Abstract
Major histocompatibility complex genes in mammals include highly polymorphic class I and class II genes that are critical for donor-recipient matching for transplantation. Dogs have served as an effective, directly translatable model for stem/progenitor cell transplantation. Previous analyses of major histocompatibility complex class I genes in dogs point to a single highly polymorphic gene, dog leukocyte antigen (DLA)-88, as an important factor in the success or failure of hematopoietic stem cell transplants. Fifty-nine DLA-88 alleles have been identified and reported so far. Here, we extend this list by presenting 13 novel DLA-88 alleles found in domestic dogs.
Keywords: allele, canine, class I, dog, dog leukocyte antigen, DLA-88, major histocompatibility complex
Introduction
Advancement of allogeneic hematopoietic stem cell transplantation (HSCT) is due largely to preclinical progress in the canine model (1–3). The typing of highly polymorphic canine major histocompatibility complex (MHC) genes, termed dog leukocyte antigen (DLA), was fundamental in the successful development of the canine model for transplantation (4,5). As in humans, the dog MHC class I and class II genes exhibit an exceptionally high degree of polymorphism (6–9). Histocompatibility matching of donor-recipient pairs for HSCT has traditionally focused on the most polymorphic DLA genes. The canine class I and class II genes have been localized to chromosome 12, with the exception of DLA-79, a non-classical class I gene located on chromosome 18 (6–9). Four canine class II genes have been identified; DLA-DQA1, DQB1 and DRB1 are polymorphic while DRA is not. Historically, DRB1 has been most commonly utilized for DLA-matching for HSCT among sibling donor-recipient pairs, due to this gene’s high level of polymorphism, close association with other DLA genes on chromosome 12, and ease of allele typing. Of the three classical class I genes, DLA-88, DLA-12 and DLA-64, DLA-88 is the most pertinent for HSCT, as it is highly polymorphic with 59 alleles reported so far (8,10–14). DLA-88 typing was added to the DLA matching protocol, when methods for routine typing of the alleles were worked out (12). DLA-79 is the second most polymorphic class I gene, but a specific association in graft rejection could not be demonstrated, as about 80 percent of the transplanted dogs were homozygous for the most common allele (15). Alleles of DLA-12 and DLA-64 are less well studied as the polymorphism was low in the initial set of dogs typed (8). In this report, we extend the list of DLA-88 alleles by presenting 13 novel alleles.
Materials and methods
Source of DNA
A total of 427 dogs were tested in this study. Random-bred litters of beagles and mini-mongrel crossbreeds including Basenji, Golden Retriever, hounds and harriers (15), were raised at the Fred Hutchinson Cancer Research Center (FHCRC), Seattle, WA. Archived samples of genomic DNA were obtained from 200 of these animals. An additional 200 blood samples included in this study came from breeders and their pups in our colony and dogs from Marshall Bioresources, USA, Ridglan Farms Inc, USA and from Phoenix Laboratories, Edmonds, WA. In addition, DNA from 27 dogs that were typed by reference strand-mediated conformation analysis and chosen to likely contain new DLA-88 alleles were included in this screen. All experiments were approved by Institutional Animal Care and Use Committee of the FHCRC. Standard care for dogs was provided as described previously (16, 17).
