Abstract
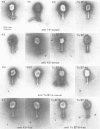
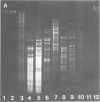
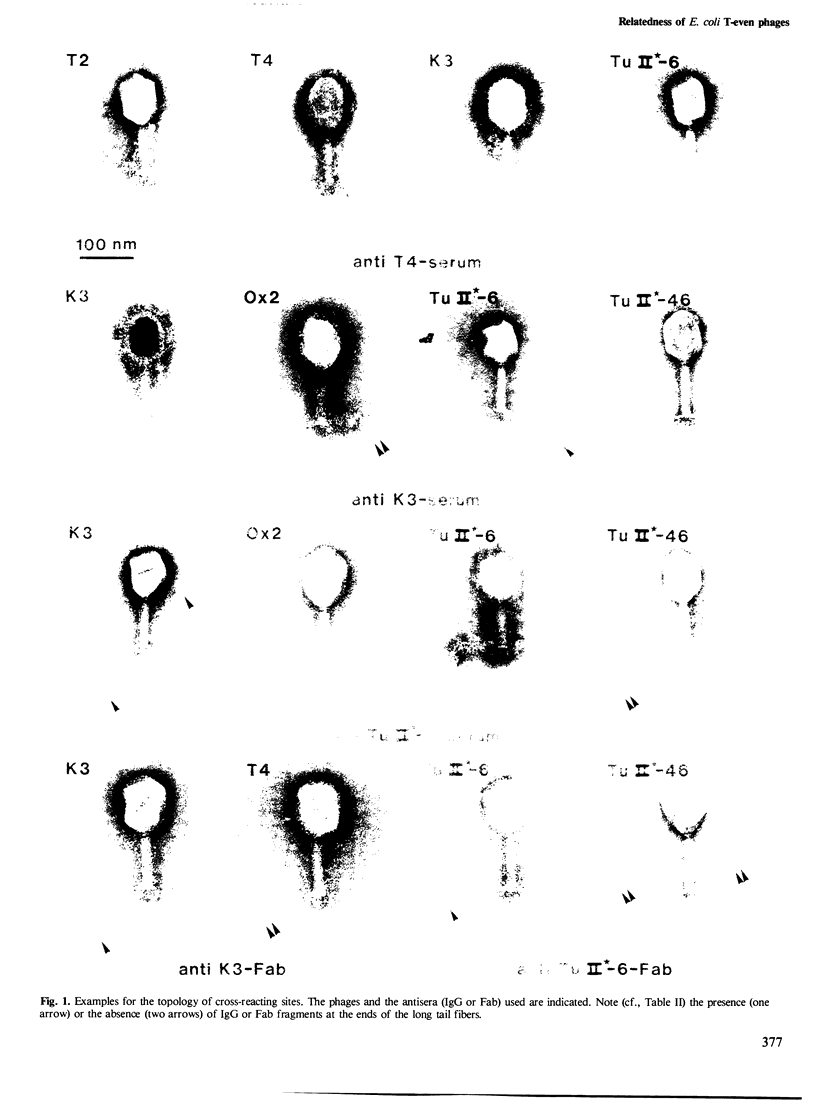
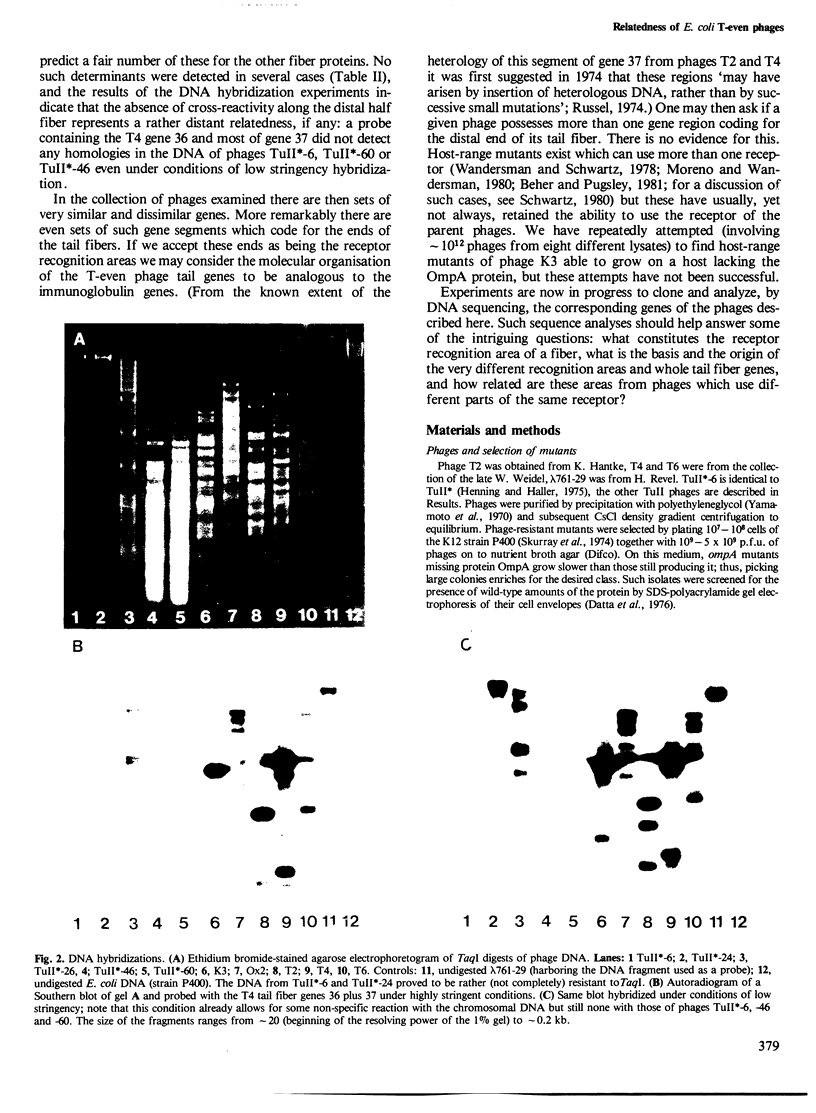
The relatedness of a series of T-even like phages which use the Escherichia coli outer membrane protein OmpA as a receptor, and the classical phages T2, T4 and T6 has been investigated. Immunoelectron microscopy and the pattern of phage resistance in bacterial mutants revealed that: (i) phages of this morphology do not necessarily cross-react serologically; (ii) phages using different receptors may bind heterologous IgG everywhere except to the tip (comprising approximately 10% of one fiber polypeptide) of the long tail fibers; (iii) cross-reacting OmpA-specific phages may bind heterologous IgG only to the tip of these fibers: (iv) OmpA-specific phages not cross-reacting at the tip of the tail fibers use different receptor sites on the protein. Absence of cross-reactivity appears to reflect high degrees of dissimilarity. A DNA probe consisting of genes encoding the two most distal tail fiber proteins of T4 detected homologies only in DNA from phages serologically cross-reacting at this fiber. Even under conditions of low stringency, allowing the formation of stable hybrids with almost 30% base mismatch, no such homologies could be found in serologically unrelated phages. Thus, in the collection of phages examined, there are sets of very similar and very dissimilar tail fiber genes and even of such gene segments.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ADAMS M. H. Classification of bacterial viruses: characteristics of the T5 species and of the T2, C16 species. J Bacteriol. 1952 Sep;64(3):387–396. doi: 10.1128/jb.64.3.387-396.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achtman M., Willetts N., Clark A. J. Beginning a genetic analysis of conjugational transfer determined by the F factor in Escherichia coli by isolation and characterization of transfer-deficient mutants. J Bacteriol. 1971 May;106(2):529–538. doi: 10.1128/jb.106.2.529-538.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckendorf S. K., Kim J. S., Lielausis I. Structure of bacteriophage T4 genes 37 and 38. J Mol Biol. 1973 Jan;73(1):17–35. doi: 10.1016/0022-2836(73)90156-3. [DOI] [PubMed] [Google Scholar]
- Beckendorf S. K. Structure of the distal half of the bacteriophage T4 tail fiber. J Mol Biol. 1973 Jan;73(1):37–53. doi: 10.1016/0022-2836(73)90157-5. [DOI] [PubMed] [Google Scholar]
