Abstract
CRM1 (chromosome maintenance region 1, Exportin 1) binds to nuclear export signals and is required for nucleocytoplasmic transport of a large variety of proteins and RNP complexes. Leptomycin B (LMB), the first specific inhibitor of CRM1 identified, binds covalently to cysteine 528 in the nuclear export signal binding region of CRM1 leading to the inhibition of protein nuclear export. Although the biochemical mechanisms of action of CRM1 inhibitors such as LMB are well studied, the subcellular effects of inhibition on CRM1 are unknown. We have found that LMB causes CRM1 to redistribute from the nucleus to the cytoplasm in A549 cells. A significant decrease in nuclear CRM1 coupled with an increase in cytoplasmic CRM1 was sustained for up to 4 hours, while there was no change in total CRM1 protein in fractionated cells. Cells expressing an LMB insensitive HA-tagged CRM1-C528S protein were unaffected by LMB treatment, whereas HA-tagged wildtype CRM1 redistributed from the nucleus to the cytoplasm with LMB treatment, similar to endogenous CRM1. GFP-tagged CRM1 protein microinjected into the cytoplasm of A549 cells distributed throughout the cell in untreated cells remained primarily cytoplasmic in LMB-treated cells. Upon nuclear microinjection, GFP-CRM1 translocated to and accumulated in the cytoplasm of LMB-treated cells. Thus, LMB binds to CRM1 and causes its redistribution to the cytoplasm by inhibiting its nuclear import. Decreasing the nuclear availability of CRM1 likely contributes to the accumulation of CRM1 cargo proteins in the nucleus, suggesting a new mechanism of action for LMB.
Keywords: Chromosome maintenance region 1, karyopherin, LMB, exportin, nucleocytoplasmic localization, protein nuclear export
Introduction
Nuclear pore complexes (NPC) distributed throughout the nuclear envelope allow the regulated exchange of macromolecules (e.g. proteins, mRNA, tRNA) [1]. Nucleocytoplasmic transport of proteins through the NPC is primarily mediated by the karyopherin family of transport receptors that recognize nuclear localization signals (NLS) for nuclear import or nuclear export signals (NES) for nuclear export within the cargo protein’s sequence [2]. These karyopherins include nuclear import factors (importins), nuclear export factors (exportins), and bidirectional transporters [3]. CRM1 functions as the canonical nuclear protein export factor in the cell that recognizes the classical leucine-rich NES sequence on proteins [4,5]. Among nuclear export factors, CRM1 has the broadest range of substrates which include transcription factors, cell cycle regulators, and RNP complexes [4,6]. In the nucleus, RanBP3 acts as co-factor that stimulates the recognition of cargo by CRM1 and stabilizes the trimeric export complex of CRM1-RanGTP-cargo [7,8]. Once the CRM1 export complex is formed it is translocated through the NPC into the cytoplasm where the complex is disassembled by RanGAP and the co-factor RanBP1 [9].
CRM1 has been observed to play a role in inflammatory diseases, viral infections, wound healing, and most notably in cancer [10]. The overexpression of CRM1 seen in many different types of cancers is correlated with increased metastasis, histological grade, tumor size, and decreased survival [11,12]. Although there is extensive research on cellular processes that involve CRM1 activity, there is a lack of research on the molecular mechanisms of the cellular regulation of CRM1 itself. Immunelectron microscopy shows CRM1 concentrated around the nuclear envelope on both the cytoplasmic and nuclear faces [13]. Altering its localization would likely be an important mechanism in the cellular regulation of the protein.
LMB is a 540 Da polyketide composed of an unsaturated, branched fatty acid chain with a terminal α,β-unsaturated lactone ring [14]. LMB binds covalently and irreversibly to cys528 of CRM1 [14]. It has been used as a tool to study the cellular functions of CRM1 and to identify CRM1 cargo proteins. Structural analysis revealed that following the covalent modification of cys528 by the lactone ring of LMB, the lactone ring was hydrolyzed to a hydroxy acid, resulting in extensive electrostatic interactions between LMB and residues in the NES-binding groove, increasing the area of coverage of the NES-binding site and making the adduct virtually irreversible [14]. Interestingly, our lab has determined that inhibition of CRM1 by LMB also causes nucleocytoplasmic redistribution of CRM1 itself from the nucleus to the cytoplasm.
Material and Methods
Plasmids
Human CRM1 cDNA was PCR amplified using pET-His-CRM1-HMK (gift from Jorgen Kjems, University of Aarhus) as a template and cloned into the SspI site of pET-His-GFP (plasmid 29663 Addgene, Cambridge, MA) to create pET-His-GFP-CRM1 and verified by sequencing. Plasmids pHA-CRM1-WT and pHA-CRM1-C528S were a gift from Ralph H. Kehlenbach (University of Gottingen, Germany)[15].
