Prevalence of resistance to studied antibiotics in Escherichia coli isolates found in hospital wastewater.
| Studied Antibiotics | Rural Hospital (n = 158) | Urban Hospital (n = 107) | Both Hospitals (n = 265) Overall (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| WBT (%) n = 84 | WAT (%) n = 74 | p-Value | Overall (%) | WBT (%) n = 60 | WAT (%) n = 47 | p-Value | Overall (%) | ||
| Amoxicillin/clavulanic acid | 51 | 24 | 0.001 * | 39 | 28 | 19 | 0.36 | 24 | 33 |
| Ceftazidime | 42 | 36 | 0.52 | 39 | 32 | 23 | 0.40 | 28 | 35 |
| Ceftriaxone | 55 | 42 | 0.11 | 49 | 45 | 32 | 0.23 | 39 | 45 |
| Ciprofloxacin | 25 | 35 | 0.22 | 30 | 23 | 17 | 0.50 | 21 | 26 |
| Co-trimoxazole | 86 | 53 | <0.001 * | 70 | 80 | 60 | 0.03 * | 71 | 70 |
| Fosfomycin | 1 | 3 | 0.60 | 2 | 15 | 0 | 0.005 * | 8 | 4 |
| Gentamycin | 51 | 31 | 0.02 * | 42 | 33 | 23 | 0.30 | 29 | 37 |
| Imipenem | 1 | 0 | 1.00 | 1 | 0 | 0 | N/A | 0 | 1 |
| At least one antibiotic | 94 | 74 | 0.001 * | 85 | 88 | 68 | 0.02 * | 79 | 83 |
| MDR | 44 | 26 | 0.02 * | 35 | 32 | 21 | 0.30 | 27 | 32 |
MDR: multidrug resistance; N/A: not available; WBT: wastewater before treatment; WAT: wastewater after treatment. * Differences in prevalence of resistant Escherichia coli strains isolated from WBT and WAT are significant.
In the urban hospital, 79% of E. coli isolates were resistant to at least one of the studied antibiotics (WBT = 88%; WAT = 68%). Co-trimoxazole resistance was again most common, with resistance found in 71% of isolates (WAT = 80%; WAT = 60%), followed by ceftriaxone resistance (39%) (WBT = 45%; WAT = 32%). Resistance to gentamicin and ceftazidime was found in 29% and 28% of isolates, respectively, followed by amoxicillin/clavulanic acid (24%) and ciprofloxacin (21%). Fosfomycin resistance was least common, as it was detected in only 8% of isolates. MDR was found in 27% of isolates (WBT = 32%; WAT = 21%). Prevalence of resistance to the studied antibiotics in E. coli isolates from WAT was lower than WBT. The differences were statistically significant for co-trimoxazole, fosfomycin, and resistance to at least one studied antibiotic (Table 1).
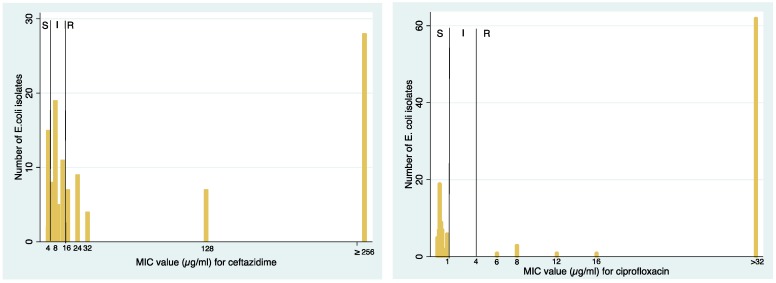
The distribution of MIC values for ceftazidime and ciprofloxacin susceptibility testing is presented in Figure 1. The number of E. coli isolates with high MIC values is large compared to the number of isolates with lower MIC values, indicating high levels as well as high proportions of resistance.
Figure 1.
Distribution of minimum inhibitory concentration (MIC) values for ceftazidime and ciprofloxacin susceptibility testing.
When applying ECOFF values, we found decreased susceptibility to amoxicillin/clavulanic acid (45% of isolates), ceftazidime (39% of isolates), ceftriaxone (48% of isolates), ciprofloxacin (29% of isolates), and imipenem (2% of isolates).
MDR patterns are presented in Table 2 with identified antibiotic combinations. MDR to six out of eight studied antibiotics was found in 25 isolates (10%).
Table 2.
