Abstract
Objective
Lower body fat is associated with diminishing cardiometabolic risk. Physiological differences between gluteofemoral and abdominal subcutaneous adipocyte functions are known, but the molecular basis for depot differences in adipocyte function is poorly understood. Our objective is to identify depot-differences in microRNA expression in human abdominal and gluteofemoral subcutaneous adipose tissues and their implication in gene regulation.
Methods
Abdominal and gluteofemoral adipose tissue aspirates obtained from 18 participants (9M/9F, age 30±1.5 years, BMI 27.3.±1.23 kg/m2) analyzed for miRNA expression profiles by next-generation DNA sequencing. The raw reads were mapped to miRBase v17 and differentially expressed miRNAs were confirmed by qRT-PCR. The hsa-mimic-miR196a was transfected into cultured abdominal preadipocytes isolated from 5 obese women. Target gene expression was evaluated by RT-qPCR.
Results
Among the 640 miRNAs detected in adipose tissue; miR196a2, miR196a1, miR196b and miR204 showed a higher expression in gluteofemoral (fold change=2.7, 2.3, 1.7 and 2.3 respectively) independent of the sex. Bioinformatic analyses and human primary preadipocyte transfection with miR196 suggested that the differentially expressed miRNAs could modulate directly or indirectly Homeobox (HOX) genes expression.
Conclusion
MiR196 gene family could play an important role in the regulation of HOX gene expression in subcutaneous AT and then in fat distribution variation.
Key terms: Adipose tissue distribution, microRNA, Gluteal adipose tissue, HOX genes
Introduction
Overweight and obesity presents a worldwide clinical and public health burden and is correlated with an increased risk of cardiometabolic diseases, cancer, and mortality (1, 2). In obese individuals, triglycerides are stored in an expanded adipose tissue leading to inflammation, altered adipokine secretion and inflexible lipid metabolism. However, not all obese subjects have the same risk of developing these complications. Multiple correlation studies have shown that central obesity, characterized by triglyceride accumulation in visceral and abdominal subcutaneous depots, is closely associated with the development of cardiovascular disease (CVD) and type 2 diabetes (T2D) (3, 4). Inversely, lower body defined as fat accumulation in the gluteofemoral (GF) region and typically observed in premenopausal women, appears to be protective and is paradoxically associated with improved metabolic and cardiovascular profiles (3, 5–7). GF body fat distribution plays then a key protective role in metabolic disorders associated with overweight/obesity and, therefore, understanding the mechanisms regulating its expansion is an important challenge. Excellent reviews showing basic physiological differences in abdominal and GF adipocytes have been recently published, mainly focusing on gender specificity (8–12). However, we currently do not understand the underlying molecular basis for depot differences, and the molecular mechanisms that control the development and function of the different subcutaneous adipose tissue.
At the transcriptional level, previous transcriptome profiling in our laboratory found that genes in the HOX family were differentially expressed in abdominal and GF subcutaneous adipose tissue in both sexes with HOXA3, HOXA5, HOXB8 and HOXC8 downregulated in GF depot (13). Differences were present in both isolated adipocytes and stroma vascular fraction. Homeobox genes make up a network of transcription factors controlling embryonal development as well as crucial functions of adult eukaryotic cells. Their role in metabolic diseases was discussed in the excellent review of Procino A et al (14). The HOX code of different depots suggest the involvement of the HOX network in transcriptional regulation of human adipogenesis and adipose tissue function. Their specific role in fat development and their regulation are still under investigation.
MiRNAs represent the most abundant regulators of gene expression in the human genome. Through their influence on target mRNAs, miRNAs are involved in numerous physiologic and pathological processes and in particular in many functional aspects of adipocyte differentiation (15) and potentially contribute to the pathogenesis of obesity (16), (17). Some of the more interesting genomic locations of miRNA genes include those in the Hox clusters where they can affect the translational repression of the HOX genes but could also mediate degradation of the target mRNA (18).
As part of a broader exploration to identify the molecular mechanisms underlying the fundamental differences in GF and abdominal subcutaneous adipose tissue, we hypothesized that a differential miRNA profile exists between abdominal and GF adipose tissue depots. We employed a new DNA sequencing technology to identify miRNAs present in abdominal and GF subcutaneous human adipose tissue. This approach allows the sequencing and direct quantification of all the miRNAs independent of any a priori information on what miRNAs may exist in the sample and should complement the previous work of Rantalainen M et al who analyzed global miRNA expression in gluteal and abdominal adipose tissue in human subjects using an eQTL method (19). To test this hypothesis, we performed next generation sequencing of small RNAs (size<45bp) and miRNAs from GF and abdominal subcutaneous adipose tissue. In order to determine an intrinsic regulator of fat development not under the influence of sex hormones, we decided to focus on miRNA differentially expressed between depots independent of the gender. Because in silico analysis indicated that Hox cluster-embedded miRNAs preferentially target Hox mRNAs (20), we made the assumption that some of these miRNAs target specifically depot specific HOX genes expression. We explored in silico their potential interactions with HOX gene expression which we previously identified as differentially expressed in abdominal and gluteal adipose tissue (13). Finally, we tested these potential interactions in vitro by transfecting primary human preadipocytes with a pre-miRNA that mimic endogenous precursor miR196a.
Methods
Twenty one men and 14 women were recruited according to the criteria described in (13). To participate, volunteers needed to be between the age of 18 and 40 with percent body fat of 20–50%. At screening, volunteers were excluded for significant medical illness, glucocorticoid use, smoking, or substance/alcohol abuse, and women needed to have normal menstrual cycles. Oral contraceptive pills were not allowed. Men and female matched for age, ethnicity and BMI. According the tissue availability, we selected nine men and nine women for the conduct of our sub-study. The characteristics of the sub-study population are presented in Table 1.