Cell culture
A549 cells (#CCL-185; American Type Culture Collection, Rockville, MD) were grown in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum and 1× anitmycotic/antibiotic solution (Invitrogen, Carlsbad, CA). LMB (LC Laboratories, Woburn, MA) was diluted in ethanol and used at a final concentration of 50 nM.
Whole cell lysates and subcellular fractionation
Cells were lysed with 2X Laemmli buffer (125 mM Tris HCl pH 6.8, 4% sodium dodecyl sulfate (SDS), 20% glycerol, 10% β-mercaptoethanol, 0.1% bromophenol blue) diluted to 1X with phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 8 mM Na2HPO4, and 2 mM KH2PO4), passed several times through a 28 G × ½ inch syringe, and boiled for 5 minutes. All treatments were done in triplicate plates and repeated at least three times.
For fractionation, A549 cells were grown to confluence and treated with or without LMB for the amount of time specified. Post drug treatment, the cells were washed with ice cold PBS and resuspended in 400 μl of buffer A (10 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA (ethylene glycolbis(β-aminoethyl ether)-N,N,N′,N′-tetraacetic acid), and 1.0 mM DTT) with and incubated on ice for 20 minutes before adding 25 μl of 10% NP-40. All buffers contained 1X protease inhibitor cocktail (Roche cOmplete EDTA-free; Sigma Aldrich, St. Louis, MO). The cells were vortexed vigorously and centrifuged to collect the cytoplasmic fraction. The nuclear pellet was resuspended in 100 μl of buffer C (20 mM HEPES, 0.4 mM NaCl, 1.0 mM EDTA, 1.0 mM EGTA, and 1.0 mM DTT), incubated on ice for 30 minutes, homogenized using a 28 G × ½ inch syringe, and centrifuged to collect the nuclear fraction. Nuclear and cytoplasmic lysates were diluted with 2× Laemmli buffer and boiled for 5 minutes. All treatments were done in triplicate plates and repeated at least three times.
Western Blotting
SDS-polyacrylamide gel electrophoresis was conducted using either equal volumes for total cell lysates or equal micrograms (10 μg) for nuclear and cytoplasmic lysates. Membranes were blocked with 5% non-fat dairy milk in PBS-T (phosphate buffered saline, 0.1% Tween-20) and immunoblotted with antibodies against CRM1 (1:2000; 611832, BD Biosciences, Franklin Lakes, New Jersey), super oxide dismutase (SOD; 1: 10,000; sc-11407, Santa Cruz Biotechnology, Santa Cruz, CA), and TATA-Binding Protein (TBP; 1: 10,000 ab818, Abcam, Cambridge, UK) concurrently. Blots were reacted with secondary anti-mouse (1:5,000) and anti-rabbit (1: 5,000) HRP conjugated secondary antibodies (554002, BD Biosciences, Franklin Lakes, New Jersey; 70745S, Cell Signaling, Danvers, MA) and SuperSignal® West Dura Extended Duration chemiluminescent substrate (Thermo Fisher Scientific, Waltham, MA) was used to detect the proteins on the western blot. Films were scanned for densitometry using Image J.
Immunofluorescence microscopy
A549 cells were grown on coverslips with or without 50 nM LMB then washed with PBS and fixed with 50/50 methanol/acetone for 10 minutes at −20°C. Cells were blocked with 3% bovine serum albumin in PBS for 1 hour at room temperature, and incubated overnight at 4°C in 3% BSA in PBS containing antibodies against CRM1 (1: 200). The coverslips were washed with PBS and incubated for 1 hour at room temperature with goat anti-mouse secondary antibody conjugated with AlexaFluor 488 (1: 500; Abcam, Cambridge, UK) diluted in 3% BSA in PBS. Coverslips were washed with PBS, and mounted with DABCO (1,4-diazabicyclo [2.2.2] octane) mounting media containing DAPI (4′,6-diamidino-2-phenylindole).
For immunofluorescence images of His-GFP-CRM1, 2 hours post-microinjection, cells were washed with PBS and fixed with 4% paraformaldehyde solution for 10 minutes at room temperature. Coverslips were then washed with PBS and mounted with DAPI/DABCO.