Multidrug resistance patterns in Escherichia coli isolates found in hospital wastewater (the number of MDR isolates having the respective pattern).
| MDR Pattern | Rural Hospital (n = 158) | Urban Hospital (n = 107) | ||
|---|---|---|---|---|
| WBT | WAT | WBT | WAT | |
| CTX + CIP + SXT | 0 | 0 | 0 | 2 |
| CTX + GEN + SXT | 7 | 0 | 3 | 0 |
| AMC + GEN + SXT | 0 | 0 | 1 | 0 |
| GEN + CIP + SXT | 0 | 4 | 7 | 1 |
| CAZ + CTX + AMC + CIP | 2 | 0 | 0 | 0 |
| CAZ + CTX + AMC + SXT | 2 | 0 | 0 | 2 |
| CAZ + CTX + GEN + SXT | 6 | 0 | 1 | 0 |
| CAZ + CTX + CIP + SXT | 0 | 0 | 0 | 1 |
| CAZ + CTX + SXT + FOM | 1 | 0 | 0 | 0 |
| CTX + GEN + CIP + SXT | 4 | 1 | 2 | 0 |
| AMC + GEN + CIP + SXT | 0 | 1 | 0 | 1 |
| CAZ + CTX + GEN + CIP + SXT | 2 | 2 | 3 | 0 |
| CAZ + CTX + AMC + CIP + SXT | 0 | 1 | 0 | 0 |
| CTX + GEN + CIP + SXT + FOM | 0 | 1 | 0 | 0 |
| CTX + AMC + GEN + CIP + SXT | 1 | 0 | 0 | 0 |
| IMP + CAZ + CTX + AMC + SXT | 1 | 0 | 0 | 0 |
| CAZ + CTX + AMC + GEN + CIP + SXT | 11 | 9 | 1 | 3 |
| CAZ + CTX + AMC + GEN + CIP + FOM | 0 | 0 | 1 | 0 |
| Total (%) | 37 (44%) | 19 (26%) | 19 (32%) | 10 (21%) |
| 56 (35%) | 29 (27%) | |||
MDR: multidrug resistance; WBT: wastewater before treatment; WAT: wastewater after treatment; AMC: amoxicillin/clavulanic acid; CAZ: ceftazidime; CIP: ciprofloxacin; CTX: ceftriaxone; FOM: fosfomycin; GEN: gentamicin; IMP: imipenem; SXT: trimethoprim/sulfamethoxazole (co-trimoxazole).
3.2. ESBL-Producing E. coli, ESBL, and Quinolone Resistance Genes
In the rural hospital, 76 E. coli isolates were ESBL-producing (48%). Among them, blaTEM was detected in 97% of isolates and blaCTX-M in 76%. Both blaCTX-M and blaTEM were detected in 75% of isolates. Quinolone-resistance gene (qepA) was detected in 72% of ciprofloxacin-resistant isolates. All three genes were detected in 51% of ciprofloxacin-resistant isolates (Table 3).
Table 3.
Genetic analysis of extended-spectrum beta-lactamase (ESBL)-producing and ciprofloxacin-resistant Escherichia coli strains found in hospital wastewater.
| Genetic Analysis | Rural Hospital | Urban Hospital | Both Hospitals | ||||
|---|---|---|---|---|---|---|---|
| WBT n (%) | WAT n (%) | Overall n (%) | WBT n (%) | WAT n (%) | Overall n (%) | Overall n (%) | |
| ESBL-producing | 45 (54) | 31 (42) | 76 (48) | 27 (45) | 12 (26) | 39 (36) | 115 (43) |
| blaCTX-M | 29 (64) | 29 (94) | 58 (76) | 14 (52) | 2 (17) | 16 (41) | 74 (64) |
| blaTEM | 44 (98) | 30 (97) | 74 (97) | 27 (100) | 10 (83) | 37 (95) | 111 (97) |
| blaCTX-M + blaTEM | 29 (64) | 28 (90) | 57 (75) | 14 (52) | 2 (17) | 16 (41) | 73 (63) |
| Ciprofloxacin resistance | 21 (25) | 26 (35) | 47 (30) | 14 (23) | 8 (17) | 22 (21) | 69 (26) |
| qepA | 14 (67) | 20 (77) | 34 (72) | 12 (86) | 7 (88) | 19 (86) | 53 (77) |
| qepA + blaCTX-M + blaTEM | 13 (62) | 11 (42) | 24 (51) | 6 (43) | 2 (25) | 8 (36) | 32 (46) |
WAT: wastewater after treatment; WBT: wastewater before treatment.
In the urban hospital, 39 E. coli isolates were ESBL-producing strains (36%). Among them, blaTEM was detected in 95% of isolates and blaCTX-M in 41%. Both blaCTX-M and blaTEM were detected in 41% of isolates. Quinolone-resistance gene (qepA) was detected in 86% of ciprofloxacin-resistant isolates. All three genes, blaTEM, blaCTX-M, and qepA, were detected in 36% of ciprofloxacin-resistant isolates (Table 3).
