Abstract
Genetically engineered bacteria can be used for a wide range of applications, from monitoring environmental toxins to studying complex communication networks in the human digestive system. Although great strides have been made in studying single strains of bacteria in well-controlled microfluidic environments, there remains a need for tools to reliably control and measure communication between multiple discrete bacterial populations. Stable long-term experiments (e.g., days) with controlled population sizes and regulated input (e.g., concentration) and output measurements can reveal fundamental limits of cell-to-cell communication. In this work, we developed a microfluidic platform that utilizes a porous monolith to reliably and stably partition adjacent strains of bacteria while allowing molecular communication between them for several days. We measured small molecule production by the bacterial populations in response to stimuli using analytical chemistry methods and measured fluorescent output. The results are compared with communication and diffusion delay models. This porous monolith microfluidic system enables bacterial cell-to-cell communication assays with dynamic control of inputs, relatively long-term experimentation with no cross contamination, and stable bacterial population size. This system can serve as a valuable tool in understanding bacterial communication and improving biosensor design capabilities.
I. INTRODUCTION
The human gastrointestinal (GI) tract maintains a large and diverse microbiome that plays a vital role in maintaining human health.1 Gut microbiota are involved in essential processes including immune system development, nutrient synthesis, epithelial renewal, and antimicrobial secretion.2 Dysregulation of GI tract bacterial populations has detrimental inflammatory and metabolic effects, leading to cardiovascular, neurodegenerative, and autoimmune disorders.3 Although great advancements have been made in mapping the genetic diversity of this microbiome, we still lack a fundamental, scientific understanding of how this complex community of over 1000 bacterial species and 100 trillion individual cells in the human GI tract is able to maintain homeostasis within a living organism.4
Current hypotheses suggest that GI tract bacterial species survey their microbiome and regulate their symbiotic relationships in response to stimuli such as pH, temperature, and molecular signals.5 While the vast complexity of these species and stimuli is still overwhelming, isolating and genetically engineering a few dominant species within a controllable, engineered environment (e.g., a microfluidic device) to monitor and perturb their molecular signals may yield fundamental insights into the network dynamics within the human gut. Beyond elucidating the fundamental mechanisms in these physiological systems, understanding these communication signals could enable us to modulate, regulate, and even accelerate communication for both human health and engineered bacterial biosensors (e.g., Refs. 6–8).
Microfluidics can provide control over experimental conditions such as population size, temperature, nutrient levels, and stimulus. As such they have been used for a wide variety of cell studies,9 yet stable isolation of different populations while allowing molecular communication remains challenging. Isolation, or partitioning, has been achieved through a micro-ratchet structure,10 an alumina matrix filter,11 and a chitosan membrane.12,13 Although the micro-ratchet structure10 was able to culture isolated populations of bacteria on chip, cross-contamination limited experiments to 8 h—the duration of a single signaling event with insufficient time to stabilize population size (24 h). The alumina matrix filter11 successfully kept populations isolated without cross-contamination, but the chambers were very large, sealed off, and had no flow, thus precluding dynamic experimentation with control of stimulus, population size, nutrients, and removal of excess bacteria. The work done by Luo et al.12,13 made progress culturing populations of bacteria in an alginate gel, isolating them using a chitosan membrane. Flow rates were necessarily extremely limited, and after 15 h, bacteria outgrew the gel. Further, the alginate gel used to mimic the biofilm could potentially have deleterious effects on bacterial growth and communication. In this work, we developed a microfluidic platform that utilizes a porous monolith to reliably and stably partition adjacent strains of bacteria while allowing communication between them for several days. This microfluidic system allows for continual monitoring of output using fluorescent microscopy as well as collection of effluent for offline small molecule quantification.
II. METHODS
A. Bacteria and communication network architecture
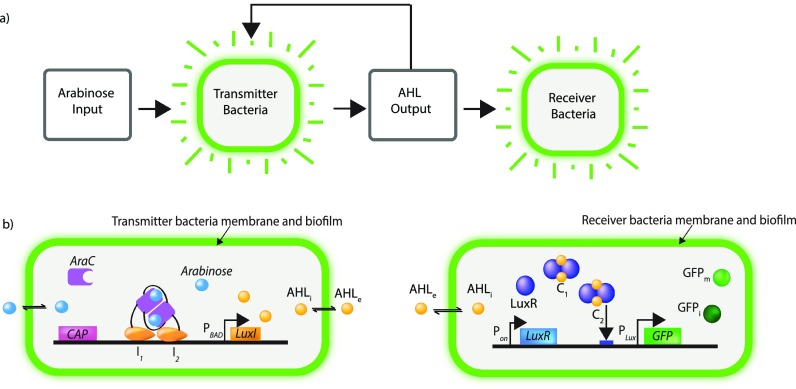
Two distinct bacterial strains with different functions were engineered for this work: a transmitter and a receiver. In response to an inducer from its surroundings, the transmitter cells emit chemical signal molecules to the receiver cells, which in turn report via fluorescent output. Figure 1 depicts this bacterial communication network and shows the bacterial strains in detail. As shown in Fig. 1(a), arabinose is the inducer (Arabinose Input) for the transmitter, which produces the chemical signal N-Hexanoyl-L-homoserine lactone (AHL) (AHL output). The AHL is then the input for the receiver bacteria. As we have engineered them, both the transmitter and receiver bacteria produce green fluorescent protein (GFP) as a visual output that can be imaged using fluorescent microscopy.