Table 1.
Clinical parameters of subjects (mean ± standard deviation)
| Clinical parameters | Female subjects (n=9) | Male subjects (n=9) | p value |
|---|---|---|---|
| Age (years) | 29 ± 2.3 | 31 ± 1.9 | NS* |
| Adiposity markers | |||
| BMI (kg/m2) | 28.2 ± 2.26 | 26.4 ± 1.06 | NS |
| Body weight (kg) | 76.5 ± 5.46 | 85.8 ± 4.46 | NS |
| Waist to hip ratio | 0.84 ± 0.02 | 0.89 ± 0.03 | NS |
| Total Fat mass (kg) | 29.2 ± 4.42 | 20.7 ± 2.89 | NS |
| Total Lean mass (kg) | 46.8 ± 2.37 | 63.9 ± 2.10 | 0.0002 |
| Fat mass % | 36.8 ± 3.51 | 23.3 ± 2.25 | 0.01 |
| Plasma Glucose homeostasis | |||
| Glucose (mg/dL) | 90.8 ± 3.81 | 90.3 ± 2.55 | NS |
| Insulin (μU/mL) | 9.46 ± 2.75 | 7.83 ± 2.39 | NS |
| Plasma lipid homeostasis | |||
| Total cholesterol (mg/dL) | 172.3 ± 7.58 | 170.3 ± 10.0 | NS |
| Triglycerides (mg/dL) | 83.2 ± 15.9 | 76.3 ± 6.64 | NS |
| HDL cholesterol (mg/dL) | 56.4 ± 5.59 | 47.8 ± 3.04 | NS |
| LDL cholesterol (mg/dL) | 99.3 ± 5.64 | 107.2 ± 7.75 | NS |
NS: not significant
After an overnight fast blood samples were collected, accessioned, aliquoted, frozen and then shipped to the Pennington Biomedical Research Center Clinical Chemistry Laboratory for analysis. Body composition was measured by dual-energy X-ray absorptiometry (DEXA) using a single brand of the instrument (Hologic QDR 4500A, Hologic, Waltham, MA).
Adipose tissue biopsies were collected and total RNA was isolated as described in (13).
MicroRNA sequencing
We used the SOLiD™ Total RNA-Seq Kit for preparing small RNA libraries from abdominal and gluteal adipose tissue and the SOLiD V4 next generation sequencing instrument to identify the microRNAs. Briefly, the small RNA (20 to 45nt region) was enriched from total RNA using FlashPage gel and were concentrated by precipitating overnight with sodium acetate and ethanol. The small RNA was used as input for library preparation as described in the protocol “SOLiD Small RNA Expression Kit protocol” (part#4399434). The detection method was based on sequencing by ligation. The 40 libraries were multiplexed, loaded onto two full slides and 35bp reads were generated from the SOLiD V4 instrument.
MicroRNA sequence data analysis
The raw SOLiD reads contained part of the adapter sequences; as the SOLiD reads (35nt) were longer than the most of the expected length of mature miRNAs. The “cutadapt” program was first applied to trim the adapters from the reads (21). Trimmed reads which were shorter than 15 nt were discarded. Subsequently, reads that matched RNA contaminants such as tRNA, rRNA, and DNA repeats were filtered out. The remained trimmed reads were mapped to the human genome (hg19) (UCSC genome browser) and to the known mature miRNAs (miRBase, release 17) using the Bowtie aligner (22) with parameters (-n 0 –l; 16 –e; 100 –m 5; –best). The mature miRNA coordinates in hg19 were based on miRBase release 17 (23). The raw expression values (read counts) were obtained by summing the number of uniquely mapped reads to mature miRNA or to mature miRNA coordinates. Normalized expression values were obtained using RPKM approach (24). Custom Perl scripts were developed to summarize the raw read counts and normalize the expression values.
Statistical analysis of the miRNA sequencing data
Genes with zero read counts across all samples were filtered from the data base. Box-plots and M-A plots were applied to evaluate the data quality for each replicate. Two samples which showed extremely low counts were excluded. After cleaning the data, the read counts were logarithmically (base 2) transformed and then used for downstream analysis. Accounting for the high proportion of zero and low counts of each sample, the upper-quartile (3rd) normalization method was applied to remove any impacts of technical factors and sequencing depth among samples (25). Specifically, a negative binomial general linear model was used to detect the statistical differential genes between two depots. The raw P values generated from multiple comparisons were adjusted by FDR using the linear step-up method of Benjamini Hochberg. 2640 hypothesis have been tested. A miRNA was considered to be significantly different between two depots if the FDR-adjusted P-value was less than 0.05 and Fold Change of expression was greater than 1.3 in either direction. All statistical analyzes were performed in SAS (V9.2).
Confirmatory miRNA gene expression analyses
MiRNAs were reverse transcribed by TaqMan™ MicroRNA reverse transcription kit (Applied Biosystems). MiRNA expression was assessed by real time-PCR using a ViiA7 sequence detection system (Life Technologies) and Taqman technology suitable for relative miRNA expression quantification using the following parameters: one cycle of 95°C for 10 minutes, followed by 40 cycles at 95°C for 10 seconds and 60°C for 1 minute. For all assays, the RNU48 gene was used as internal control. All expression data were normalized by dividing the target gene by the internal control.
Human adipocytes and stromal vascular fraction isolation
Adipocytes and stromal-vascular fractions were isolated by collagenase digestion with 1 mg/ml collagenase type 1 (Worthington Biochemical, Lakewood Township, NJ) in Hanks’ balanced salt solution shaken for 2h at 37°C and used for RNA extraction.