4. Discussion
Our novel findings show that, in Vietnam, bacteria resistant to commonly used antibiotics along with genes coding for resistance are present in hospital wastewater, even after treatment. Prior to our study, Duong et al. examined E. coli resistance to ciprofloxacin and norfloxacin in hospital wastewater of another Hanoian hospital and reported that E. coli strains isolated from WAT samples were susceptible [22]. In their study, water samples were collected over two days using grab sampling and only 15 E. coli isolates, including three isolates from treated water samples, were tested. Conversely, in our study, water samples were collected every month over one year using continuous sampling and a total of 265 E. coli isolates were tested.
In both hospitals, E. coli isolates were most resistant to co-trimoxazole (around 70% of isolates). Previous reports show comparatively lower prevalence rates. In 2004–2005, an Indian study indicated that 55% of enteric bacteria found in hospital wastewater were resistant to co-trimoxazole [23]. A study from Poland showed that 20% of E. coli isolates from hospital wastewater collected before 2013 were resistant to co-trimoxazole [24]. Co-trimoxazole is a combination of sulfamethoxazole and trimethoprim. Sulfamethoxazole, one of the first antibiotics to be developed, was put into clinical use in 1935 and trimethoprim was first used in 1962. The two antibiotics first started to be used in combination in 1968. Over the past decades, its extensive use in clinical settings to treat a variety of bacterial infections, such as urinary tract infections and respiratory tract infections, which was also the case in the studied hospitals, might explain the high occurrence of co-trimoxazole resistance [25]. Moreover, both sulfamethoxazole and trimethoprim are not readily degradable and their residues found every month over the studied period in the same hospital wastewater could favour the development of co-trimoxazole resistance in the bacteria [4].
Cephalosporin resistance was also found in higher proportions than other antibiotics investigated. Similar prevalence rates of cephalosporin resistant bacteria in hospital effluent were shown by Chagas et al. [26]. Resistance mechanisms to second and third generation cephalosporins differ, with ESBLs being the most important [27]. ESBL enzymes are capable of hydrolysing and inactivating beta-lactam antibiotics and are often plasmid-mediated [28]. The plasmid genes encoding ESBLs can be transferred between different bacterial strains (horizontal gene transfer), facilitating easy spread of antibiotic resistance within as well as between species. Moreover, plasmid-encoded ESBL-producing bacteria can show co-resistance to quinolones, aminoglycosides, and sulfonamides [29]. Consequently, infections caused by ESBL-producing bacterial strains can be difficult to treat due to the restricted amount of antibiotics left for successful treatment.
ESBLs were first isolated in the 1980s [30]. In a study conducted in an urban and rural hospital in central India, Chandran et al. reported a very high prevalence of ESBL-producing E. coli in the hospital wastewater (96%) [31]. In our study, the prevalence was around 40%, lower than the aforementioned study but relatively higher than the figures reported by Diwan et al. (25%), Abdulhaq et al. (25%), and Korzeniewska et al. (37%) [22,32,33]. Among genes coding for ESBL production, TEM and CTX-M are the most common [34]. Our findings indicate the presence of blaCTX-M and blaTEM in ESBL-producing strains, with the blaTEM gene being predominant. This is in accordance with the findings of Varela et al., where blaTEM was found to be the most prevalent ESBL-encoding gene, followed by blaCTX-M [35]. In contrast, Chandran et al. reported higher prevalence of blaCTX-M than blaTEM in hospital wastewater in India [31]. However, not all the blaTEM genes are responsible for ESBL, and in our study, we were not able to do further sequencing to show the frequency of the ESBL blaTEM. In addition, according to the PCR protocol used, the detected blaCTX-M were restricted to CTX-M group 1 including CTX-M-1, CTX-M-3, and CTX-M-15. Ciprofloxacin resistance was detected in our samples, with gene qepA coding for high proportions of the resistant strains. Similar to ESBL-coding genes blaCTX-M and blaTEM, quinolone resistance gene qepA is plasmid-mediated and capable of horizontal gene transfer [36]. Of note is that co-existence of blaCTX-M, blaTEM, and qepA was detected, genetically proving co-resistance in the bacterial strains. In our study, the prevalence of MDR found phenotypically was around 35% with the detection of co-resistance to six out of eight of the studied antibiotics.