FIG. 1.
Bacterial (a) network flow chart, with (b) detailed representations of the transmitter bacterial and receiver bacterial strains showing their signaling pathways.
For both bacterial strains, we utilized genetic features of a marine symbiotic bacterium Vibrio fischeri (V. fischeri) that possesses a quorum sensing system called the LuxIR circuit. In the native system, the LuxI enzyme catalyzes the generation of the AHL signaling molecule, and the LuxR receptor protein upon binding AHL can serve as a transcription factor that activates the Plux promoter that controls production of a visual output, light. To confer communication properties to the Escherichia coli (E. coli) strains used in this work, both receiver and transmitter bacteria were transformed with plasmids constructed with components from V. fischeri. The arabinose input and AHL output [Fig. 1(a)] plasmid construction was developed using additional E. coli genetic components as described by Schleif.14
The receiver bacterial development [Fig. 1(b)], testing, and modeling have been reported in our previous work15 but will be briefly mentioned here. In the receiver bacterial construction, AHL diffuses across the membrane while a constitutive promoter (Pon) drives the expression of the luxR gene. AHLe represents AHL external to the cell, while AHLi is internal AHL. LuxR forms a dimer with the AHL to form a complex. That complex then activates the lux promoter (PLux), which in this system results in expression of the green fluorescent protein (GFP) gene carrying a degradation tag. In its immature state (GFPi), it is not fluorescent. Once the GFP reaches its mature state (GFPm), it becomes fluorescent. GFP production causes the receiver bacteria to fluoresce in response to AHL accumulation.
The transmitter bacteria feature both “off” (non-fluorescent) and “on” [fluorescent, shown in Fig. 1(b)] states, triggered by the presence of an arabinose inducer. Without the presence of arabinose in the cell (“off”), an AraC protein dimer is bound to the PBAD promoter DNA for the luxI gene and represses AHL production by preventing access by RNA polymerase. When arabinose is present and transported across the bacterial membrane, it can bind to AraC (“on”) and induces a change in AraC protein conformation. This leads to the release of AraC from the PBAD promoter, “de-repressing” it so that RNA polymerase can express the luxI gene to synthesize AHL. The AHL produced in response to arabinose can diffuse out of the transmitter bacteria into the microfluidic chamber for reception by the receiver bacteria. To facilitate monitoring of AHL production by the transmitter bacteria fluorescently, we also integrated the AHL-receptive GFP gene network within the transmitters so that they can also receive self-produced AHL Output and express GFP in response [Fig. 1(a)].
B. Acyl homoserine lactone (AHL) quantification
Quantification of the AHL output of the transmitter bacteria is necessary to understand the entire communication network and to model the events depicted in Fig. 1(a). AHL is traditionally measured using a bacterial bioassay,16 but this method only yields order of magnitude concentration resolution. To improve the quantification to micro-molar resolution, we adapted the methods used by Kannappan et al.,17,18 after collecting effluent media (360 μl for every hour of experimentation) containing AHL from the transmitters isolated on the microfluidic chip.
To extract the AHL from the media, the solution was centrifuged for 10 min at 10,000 rpm in a sterile tube and the supernatant was collected. The supernatant was mixed with ethyl acetate (2:1 ratio), incubated at room temperature for 15 min, and transferred to a separatory funnel for 5 min. The organic layer was collected in a sterile container, and the aqueous layer was extracted. The separatory step was repeated twice.
To separate the AHL from the ethyl acetate, a rotary evaporator was used. With the solution in a 100 ml round bottom flask submerged in 37 °C water, the pressure of the rotary evaporator system was slowly lowered until the pressure reached 80 mbar and then left until all solvent had evaporated. The AHL was re-suspended in 15% methanol and stored (−60 °C) for analysis using liquid chromatography-mass spectrometry (LC-MS, using ThermoFisher Scientific LTQ Orbitrap XL ETD), with a detection threshold of 1 μM.
To determine whether AHL extraction and quantification could be performed with high yield, linearly, we constructed a set of calibration curves. We compared the AHL quantification achieved through the extraction from media (spiked with known concentrations of AHL) to AHL in 15% methanol (control). For both test and control, AHL concentrations were as follows: 100 μM, 75 μM, 50 μM, 25 μM, 10 μM, and 1 μM. The controls were repeated seven times and the extracts were repeated six times, with averages shown.
C. Microfluidic chip design and fabrication
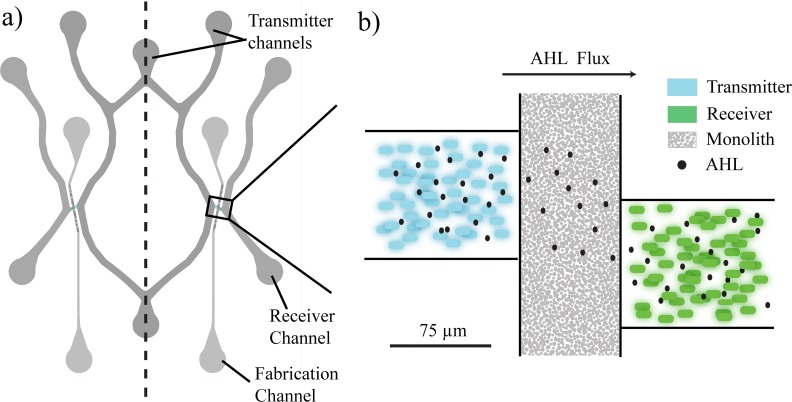
The microfluidic device design is shown in Fig. 2(a), with a detailed view of the “communication zone” featuring the porous monolith and trapping chambers shown in Fig. 2(b). The line of symmetry [Fig. 2(a)] enables two simultaneous communication experiments to be conducted. To optimize the flow channel and chamber design to maximize the AHL concentration in the receiver chamber while minimizing clogging, we used finite element analysis (COMSOL) modeling.