Preadipocytes culture and miRNA precursor transfection
Human preadipocytes were isolated from obese women’s abdominal adipose tissue biopsy and cultured as described in (13). One week after seeding, 5nM of hsa-miR-196a-5p mimic (mimics the all miR-196 family including miR196a1, miR196a2 and miR196b) or negative control (Ambion, ThermoFisher Scientific, USA) were transfected into abdominal preadipocytes using lipofectamine RNAiMax (Life Technologies, USA) for 72h. At the end of the transfection, the number of viable cells was measured using the RealTime Glo MT Cell Viability Assay (Promega Corp., Madison, WI). Experiments were conducted in cells isolated from 5 different donors (age=30±6.9 years; BMI=34.6±2.22 kg/m2; WHR=0.91±0.04) and experiments were performed in duplicate.
Bioinformatics analysis: Pathways and Structural Equation Models (SEM) mapping
The pathway analysis was performed with IPA (Ingenuity Pathway Analysis), using a new microRNA target filter functionality which enables prioritization of experimentally validated and predicted mRNA targets. Data input were the HOX genes and the microRNAs differentially expressed between both depots. The network pathways and biofunctions were tested by Fisher exact test. All significant differential genes (mRNA and mirRNA) were highlighted with green (Abdominal/GF>1) and red (GF/Abdominal>1) in Supplemental data Figure 1 and Figure 2.
A modified SEM algorithm was applied to construct the co-expression network of miRNA and mRNA. It included two stages: P-stage and S-stage. In the P-stage, Partial Least Square regression (PLS) was used to select a cluster of new genes with high correlations to target mRNA. Then in S-stage, Structural Equation Models (SEM) were performed to build relationships between this cluster of miRNA genes to target mRNA based on multivariate multi-layer linear regression coefficients. This two-stage SEM analysis was performed in SAS (V9.2) (30).
Results
Identification of miRNAs differentially expressed in the gluteofemoral depot
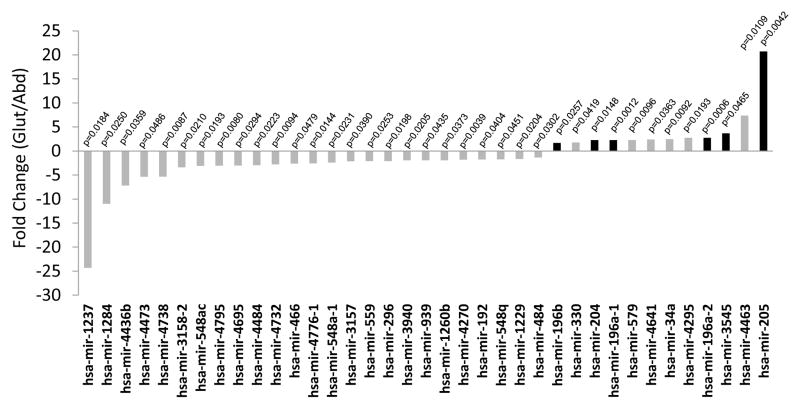
Abdominal and GF adipose tissue aspirates obtained from premenopausal women and age- and BMI-matched men were used for identifying miRNA by ABI solid sequencing technology (LifeTech SOLID v4). Their demographics and clinical characteristics are shown in Table 1. Among the 1733 known microRNA, 640 were present in the subcutaneous adipose tissue, defined as present only in an abdominal depot or only in a GF depot or both depots. Thirty seven miRNA (2.1%) were differentially expressed between abdominal and GF adipose tissue, 25 being significantly down-regulated in GF adipose tissue whereas 12 were significantly up-regulated. Figure 1 represents the fold change (FC) of each 37 miRNA differentially expressed (p-value < 0.05), with a maximal FC of 24.3 and a vast majority of miRNA (n=30) showing an FC between 1.37 and 3.40.
Figure 1. MicroRNAs differentially expressed between Abdominal and Gluteal subcutaneous adipose tissue in both sexes (FDR p<0.05).
Fold change is the ratio between abdominal and gluteal expression. Black bars indicate microRNA selected for qRT-PCR validation. Fold changes were calculated by dividing the average of the means of miRNA copies number in abdominal depot by the average of the means of miRNA copies number in gluteofemoral depot.
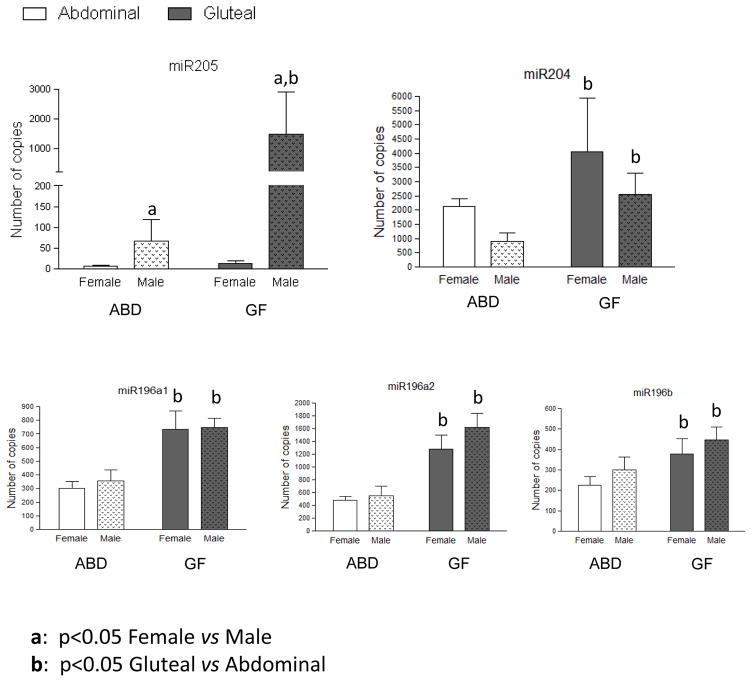
Because the expansion of lower-body subcutaneous depots is associated with protection from impairments in glucose-insulin homeostasis and hypertriglyceridemia, we chose to focus on this depot and selected the miRNA up regulated in the GF depot, making the hypothesis that they have a role in GF tissue development. Among these genes only miR196, miR203, miR204, miR205 and miR3545 showed an expression detectable by qPCR. MiR196 family genes also showed a very interesting genomic location, in the middle of HOX gene cluster as represented in Figure 4. miR205, miR204 and three isoforms of miR196 (miR196a1, miR196a2 and miR196b) showed an increase in GF adipose tissue (FC 21, 2.3, 2.3, 2.7 and 1.7 respectively, Figure 1) without distinction between males and females. No significant difference appeared for these four genes between female and male no matter the depot (Figure 2). Inversely, miR205 was highly expressed in male compared to female adipose tissue in both depots, FC 12.4 for abdominal depot and 154 for GF depot (Figure 2).