Fosfomycin-resistance and imipenem-resistance were also detected among the E. coli isolates in our study. Fosfomycin has broad activity against Gram-negative and some Gram-positive bacteria. In some countries, it is recommended as one of the first-line drugs to treat uncomplicated urinary tract infections because of increasing E. coli resistance towards other commonly used antibiotics, such as ciprofloxacin and co-trimoxazole [37]. Imipenem, the first carbapenem developed, is used to treat infections caused by β-lactamase-producing bacteria and should be saved to treat infections not readily treated by other antibiotics [38]. High prevalence of carbapenem-resistance in clinical isolates in Vietnamese hospitals has been reported [39]. The detection of resistance to these last-line antibiotics in bacterial isolates from hospital wastewater is of concern, since this can contribute to the spread of resistance among bacterial populations in the environment.
Although there is increasing evidence of the occurrence of antibiotic resistant bacteria in the environment, there are no standardized methods for antibiotic susceptibility testing which are directly applicable to environmental samples so far [2]. Epidemiological cut-off values developed by the European Committee on Antimicrobial Susceptibility Testing (EUCAST) can be used for the interpretation of antibiotic resistance in environmental bacteria. The ECOFF values separate bacteria with acquired resistance mechanisms (non-wild type) from the wild type population (having no resistance) [15]. We found in general decreased susceptibility in the E. coli isolates when using ECOFF values.
The role of hospitals in the environmental release of antibiotic-resistant bacteria and ARGs has been demonstrated and has become a growing concern for public health [40,41,42,43,44,45,46,47]. Hospital WWTPs can harbour antibiotic-resistant bacteria and ARGs [48,49,50]. Antibiotic residues in WWTPs can favour the development of antibiotic resistance due to the selective pressure placed on bacteria. In our previous study, we found that antibiotic concentrations in wastewater collected from hospital WWTPs were often higher than the reported predicted no-effect concentrations for resistance selection as well as the minimum selective concentrations, meaning that the selection of antibiotic-resistant bacteria can occur [4]. Published studies have reported the enrichment of antibiotic-resistant E. coli in WWTPs [51,52]. Our findings show significant reductions as well as the enrichment of antibiotic-resistant E. coli in WAT. Resistance to at least one studied antibiotic in the E. coli isolates from WAT was still detected in high proportions. Consequently, certain amounts of antibiotic-resistant bacteria, along with ARGs, are released into the ambient aquatic environment. They can then enter water bodies used for agriculture, irrigation, or household purposes, which poses a threat to public health. The problem can be aggravated if hospitals do not have WWTPs and the wastewater is discharged directly into the environment, which is common practice in Vietnam as well as many other low- and middle-income countries [53]. It has been shown that the prevalence of antibiotic-resistant bacteria is significantly reduced by advanced wastewater treatment processes such as ozone, UV, and ultrafiltration [54,55]. However, even in such advanced plants, resistant bacteria are not completely removed, therefore, hospitals must invest in effective WWTPs with treatment processes that completely eliminate antibiotic-resistant bacteria.
It is plausible that the dissemination of antibiotic-resistant bacteria and ARGs in the environment can result in their transmission to humans [56], however, direct evidence for this is very scarce [57]. So far, the strongest evidence available has shown the genetic similarities between human-related bacterial strains and environmental isolates collected at exposure-relevant sites [58]. Further studies are needed to identify links between the discharge of antibiotic-resistant bacteria by hospital WWTPs, their occurrence in the ambient environment, and their acquisition by humans via environmental exposure.
Our study has some limitations. Importantly, the prevalence of antibiotic resistance in E. coli isolates presented here might not be representative for the whole E. coli population in the hospital wastewater because of the limited number of E. coli isolates from each water sample. Moreover, due to financial constraints, we were not able to study more antibiotics and ARGs than what we have done. Screening for blaSHV gene, which is also common in ESBL-producing E. coli, and genes coding for imipenem resistance would make the study more comprehensive. We were also not able to do sequencing for blaTEM genes to show the frequency of the genes encoding for ESBL. Another limitation is that data from the urban hospital were unavailable for three months, as sampling could not be carried out due to construction work at the hospital. Furthermore, a detailed description of the functioning of the WWTPs was not available for us.
5. Conclusions
High prevalence of antibiotic resistance and ARGs were detected in E. coli isolates from hospital wastewater both before and even after wastewater treatment. There is a need for inclusion and development of hospital WWTPs which are effective at eliminating antibiotic-resistant bacteria and ARGs. Further studies are needed to identify links between the discharge of antibiotic-resistant bacteria by hospital WWTPs, their occurrence in the nearby environment, and their acquisition by humans when exposed in the environment.
Acknowledgments
This work was supported by the Swedish International Development Cooperation Agency (Sida) (grant number SWE-2010-50) and the Vietnamese Government Scholarship for doctoral study of the first author.