Real Time PCR
Total RNA was isolated from cells using the RNeasy Plus mini kit (Qiagen, Hilden, Germany) and reverse-transcribed into cDNA using the GoScript Reverse Transcriptase system (Promega, Madison, WI). One μg of RNA was used per reaction. cDNA was amplified using primers for CRM1 (5′-TCTGCAGCTATCCAAGCTAATG-3′ and 5′-GGCTCACCCAACCAGATATT-3′) and GAPDH (5′-TCGACAGTCAGCCGCATCTTCTTT-3′ and 5′-ACCAAATCCGTTGACTCCGACCTT-3′). Quantitative real-time PCR was performed using the SYBR Green qPCR kit (Bio-Rad, Hercules, CA) and quantified with the Bio-Rad CFX Connect Real-Time system (Bio-Rad, Hercules, CA). All samples were run in duplicate and the relative expression levels of CRM1, normalized to GAPDH, were calculated using the 2-∆∆CT method [16]. PCR products were confirmed by the presence of a single peak in the melting curve analysis (38). Each treatment group was done in triplicate wells for each experiment, and each experiment was conducted at least three times.
Protein Purification
His-GFP-CRM1 and His-GFP were expressed in E. coli BL21 by induction with 1 mM isopropyl-1-thio-β-D-galactopyranoside at 27°C for 4 hours and purified as described [17]. Purified eluates were dialyzed against 0.5X PBS and concentrated by ultrafiltration. Purification was followed by SDS-PAGE and western blots using antibodies against CRM1 and GFP.
Plasmids expressing a GST-NLS fusion protein was a gift from Mark Hannink at the University of Missouri-Columbia, purified as described [18], and reacted with equimolar fluorescein-5′-maleimide. The labeling reaction was quenched with 50 mM β-mercaptoethanol, and then dialyzed against PBS, and frozen at 1 mg/ml.
Microinjections
Nuclear and cytoplasmic microinjections of A549 cells were performed using an Eppendorf Femtojet microinjection system fitted onto an inverted Leica DMI 6000B microscope (Wetzlar, Germany) as described [17].
Imaging
Immunofluorescent images for His-GFP-CRM1 were acquired using a Leica DMI 6000 B inverted microscope with a 40× or 63× objective and a Hamamatsu EM-CCD camera (Hamamamtsu, Japan). Images were taken using the Openlab software suite (Perkin Elmer, Waltham, MA) and exported as TIFFs.
For confocal images of CRM1 staining, slides were imaged in the University of Rochester School of Medicine and Dentistry Confocal and Conventional Microscopy Core using an Olympus FV1000 Laser Scanning Confocal microscope (Olympus America, Center Valley, PA) with a 100X Plan-Apo (NA 1.4) oil objective with an optical zoom setting of 3 and sequential scanning option. Individual z planes were acquired using the 1024×1024 format setting with a 4 μs pixel dwell time and exported as TIFF’s.
Statistics
Data presented are means and standard deviations of one representative experiment from at least three independent experiments. The western blot results were analyzed using a two-tailed Student’s t-test. Data was considered biologically significant when p values <0.05.
Results
CRM1 redistributes in A549 cells with LMB treatment
CRM1 inhibitors such as LMB have been used to identify CRM1 cargo proteins, study mechanisms of nuclear export, and to regulate nuclear localization of proteins to investigate downstream molecular pathways [4]. To determine whether LMB affects the subcellular localization of CRM1 similar to the way it alters cargo protein localization, a time course analysis was conducted. A549 cells were treated with 50 nM LMB for 5, 15, 30, 60, and 240 minutes and lysed into nuclear and cytoplasmic fractions (Figure 1). As early as 15 minutes, LMB significantly decreased nuclear CRM1 compared to vehicle control (p<0.05; Figure 1A). The depletion of nuclear CRM1 was observed at each time point thereafter, indicating a sustained depletion of CRM1 from the nucleus with LMB treatment. Concurrently, as early as 30 minutes, LMB caused an increase in cytoplasmic CRM1 compared to vehicle control (p<0.05 Figure 1A). To determine if this observation was due directly to redistribution of CRM1 and not due to degradation and/or increased synthesis of CRM1, total cellular CRM1 was assayed by western blotting. A549 cells treated with LMB or vehicle for 4 hours showed no change in total CRM1 protein, supporting the hypothesis that LMB treatment redistributes CRM1 from the nucleus to the cytoplasm (Figure 1B). There was also no change in CRM1 mRNA levels with 4 hours of LMB treatment as determined by quantitative PCR (Figure 1C). The CRM1 cargo protein RanBP1 accumulated strongly in the nucleus by 4 hours of LMB treatment, demonstrating that LMB, at the concentration used, inhibited the nuclear protein export (data not shown).
Figure 1. LMB causes the subcellular redistribution of CRM1.