DLA-88 typing
Dog genomic DNA was isolated from whole blood using either Puregene DNA purification kit (Gentra Systems, Minneapolis, MN) or QIAamp DNA blood mini kit (Qiagen, Valencia, CA). Amplification and direct sequencing reactions and conditions were as described (12) except for the use of polymerase chain reaction (PCR) reverse primer, DLA88R57e3. The locus specific primers used for PCR were DLA88F-8e1: 5′-GCGGCGACGGCCAGTGTCCCCGGAG-3′ and DLA88R57e3 5′-GACCCTGAGTCCATATTCCCTTCC-3′. The primers used for sequencing were DLA88F-8e1, DLA88R57e3, DLA88I1F: 5′-CCCGGGCATCTCCCCCTG-3′, DLA88I2F: 5′-GAACCCGCGGGAACTCCCGGGAGG-3′, and DLA88I2R: 5′-GTGACGCCCGGACCCGGACCCTC-3′. The amplification primers were chosen based on their ability to amplify the polymorphic HVR regions of interest as a single fragment that includes DLA-88 exon 1 - exon 3, and for their inability to amplify the corresponding region from other class I genes. The forward primer maps to 8 bases upstream of the ATG start codon on exon 1 while the reverse primer maps to 57 bases downstream of exon 3. It was not possible to design a forward primer that can distinguish DLA-88 from DLA-64 in the region of interest. This handicap was remedied by designing a reverse primer, DLA88R57e3, that would anneal to DLA-88 but not DLA-64. Similarly, the same reverse primer sequence is also present in DLA-12 gene, so the specificity for DLA-88 amplification is rendered by the forward primer, DLA88F-8e1. In all cases, when possibilities of new alleles were suggested by the direct sequencing data, new amplicons were generated by high fidelity PCR as modified from methods previously described (12,13) using the same PCR primers indicated above and sequenced after cloning individual alleles. Briefly, amplification reactions (typically 30 μl) contained 100 ng of genomic DNA, 200 nM each of the primers and 200 μM each of dNTPs. Velocity DNA polymerase (0.5 μl) was used with the 5× GC buffer provided by the manufacturer (Bioline, Tauton, MA). All reactions were run in a Biorad C1000 Cycler (Bio-Rad, Hercules, CA) using the following amplification conditions: 96 °C for 2 min; 30 cycles of 96 °C for 30 s, 65 °C for 15 s, and 72 °C for 60 s; and 68 °C for 5 min. PCR products analyzed by electrophoresis on a 1 % agarose gel produced a sharp band of about 1.1 kb. The DNA fragments were cloned in pCR2.1-TOPO vector using the TOPO TA Cloning Kit (Invitrogen). Plasmid insert DNA from multiples clones were sequenced to identify the alleles. Plasmid sequence-derived primers (TopoF1: 5′-CAGCTATGACCATGATTACGCCAAGC -3′ and TopoR1 5′-GCCAGGGTTTTCCCAGTCACGACG -3′) and DLA-88-specific primers indicated above were used for sequencing the plasmid insert fragments. In all cases, whether by direct sequencing or plasmid-cloned sequencing, the polymorphic regions of interest were sequenced in both directions, reactions were run in the FHCRC sequencing core facility and a custom-built software (developed in-house) was used to resolve the combinations of alleles.
New allele discovery and designation
For all sequences, the intron-exon boundaries were identified by aligning with previously described DLA-88 genomic sequence (8,18); the derived amino acid sequence of the new alleles were aligned with those of closely related known alleles and the reference allele DLA-88*001:01 using ClustalW2 (19) [http://www.ebi.ac.uk/clustalw2/]. All new alleles were verified by cloning and sequencing multiple independent PCR products, using DNA derived from at least 2 dogs. Only alleles encountered in multiple animals and repeatedly confirmed by multiple independent PCR and sequencing reactions were considered for new allele designations. New alleles found in our breeder population were also confirmed in pups after breeding. The allele identification process met the published conventions of the International Society of Animal Genetics (ISAG) Comparative MHC Nomenclature Committee and the new allele designations were assigned in consultation with the curator of the canine DLA database, Dr. L. J. Kennedy (20, 21). A rooted phylogenetic tree was derived using a web based software program at phylogeny.fr in the ‘One Click’ mode (22). Amino acid sequences of all known DLA-88 alleles along with that of HLA-A*01:01 as an out-group were used. The ‘One Click’ mode by default uses a set of reliable, widely used programs: MUSCLE for multiple alignment, Gblocks for automatic alignment curation, PhyML for tree building and TreeDyn for tree drawing.