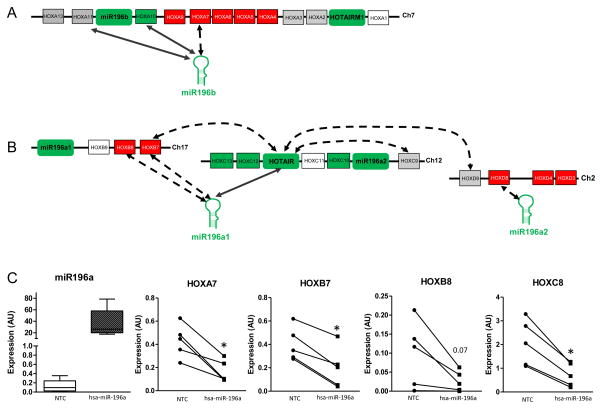
Figure 4. Relationships between selected microRNAs and HOX gene expression.
The relationships of the differentially expressed HOX genes, miR196a, miR196band miR204 were determined using SEM (see methods for details) to build a co-expression network HOX genes are represented by numbered boxes (not to scale). Genes upregulated in gluteal depot as compared to abdominal are in green; genes down regulated in gluteal depot are in red. Arrows indicate a positive correlation, dashed arrows indicate negative correlation. A: miR196a2 and miR196b showed interactions with HOX A cluster on chromosome 7. B: miR196a1/a2 and miR204 showed interactions with HOX B cluster on chromosome 17, HOX C cluster on chromosome 12 and HOX D cluster on the chromosome 2. C: Expression of HOXA7, HOXB7, HOXB8 and HOXC8 in miR196a transfected abdominal preadipocytes. Bar graph represents mean and SEM of relative expression of miR-196a at 72hrs post transfection. HOX genes relative expression was plotted for each participant, circle represent cells transfected by hsa-Control, square represent cells transfected by hsa-miR196a. Gene expression was normalized to Cyclophilin A. * paired t-test p<0.01. NTC: Negative control
Figure 2. miRSequencing quantification for three selected microRNAs differentially expressed between Abdominal (white bars) and Gluteal (grey bars) subcutaneous adipose tissue split by sex.
Data are presented as mean ± SEM of females (n=9) and males (n=9). GF: Gluteofemoral depot – Abd: Abdominal depot
miR203a and miR3545 (or miR203b), located at the same region on chromosome 14, were specifically increased in GF tissue compared to abdominal depot only in males (5 and 6 fold higher, respectively, Figure 2) with no difference in female adipose tissues. These two miRNAs showed a significant increase in male GF depot compared to female GF depot (12.7 and 13.6 fold increased respectively, Figure 2), with no difference between both sexes in the abdominal depot.
qRT-PCR confirmation
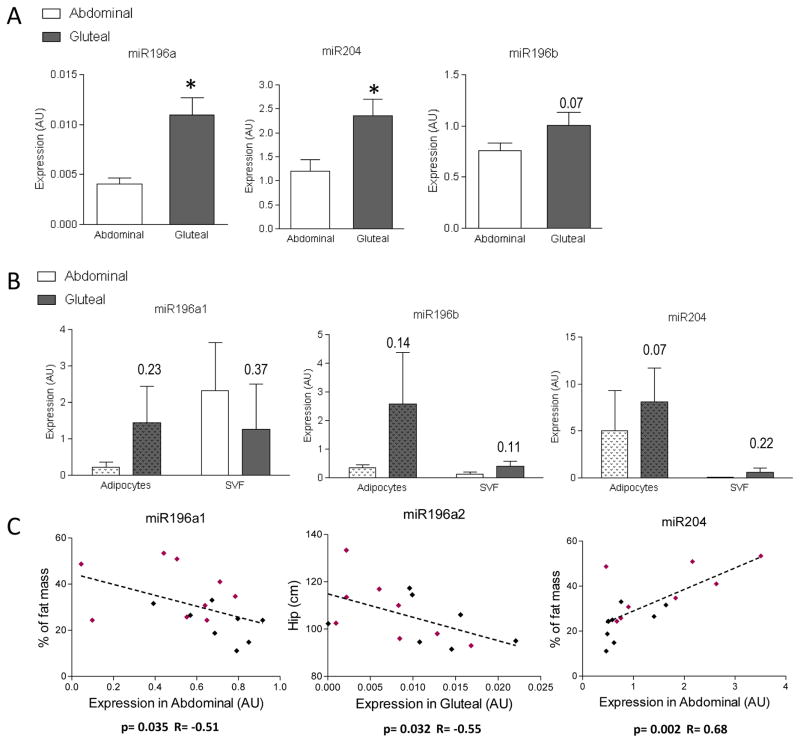
To confirm these sequence based differences, we performed qRT-PCR on the same isolated RNA using specific primers for the corresponding mature form of miRNAmiR196a (primers recognizing miR196a-1 and miR196a-2) and miR204 were significantly higher expressed in the GF depot compared to the abdominal depot (Figure 3A). We observed the same tendency for miR196b (p=0.07) but no difference detected between depots for miR205, miR203 or miR3545.