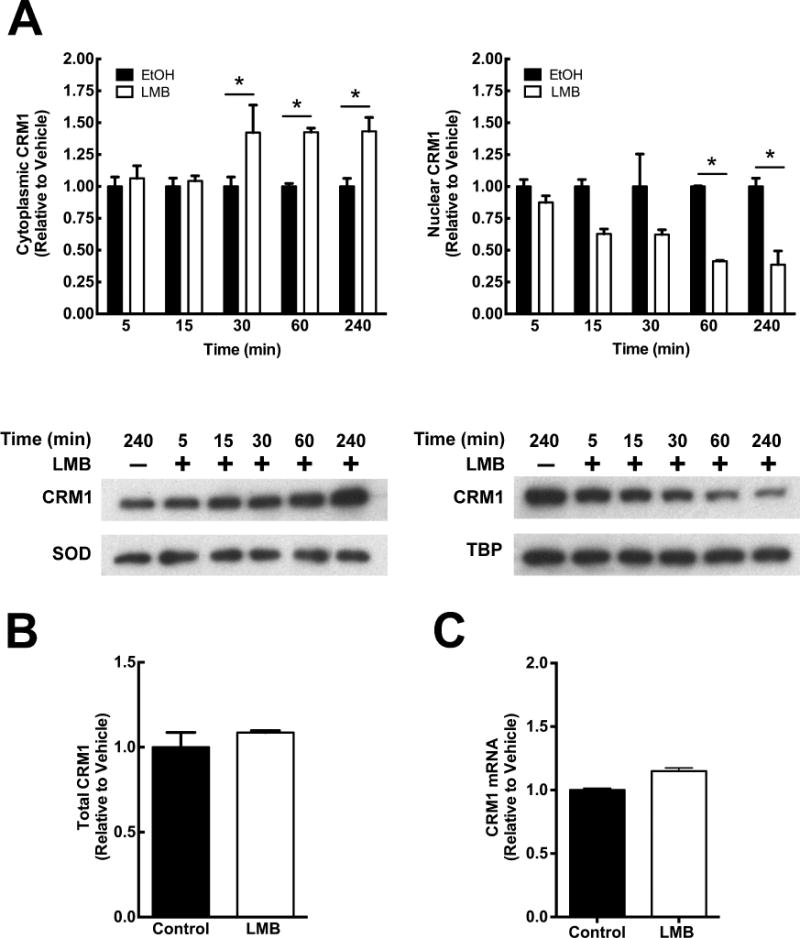
(A) Time course. A549 cells were treated with LMB (50 nM) or vehicle control and lysed into nuclear and cytoplasmic fractions. Cytoplasmic and nuclear CRM1 were normalized to SOD and TBP, respectively, and represented as fold change compared to vehicle control. (B) Total CRM1 does not change with LMB. Western blots of total cellular lysates from cells treated for 4 hours with 50 nM LMB or vehicle were probed for CRM1 and SOD. Densitometric analysis of CRM1 normalized to SOD is represented as fold change compared to vehicle control. (C) LMB does not change expression of CRM1. CRM1 mRNA was analyzed from cells treated with LMB (50 nM) or vehicle for 4 hours by qPCR and represented as fold change above control. Data is representative of three independent experiments. *, p<0.05. Error bars represent mean +/− SD.
To determine whether CRM1 is redistributed broadly and diffusely throughout the cytoplasm or in particular loci, confocal imaging was used. CRM1 immunofluorescence in A549 cells 4 hours post LMB treatment validated the western blot findings and showed an increase in diffuse cytoplasmic staining of CRM1 and an overall decrease in nuclear CRM1 compared to vehicle control (Figure 2). Interestingly, staining of CRM1 protein is concentrated at the nuclear envelope in cells treated with LMB compared to control cells (Figure 2). Taken together, this suggests that LMB causes the redistribution of CRM1 from the nucleus to the cytoplasm.
Figure 2. LMB affects the subcellular localization of CRM1.