Author Contributions
Cecilia Stålsby Lundborg, Nguyen Thi Kim Chuc, Pham Thi Lan, Nguyen Quynh Hoa, Ashok J. Tamhankar, and Vishal Diwan conceived and designed the experiments; La Thi Quynh Lien, Nguyen Thi Minh Thoa, and Pham Hong Nhung performed the experiments; La Thi Quynh Lien and Cecilia Stålsby Lundborg in collaboration with all co-authors analysed the data and contributed with reagents/materials/analysis tools; La Thi Quynh Lien wrote the first draft of the manuscript; and all authors further contributed in writing the manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Williams M.R., Stedtfeld R.D., Guo X., Hashsham S.A. Antimicrobial resistance in the environment. Water Environ. Res. 2016;88:1951–1967. doi: 10.2175/106143016X14696400495974. [DOI] [PubMed] [Google Scholar]
- 2.Berendonk T.U., Manaia C.M., Merlin C., Fatta-Kassinos D., Cytryn E., Walsh F., Burgmann H., Sorum H., Norstrom M., Pons M.N., et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015;13:310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 3.Verlicchi P., Al Aukidy M., Galletti A., Petrovic M., Barceló D. Hospital effluent: Investigation of the concentrations and distribution of pharmaceuticals and environmental risk assessment. Sci. Total Environ. 2012;430:109–118. doi: 10.1016/j.scitotenv.2012.04.055. [DOI] [PubMed] [Google Scholar]
- 4.Lien L.T., Hoa N.Q., Chuc N.T., Thoa N.T., Phuc H.D., Diwan V., Dat N.T., Tamhankar A.J., Lundborg C.S. Antibiotics in Wastewater of a Rural and an Urban Hospital before and after Wastewater Treatment, and the Relationship with Antibiotic Use-A One Year Study from Vietnam. Int. J. Environ. Res. Public Health. 2016;13:558. doi: 10.3390/ijerph13060588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guenther S., Ewers C., Wieler L.H. Extended-Spectrum Beta-Lactamases Producing E. coli in Wildlife, yet Another Form of Environmental Pollution? Front. Microbiol. 2011;2:246. doi: 10.3389/fmicb.2011.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drieux L., Haenn S., Moulin L., Jarlier V. Quantitative evaluation of extended-spectrum beta-lactamase-producing Escherichia coli strains in the wastewater of a French teaching hospital and relation to patient strain. Antimicrob. Resist. Infect. Control. 2016;5:9. doi: 10.1186/s13756-016-0108-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.World Health Organization (WHO) Antibiotic Resistance: Global Report on Surveillance 2014. WHO; Geneva, Switzerland: 2015. [Google Scholar]
- 8.Chandy S.J., Naik G.S., Balaji V., Jeyaseelan V., Thomas K., Lundborg C.S. High cost burden and health consequences of antibiotic resistance: The price to pay. J. Infect. Dev. Ctries. 2014;8:1096–1102. doi: 10.3855/jidc.4745. [DOI] [PubMed] [Google Scholar]
- 9.Alsan M., Schoemaker L., Eggleston K., Kammili N., Kolli P., Bhattacharya J. Out-of-pocket health expenditures and antimicrobial resistance in low-income and middle-income countries: An economic analysis. Lancet Infect. Dis. 2015;15:1203–1210. doi: 10.1016/S1473-3099(15)00149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.World Health Organization (WHO) Global Action Plan on Antimicrobial Resistance. WHO; Geneva, Switzeland: 2015. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization (WHO) Worldwide Country Situation Analysis: Response to Antimicrobial Resistance. WHO; Geneva, Switzerland: 2015. [Google Scholar]
- 12.Velez R., Sloand E. Combating antibiotic resistance, mitigating future threats and ongoing initiatives. J. Clin. Nurs. 2016;25:1886–1889. doi: 10.1111/jocn.13246. [DOI] [PubMed] [Google Scholar]
- 13.Roca I., Akova M., Baquero F., Carlet J., Cavaleri M., Coenen S., Cohen J., Findlay D., Gyssens I., Heuer O.E., et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015;6:22–29. doi: 10.1016/j.nmni.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.General Assembly of the United Nations. [(accessed on 7 November 2016)];PRESS RELEASE: High-Level Meeting on Antimicrobial Resistance. Available online: http://www.un.org/pga/71/2016/09/21/press-release-hl-meeting-on-antimicrobial-resistance/
- 15.Hocquet D., Muller A., Bertrand X. What happens in hospitals does not stay in hospitals: Antibiotic-resistant bacteria in hospital wastewater systems. J. Hosp. Infect. 2016;93:395–402. doi: 10.1016/j.jhin.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 16.Talaro K.P., Talaro A. Foundations in Microbiology. 4th ed. The McGraw−Hill Companies; Boston, MA, USA: 2002. pp. 799–813. [Google Scholar]
- 17.Cinical and Laboratory Standards Institute (CLSI) Performance Standards for Antimicrobial Disk Susceptibility Tests. Approved Standard-Eleventh Edition. CLSI; Wayne, PA, USA: 2012. [Google Scholar]
- 18.EUCAST Antimicrobial Wild Type Distributions of Microorganisms. [(accessed on 2 December 2016)]; Available online: http://mic.eucast.org/Eucast2/SearchController/search.jsp?action=performSearch&BeginIndex=0&Micdif=dif&NumberIndex50&Antib=−1&Specium=162.