Figure 3. Expression of selected microRNAs in sc adipose tissue (qRT-PCR) in male and female combined.
A: Confirmation of the miRNA sequencing data by qPCR for miR196a, miR196b and miR204. B: Expression of selected microRNAs trend to be depot specific only in adipocytes. Data are presented as mean ± SEM of n=16 paired abdominal and gluteal tissue samples, n between 2 and 4 for isolated adipocytes and stromal-vascular fractions. * P<0.05 compared with abdominal depot, paired t-test. C: Spearman correlations between miRNA expression in specific depot and clinical data for n=9 women (pink) and n=9 men (black).
To gain insight into the causes of the differences in miRNA expression, we analyzed the association of selected mature miRNA of interest with clinical/anthropometric variables. This analysis was performed with gene expression data in both depots. As shown in Figure 3C, the correlations between % body fat, waist circumference and the expression of miR196a1 and miR204 were found only in in the abdominal depot. Inversely, the correlation between hip circumference and miR196a2 expression were observed only in gluteofemoral depot. The correlation between miR204 expression and % body fat was found only in female (p=0.047 R=0.67) but not in male only (Figure 3C).
A second independent group of healthy participants (n=2F/2M; age=27±1.84 years; BMI=28.4±2.53 kg/m2; WHR=0.87±0.04) was recruited to address the question whether the differential expression of the miRNA previously observed is due to alterations in adipocytes and/or the stromal vascular fraction (SVF). As shown in Figure 3B, miR196a, miR196b and miR204 are expressed in both collagenase isolated adipocyte and SVF. Depot differences were apparent in both fractions (except for miR196a in SVF). The tendency was nevertheless more apparent in adipocyte fractions (Figure 3B). For these selected miRNA, expression differences observed in adipose tissues were not maintained in culture after differentiation of human preadipocytes (data not shown), i.e. were not cell-autonomous.
Identification of miRNA-HOX gene interactions by pathway analysis and Structural Equations Model
We recently identified several HOX genes downregulated in the GF depot of both sexes, and we hypothesized that these genes might play a role in the inter-individual variation in fat distribution. To identify gene networks connected to specific miRNAs that was higher in gluteal adipose tissue, we performed Ingenuity Pathway Analysis (IPA). These analyses regrouped the miRNA differentially expressed between depots into 2 different modules (Supplemental data). “Cellular development, tissue development, cellular growth and proliferation” were identified as the primary functions for genes implicated in the first network of genes. Interestingly, pathway analysis highlighted a direct interaction between miR204 and HOXA10, a mRNA that we previously identified strongly upregulated in GF male and female adipose tissue (13).
The second module established a relationship between miR196a/b and a different set of HOX genes (Supplemental data Figure 2), characterized by direct interactions between miR196a/b and HOX8 genes (HOXB8, HOXC8, HOXD8) three mRNAs that are downregulated in GF depots. IPA also identified direct interaction between miR196a/b and HOXA7 and indirect interactions with HOXB7.
We then performed correlation analysis using Structural Equations Model (SEM) described in details in the Method section and in (26). The HOX genes expression data, previously obtained by transcriptome microarray experiment with the same group of subjects (13), was loaded into the model along with each miRNA. To study only the relationships between genes showing a differential expression between both depots, the correlations were established between the ratio of expression in abdominal and in GF. We identified gene expression correlations between miR196b and HOXA7, HOXA10 and HOXA11, 3 HOX genes located adjacent to each other on chromosome 7 (Table 2 and Figure 4A). miR196a2 gene expression was negatively correlated with HOXD8. miR196a1 expression was correlated with HOXB7, HOXB8 and HOTAIR (HOX antisense intergenic RNA), a well-studied lncRNA and previously identified as a regulator of adipogenesis by our labs (27). Finally, miR204 expression was positively correlated with HOXC10 expression (Table 2 and Figure 4B).
Table 2.
Correlations between miRNA expression and HOX genes expression
| HOXA7 | HOXA9 | HOXA10 | HOXA11 | HOXB7 | HOXB8 | HOXC9 | HOXC10 | HOXD8 | |
|---|---|---|---|---|---|---|---|---|---|
| miR196a1 | β=−0.7404 P<0.01 |
β=−0.6149 P<0.01 |
|||||||
| miR196a2 | β=−0.5221 P<0.05 |
||||||||
| miR196b | β=0.5013 P<0.05 |
β=−0.5074 P<0.05 |
β=0.5389 P<0.05 |
||||||
| miR204 | β=0.5757 P<0.05 |
miR196 decreases HOX genes expression in human preadipocytes
To determine whether miR196a can also influence the expression of mRNA in human adipose tissue we transfected human abdominal preadipocytes with the corresponding miRNA mimics as described in Material and Methods. The list of the highest-scoring putative gene targets as predicted by TargetScan (http://www.targetscan.org/) for the miR-196 family is shown in Table S1. This analysis predicted that some HOX genes are putative targets of miR-196. Among them HOXA5, HOXA9, HOXB8, and HOXC8, all of which have been shown to be upregulated in the abdominal depot in both sexes, whereas HOXB7 was shown upregulated in abdominal depot only in males (13). Cells transfection resulted in an increase of miR196 expression between 100 to 900 fold and a reduction of 3, 3.5, 3.4 and 5 times of HOXA7, HOXB7, HOXC8 and HOXB8 expression, respectively. Expression of other HOX genes (such as HOXA5 and HOXA9) or HOTAIR were not influenced by miR196 modulation (data not shown). Interestingly we did not observe a modification of gene expression when we modulated miR196a expression in the corresponding GF preadipocytes where expression was already high (data not shown).