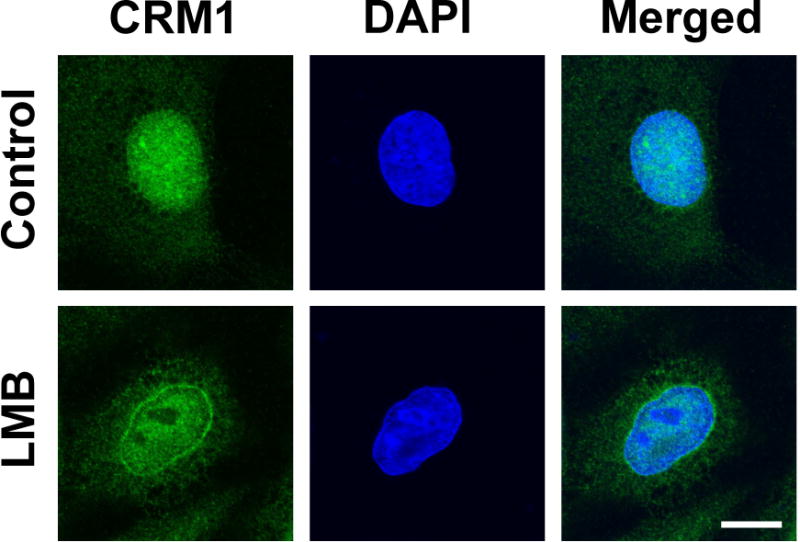
A549 cells treated with 50 nM LMB or vehicle for 4 hours, fixed, stained for CRM1 (green), and treated with DAPI to stain the nucleus (blue). Cells were imaged by confocal microscopy. Bar = 10 μm.
LMB does not affect localization of mutant CRM1-C528S
To determine whether direct binding of LMB to cys528 on CRM1 is necessary for the redistribution of CRM1 from the nucleus to the cytoplasm, A549 cells were transfected with plasmids expressing either HA-tagged wildtype (wt) CRM1, or HA-tagged CRM1-C528, a mutant that cannot bind LMB and is recused from the inhibitory effect of LMB on nuclear export [19]. Twenty-four hours post-transfection, cells were treated with LMB or vehicle for 1 hour and harvested for nucleocytoplasmic fractionation analysis. The nuclear and cytoplasmic localization of HA-CRM1-C528 mutant was unaffected by LMB treatment, while LMB caused redistribution of HA-wtCRM1 in the same manner as endogenous CRM1 (Figure 3). These findings suggest that LMB binding to cys528 is necessary for the redistribution of CRM1 from the nucleus to the cytoplasm.
Figure 3. LMB has no effect on the subcellular localization of CRM1-C528S.
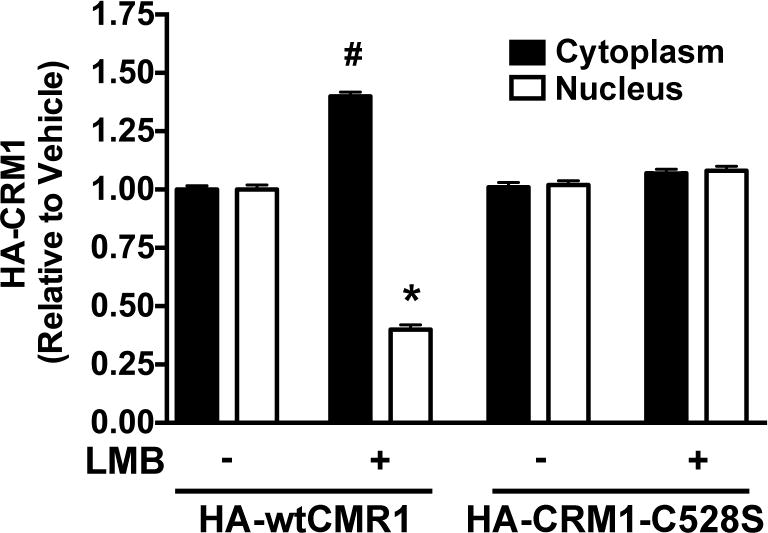
A549 cells were transiently transfected with either pHA-wtCRM1 or pHA-CRM1-C528S and 24 hours later treated with or without 50 nM LMB for 1 hour. Western blots of cytoplasmic and nuclear fractions of cell lysates were probed for HA, SOD, and TBP. Densitometric analysis of cytoplasmic and nuclear HA-tagged CRM1 (wt or mutant) was normalized to SOD and TBP respectively. Data represents fold change of HA-tagged wtCRM1 and HA-tagged CRM1-C528S compared to vehicle control within each fraction. * p<0.05 compared to cytoplasmic control. # p<0.05 compared to nuclear control. Error bars represent mean +/− SD. Data is representative of three independent experiments.