- 19.Dallenne C., Da Costa A., Decre D., Favier C., Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J. Antimicrob. Chemother. 2010;65:490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 20.Le T.M., Baker S., Le T.P., Cao T.T., Tran T.T., Nguyen V.M., Campbell J.I., Lam M.Y., Nguyen T.H., et al. High prevalence of plasmid-mediated quinolone resistance determinants in commensal members of the Enterobacteriaceae in Ho Chi Minh City, Vietnam. J. Méd. Microbiol. 2009;58:1585–1592. doi: 10.1099/jmm.0.010033-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magiorakos A.P., Srinivasan A., Carey R.B., Carmeli Y., Falagas M.E., Giske C.G., Harbarth S., Hindler J.F., Kahlmeter G., Olsson-Liljequist B., et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012;18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 22.Duong H.A., Pham N.H., Nguyen H.T., Hoang T.T., Pham H.V., Pham V.C., Berg M., Giger W., Alder A.C. Occurrence, fate and antibiotic resistance of fluoroquinolone antibacterials in hospital wastewaters in Hanoi, Vietnam. Chemosphere. 2008;72:968–973. doi: 10.1016/j.chemosphere.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 23.Alam M.Z., Aqil F., Ahmad I., Ahmad S. Incidence and transferability of antibiotic resistance in the enteric bacteria isolated from hospital wastewater. Braz. J. Microbiol. 2013;44:799–806. doi: 10.1590/S1517-83822013000300021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Korzeniewska E., Korzeniewska A., Harnisz M. Antibiotic resistant Escherichia coli in hospital and municipal sewage and their emission to the environment. Ecotoxicol. Environ. Saf. 2013;91:96–102. doi: 10.1016/j.ecoenv.2013.01.014. [DOI] [PubMed] [Google Scholar]
- 25.Huovinen P. Resistance to trimethoprim-sulfamethoxazole. Clin. Infect. Dis. 2001;32:1608–1614. doi: 10.1086/320532. [DOI] [PubMed] [Google Scholar]
- 26.Chagas T.P., Seki L.M., Cury J.C., Oliveira J.A., Davila A.M., Silva D.M., Asensi M.D. Multiresistance, beta-lactamase-encoding genes and bacterial diversity in hospital wastewater in Rio de Janeiro, Brazil. J. Appl. Microbiol. 2011;111:572–581. doi: 10.1111/j.1365-2672.2011.05072.x. [DOI] [PubMed] [Google Scholar]
- 27.Rawat D., Nair D. Extended-spectrum β-lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis. 2010;2:263–274. doi: 10.4103/0974-777X.68531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bush K. Characterization of beta-lactamases. Antimicrob. Agents Chemother. 1989;33:259–263. doi: 10.1128/AAC.33.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva J., Castillo G., Callejas L., Lopez H., Olmos J. Frequency of transferable multiple antibiotic resistance amongst coliform bacteria isolated from a treated sewage effluent in Antofagasta, Chile. Electron. J. Biotechnol. 2006;5:533–540. doi: 10.2225/vol9-issue5-fulltext-7. [DOI] [Google Scholar]
- 30.Brun-Buisson C., Legrand P., Philippon A., Montravers F., Ansquer M., Duval J. Transferable enzymatic resistance to third-generation cephalosporins during nosocomial outbreak of multiresistant Klebsiella pneumoniae. Lancet. 1987;2:302–306. doi: 10.1016/S0140-6736(87)90891-9. [DOI] [PubMed] [Google Scholar]
- 31.Chandran S.P., Diwan V., Tamhankar A.J., Joseph B.V., Rosales-Klintz S., Mundayoor S., Lundborg C.S., Macaden R. Detection of carbapenem resistance genes and cephalosporin, and quinolone resistance genes along with oqxAB gene in Escherichia coli in hospital wastewater: A matter of concern. J. Appl. Microbiol. 2014;117:984–995. doi: 10.1111/jam.12591. [DOI] [PubMed] [Google Scholar]
- 32.Diwan V., Chandran S.P., Tamhankar A.J., Lundborg C.S., Macaden R. Identification of extended-spectrum -lactamase and quinolone resistance genes in Escherichia coli isolated from hospital wastewater from central India. J. Antimicrob. Chemother. 2012;67:857–859. doi: 10.1093/jac/dkr564. [DOI] [PubMed] [Google Scholar]