Discussion
Since their discovery, miRNAs have attracted considerable interest, and the volume of research is growing exponentially. Their importance in adipose tissue formation came first from the observation that inhibition of Drosha and Dicer, critical miRNA-processing enzymes, in human mesenchymal stem cells inhibited the differentiation of these cells into adipocytes (28). Since then, several global profiling studies have been performed to identify new miRNA candidates implicated in adipose tissue development and biology (29–31).
In the present study, we hypothesized that miRNA might be differentially expressed in abdominal and GF adipose tissue and that they could play a role in adipose tissue distribution. Rantalainen et al analyzed global miRNA expression in gluteal and abdominal adipose tissue in human subjects using an eQTL method (19). Among all miRNA identified with a higher expression in gluteal adipose tissue three are in common with our results: miR-196a, miR-196b, and miR-204. Our actual method using next generation sequencing presents an advantage as it detects miRNAs from a library of more than 1400 miRNAs instead of detecting defined miRNAs. We observed for the first time a specific upregulation of both isoforms miR196a1 and miR196a2, together with miR205 in the GF depot. To identify miRNA targets, we combined our miRNA data with previous mRNA data (generated using the same subjects) and performed in silico analysis (IPA and SEM). IPA revealed the central position of beta-estradiol, hormones known to influence fat distribution, in the first gene network (including miR203, miR204 and miR205), suggesting a regulation of adipose tissue miRNAs expressions by sex hormones. The effect of these molecules on miR205 and other miRNAs merit further investigation. Unfortunately in this study the low number of subjects don’t allow us to perform sex specific analysis and in order to limite the influence of sex hormones, we decided to focus on miRNAs up regulated in GF in both sexes.
Previous studies have shown interaction of some miRNA and transcription factors influencing adipose tissue biology (32, 33). Our current bioinformatics analyses showed potential interaction between miR196a/b and other molecules like KRT5, ANXA1 or GFI1, but no one known to be differentially regulated in human adipose tissue. We then focused on the potential interaction between miRNA and HOX genes that we had recently identified as potential contributor to functional and morphological differences between lower body (GF) and upper body (abdominal) subcutaneous adipose tissues (13). Interestingly we established an interaction between miR196 gene family and HOX gene expression in accordance with their genomic location (34). In silico analysis predicts that miRNAs identified in the same chromosomal region as HOX clusters preferentially target local (in cis) HOX mRNAs. Our correlation data, pathway analysis and then our in vitro experiments suggested these interactions in adipocytes. Further experiments such as knockdown of miR196 genes and other experiments will be necessary to identify their actual targets in adipocytes.
We observed that a higher expression of miR196a in gluteal depot lead to a decrease of fat development in this depot (characterized by a lower hip circumference, fig 3C). At the opposite a higher expression of miR-196a in abdominal depot seems beneficial because associated with a decrease of fat mass. We also showed that miR196a overexpression is associated with a decrease of HOXC8 expression (fig 4C), gene known to be involved in the development of brown adipose tissue, targeted tissue to combat obesity and fat development. Our data suggested that a repression of miR196a expression in gluteal depot would be a good therapeutic target to stimulate the expansion of the gluteal depot. This hypothesis merits further exploration.
The present study did not find evidence for cell-autonomous expression of all miRNAs differentially expressed in adipose tissue, suggesting that the microenvironment or the hormonal milieu in vivo may be important. Alternately, epigenetic marks may be erased in the transition to an artificial cell culture system. For example, hypoxia or inflammatory cytokines, both known to be increased in obese subjects, could influence microRNA expression in adipose tissues (37, 38).
Adipose tissue is a heterogeneous tissue where microRNAs could be found in all cells type including adipocytes, resident inflammatory cells, preadipocytes, vascular smooth muscle cells, endothelial cells, and pericytes. Moreover, the proportion of each type of cells varies among obesity or diabetes status. In the future, it will be crucial to determining which cells express each miRNA to establish their role in adipose tissue biology.
In conclusion, miR196 gene family is up regulated in GF depot compared to abdominal depot, independent of the gender. Bioinformatics analysis identified a complex system of interaction of miR196 and HOX genes, also found in vitro in human preadipocytes isolated from abdominal adipose tissue where miR196a overexpression decreased HOX genes expression, further in vitro experiments are necessary to conclude on a direct or indirect effect.
Supplementary Material
What is already known about this subject?
Lower body defined as fat accumulation in the gluteofemoral (GF) region appears to be protective and is associated with improved metabolic and cardiovascular profiles
Physiological differences between gluteofemoral (GF) and abdominal subcutaneous adipocyte functions.
MicroRNAs (miRNA) have emerged as key transcriptional and post-transcriptional modulators of gene expression.
What does this study add?
Depot-differences in microRNA expression in human abdominal and gluteofemoral subcutaneous adipose tissues.
miR196 genes family is up regulated in gluteaofemoral depot, independently of the sex.
miR196 regulates some HOX genes expression.
Acknowledgments
Funding sources: This work was supported by National Institutes of Health Grants R24DK087669, P30DK46200 and R01DK107009; the Society for Women’s Health Research (SWHR) Interdisciplinary Studies on Sex Differences Network on Metabolism; the Evans Center for Interdisciplinary Biomedical Research Affinity Research Collaborative on Sex Differences in Adipose Tissue at Boston University School of Medicine; and the Translational Research Institute for Metabolism and Diabetes.