FIG. 2.
(a) Microfluidic chip design with flow channels and adjacent trapping chambers shown (b) schematically. The bacterial populations grow in isolated chambers separated by the monolith. AHL diffuses through the porous monolith from the transmitters to the receivers, the adjacent flow channels of which provide nutrients, oxygen, and carry away excess or dead bacteria.
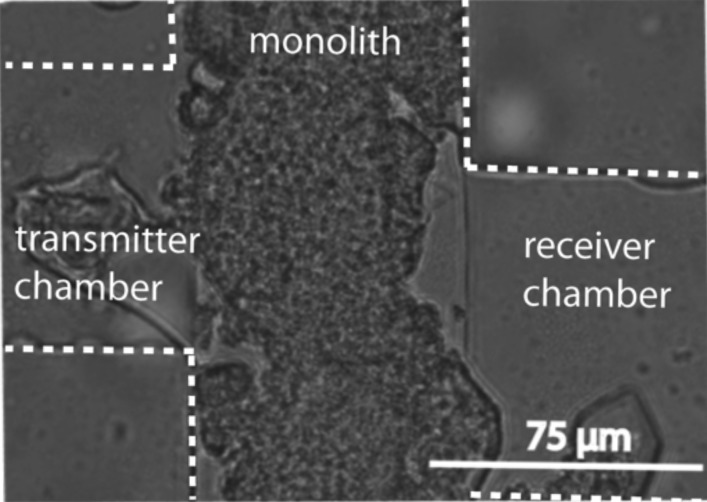
Each of the two communication zones comprises a pair of trapping chambers (100 μm × 100 μm × 5 μm tall) to isolate the bacterial populations that we have developed previously.15,19 They are separated by a porous monolith (75 μm wide, 10 μm tall). During communication experiments, Fig. 2(b) shows that the transmitter produces AHL (black dots), and the AHL is free to diffuse through the porous monolith (gray) to the receiver bacteria (green). The flow channels (250 μm wide × 10 μm high) are in direct open contact with the chambers and are used to flow receiver media and either transmitter media or transmitter stimulus as necessary to stimulate this communication. Flow was collected from the “transmitter output” port [Fig. 2(a)] for AHL quantification. The constant flow of media is critical not only for the health of the bacteria but also to remove excess bacteria to maintain constant population sizes for the duration of the experiment and across all experiments. Port separation is minimized, allowing for metal connectors for tubing without collapsing the channels. To reduce the number of ports, thus the number of tubing connections, we tried to simplify the design by combining common channels. Prior to each communication experiment, the monolith fabrication ports are used to flow monolith reagents through a channel where they are polymerized.
To fabricate the microfluidic devices, we utilized standard soft lithography20 to create a SU-8 and silicon mold. Polydimethylsiloxane (PDMS) was poured and baked onto the mold, peeled from the mold, and then plasma treated and bonded, resulting in a PDMS device bonded to a glass coverslip.
D. Monolith fabrication
Several methods of porous monolith fabrication inside PDMS microfluidics21–24 have been published, and in particular, Araya-Farias et al.24 developed an innovative single step approach—a monolith consisting of ethylene glycol methacrylate phosphate monomer and N,N′-methylene-bis-acrylamide crosslinker.
The monolith solution consists of 2‐(methacryloxy) ethyl phosphate (EGMP) monomer, N,N′-methylenebis(acrylamide) crosslinker, and 2,2-dimethoxy-2-phenylacetophenone photoinitiator dissolved in a solvent mixture of 1-dodecanol (DDC), dimethylsulfoxide (DMSO), and N,N-dimethylformamide (DMF). The solvents were mixed in proportions of 58.2% w/w DMSO, 32.2% w/w DDC, and 9.6% w/w DMF and left to sit at room temperature for ∼2 h. The monomer, crosslinker, and photoinitiator were weighed at proportions of 52.3% w/w EGMP, 40.3% w/w BAA (N,N′-methylene-bis-acrylamide), and 7.4% w/w DMPA (2,2-dimethoxy-2-phenylacetophenone) and dissolved in the solvent (to give a final mixture of 44.0% w/w DMSO, 24.3% w/w DDC, 12.8% w/w EGMP, 9.8% w/w BAA, 7.3% w/w DMF, and 1.8% w/w DMPA). Due to the oxygen-sensitive nature of free-radical polymerization, the reaction solution was degassed via nitrogen sparging while under sonication and the prepared PDMS device was subjected to a near-vacuum each for 20 min prior to injection.