- 33.Abdulhaq A., Basode V.K. Prevalence of extended-spectrum beta-lactamase-producing bacteria in hospital and community sewage in Saudi Arabia. Am. J. Infect. Control. 2015;43:1139–1141. doi: 10.1016/j.ajic.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Bush K., Jacoby G.A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother. 2010;54:969–976. doi: 10.1128/AAC.01009-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Varela A.R., Macedo G.N., Nunes O.C., Manaia C.M. Genetic characterization of fluoroquinolone resistant Escherichia coli from urban streams and municipal and hospital effluents. FEMS Microbiol. Ecol. 2015;91 doi: 10.1093/femsec/fiv015. [DOI] [PubMed] [Google Scholar]
- 36.Yamane K., Wachino J., Suzuki S., Arakawa Y. Plasmid-mediated qepA gene among Escherichia coli clinical isolates from Japan. Antimicrob. Agents Chemother. 2008;52:1564–1566. doi: 10.1128/AAC.01137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alrowais H., McElheny C.L., Spychala C.N., Sastry S., Guo Q., Butt A.A., Doi Y. Fosfomycin Resistance in Escherichia coli, Pennsylvania, USA. Emerg. Infect. Dis. 2015;21:2045–2047. doi: 10.3201/eid2111.150750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edwards J.R., Betts M.J. Carbapenems: The pinnacle of the β-lactam antibiotics or room for improvement? J. Antimicrob. Chemother. 2000;45:1–4. doi: 10.1093/jac/45.1.1. [DOI] [PubMed] [Google Scholar]
- 39.Le N.K., Wertheim H.F., Vu P.D., Khu D.T., Le H.T., Hoang B.T., Vo V.T., Lam Y.M., Vu D.T., Nguyen T.H., et al. High prevalence of hospital-acquired infections caused by gram-negative carbapenem resistant strains in Vietnamese pediatric ICUs: A multi-centre point prevalence survey. Medicine. 2016;95:e4099. doi: 10.1097/MD.0000000000004099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laffite A., Kilunga P.I., Kayembe J.M., Devarajan N., Mulaji C.K., Giuliani G., Slaveykova V.I., Pote J. Hospital Effluents Are One of Several Sources of Metal, Antibiotic Resistance Genes, and Bacterial Markers Disseminated in Sub-Saharan Urban Rivers. Front. Microbiol. 2016;7:1128. doi: 10.3389/fmicb.2016.01128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mandal S.M., Ghosh A.K., Pati B.R. Dissemination of antibiotic resistance in methicillin-resistant Staphylococcus aureus and vancomycin-resistant S. aureus strains isolated from hospital effluents. Am. J. Infect. Control. 2015;43:e87–e88. doi: 10.1016/j.ajic.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 42.Ory J., Bricheux G., Togola A., Bonnet J.L., Donnadieu-Bernard F., Nakusi L., Forestier C., Traore O. Ciprofloxacin residue and antibiotic-resistant biofilm bacteria in hospital effluent. Environ. Pollut. 2016;214:635–645. doi: 10.1016/j.envpol.2016.04.033. [DOI] [PubMed] [Google Scholar]
- 43.Roderova M., Sedlakova M.H., Pudova V., Hricova K., Silova R., Imwensi P.E., Bardon J., Kolar M. Occurrence of bacteria producing broad-spectrum beta-lactamases and qnr genes in hospital and urban wastewater samples. New Microbiol. 2016;39:124–133. [PubMed] [Google Scholar]
- 44.Santoro D.O., Cardoso A.M., Coutinho F.H., Pinto L.H., Vieira R.P., Albano R.M., Clementino M.M. Diversity and antibiotic resistance profiles of Pseudomonads from a hospital wastewater treatment plant. J. Appl. Microbiol. 2015;119:1527–1540. doi: 10.1111/jam.12936. [DOI] [PubMed] [Google Scholar]
- 45.Varela A.R., Nunes O.C., Manaia C.M. Quinolone resistant Aeromonas spp. as carriers and potential tracers of acquired antibiotic resistance in hospital and municipal wastewater. Sci. Total Environ. 2016;542:665–671. doi: 10.1016/j.scitotenv.2015.10.124. [DOI] [PubMed] [Google Scholar]
- 46.Vaz-Moreira I., Varela A.R., Pereira T.V., Fochat R.C., Manaia C.M. Multidrug Resistance in Quinolone-Resistant Gram-Negative Bacteria Isolated from Hospital Effluent and the Municipal Wastewater Treatment Plant. Microb. Drug Resist. 2016;22:155–163. doi: 10.1089/mdr.2015.0118. [DOI] [PubMed] [Google Scholar]