The authors would like to thank the SWHR network and the members of the SWHR group, Jennifer Lovejoy, Nori Geary, Joel Elmquist, Philipp Scherer, Randy Seeley, and Richard Simerly, for their support in the origins of this research. The authors would like to thank the personnel of the genomic core at Sanford-Burnham Prebys Medical Discovery Institute, Orlando Florida, who performed the microRNA sequencing and more particularly Subramaniam Shyamala Govindarajan.
List of abbreviations in alphabetical order
- FDR
False discovery rate
- FFA
Free fatty acids
- GF
Gluteofemoral
- HOX
Homeobox
- IPA
Ingenuity pathway analysis
- lncRNA
Long non-coding RNA
- miRNA
MicroRNA
- nt
nucleotide
- SEM
Structural equations model
- SVF
Stroma vascular fraction
- T2D
Type 2 Diabetes
- WHR
Waist to hip ratio
Footnotes
Disclosure: The authors have no conflicts of interest and nothing to disclose
Clinical Trial Registration Number: NCT00704197, NCT01745471 and NCT0174545471
Study approved by Florida Hospital Institutional Review Board
List of references
- 1.Kopelman PG. Obesity as a medical problem. Nature. 2000;404(6778):635–43. doi: 10.1038/35007508. Epub 2000/04/15. [DOI] [PubMed] [Google Scholar]
- 2.Uchegbu EC, Kopelman PG. Cardiovascular risks in obesity. J Endocrinol Invest. 2002;25(10):915–8. doi: 10.1007/BF03344056. Epub 2003/01/02. [DOI] [PubMed] [Google Scholar]
- 3.Yusuf S, Hawken S, Ounpuu S, Bautista L, Franzosi MG, Commerford P, et al. Obesity and the risk of myocardial infarction in 27,000 participants from 52 countries: a case-control study. Lancet. 2005;366(9497):1640–9. doi: 10.1016/S0140-6736(05)67663-5. Epub 2005/11/08. [DOI] [PubMed] [Google Scholar]
- 4.Kissebah AH, Krakower GR. Regional adiposity and morbidity. Physiol Rev. 1994;74(4):761–811. doi: 10.1152/physrev.1994.74.4.761. Epub 1994/10/01. [DOI] [PubMed] [Google Scholar]
- 5.Canoy D, Boekholdt SM, Wareham N, Luben R, Welch A, Bingham S, et al. Body fat distribution and risk of coronary heart disease in men and women in the European Prospective Investigation Into Cancer and Nutrition in Norfolk cohort: a population-based prospective study. Circulation. 2007;116(25):2933–43. doi: 10.1161/CIRCULATIONAHA.106.673756. Epub 2007/12/12. [DOI] [PubMed] [Google Scholar]
- 6.Bigaard J, Frederiksen K, Tjonneland A, Thomsen BL, Overvad K, Heitmann BL, et al. Body fat and fat-free mass and all-cause mortality. Obes Res. 2004;12(7):1042–9. doi: 10.1038/oby.2004.131. Epub 2004/08/05. [DOI] [PubMed] [Google Scholar]
- 7.Manolopoulos KN, Karpe F, Frayn KN. Gluteofemoral body fat as a determinant of metabolic health. International journal of obesity. 2010;34(6):949–59. doi: 10.1038/ijo.2009.286. Epub 2010/01/13. [DOI] [PubMed] [Google Scholar]
- 8.Fried SK, Lee MJ, Karastergiou K. Shaping fat distribution: New insights into the molecular determinants of depot- and sex-dependent adipose biology. Obesity (Silver Spring) 2015;23(7):1345–52. doi: 10.1002/oby.21133. Epub 2015/06/10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi H, Seeley RJ, Clegg DJ. Sexual differences in the control of energy homeostasis. Frontiers in neuroendocrinology. 2009;30(3):396–404. doi: 10.1016/j.yfrne.2009.03.004. Epub 2009/04/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karastergiou K, Smith SR, Greenberg AS, Fried SK. Sex differences in human adipose tissues - the biology of pear shape. Biol Sex Differ. 2012;3(1):13. doi: 10.1186/2042-6410-3-13. doi:0.1186/2042-6410-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pramyothin P, Karastergiou K. What Can We Learn from Interventions That Change Fat Distribution? Curr Obes Rep. 2016;5(2):271–81. doi: 10.1007/s13679-016-0215-x. [DOI] [PubMed] [Google Scholar]
- 12.White UA, Tchoukalova YD. Sex dimorphism and depot differences in adipose tissue function. Biochim Biophys Acta. 2014;3:377–92. doi: 10.1016/j.bbadis.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karastergiou K, Fried SK, Xie H, Lee MJ, Divoux A, Rosencrantz MA, et al. Distinct developmental signatures of human abdominal and gluteal subcutaneous adipose tissue depots. J Clin Endocrinol Metab. 2013;98(1):362–71. doi: 10.1210/jc.2012-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Procino A, Cillo C. The HOX genes network in metabolic diseases. Cell Biol Int. 2013;37(11):1145–8. doi: 10.1002/cbin.10145. [DOI] [PubMed] [Google Scholar]
- 15.Fujimoto Y, Nakagawa Y, Shingyouchi A, Tokushige N, Nakanishi N, Satoh A, et al. Dicer has a crucial role in the early stage of adipocyte differentiation, but not in lipid synthesis, in 3T3-L1 cells. Biochemical and biophysical research communications. 2012;420(4):931–6. doi: 10.1016/j.bbrc.2012.03.110. Epub 2012/04/06. [DOI] [PubMed] [Google Scholar]
- 16.Xie H, Sun L, Lodish HF. Targeting microRNAs in obesity. Expert Opin Ther Targets. 2009;13(10):1227–38. doi: 10.1517/14728220903190707. Epub 2009/08/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Capobianco V, Nardelli C, Ferrigno M, Iaffaldano L, Pilone V, Forestieri P, et al. miRNA and Protein Expression Profiles of Visceral Adipose Tissue Reveal miR-141/YWHAG and miR-520e/RAB11A as Two Potential miRNA/Protein Target Pairs Associated with Severe Obesity. J Proteome Res. 2012 doi: 10.1021/pr300152z. Epub 2012/04/28. [DOI] [PubMed] [Google Scholar]
