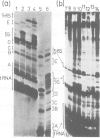
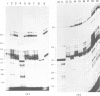
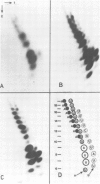
Abstract
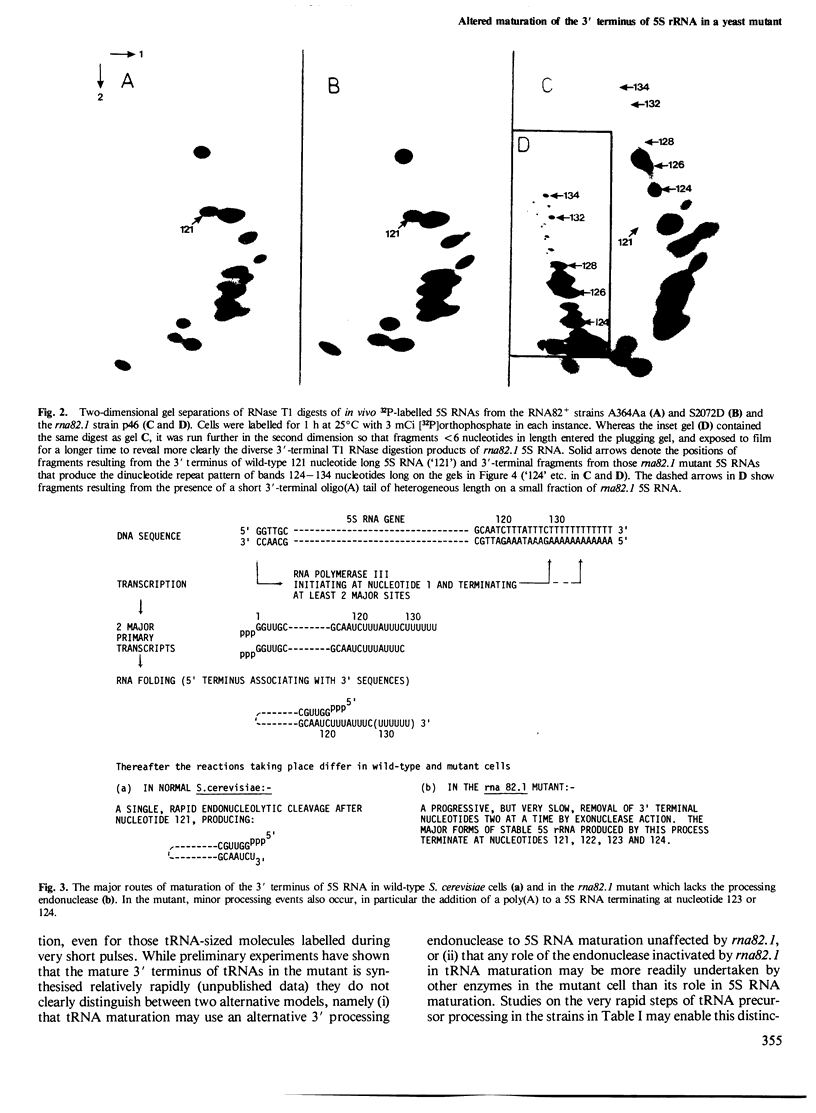
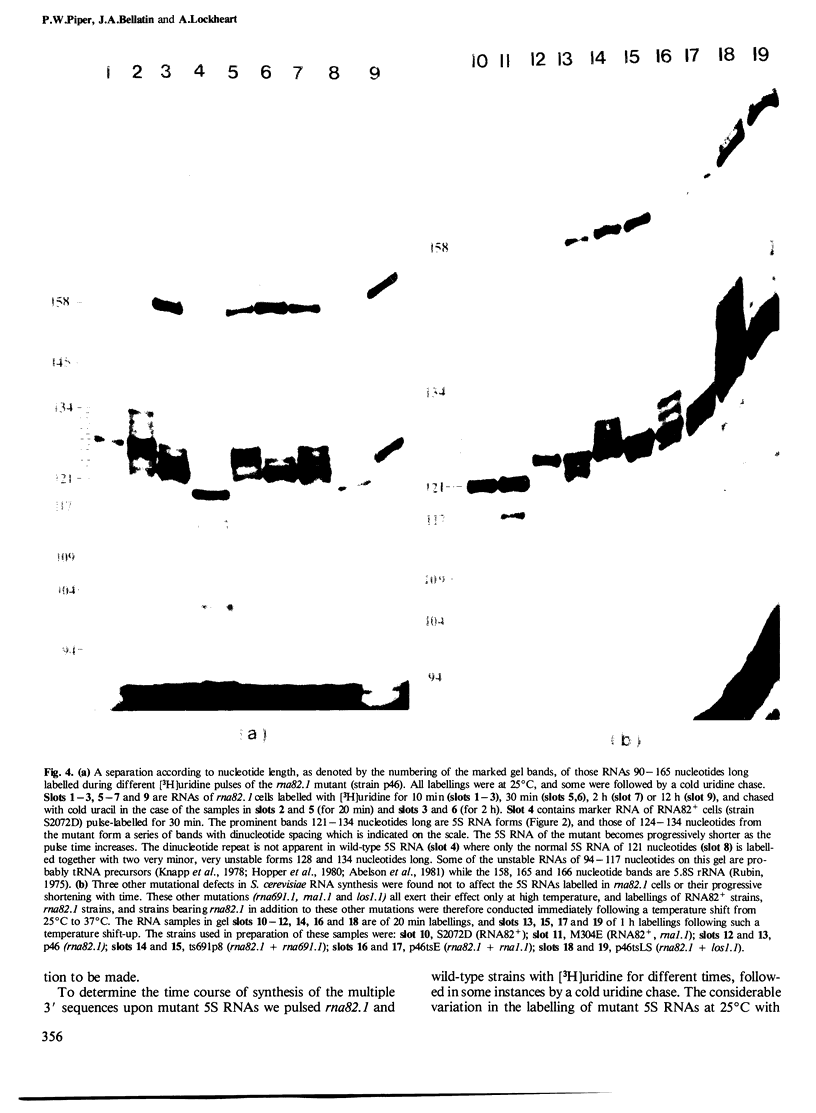
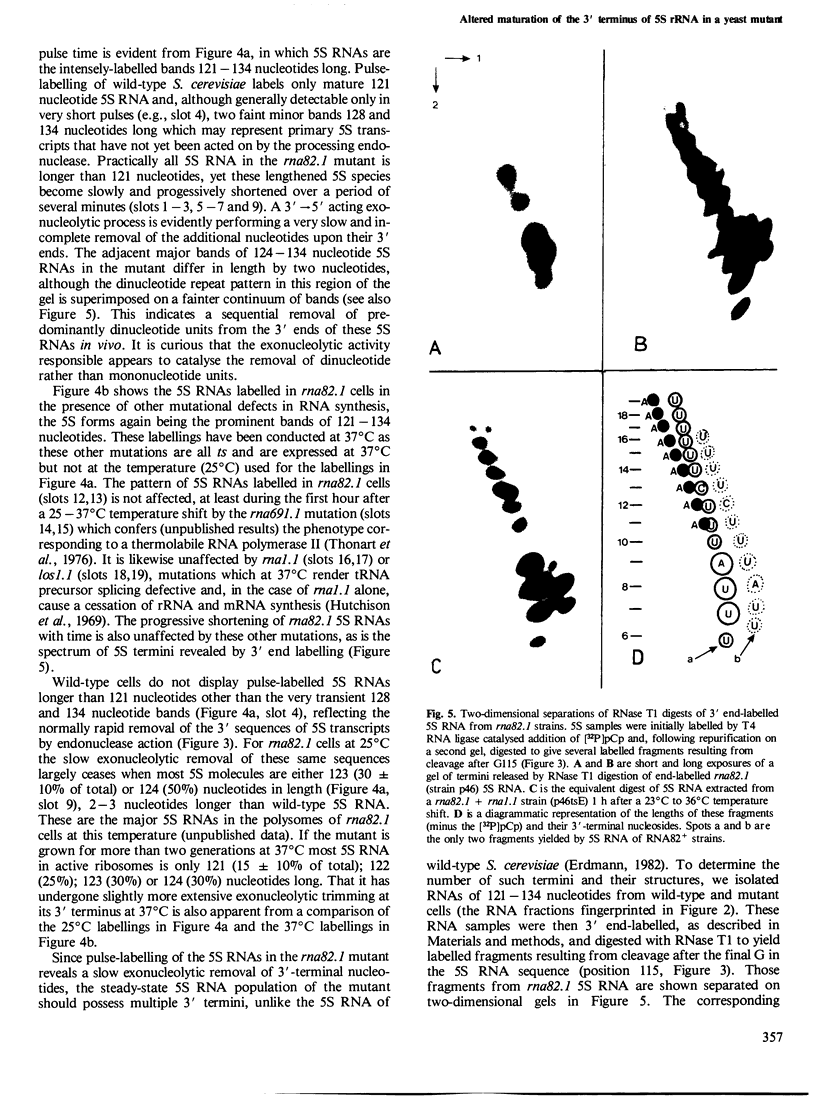
Sequences at the immediate 3' terminus of several eukaryotic primary transcripts, synthesised just before the termination of transcription, are often lost during RNA processing. The rna82.1 mutation in Saccharomyces cerevisiae appears to result in a deficiency of the endonuclease that removes such sequences from certain yeast transcripts. Some small RNAs of rna82.1 cells are a few nucleotides longer than their counterparts in wild-type S. cerevisiae. The 5S rRNAs made during very short pulse-labellings of the mutant have, relative to the mature 121 nucleotide 5S RNA of wild-type cells, an additional 7, 11 or 13 nucleotides at their 3' terminus. These 5S forms reveal sites upon 5S genes where transcription probably terminates in vivo. The extra nucleotides upon 5S RNAs in rna82.1 cells are lost very slowly by sequential removal from the 3' terminus. Through this 3'-5' exonuclease action the total 5S RNA of the mutant possesses several 3'-terminal sequences yet is mostly only 0-3 nucleotides longer than in wild-type S. cerevisiae. Just one or two of these 3'-terminal sequences serve as a substrate in vivo for a poly(A) polymerase since a small proportion of rna82.1 5S RNAs terminate in the sequence: CAAUCUUU(A)n.
Full text
PDF
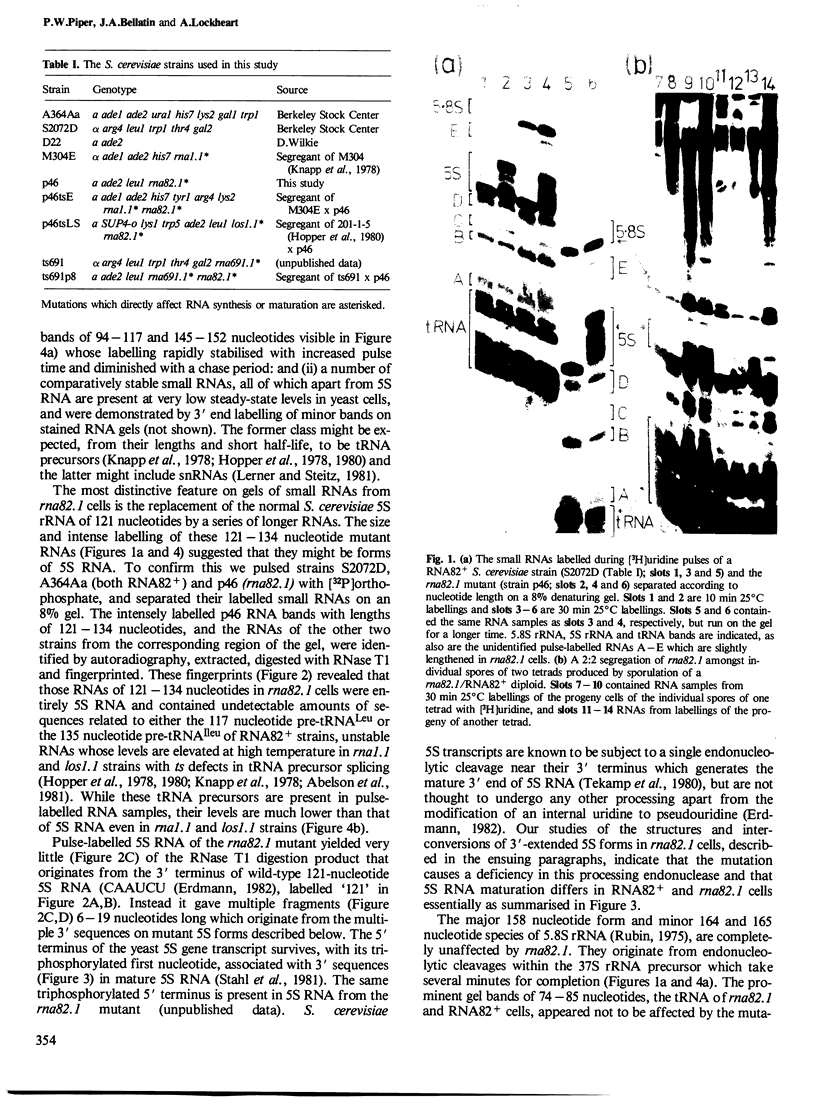