- 47.Anssour L., Messai Y., Estepa V., Torres C., Bakour R. Characteristics of ciprofloxacin-resistant Enterobacteriaceae isolates recovered from wastewater of an Algerian hospital. J. Infect. Dev. Ctries. 2016;10:728–734. doi: 10.3855/jidc.6727. [DOI] [PubMed] [Google Scholar]
- 48.Li C., Lu J., Liu J., Zhang G., Tong Y., Ma N. Exploring the correlations between antibiotics and antibiotic resistance genes in the wastewater treatment plants of hospitals in Xinjiang, China. Environ. Sci. Pollut. Res. Int. 2016;23:15111–15121. doi: 10.1007/s11356-016-6688-z. [DOI] [PubMed] [Google Scholar]
- 49.Harris S., Morris C., Morris D., Cormican M., Cummins E. Antimicrobial resistant Escherichia coli in the municipal wastewater system: Effect of hospital effluent and environmental fate. Sci. Total Environ. 2014;468–469:1078–1085. doi: 10.1016/j.scitotenv.2013.09.017. [DOI] [PubMed] [Google Scholar]
- 50.Iweriebor B.C., Gaqavu S., Obi L.C., Nwodo U.U., Okoh A.I. Antibiotic susceptibilities of enterococcus species isolated from hospital and domestic wastewater effluents in alice, eastern cape province of South Africa. Int. J. Environ. Res. Public Health. 2015;12:4231–4246. doi: 10.3390/ijerph120404231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Galvin S., Boyle F., Hickey P., Vellinga A., Morris D., Cormican M. Enumeration and Characterization of Antimicrobial-Resistant Escherichia coli Bacteria in Effluent from Municipal, Hospital, and Secondary Treatment Facility Sources. Appl. Environ. Microbiol. 2010;76:4772–4779. doi: 10.1128/AEM.02898-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferreira da Silva M., Vaz-Moreira I., Gonzalez-Pajuelo M., Nunes O.C., Manaia C.M. Antimicrobial resistance patterns in Enterobacteriaceae isolated from an urban wastewater treatment plant. FEMS Microbiol. Ecol. 2007;60:166–176. doi: 10.1111/j.1574-6941.2006.00268.x. [DOI] [PubMed] [Google Scholar]
- 53.Rizzo L., Manaia C., Merlin C., Schwartz T., Dagot C., Ploy M.C., Michael I., Fatta-Kassinos D. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: A review. Sci. Total Environ. 2013;447:345–360. doi: 10.1016/j.scitotenv.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 54.Nagulapally S.R., Ahmad A., Henry A., Marchin G.L., Zurek L., Bhandari A. Occurrence of ciprofloxacin-, trimethoprim-sulfamethoxazole-, and vancomycin-resistant bacteria in a municipal wastewater treatment plant. Water Environ. Res. 2009;81:82–90. doi: 10.2175/106143008X304596. [DOI] [PubMed] [Google Scholar]
- 55.Rosenberg Goldstein R.E., Micallef S.A., Gibbs S.G., George A., Claye E., Sapkota A., Joseph S.W., Sapkota A.R. Detection of vancomycin-resistant enterococci (VRE) at four U.S. wastewater treatment plants that provide effluent for reuse. Sci. Total Environ. 2014;466–467:404–411. doi: 10.1016/j.scitotenv.2013.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maheshwari M., Yaser N.H., Naz S., Fatima M., Ahmad I. Emergence of ciprofloxacin-resistant extended-spectrum beta-lactamase-producing enteric bacteria in hospital wastewater and clinical sources. J. Glob. Antimicrob. Resist. 2016;5:22–25. doi: 10.1016/j.jgar.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 57.Huijbers P.M., Blaak H., de Jong M.C., Graat E.A., Vandenbroucke-Grauls C.M., de Roda Husman A.M. Role of the Environment in the Transmission of Antimicrobial Resistance to Humans: A Review. Environ. Sci. Technol. 2015;49:11993–12004. doi: 10.1021/acs.est.5b02566. [DOI] [PubMed] [Google Scholar]
- 58.Hower S., Phillips M.C., Brodsky M., Dameron A., Tamargo M.A., Salazar N.C., Jackson C.R., Barrett J.B., Davidson M., Davis J., et al. Clonally related methicillin-resistant Staphylococcus aureus isolated from short-finned pilot whales (Globicephala macrorhynchus), human volunteers, and a bayfront cetacean rehabilitation facility. Microb. Ecol. 2013;65:1024–1038. doi: 10.1007/s00248-013-0178-3. [DOI] [PubMed] [Google Scholar]