Results
Identification of new DLA-88 alleles
All sequences obtained were found to be specific DLA-88 sequences, and not derived from other class I gene fragments as revealed by sequence analysis of gene-specific variations within polymorphic exons, as well as intron 1 and intron 2. Fifty of the 59 previously published alleles were identified among the dogs tested. In addition, thirteen novel alleles were also discovered. The nucleotide sequences for these alleles are recorded in the GenBank database, including complete intron 2 sequences. Table I lists the new alleles along with GenBank accession numbers, and source (laboratory and breed) of the dogs carrying those alleles. Seven of the new alleles were found within the relatively small group of dogs of diverse breeds pre-screened by reference strand-mediated conformation analysis, and these alleles accounted for 26 of the 54 allele identifications in this group.
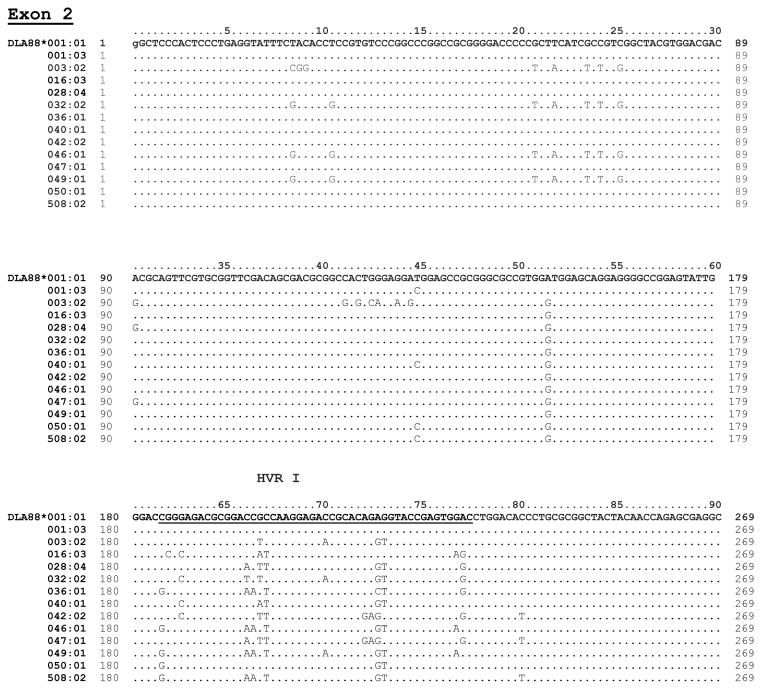
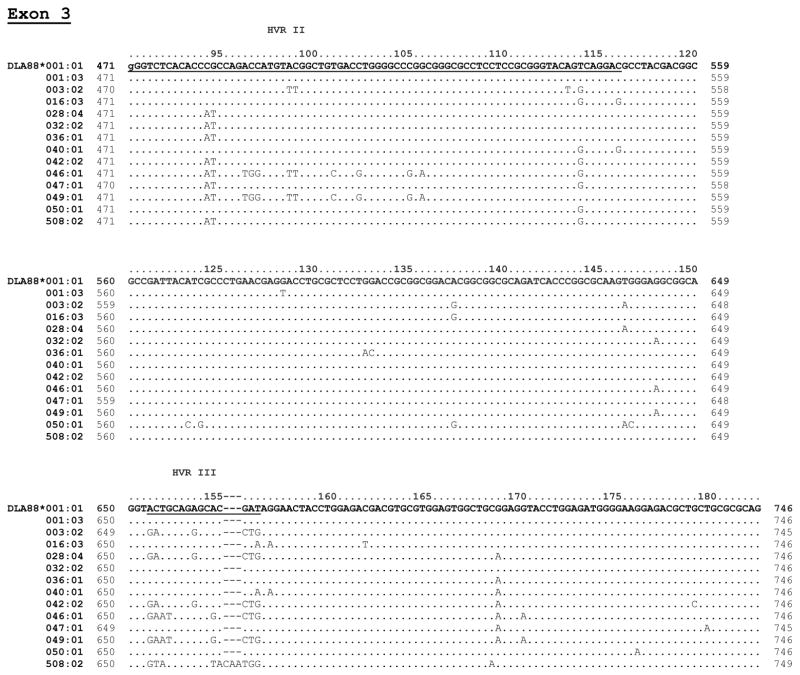
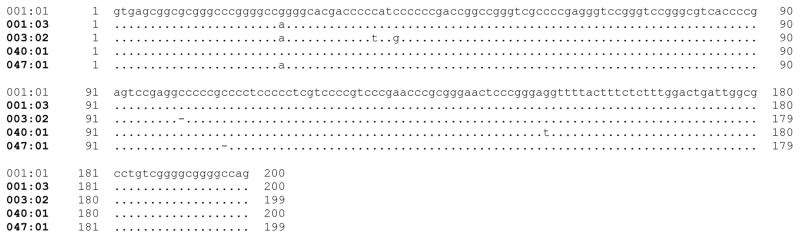
Figure 1 shows the nucleotide sequences of the 13 alleles aligned with reference allele, DLA-88*001:01. The hyper-variable regions (HVR I–III) and variations between the alleles in the polymorphic exons 2 and exon 3 are indicated. Nine of the 13 alleles had the same intron 2 sequence as the most common 200 bp intron 2 found in the reference allele 001:01 and most other alleles. However, 4 alleles varied within intron 2 from the canonical 001:01 allele, each having 1–3 base substitutions (Figure 2). Alleles 003:02 and 047:01, each also carried a single base deletion.
Figure 1. Nucleotide sequence alignment of the new DLA-88 alleles.
Nucleotide sequence of exons 2 and 3 for the 13 new alleles are shown aligned to the reference allele DLA-88*001:01. The nucleotide position numbers indicated start from the first base of exon 2. The exon 2 alignment starts with a lowercase ‘g’ from exon 1 to provide full codon context. The codon numbers are indicated