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andrew C., Hopper A. K., Hall B. D. A yeast mutant defective in the processing of 27S r-RNA precursor. Mol Gen Genet. 1976 Feb 27;144(1):29–37. doi: 10.1007/BF00277300. [DOI] [PubMed] [Google Scholar]
- Bayev A., Georgiev O. I., Hadjiolov A. A., Nikolaev N., Skryabin K. G., Zakharyev V. M. The structure of the yeast ribosomal RNA genes. 3. Precise mapping of the 18 S and 25 S rRNA genes and structure of the adjacent regions. Nucleic Acids Res. 1981 Feb 25;9(4):789–799. doi: 10.1093/nar/9.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenmeier E. H., Brown D. D., Jordan E. A nuclear extract of Xenopus laevis oocytes that accurately transcribes 5S RNA genes. Cell. 1978 Nov;15(3):1077–1086. doi: 10.1016/0092-8674(78)90291-x. [DOI] [PubMed] [Google Scholar]
- Chamberlain J. P. Fluorographic detection of radioactivity in polyacrylamide gels with the water-soluble fluor, sodium salicylate. Anal Biochem. 1979 Sep 15;98(1):132–135. doi: 10.1016/0003-2697(79)90716-4. [DOI] [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujardin G., Jacq C., Slonimski P. P. Single base substitution in an intron of oxidase gene compensates splicing defects of the cytochrome b gene. Nature. 1982 Aug 12;298(5875):628–632. doi: 10.1038/298628a0. [DOI] [PubMed] [Google Scholar]
- England T. E., Bruce A. G., Uhlenbeck O. C. Specific labeling of 3' termini of RNA with T4 RNA ligase. Methods Enzymol. 1980;65(1):65–74. doi: 10.1016/s0076-6879(80)65011-3. [DOI] [PubMed] [Google Scholar]
- Feunteun J., Jordan B. R., Monier R. Study of the maturation of 5 s RNA precursors in Escherichia coli. J Mol Biol. 1972 Oct 14;70(3):465–474. doi: 10.1016/0022-2836(72)90553-0. [DOI] [PubMed] [Google Scholar]