LMB decreases the translocation of CRM1 from the cytoplasm into the nucleus
The nucleocytoplasmic redistribution of CRM1 caused by LMB suggests that the mechanisms of nuclear translocation may be affected by the inhibition of CRM1 with LMB. To determine if LMB impairs the import of cytoplasmic CRM1 into the nucleus, cells were cytoplasmically microinjected with affinity purified His-GFP-CRM1. The cells were treated with either LMB or vehicle (ethanol) for 1 hour prior to injections and maintained with or without LMB for 1 hour post-microinjection prior to fixation and visualization. In the absence of LMB, His-GFP-CRM1 was evenly distributed throughout the cell in all of the cells observed (Figure 4A), demonstrating that the purified His-GFP-CRM1 has the ability to move import into the nucleus following injection into the cytoplasm. However, in the presence of LMB, the cytoplasmically injected His-GFP-CRM1 remained almost entirely cytoplasmic in all of the cells observed compared to control cells (Figure 4A). This suggests that the nuclear import of His-GFP-CRM1 is blocked by LMB. To ensure that the blocked subcellular localization of His-GFP-CRM1 caused by LMB is specific to CRM1, cells were microinjected with His-GFP in the presence of LMB or vehicle control. His-GFP distributed evenly throughout the cytoplasm and nucleoplasm in both vehicle and LMB treated cells (Figure 4C). Additionally, to ensure that canonical nuclear import is not affected by LMB in these experiments, a purified GST-NLS fusion protein conjugated to FITC was injected into the cytoplasm of cells treated with or without LMB for one hour. As shown in figure 4D, FITC-GST-NLS accumulated in the nucleus either in the presence or absence of LMB, suggesting that LMB has no observable effect on canonical importin α/β-mediated nuclear import. These results suggest that LMB impeded the translocation of CRM1 from the cytoplasm into the nucleus.
Figure 4. LMB inhibits nuclear import of CRM1.
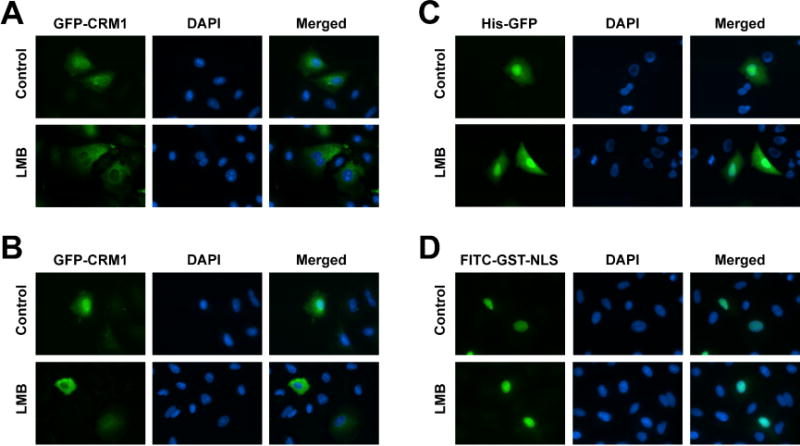
A549 cells were plated on coverslips and pre-treated with vehicle or 50 nM LMB, and either (A) cytoplasmically or (B) nuclear microinjected with purified His-GFP-CRM1 or cells were cytoplasmically microinjected with (C) purified His-GFP or (D) FITC-GST-NLS. Cells were then incubated with or without LMB and fixed 1 hour later for imaging by fluorescence microscopy. Representative images are shown. Bar = 25 μm.
To determine whether LMB could also affect the movement of nuclear CRM1 to the cytoplasm, affinity purified His-GFP-CRM1 was microinjected into the nuclei of cells that were pre-treated with LMB or vehicle (ethanol) for 1 hour and maintained in the same media throughout and after the injection process. One hour later, cells were fixed and the distribution of His-GFP-CRM1 was visualized. In the control group, His-GFP-CRM1 injected into the nucleus had distributed throughout the cytoplasm and the nucleoplasm by 1-hour post-microinjection in all of the cells observed (Figure 4B). In the presence of LMB, His-GFP-CRM1 had almost entirely redistributed from the nucleus into the cytoplasm in all of the microinjected cells (Figure 4B). This suggests that LMB does not inhibit the nuclear export of CRM1 but does prevent its nuclear import.
Discussion
CRM1 binds cargo proteins within the nucleoplasm and transports them to the cytoplasmic side of NPC. Inhibition of CRM1 nuclear export activity has been largely assumed to cause a nuclear accumulation of CRM1; by blocking the NES binding site of CRM1, CRM1 would be confined to the nucleus, unable to bind to cargo proteins. This would thereby increase the fraction of total cellular CRM1 in the nucleus. However, here we report that in cells treated with LMB, the opposite effect was observed: LMB binding caused a significant redistribution of CRM1 from the nucleus into the cytoplasm. This redistribution of CRM1 was rapid and required the direct binding of LMB to CRM1 since the HA-CRM1-C528S mutant showed no observable change in cytoplasmic and nuclear localization in the presence of LMB. Furthermore, microinjection of exogenously expressed CRM1-GFP into the cytoplasm or nucleus revealed that LMB inhibited nuclear import of CRM1 to cause the redistribution of nuclear CRM1 into the cytoplasm.