Injection was done manually by using a syringe quickly so as to reduce the solution's exposure to open air after the reaction mixture was removed from the nitrogen stream. The device was filled entirely with monolith solution (approximately 0.1 ml). By injecting through only one monolith fabrication port on each side (of the symmetrical device), we reduced the chance of trapped bubbles in the microfluidic device.
We advanced the monolith fabrication technique of Araya-Farias et al. by lithographically patterning the monolith with micrometer resolution. Each device was fitted with a 75 μm thick stainless steel sheet that was laser cut (Stricklin Company) and masked with tape such that the monolith channel of the device is exposed to incident light while masking the flow channels and chambers. The device with mask was irradiated at 1.5 mW/cm2, 365 nm light (in a UL-2000L Crosslinker) for 12 min. Following irradiation, the device was gently flushed with 30% v/v methanol in deionized water to remove unreacted monomers and stored in deionized water at 4 °C.
Following lithography and flushing, all monolith fabrication ports were plugged with 25 gauge stainless steel wire to prevent further air/fluid flow. The chip was flushed with deionized (DI) water and then filled with LB (lysogeny broth) (2XYT Sigma Aldrich) media containing ampicillin (20 μM) via a syringe (BD, Luer-Lok) and placed under a UV light in a biosafety cabinet hood for sterilization.
E. Bacterial communication
Bacteria were initially injected into the device through, respectively, the receiver media port and the transmitter media port [see Fig. 2(a)] using a syringe (BD, Luer-Lok), filling their respective channels entirely. Excess bacteria were flushed from the channels, leaving bacterial populations in the chambers. On a heated microscope stage (30 °C), bacteria were allowed to populate the chambers for 24 h to reach capacity (∼105/chamber), while the chamber was supplied with LB at 100 μl/h.
Once the bacteria had filled the chamber and the biofilm had formed protecting the colony from shear stress, the flow rate was increased to 360 μl/h. For the transmitter bacteria, which is supplied by two ports [media port and stimulus port in Fig. 2(a)], one syringe was used for LB (at 350 μl/h), while the second (10 μl/h) was used to deliver arabinose. An “arabinose pulse” is the duration and concentration over which this arabinose inducer is delivered to the transmitter bacterial chamber. Pulse durations of 50 min and concentrations of 20 μM that we have previously optimized for stimulus15,25 were used. Fluorescent images were captured once in every 10 min at 20× magnification and post-processed using MATLAB. The intensity of the pixels within the bacterial chambers was averaged and the background fluorescence was subtracted, yielding relative fluorescence [arbitrary units (AU)]. Tygon tubing and syringe pumps (Harvard Apparatus) were used for connections and flow. After approximately two days of growth and experimentation, experiments were terminated due to bacterial growth issues (e.g., clogging).
Experimentally, the duration to fluorescence after induction was determined to be the difference between the time start of the induction pulse and the time at which the fluorescence exceeded 10% of maximum fluorescence.
III. RESULTS
A. AHL quantification
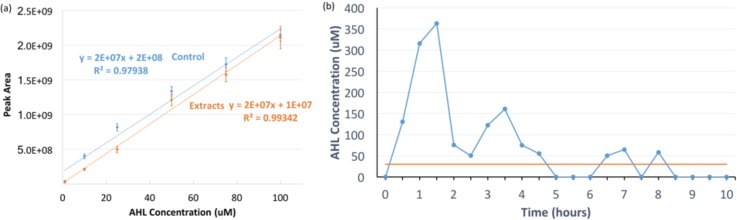
The results comparing AHL quantification from media to controls are shown in Fig. 3(a). The linear fit and the corresponding R2 values are displayed. The AHL retention efficiency of the extraction method on average was above 90% for the range of concentrations measured (1–100 μM). Thus, we successfully extracted AHL from the media, as compared to controls in methanol) using analytical chemistry tools, and the response to our quantification method is relatively linear. The resulting calibration curve between the mass spectrometry “peak area” and AHL concentration enabled us to subsequently determine the AHL concentration in the transmitter chambers during communication experiments.
FIG. 3.
(a) Comparison of AHL quantification (Mass spectrometry peak area) achieved through the extraction from media (spiked with known concentrations of AHL) to AHL in 15% methanol (control). For both test and control, AHL concentrations were as follows: 100 μM, 75 μM, 50 μM, 25 μM, 10 μM, and 1 μM. Error bars represent standard deviation. Dashed lines represent a linear fit, and the corresponding R2 values are displayed. (b) Transmitter AHL output as a function of time. The output was collected in 30 min intervals, extracted, and quantified using LC-MS. Values were then compared to extract the calibration curve to obtain AHL concentrations. The orange line indicates AHL to GFP circuit saturation.
Using the microfluidic device loaded with transmitter bacteria, we collected media from the “transmitter output” port [Fig. 2(a)] using a syringe pump and Tygon tubing with an opposite flow rate to the input ports. We extracted and analyzed the AHL concentration in the trapping chambers at 30 min intervals and correlated it with the bacterial fluorescent response. These results are essential in understanding the communication between transmitter and receiver bacteria.
The transmitter AHL output over a 10 h experiment was collected every 30 min. This was repeated 8 times and the time segmented outputs were combined over the 8 experiments. The results are shown in Fig. 3(b). The AHL concentration was determined by taking the area under the curve of the mass spectrometry data, correcting for volume changes, and comparing to the extraction calibration curve of Fig. 3(a) using the “extracts” linear fit equation.