- Gegenheimer P., Apirion D. Processing of procaryotic ribonucleic acid. Microbiol Rev. 1981 Dec;45(4):502–541. doi: 10.1128/mr.45.4.502-541.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., Banks F. A yeast mutant which accumulates precursor tRNAs. Cell. 1978 Jun;14(2):211–219. doi: 10.1016/0092-8674(78)90108-3. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Furukawa A. H., Pham H. D., Martin N. C. Defects in modification of cytoplasmic and mitochondrial transfer RNAs are caused by single nuclear mutations. Cell. 1982 Mar;28(3):543–550. doi: 10.1016/0092-8674(82)90209-4. [DOI] [PubMed] [Google Scholar]
- Hopper A. K., Schultz L. D., Shapiro R. A. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell. 1980 Mar;19(3):741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- Hutchison H. T., Hartwell L. H., McLaughlin C. S. Temperature-sensitive yeast mutant defective in ribonucleic acid production. J Bacteriol. 1969 Sep;99(3):807–814. doi: 10.1128/jb.99.3.807-814.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq B., Jourdan R., Jordan B. R. Structure and processing of precursor 5 S RNA in Drosophila melanogaster. J Mol Biol. 1977 Dec 15;117(3):785–795. doi: 10.1016/0022-2836(77)90069-9. [DOI] [PubMed] [Google Scholar]
- Knapp G., Beckmann J. S., Johnson P. F., Fuhrman S. A., Abelson J. Transcription and processing of intervening sequences in yeast tRNA genes. Cell. 1978 Jun;14(2):221–236. doi: 10.1016/0092-8674(78)90109-5. [DOI] [PubMed] [Google Scholar]
- Lerner M. R., Steitz J. A. Snurps and scyrps. Cell. 1981 Aug;25(2):298–300. doi: 10.1016/0092-8674(81)90047-7. [DOI] [PubMed] [Google Scholar]