- 18.Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304(5670):594–6. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- 19.Rantalainen M, Herrera BM, Nicholson G, Bowden R, Wills QF, Min JL, et al. MicroRNA expression in abdominal and gluteal adipose tissue is associated with mRNA expression levels and partly genetically driven. PloS one. 2011;6(11):e27338. doi: 10.1371/journal.pone.0027338. Epub 2011/11/22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yekta S, Tabin CJ, Bartel DP. MicroRNAs in the Hox network: an apparent link to posterior prevalence. Nature reviews Genetics. 2008;9(10):789–96. doi: 10.1038/nrg2400. Epub 2008/09/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011 [Google Scholar]
- 22.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. Epub 2009/03/06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39(Database issue):D152–7. doi: 10.1093/nar/gkq1027. Epub 2010/11/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5(7):621–8. doi: 10.1038/nmeth.1226. Epub 2008/06/03. [DOI] [PubMed] [Google Scholar]
- 25.Vague J. The degree of masculine differentiation of obesities: a factor determining predisposition to diabetes, atherosclerosis, gout, and uric calculous disease. Am J Clin Nutr. 1956;4(1):20–34. doi: 10.1093/ajcn/4.1.20. *OBESITY/complications. [DOI] [PubMed] [Google Scholar]
- 26.Xie JBPM. Covariance Structure Models for Gene Expression Microarray Data. Structural Equation Modeling. 2003;10(4):566–82. [Google Scholar]
- 27.Fontbonne A, Thibult N, Eschwege E, Ducimetiere P. Body fat distribution and coronary heart disease mortality in subjects with impaired glucose tolerance or diabetes mellitus: the Paris Prospective Study, 15-year follow-up. Diabetologia. 1992;35(5):464–8. doi: 10.1007/BF02342445. [DOI] [PubMed] [Google Scholar]
- 28.Van Harmelen V, Rohrig K, Hauner H. Comparison of proliferation and differentiation capacity of human adipocyte precursor cells from the omental and subcutaneous adipose tissue depot of obese subjects. Metabolism. 2004;53(5):632–7. doi: 10.1016/j.metabol.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 29.Karpe F, Pinnick KE. Biology of upper-body and lower-body adipose tissue--link to whole-body phenotypes. Nat Rev Endocrinol. 2015;11(2):90–100. doi: 10.1038/nrendo.2014.185. [DOI] [PubMed] [Google Scholar]
- 30.Tchkonia T, Thomou T, Zhu Y, Karagiannides I, Pothoulakis C, Jensen MD, et al. Mechanisms and metabolic implications of regional differences among fat depots. Cell Metab. 2013;17(5):644–56. doi: 10.1016/j.cmet.2013.03.008. Epub Apr 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinnick KE, Nicholson G, Manolopoulos KN, McQuaid SE, Valet P, Frayn KN, et al. Distinct developmental profile of lower-body adipose tissue defines resistance against obesity-associated metabolic complications. Diabetes. 2014;63(11):3785–97. doi: 10.2337/db14-0385. [DOI] [PubMed] [Google Scholar]
- 32.Karbiener M, Fischer C, Nowitsch S, Opriessnig P, Papak C, Ailhaud G, et al. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochemical and biophysical research communications. 2009;390(2):247–51. doi: 10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- 33.Qin L, Chen Y, Niu Y, Chen W, Wang Q, Xiao S, et al. A deep investigation into the adipogenesis mechanism: profile of microRNAs regulating adipogenesis by modulating the canonical Wnt/beta-catenin signaling pathway. BMC Genomics. 2010;11(320):1471–2164. doi: 10.1186/1471-2164-11-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Woltering JM, Durston AJ. MiR-10 represses HoxB1a and HoxB3a in zebrafish. PloS one. 2008;3(1):e1396. doi: 10.1371/journal.pone.0001396. Epub 2008/01/03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tanzer A, Amemiya CT, Kim CB, Stadler PF. Evolution of microRNAs located within Hox gene clusters. J Exp Zool B Mol Dev Evol. 2005;304(1):75–85. doi: 10.1002/jez.b.21021. Epub 2005/01/12. [DOI] [PubMed] [Google Scholar]
- 36.Popovic R, Riesbeck LE, Velu CS, Chaubey A, Zhang J, Achille NJ, et al. Regulation of mir-196b by MLL and its overexpression by MLL fusions contributes to immortalization. Blood. 2009;113(14):3314–22. doi: 10.1182/blood-2008-04-154310. Epub 2009/02/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mace TA, Collins AL, Wojcik SE, Croce CM, Lesinski GB, Bloomston M. Hypoxia induces the overexpression of microRNA-21 in pancreatic cancer cells. The Journal of surgical research. 2013;184(2):855–60. doi: 10.1016/j.jss.2013.04.061. Epub 2013/06/04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu L, Chen L, Shi CM, Xu GF, Xu LL, Zhu LL, et al. MiR-335, an Adipogenesis-Related MicroRNA, is Involved in Adipose Tissue Inflammation. Cell biochemistry and biophysics. 2013 doi: 10.1007/s12013-013-9708-3. Epub 2013/06/27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.