Figure 3(b) illustrates that the AHL concentration first increases in response to the 50 min pulse of 20 μM arabinose and then begins decreasing. Notably, we found in previous experiments (not shown) that the saturation of the AHL to GFP circuit occurs around 30 μM of AHL (shown in orange). Thus, concentrations above 30 μM do not elicit additional GFP production, due to the limited amount of LuxR in the bacteria. Without any free LuxR, AHL cannot form a dimer. The variation of the concentration level over time is likely due to AHL loss during the extraction process. Most importantly, the results show the ability to quantify the AHL production of the bacteria, a vital capability for future modeling work.
B. Modeling
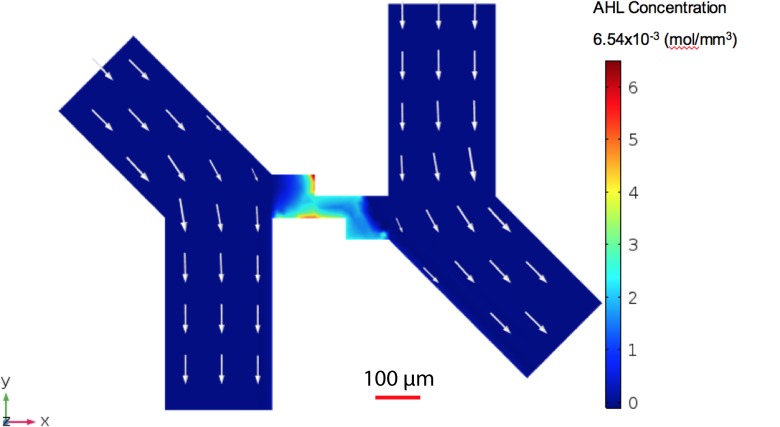
The microfluidic device design shown in Fig. 2(a) is intended to enable long term experimentation by minimizing areas of low velocity flow in/near the trapping chambers that could lead to errant bacterial growth while maximizing diffusion of AHL from the transmitter to receiver chamber. This required optimizing the angular orientation of the flow channels, which was performed in COMSOL. As shown in Fig. 4, the inlet to the trapping chamber is oriented at 45° relative to the outlet. This design resulted in 30.6% of AHL produced by the transmitters reaching the receiver bacterial population. While there were relatively low velocity locations in this design, there were no stagnation points that would lead to unwanted bacterial growth. Thus, the device features this angular orientation. Although this model neglects the effects of the bacteria in the trapping chambers, AHL loss is minimal compared to channel AHL loss.
FIG. 4.
Two-node microfluidic chip COMSOL modeling. The arrows represent the velocity fields of the media and the color intensity represents the AHL concentration (blue low to red high). The communication zone is in the center, comparable to Fig. 2(b).
C. Microfabricated chip and monolith
A representative image of the “communication zone” as fabricated is shown with the central monolith region in Fig. 5, with channel and chamber edges highlighted for comparison to Fig. 2(b). The image shows monolith formation in a chip filled with DI (deionized) water. The monolith creates a barrier between the two trapping chambers to prevent bacteria traveling from one side to the other and cross contaminating the populations, while still allowing AHL to diffuse across. Although there is some monolith debris in the trapping chamber, it is minimal and still allows the bacteria to create a stable population.
FIG. 5.
Monolith formation in the bacterial communication microfluidic device.
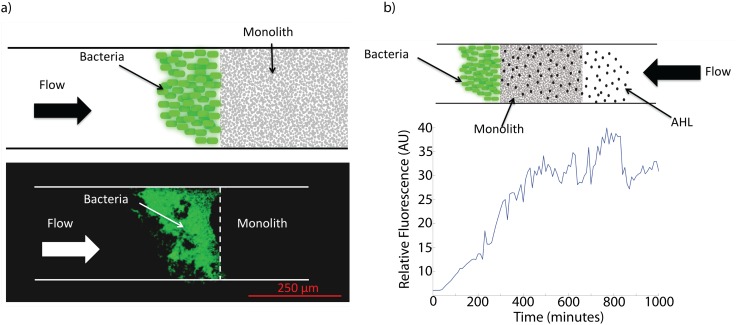
We performed a set of experiments in a simple straight channel (250 μm × 10 μm) with a porous monolith plug fabricated as described to ensure that the monolith, with its inherent pore size, could successfully restrict bacterial growth while allowing AHL flow. Figure 6(a) shows the results for a bacterial strain producing GFP injected on one side of the monolith plug. These results confirm that the bacteria could not pass through the pores in the monolith and that the monolith was well attached to the device under flow.
FIG. 6.
(a) (top) Schematic and (bottom) photograph demonstrating a plug of monolith inside a straight channel (250 μm × 10 μm). Bacteria producing GFP are introduced to the left side of the plug and do not pass through the stable monolith, while media flow does. (b) (top) Flow is reversed with AHL simulant for receiver bacteria flowing from the right, (bottom) Receiver fluorescent response in time to AHL, showing successful passage of AHL molecules with the media through the monolith.
Reversing the flow with AHL in the media enabled us to determine whether AHL could successfully pass through the monolith and stimulate GFP production receiver bacteria. This induction represents a step function. The results are shown in Fig. 6(b). As the receivers begin producing GFP, the time domain signal of relative fluorescence increases to a plateau, as expected.