above the reference allele sequence. Nucleotide position 270 from exon 2 is included as the first position (in lower case ‘g”) along with the exon 3 sequence. The exon 3 sequence numbering is also indexed from the first base of exon 2 and continues through the intron 2 sequence. The new alleles are presented in sequential order with the differences from the reference allele indicated. The HVR I, II and III are in bold and underlined.
Figure 2. Alignment of atypical DLA-88 introns.
New alleles with atypical intron 2 are shown aligned with the reference allele (DLA-88*001:01) intron 2 sequence. The numbering pertains to the full intron 2 sequence. All other alleles reported in this paper have an intron 2 sequence that is identical to the 200 bp intron 2 of the reference allele DLA-88*001:01.
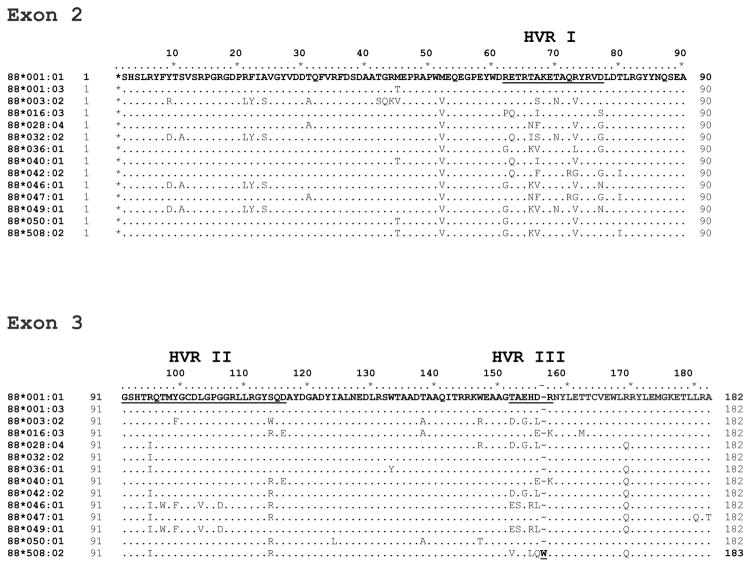
Figure 3 shows the alignment of the amino acid sequence of exons 2 and 3 of the new alleles with the reference allele, 001:01. Most amino acid substitutions are seen in positions known to be polymorphic and the majority of these changes are seen within the three HVR. Allele 001:03 is a minor variant of the reference allele 001:01, with a single amino acid change, Thr instead of Met at position 45 in exon 2, which is outside the HVR. A new variant amino acid position is reported here that was not previously seen: Ser153 in alleles 046:01 and 049:01. The alignment of amino acid sequences of the new DLA-88 alleles along with all the other published alleles in a clustered arrangement as well as in their numerical order, as generated by ClustalW (18) are included in the supplementary information (supplementary fig. 1). A phylogenetic tree derived from this alignment analysis is also provided (supplementary fig. 2). A summary of the known DLA-88 alleles’ nomenclature and their primary references is also included (supplementary table 1). The nucleotide and amino acid sequences of all these alleles are depicted in FASTA format in supplementary figure 3.