Most published studies have focused on the formation of the trimeric CRM1-RanGTP-NES export complex and its nuclear export through interactions with specific nucleoporins within the NPC [15,20,21]. By contrast, very little is known about the nuclear import of free CMR1 once the complex has dissociated on the cytoplasmic face of the NPC. During the terminal steps of nuclear export, the trimeric CRM1-RanGTP-NES export complex binds to the cytoplasmic facing nucleoporins Nup358 (RanBP2) and Nup214 [20,22,23]. Depletion studies have shown that Nup214 is necessary for the export of several known CRM1 cargo proteins [21]. Nup214, which strongly associates with Nup88, is a major component of the cytoplasmic filaments on the NPC and likely creates a docking site in the cytoplasm for CRM1 bound to its NES substrate [20]. Interestingly, it has been shown previously that CRM1 bound to LMB can still form a complex with Nup214 and RanGTP, suggesting that the NES binding site is not needed for this interaction [21]. The docking of CRM1 to these nucleoporins on the cytoplasmic side of the NPC allows for the hydrolysis of RanGTP to RanGDP by RanGAP, releasing the cargo protein into the cytoplasm and enabling CRM1 to return to the nucleus [24]. However, the role of these nucleoporins in CRM1 nuclear import is unclear.
NES sequences form a weak bond with CRM1, allowing for efficient release of cargo protein into the cytoplasm [6]. When a synthetic NES peptide with high affinity to CRM1 that prevented complex dissociation was fused to GFP and overexpressed, it caused the nuclear accumulation of CRM1 on the cytoplasmic face of the nuclear envelope [25]. This accumulation was LMB sensitive and dependent on CRM1 binding to Nup358, suggesting that the inefficient disassembly of the CRM1-NES complex may sequester CRM1 to the cytoplasmic face of the NPC and prevent recycling of CRM1 back into the nucleus [25]. The phenotype of LMB-bound CRM1 observed here is strikingly similar to that of the non-dissociable NES. In both cases, the NES binding pocket of CRM1 is occupied or blocked and suggests that an open conformation could be necessary for import of the free CRM1. While crystalographic analysis of the trimeric export complex bound to an FG repeat peptide identified three domains on CRM1 that form contacts with the FG repeats, none of them are coincident with the NES binding pocket [26,27]. However, binding of LMB or NES cargo to CRM1 does induce conformational changes that could alter interactions with the NPC, to prevent nuclear import of the LMB-CRM1 or high affinity NES-CRM1 proteins. Taken together, our results could suggest that unless the CRM1 NES binding pocket is open/unbound, nuclear re-import of free cytoplasmic CRM1 cannot occur.
Supplementary Material
Acknowledgments
We thank Gillian Schiralli-Lester and Xin Lin for insightful discussions and Rosemary Norman for technical assistance.
Funding: This work was supported by the National Institutes of Health [EB9903, ES07026, and center Grant ES01247].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Biol. 2010;11:490–501. doi: 10.1038/nrm2928. [DOI] [PubMed] [Google Scholar]
- 2.Stewart M. Structural basis for the nuclear protein import cycle. Biochem Soc Trans. 2006;34:701–704. doi: 10.1042/BST0340701. [DOI] [PubMed] [Google Scholar]
- 3.Xu D, Farmer A, Chook YM. Recognition of nuclear targeting signals by Karyopherin-beta proteins. Curr Opin Struct Biol. 2010;20:782–790. doi: 10.1016/j.sbi.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 2007;17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 5.Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 6.Kutay U, Guttinger S. Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol. 2005;15:121–124. doi: 10.1016/j.tcb.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 7.Petosa C, Schoehn G, Askjaer P, Bauer U, Moulin M, Steuerwald U, Soler-Lopez M, Baudin F, Mattaj IW, Muller CW. Architecture of CRM1/Exportin1 suggests how cooperativity is achieved during formation of a nuclear export complex. Mol Cell. 2004;16:761–775. doi: 10.1016/j.molcel.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 8.Lindsay ME, Holaska JM, Welch K, Paschal BM, Macara IG. Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J Cell Biol. 2001;153:1391–1402. doi: 10.1083/jcb.153.7.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kehlenbach RH, Dickmanns A, Kehlenbach A, Guan T, Gerace L. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J Cell Biol. 1999;145:645–657. doi: 10.1083/jcb.145.