D. Bacterial communication
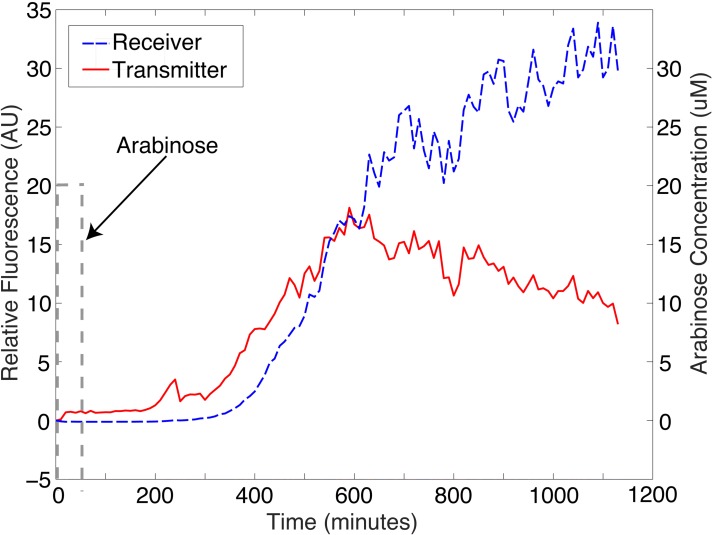
With both receiver and transmitter chambers populated with respective bacteria and nourished by media, we delivered a 50 min pulse of 20 μM arabinose to the transmitter channel. The results are shown in Fig. 7. The fluorescence response of the transmitters is first observed, followed shortly thereafter by the fluorescence response of the receiver population.
FIG. 7.
Bacterial communication results. A 50 min pulse of 20 μM arabinose was given to the transmitter bacteria. The transmitters produced AHL which in turn caused them to produce GFP. This AHL also diffuses across the porous monolith to the receiver bacterial population which in response, after a delay, produced GFP. The right axis is for Arabinose (shown as a dashed gray line).
To analyze these results, we first examined whether the delay between the transmitter response and the receiver response is as expected. This delay is governed primarily by diffusion of the small AHL molecules through the monolith. We neglected the convective forces and biofilm inhibition in these calculations.
Fick's law of diffusion utilizes a diffusion coefficient that depends on the size and shape of the molecule, interaction with the solvent, and viscosity of the solvent. This diffusion coefficient works when considering an unobstructed medium that the molecule is diffusing through. Since the AHL passes through a porous monolith, it was necessary to calculate an effective diffusion coefficient for the slowing effect the monolith would cause. This effective diffusion coefficient considers the size of the molecule and the size of the pores. The effective diffusivity (De) in porous medium is
| (1) |
where D is the diffusion coefficient in gas or liquid filling the pores (m2 s−1), εt is the porosity available for the transport (dimensionless), δ is the constrictivity (dimensionless), and τ is the tortuosity (dimensionless). Porosity is a measure of the void (i.e., “empty”) spaces in a material and is a fraction of the volume of voids over the total volume measured between 0 and 1. Constrictivity is the ratio of the diameter of the diffusing particle to the pore diameter, and tortuosity is a property of curve being tortuous inclusive of range 1–2. Since the tortuosity cannot be determined, the two extremes were examined.
To calculate the constrictivity, we first needed to establish a radius for the AHL molecule. The Stokes radius, or a hydrodynamic radius, is the radius of a hard sphere that diffuses at the same rate as the solute of interest, or
| (2) |
where D is the diffusion coefficient in gas or liquid filling the pores (m2 s−1), T is the temperature (K), η is the viscosity of the media, and kB is the Boltzmann constant. The values listed Table I were used to calculate the 3.88 nm AHL radius. The constrictivity was then calculated by taking the ratio of the AHL radius to the monolith pore size and was found to be 0.06. This was finally used to calculate the effective diffusivity of AHL of 1.335 μm2/s. All variables and their respective sources, when applicable, are shown in Table I.
TABLE I.
Coefficients and calculated values for AHL diffusion through the porous monolith. Where applicable, a source is provided and blank units are equated to dimensionless parameters.
| Symbol | Name | Value | Units | Source |
|---|---|---|---|---|
| D | AHL diffusion coefficient | 43 | μm2/s | 26 |
| εT | Porosity | 0.52 | 24 | |
| RM | Monolith pore size | 65 | Nm | 24 |
| RH | Stokes radius | 3.88 | Nm | Calculated |
| KB | Boltzmann constant | 1.38 × 10−23 | m2 kg/s2 K1 | |
| T | Temp | 303.15 | K | |
| Η | Viscosity of media | 8.01 × 10−07 | m2/s | Water |
| Δ | Constrictivity | 0.060 | Calculated | |
| τ | Tortuosity | 1–2 | Estimated | |
| DE | Effective diffusion coefficient | 1.335 | μm2/s | Calculated |
| X | Monolith width | 75 | μm | Designed |
| T | Diffusion time through Monolith | 70.22 | min | Calculated |
The distance the AHL will travel through the monolith between the transmitters and the receivers is the width of the monolith fabrication channel, 75 μm. This distance combined with the effective diffusivity leads to the calculation of the diffusion times of 70.22 min (when tortuosity = 1) and 139.76 min (when tortuosity = 2). This means that we expect the receiver bacteria to begin fluorescing approximately 70 min after the transmitter.