Figure 3. Amino acid sequence alignment of the new DLA-88 alleles.
The amino acid sequence of 13 new alleles, in bold, are shown aligned with the reference allele DLA-88*001:01. The amino acid sequence starts with an asterisk ‘*’ at position 1 to indicate the partial glycine codon from exon 2. The three hypervariable regions (HVR) are indicated. The amino sequences of the new alleles are presented in sequential order with the amino acid substitutions from the reference allele indicated. The HVR I, II and III are in bold and underlined.
Conclusion
In this report, we extend the list of known DLA-88 alleles by presenting 13 additional alleles discovered during retrospective analyses of archival canine donor/recipient genomic DNA samples and new canine DNA samples from many breeds earmarked as likely to contain new alleles. Since the animals tested in this study were often related, and no attempt was made to randomize the study with respect to breed or family, no estimates of allele frequency should be inferred from this data. The true extent of diversity of the DLA genes in canines, particularly of the class I genes, is still not known. Ongoing studies include a diversity survey to genotype and sequence DLA-88 in numerous dog breeds. A detailed characterization of haplotype diversity of DLA genes among breeds will:
Allow a donor registry for clinical transplantation in dogs.
Help focus MHC-linked disease association studies.
Provide a more comprehensive understanding of the MHC diversity in dogs.
Promote further study of the CTL response in dogs.
Enhance the preclinical canine transplantation model, which predicts for engraftment and graft-versus-host-disease after HSCT in humans.
Supplementary Material
Supplementary Figure 1. The amino acid sequence alignment of DLA-88 alleles.
The amino sequences of all published DLA-88 alleles are presented in (A) the clustered alignment as generated by the ClustalW (1) or (B) in the numerical order. The amino acid sequence of the same reference allele is also shown aligned with other class I reference alleles for DLA-12, DLA-64, and DLA-79. Human HLA*A01:01:01 allele is included as an outgroup. Variations between the alleles in the polymorphic exons 2 and exon 3 including within the hyper-variable regions (HVR I–III; underlined) are indicated.
Supplementary Figure 2. Phylogenetic tree derived from the amino acid sequences of exons 2 and 3 of DLA-88 alleles using ClustalW program (1).
A rooted phylogenetic tree derived from the amino acid sequences of exons 2 and 3 of all published DLA-88 alleles and their relationship to the DLA-88 reference allele, with HLA-A*01:01:01 as an outgroup. The sequences indicated with suffixes RB, RH, RJ and RL are from the GenBank submissions of Ross et al., but do not have a finalized ISAG designation yet. A web based program at Phylogeny.fr was used in the ‘One Click’ mode (see Materials and Methods for details). Use of advanced options with 100 replicates in a bootstrap analysis gave the same result. Use of exon 2 and 3 nucleotide sequences instead of amino acid sequences from the same data set resulted in a similar tree. The scale bar units refer to number of substitutions per site.
Supplementary Figure 3. The Nucleotide and amino acid sequences of DLA-88 alleles.
The nucleotide sequences of all published DLA-88 alleles are in FASTA format. The GenBank accession numbers, exonic (upper case) and the intronic sequences (lower case & underlined, when available) (A) and amino acid sequences derived from the polymorphic exons 2 and 3 (B).