- Lockard R. E., Alzner-Deweerd B., Heckman J. E., MacGee J., Tabor M. W., RajBhandary U. L. Sequence analysis of 5'[32P] labeled mRNA and tRNA using polyacrylamide gel electrophoresis. Nucleic Acids Res. 1978 Jan;5(1):37–56. doi: 10.1093/nar/5.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Tizard R., Skryabin K. G., Gilbert W. Promotor region for yeast 5S ribosomal RNA. Nature. 1977 Jun 16;267(5612):643–645. doi: 10.1038/267643a0. [DOI] [PubMed] [Google Scholar]
- Petes T. D. Molecular genetics of yeast. Annu Rev Biochem. 1980;49:845–876. doi: 10.1146/annurev.bi.49.070180.004213. [DOI] [PubMed] [Google Scholar]
- Phillips S. L., Tse C., Serventi I., Hynes N. Structure of polyadenylic acid in the ribonucleic acid of Saccharomyces cerevisiae. J Bacteriol. 1979 May;138(2):542–551. doi: 10.1128/jb.138.2.542-551.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle J. R. Methods for avoiding proteolytic artefacts in studies of enzymes and other proteins from yeasts. Methods Cell Biol. 1975;12:149–184. doi: 10.1016/s0091-679x(08)60956-5. [DOI] [PubMed] [Google Scholar]
- Rosbash M., Harris P. K., Woolford J. L., Jr, Teem J. L. The effect of temperature-sensitive RNA mutants on the transcription products from cloned ribosomal protein genes of yeast. Cell. 1981 Jun;24(3):679–686. doi: 10.1016/0092-8674(81)90094-5. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. Preparation of RNA and ribosomes from yeast. Methods Cell Biol. 1975;12:45–64. doi: 10.1016/s0091-679x(08)60951-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Selker E. U., Yanofsky C., Driftmier K., Metzenberg R. L., Alzner-DeWeerd B., RajBhandary U. L. Dispersed 5S RNA genes in N. crassa: structure, expression and evolution. Cell. 1981 Jun;24(3):819–828. doi: 10.1016/0092-8674(81)90107-0. [DOI] [PubMed] [Google Scholar]