The time between the transmitter and receiver fluorescent start was found to be 78 min. The correspondence between the model and experiment further supports our assumption that convection and biofilm effects are small.
In addition to the time between transmitter and receiver fluorescent start, the amplitude of the fluorescence was examined. Previously, we developed a mathematical model for the receivers that predicted the fluorescence output given the AHL input,15 and in reverse, we were able to determine the AHL input given the fluorescence output.6 Inputting the fluorescent response of the receivers into the reverse model, we found that the AHL concentration level was approximately 8.5 μM. With the initial Comsol modeling, we predicted that 30% of the AHL would diffuse from transmitters to receivers. With the addition of the monolith fabrication channel in the final design, shown in (Fig. 2), there was further AHL loss due to diffusion into the fabrication channel. The final expected AHL percentage that reached the receiver population was 10%. As previously shown (Fig. 3), the transmitters produce an AHL concentration between 50 μM and 360 μM. The 8.5 μM AHL determined from experimental results fall in the expected range of 5 μM and 36 μM AHL. Although the experiment ends at 1200 min (20 h), we plan on running extended experiments in future and allowing both bacterial populations to return to the off state. Despite this, we already surpassed previous attempt's two node experimental times, with a 24-h growth period followed by 20-h communication period.
IV. CONCLUSION
A design and implementation of porous monolith microfluidics for bacterial cell-to-cell communication assays was presented. Using genetically engineered “transmitter” bacteria and “receiver” bacteria that are isolated and cultured in stable populations in adjacent chambers in the microfluidic device, separated by a 75 μm thick porous monolith, we have demonstrated successful communication between them through a “pulse of arabinose (20 μM concentration, 50 min long) which is transduced to AHL and excites fluorescence in both the transmitter and receiver populations. The delay and amplitude of the receiver response are 78 min and 30 AU, which agree well with models. Experiments were conducted for 44 h, longer than other reported work, showing that the design is stable for relatively long duration experiments.
The channels have angular orientation optimized with finite element analysis to maximize communication and minimize errant bacterial growth. We also present a method for quantifying the intermediate AHL output of the transmitters for complete communication measurement to support future modeling efforts, showing good linearity across relevant concentrations of AHL and 90% yield. Although there is a 10% loss, the presented methods are quantitative vs the gold standard bioassay, which is more qualitative. The main factor in AHL loss occurs during the extraction process and AHL degradation. Due to the length of the experiment, collected AHL samples cannot be processed the same day and must be frozen before evaluated. Future development to further tune the extraction methods is under way. Advancements in lithographically defined monolith fabrication in microfluidic channels have been presented, opening this microfabrication approach for further use.
The lithographically patterned monolith in microfluidic devices is also amenable to more complex arrangements (e.g., multiple populations) and may thus be suitable for studying complex signals such as the microenvironment of the human GI tract in order to enable a fundamental understanding of the intercellular signaling between these bacterial populations. The porous monolith microfluidic system enables bacterial cell-to-cell communication assays with dynamic control of inputs, relatively long-term experimentation with no cross contamination, and stable bacterial population size and serves as a valuable tool in understanding bacterial communication in a controllable environment and improving biosensor design capabilities.
ACKNOWLEDGMENTS
C.R.F, B.K.H., and C.M.A. are grateful for funding by the National Science Foundation (CISE 1110947). C.M.A. acknowledges the National Science Foundation Graduate Fellowship Program, without which this work would not have been possible. L.H. is grateful for the National Science Foundation's National Nanotechnology Infrastructure Network (NNIN) Summer Research Experience for Undergraduates (REU) program that funded her participation in this research, and J.E.P. and S.E. are thankful for the Georgia Tech President's Undergraduate Research Award and Undergraduate Research Opportunities Program that supported their contributions.
The authors declare no competing financial interest or conflict of interest in this work.