Supplementary Table 1. All known DLA-88 Alleles. List of DLA-88 alleles to date indicating alternative names, GenBank accession numbers and primary publication references**.
Table 1.
New DLA-88 Alleles and Number of Dogs Typed by Breed
| DLA-88 Allele | Accession number | No. of dogs with allele | Source of sample, Number of dogs and Breed type |
|---|---|---|---|
| 001:03 | KR818710 | 9 | FH: 4 Papillion, 5 Schipperke |
| 003:02 | KF911090 | 6 | FH: 3 Hound mix LK: 1 each of Hovawart, Newfoundland, Shetland Sheep Dog |
| 016:03 | KF939645 | 11 | FH: 1 Beagle, 3 Beagle-Mongrel LK: 1 Doberman, 2 Great Dane, 1 Japanese Akita, 1 Newfoundland, 2 West Highland White terrier |
| 028:04 | KF939646 | 10 | FH: 4 Beagle, 2 Beagle-Golden Retriever, 1 Beagle-Mongrel, 1 Hound mix LK: 1 Cairn Terrier, 1 Hovawart |
| 032:02 | KF911096 | 7 | FH: 2 Hound mix, 5 Beagle-Mongrel |
| 036:01 | KF911094 | 7 | FH: 2 Hound mix, 5 Beagle-Mongrel |
| 040:01 | KR818708 | 4 | LK: 2 Great Dane, 2 Newfoundland |
| 042:02 | KT337312 | 2 | FH: 2 Hound mix |
| 046:01 | KR818709 | 4 | LK: 1 Japanese Akita, 3 West Highland White Terrier |
| 047:01 | KF939647 | 10 | FH: 5 Beagle, 2 Beagle-Golden Retriever, 1 Hound mix, 1 Beagle-Mongrel LK: 1 Hovawart |
| 049:01 | KF911093 | 7 | FH: 1 Beagle, 3 Hound mix, 3 Mongrel |
| 050:01 | KF911095 | 2 | FH: 2 Beagle |
| 508:02 | KF911092 | 11 | FH: 1 Beagle, 10 Hound mix |
The number of dogs and the type of dog breed for each allele analyzed. This table does not represent allele frequencies in these different breeds. The nucleotide sequences of these new DLA-88 alleles are recorded in GenBank, and accession numbers are listed. The laboratory source of the dog blood samples is FH for the Fred Hutchinson Cancer Research Center Seattle, WA and LK for LJ Kennedy, Manchester, UK.
Acknowledgments
The authors wish to thank Patrice Stroup for help with the archival DNA samples. Sue Carbonneau, Helen Crawford and Bonnie Larson are acknowledged and thanked for their assistance with the manuscript preparation.
Funding support information: The authors are grateful for research funding from the National Institutes of Health, Bethesda, MD, grants P01 CA078902, P30 DK056465, U54 DK106829 as well as The Storb Endowment. This project was supported by the AKC Canine Health Foundation Oak grant 01771. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the views of the AKC Canine Health Foundation nor the official views of the National Institutes of Health nor its subsidiary Institutes and Centers.
Abbreviations
- DLA
dog leukocyte antigen
- FHCRC
Fred Hutchinson Cancer Research Center
- HSCT
hematopoietic stem cell transplantation
- HLA
human leukocyte antigen
- HVR
Hyper-variable region
- MHC
Major histocompatibility complex
- PCR
polymerase chain reaction
Footnotes
Conflicts of Interest (None)
The authors have no financial relationships that can present a potential conflict of interest in these studies.
Authors’ contributions:
Gopalakrishnan M Venkataraman: Designed the study, performed the experiments, analyzed data and co-authored the manuscript.
Lorna J. Kennedy: Provided valuable canine samples and edited the manuscript.
Marie-Térèse E. Little: Analyzed data and co-authored the manuscript.
Scott S. Graves: Supervised dog leukocyte typing core.
Beverly Torok-Storb: Provided scientific and laboratory support and edited the manuscript.