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun Q, Chen X, Zhou Q, Burstein E, Yang S, Jia D. Inhibiting cancer cell hallmark features through nuclear export inhibition. Signal Transduction And Targeted Therapy. 2016;1:10. doi: 10.1038/sigtrans.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kojima K, Kornblau SM, Ruvolo V, Dilip A, Duvvuri S, Davis RE, Zhang M, Wang Z, Coombes KR, Zhang N, Qiu YH, Burks JK, Kantarjian H, Shacham S, Kauffman M, Andreeff M. Prognostic impact and targeting of CRM1 in acute myeloid leukemia. Blood. 2013;121:4166–4174. doi: 10.1182/blood-2012-08-447581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van der Watt PJ, Zemanay W, Govender D, Hendricks DT, Parker MI, Leaner VD. Elevated expression of the nuclear export protein, Crm1 (exportin 1), associates with human oesophageal squamous cell carcinoma. Oncol Rep. 2014;32:730–738. doi: 10.3892/or.2014.3231. [DOI] [PubMed] [Google Scholar]
- 13.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. Embo J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Q, Carrasco YP, Hu Y, Guo X, Mirzaei H, Macmillan J, Chook YM. Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1. Proc Natl Acad Sci U S A. 2013;110:1303–1308. doi: 10.1073/pnas.1217203110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kehlenbach RH, Assheuer R, Kehlenbach A, Becker J, Gerace L. Stimulation of nuclear export and inhibition of nuclear import by a Ran mutant deficient in binding to Ran-binding protein 1. J Biol Chem. 2001;276:14524–14531. doi: 10.1074/jbc.M011087200. [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Miller AM, Dean DA. Cell-specific nuclear import of plasmid DNA in smooth muscle requires tissue-specific transcription factors and DNA sequences. Gene Ther. 2008;15:1107–1115. doi: 10.1038/gt.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sachdev S, Bagchi S, Zhang DD, Mings AC, Hannink M. Nuclear import of IkappaBalpha is accomplished by a ran-independent transport pathway. Mol Cell Biol. 2000;20:1571–1582. doi: 10.1128/mcb.20.5.1571-1582.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilliard M, Frohnert C, Spillner C, Marcone S, Nath A, Lampe T, Fitzgerald DJ, Kehlenbach RH. The anti-inflammatory prostaglandin 15-deoxy-delta(12,14)-PGJ2 inhibits CRM1-dependent nuclear protein export. J Biol Chem. 2010;285:22202–22210. doi: 10.1074/jbc.M110.131821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernad R, van der Velde H, Fornerod M, Pickersgill H. Nup358/RanBP2 Attaches to the Nuclear Pore Complex via Association with Nup88 and Nup214/CAN and Plays a Supporting Role in CRM1-Mediated Nuclear Protein Export. Molecular and Cellular Biology. 2004;24:2373–2384. doi: 10.1128/MCB.24.6.2373-2384.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hutten S, Kehlenbach RH. Nup214 is required for CRM1-dependent nuclear protein export in vivo. Mol Cell Biol. 2006;26:6772–6785. doi: 10.1128/MCB.00342-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walde S, Thakar K, Hutten S, Spillner C, Nath A, Rothbauer U, Wiemann S, Kehlenbach RH. The nucleoporin Nup358/RanBP2 promotes nuclear import in a cargo- and transport receptor-specific manner. Traffic. 2012;13:218–233. doi: 10.1111/j.1600-0854.2011.01302.x. [DOI] [PubMed] [Google Scholar]
- 23.Ritterhoff T, Das H, Hofhaus G, Schroder RR, Flotho A, Melchior F. The RanBP2/RanGAP1*SUMO1/Ubc9 SUMO E3 ligase is a disassembly machine for Crm1-dependent nuclear export complexes. Nat Commun. 2016;7:11482. doi: 10.1038/ncomms11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Floer M, Blobel G. Putative reaction intermediates in Crm1-mediated nuclear protein export. J Biol Chem. 1999;274:16279–16286. doi: 10.1074/jbc.274.23.16279. [DOI] [PubMed] [Google Scholar]
- 25.Engelsma D, Bernad R, Calafat J, Fornerod M. Supraphysiological nuclear export signals bind CRM1 independently of RanGTP and arrest at Nup358. EMBO J. 2004;23:3643–3652. doi: 10.1038/sj.emboj.7600370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monecke T, Dickmanns A, Ficner R. Allosteric control of the exportin CRM1 unraveled by crystal structure analysis. FEBS J. 2014;281:4179–4194. doi: 10.1111/febs.12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Port SA, Monecke T, Dickmanns A, Spillner C, Hofele R, Urlaub H, Ficner R, Kehlenbach RH. Structural and Functional Characterization of CRM1-Nup214 Interactions Reveals Multiple FG-Binding Sites Involved in Nuclear Export. Cell Rep. 2015;13:690–702. doi: 10.1016/j.celrep.2015.09.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