- Silberklang M., Gillum A. M., RajBhandary U. L. Use of in vitro 32P labeling in the sequence analysis of nonradioactive tRNAs. Methods Enzymol. 1979;59:58–109. doi: 10.1016/0076-6879(79)59072-7. [DOI] [PubMed] [Google Scholar]
- Stahl D. A., Luehrsen K. R., Woese C. R., Pace N. R. An unusual 5S rRNA, from Sulfolobus acidocaldarius, and its implications for a general 5S rRNA structure. Nucleic Acids Res. 1981 Nov 25;9(22):6129–6137. doi: 10.1093/nar/9.22.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekamp P. A., Garcea R. L., Rutter W. J. Transcription and in vitro processing of yeast 5 S rRNA. J Biol Chem. 1980 Oct 10;255(19):9501–9506. [PubMed] [Google Scholar]
- Thonart P., Bechet J., Hilger F., Burny A. Thermosensitive mutations affecting ribonucleic acid polymerases in Saccharomyces cerevisiae. J Bacteriol. 1976 Jan;125(1):25–32. doi: 10.1128/jb.125.1.25-32.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela P., Bell G. I., Masiarz F. R., DeGennaro L. J., Rutter W. J. Nucleotide sequence of the yeast 5S ribosomal RNA gene and adjacent putative control regions. Nature. 1977 Jun 16;267(5612):641–643. doi: 10.1038/267641a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Seifart K. H. Heterogeneity in the 3'-terminal sequence of ribosomal 5S RNA synthesized by isolated HeLa cell nuclei in vitro. Biochemistry. 1978 Feb 7;17(3):457–461. doi: 10.1021/bi00596a013. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]