Michael A. Harkey: Co-authored and edited the manuscript.
Rainer Storb: Designed the study and edited the manuscript.
References
- 1.Thomas ED, Storb R. The development of the scientific foundation of hematopoietic cell transplantation based on animal and human studies. In: Thomas ED, Blume KG, Forman SJ, editors. Hematopoietic Cell Transplantation, Second Edition. Boston: Blackwell Science; 1999. pp. 1–11. [Google Scholar]
- 2.Baron F, Storb R, Little M-T. Hematopoietic cell transplantation: five decades of progress (Review) Archives of Medical Research. 2003;34:528–544. doi: 10.1016/j.arcmed.2003.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Little M-T, Storb R. History of haematopoietic stem-cell transplantation. Nature Reviews Cancer. 2002;2:231–238. doi: 10.1038/nrc748. [DOI] [PubMed] [Google Scholar]
- 4.Epstein RB, Storb R, Ragde H, Thomas ED. Cytotoxic typing antisera for marrow grafting in littermate dogs. Transplantation. 1968;6:45–58. doi: 10.1097/00007890-196801000-00005. [DOI] [PubMed] [Google Scholar]
- 5.Storb R, Rudolph RH, Thomas ED. Marrow grafts between canine siblings matched by serotyping and mixed leukocyte culture. J Clin Invest. 1971;50:1272–1275. doi: 10.1172/JCI106605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnett RC, DeRose SA, Wagner JL, Storb R. Molecular analysis of six dog leukocyte antigen class I sequences including three complete genes, two truncated genes, and one full-length processed gene. Tissue Antigens. 1997;49:484–495. doi: 10.1111/j.1399-0039.1997.tb02783.x. [DOI] [PubMed] [Google Scholar]
- 7.Burnett RC, Geraghty DE. Structure and expression of a divergent canine class I gene. J Immunol. 1995;155:4278–4285. [PubMed] [Google Scholar]
- 8.Graumann MB, DeRose SA, Ostrander EA, Storb R. Polymorphism analysis of four canine MHC class I genes. Tissue Antigens. 1998;51:374–381. doi: 10.1111/j.1399-0039.1998.tb02976.x. [DOI] [PubMed] [Google Scholar]
- 9.Ostrander EA, Galibert EF, Mellersh CS. Linkage and radiation hybrid mapping in the canine genome. In: Ruvinsky A, Sampson J, editors. The Genetics of the Dog. Oxon, UK: CABI Publishing; 2001. pp. 329–370. [Google Scholar]
- 10.Wagner JL, Creer SA, Storb R. Dog class I gene DLA-88 histocompatibility typing by PCR-SSCP and sequencing (Brief communication) Tissue Antigens. 2000;55:564–567. doi: 10.1034/j.1399-0039.2000.550607.x. [DOI] [PubMed] [Google Scholar]
- 11.Hardt C, Ferencik S, Tak R, Hoogerbrugge PM, Wagner V, Grosse-Wilde H. Sequence-based typing reveals a novel DLA-88 allele, DLA-88*04501, in a beagle family. Tissue Antigens. 2006;67:163–165. doi: 10.1111/j.1399-0039.2006.00497.x. [DOI] [PubMed] [Google Scholar]
- 12.Venkataraman GM, Stroup P, Graves SS, Storb R. An improved method for dog leukocyte antigen 88 typing and two new major histocompatibility complex class I alleles, DLA-88*01101 and DLA-88*01201. Tissue Antigens. 2007;70:53–57. doi: 10.1111/j.1399-0039.2007.00839.x. [DOI] [PubMed] [Google Scholar]
- 13.Tsai KL, Starr-Moss AN, Venkataraman GM, et al. Alleles of the major histocompatibility complex play a role in the pathogenesis of pancreatic acinar atrophy in dogs. Immunogenetics. 2013;65:501–509. doi: 10.1007/s00251-013-0704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross P, Buntzman AS, Vincent BG, et al. Allelic diversity at the DLA-88 locus in Golden Retriever and Boxer breeds is limited. Tissue Antigens. 2012;80:175–183. doi: 10.1111/j.1399-0039.2012.01889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venkataraman GM, Geraghty D, et al. Canine DLA-79 gene: an improved typing method, identification of new alleles and its role in graft rejection and graft-versus-host disease. Tissue Antigens. 