References
- 1. Kotula J. W. et al. , “ Programmable bacteria detect and record an environmental signal in the mammalian gut,” Proc. Natl. Acad. Sci. U.S.A. 111(13), 4838–4843 (2014). 10.1073/pnas.1321321111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sun J. and Chang E. B., “ Exploring gut microbes in human health and disease: Pushing the envelope,” Genes Dis. 1(2), 132–139 (2014). 10.1016/j.gendis.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. West C. E. et al. , “ The gut microbiota and inflammatory noncommunicable diseases: Associations and potentials for gut microbiota therapies,” J. Allergy Clin. Immunol. 135(1), 3–14 (2015). 10.1016/j.jaci.2014.11.012 [DOI] [PubMed] [Google Scholar]
- 4. Guinane C. M. and Cotter P. D., “ Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ,” Ther. Adv. Gastroenterol. 6(4), 295–308 (2013). 10.1177/1756283X13482996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Miller M. B. and Bassler B. L., “ Quorum sensing in bacteria,” Annu. Rev. Microbiol. 55, 165–199 (2001). 10.1146/annurev.micro.55.1.165 [DOI] [PubMed] [Google Scholar]
- 6. Jian Y. et al. , “ nanoNS3: A network simulator for bacterial nanonetworks based on molecular communication,” Nano Commun. Networks 12, 1–11 (2017). 10.1016/j.nancom.2017.01.004 [DOI] [Google Scholar]
- 7. Bicen A. O. et al. , “ Efficient sampling of bacterial signal transduction for detection of pulse-amplitude modulated molecular signals,” IEEE Trans. Biomed. Circuits Syst. 9(4), 505–517 (2015). 10.1109/TBCAS.2015.2465182 [DOI] [PubMed] [Google Scholar]
- 8. Akyildiz I. et al. , “ Monaco: Fundamentals of molecular nano-communication networks,” IEEE Wireless Commun. 19(5), 12–18 (2012). 10.1109/MWC.2012.6339467 [DOI] [Google Scholar]
- 9. Nilsson J. et al. , “ Review of cell and particle trapping in microfluidic systems,” Anal. Chim. Acta 649(2), 141–157 (2009). 10.1016/j.aca.2009.07.017 [DOI] [PubMed] [Google Scholar]
- 10. Park S. et al. , “ Microfabricated ratchet structure integrated concentrator arrays for synthetic bacterial cell-to-cell communication assays,” Lab Chip 12(20), 3914–3922 (2012). 10.1039/c2lc40294g [DOI] [PubMed] [Google Scholar]
- 11. Nagy K. et al. , “ Interaction of bacterial populations in coupled microchambers,” Chem. Biochem. Eng. Q. 28(2), 225–231 (2014). 10.15255/CABEQ.2013.1934 [DOI] [Google Scholar]
- 12. Luo X. et al. , “ Biofabrication of stratified biofilm mimics for observation and control of bacterial signaling,” Biomaterials 33(20), 5136–5143 (2012). 10.1016/j.biomaterials.2012.03.037 [DOI] [PubMed] [Google Scholar]
- 13. Luo X. et al. , “ Distal modulation of bacterial cell-cell signalling in a synthetic ecosystem using partitioned microfluidics,” Lab Chip 15(8), 1842–1851 (2015). 10.1039/C5LC00107B [DOI] [PubMed] [Google Scholar]
- 14. Schleif R., “ AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action,” FEMS Microbiol. Rev. 34(5), 779–796 (2010). 10.1111/j.1574-6976.2010.00226.x [DOI] [PubMed] [Google Scholar]
- 15. Austin C. M. et al. , “ Modeling and validation of autoinducer-mediated bacterial gene expression in microfluidic environments,” Biomicrofluidics 8(3), 034116 (2014). 10.1063/1.4884519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steindler L. and Venturi V., “ Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors,” FEMS Microbiol. Lett. 266(1), 1–9 (2007). 10.1111/j.1574-6968.2006.00501.x [DOI] [PubMed] [Google Scholar]
- 17. Kannappan S. et al. , “ Extraction and detection of quorum sensing N-acyl homoserine lactones from shrimp pathogen Vibrio harveyi and antagonistic effect of terrestrial plants against its growth,” African J. Microbiol. Res. 7(26), 3275–3284 (2013). 10.5897/AJMR2013.5562 [DOI] [Google Scholar]
- 18. Kim Y. W. et al. , “ MALDI-MS-based quantitative analysis for ketone containing homoserine lactones in pseudomonas aeruginosa,” Anal. Chem. 87(2), 858–863 (2015). 10.1021/ac5039362 [DOI] [PubMed] [Google Scholar]
- 19. Krishnaswamy B. et al. , “ Time-elapse communication: Bacterial communication on a microfluidic chip,” IEEE Trans. Commun. 61(12), 5139–5151 (2013). 10.1109/TCOMM.2013.111013.130314 [DOI] [Google Scholar]
- 20. McDonald J. C. et al. , “ Fabrication of microfluidic systems in poly(dimethylsiloxane),” Electrophoresis 21(1), 27–40 (2000). [DOI] [PubMed] [Google Scholar]
- 21. Hu S. W. et al. , “ Surface-directed, graft polymerization within microfluidic channels,” Anal. Chem. 76(7), 1865–1870 (2004). 10.1021/ac049937z [DOI] [PubMed] [Google Scholar]
- 22. Burke J. M. and Smela E., “ A novel surface modification technique for forming porous polymer monoliths in poly(dimethylsiloxane),” Biomicrofluidics 6(1), 016506 (2012). 10.1063/1.3693589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rohr T. et al. , “ Surface functionalization of thermoplastic polymers for the fabrication of microfluidic devices by photoinitiated grafting,” Adv. Funct. Mater. 13(4), 264–270 (2003). 10.1002/adfm.200304229 [DOI] [Google Scholar]
- 24. Araya-Farias M. et al. , “ A new strategy for simultaneous synthesis and efficient anchorage of polymer monoliths in native PDMS microchips,” Polymer 66, 249–258 (2015). 10.1016/j.polymer.2015.04.039 [DOI] [Google Scholar]
- 25. Krishnaswamy B., Austin C. M., Bardill J. P., Russakow D., Holst G. L., Hammer B. K., Forest C. R., and Sivakumar R., “ Time elapse communication for super-slow networks,” in IEEE International Conference on Communications (2013). [Google Scholar]
- 26. Schwarz-Schilling M. et al. , “ Chemical communication between bacteria and cell-free gene expression systems within linear chains of emulsion droplets,” Integr. Biol. 8(4), 564–570 (2016). 10.1039/C5IB00301F [DOI] [PubMed] [Google Scholar]