2013;81:204–211. doi: 10.1111/tan.12094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jochum C, Beste M, Zellmer E, Graves SS, Storb R. CD154 blockade and donor-specific transfusions in DLA-identical marrow transplantation in dogs conditioned with 1-Gy total body irradiation. Biol Blood Marrow Transplant. 2007;13:164–171. doi: 10.1016/j.bbmt.2006.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storb R, Yu C, Barnett T, et al. Stable mixed hematopoietic chimerism in dog leukocyte antigen-identical littermate dogs given lymph node irradiation before and pharmacologic immunosuppression after marrow transplantation. Blood. 1999;94:1131–1136. [PubMed] [Google Scholar]
- 18.Wagner JL, Sarmiento UM, Storb R. Cellular, serological, and molecular polymorphism of the class I and class II loci of the canine major histocompatibility complex. Tissue Antigens. 2002;59:205–210. doi: 10.1034/j.1399-0039.2002.590304.x. [DOI] [PubMed] [Google Scholar]
- 19.Chenna R, Sugawara H, Koike T, et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennedy LJ, Angles JM, Barnes A, et al. Nomenclature for factors of the dog major histocompatibility system (DLA), 2000: Second report of the ISAG DLA Nomenclature Committee. Tissue Antigens. 2001;58:55–70. doi: 10.1034/j.1399-0039.2001.580111.x. [DOI] [PubMed] [Google Scholar]
- 21.Ellis SA, Bontrop RE, Antczak DF, et al. ISAG/IUIS-VIC Comparative MHC Nomenclature Committee report, 2005. Immunogenetics. 2006;57:953–958. doi: 10.1007/s00251-005-0071-4. [DOI] [PubMed] [Google Scholar]
- 22.Dereeper A, Guignon V, Blanc G, et al. Phylogeny. fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–W469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. The amino acid sequence alignment of DLA-88 alleles.
The amino sequences of all published DLA-88 alleles are presented in (A) the clustered alignment as generated by the ClustalW (1) or (B) in the numerical order. The amino acid sequence of the same reference allele is also shown aligned with other class I reference alleles for DLA-12, DLA-64, and DLA-79. Human HLA*A01:01:01 allele is included as an outgroup. Variations between the alleles in the polymorphic exons 2 and exon 3 including within the hyper-variable regions (HVR I–III; underlined) are indicated.
Supplementary Figure 2. Phylogenetic tree derived from the amino acid sequences of exons 2 and 3 of DLA-88 alleles using ClustalW program (1).
A rooted phylogenetic tree derived from the amino acid sequences of exons 2 and 3 of all published DLA-88 alleles and their relationship to the DLA-88 reference allele, with HLA-A*01:01:01 as an outgroup. The sequences indicated with suffixes RB, RH, RJ and RL are from the GenBank submissions of Ross et al., but do not have a finalized ISAG designation yet. A web based program at Phylogeny.fr was used in the ‘One Click’ mode (see Materials and Methods for details). Use of advanced options with 100 replicates in a bootstrap analysis gave the same result. Use of exon 2 and 3 nucleotide sequences instead of amino acid sequences from the same data set resulted in a similar tree. The scale bar units refer to number of substitutions per site.
Supplementary Figure 3. The Nucleotide and amino acid sequences of DLA-88 alleles.
The nucleotide sequences of all published DLA-88 alleles are in FASTA format. The GenBank accession numbers, exonic (upper case) and the intronic sequences (lower case & underlined, when available) (A) and amino acid sequences derived from the polymorphic exons 2 and 3 (B).
Supplementary Table 1. All known DLA-88 Alleles. List of DLA-88 alleles to date indicating alternative names, GenBank accession numbers and primary publication references**.