- Beher M. G., Pugsley A. P. Coliphage which requires either the LamB protein or the OmpC protein for adsorption to Escherichia coli K-12. J Virol. 1981 Apr;38(1):372–375. doi: 10.1128/jvi.38.1.372-375.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop R. J., Conley M. P., Wood W. B. Assembly and attachment of bacteriophage T4 tail fibers. J Supramol Struct. 1974;2(2-4):196–201. doi: 10.1002/jss.400020214. [DOI] [PubMed] [Google Scholar]
- Datta D. B., Arden B., Henning U. Major proteins of the Escherichia coli outer cell envelope membrane as bacteriophage receptors. J Bacteriol. 1977 Sep;131(3):821–829. doi: 10.1128/jb.131.3.821-829.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta D. B., Krämer C., Henning U. Diploidy for a structural gene specifying a major protein of the outer cell envelope membrane from Escherichia coli K-12. J Bacteriol. 1976 Dec;128(3):834–841. doi: 10.1128/jb.128.3.834-841.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earnshaw W. C., Goldberg E. B., Crowther R. A. The distal half of the tail fibre of bacteriophage T4. Rigidly linked domains and cross-beta structure. J Mol Biol. 1979 Jul 25;132(1):101–113. doi: 10.1016/0022-2836(79)90498-4. [DOI] [PubMed] [Google Scholar]
- Gronenborn B., Messing J. Methylation of single-stranded DNA in vitro introduces new restriction endonuclease cleavage sites. Nature. 1978 Mar 23;272(5651):375–377. doi: 10.1038/272375a0. [DOI] [PubMed] [Google Scholar]
- Hancock R. E., Reeves P. Bacteriophage resistance in Escherichia coli K-12: general pattern of resistance. J Bacteriol. 1975 Mar;121(3):983–993. doi: 10.1128/jb.121.3.983-993.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K. Major outer membrane proteins of E. coli K12 serve as receptors for the phages T2 (protein Ia) and 434 (protein Ib). Mol Gen Genet. 1978 Aug 17;164(2):131–135. doi: 10.1007/BF00267377. [DOI] [PubMed] [Google Scholar]
- Henning U., Haller I. Mutants of Escherichia coli K12 lacking all 'major' proteins of the outer cell envelope membrane. FEBS Lett. 1975 Jul 15;55(1):161–164. doi: 10.1016/0014-5793(75)80983-5. [DOI] [PubMed] [Google Scholar]
- Henning U., Schwarz H., Chen R. Radioimmunological screening method for specific membrane proteins. Anal Biochem. 1979 Aug;97(1):153–157. doi: 10.1016/0003-2697(79)90339-7. [DOI] [PubMed] [Google Scholar]
- Henning U., Sonntag I., Hindennach I. Mutants (ompA) affecting a major outer membrane protein of Escherichia coli K12. Eur J Biochem. 1978 Dec;92(2):491–498. doi: 10.1111/j.1432-1033.1978.tb12771.x. [DOI] [PubMed] [Google Scholar]
- Howley P. M., Israel M. A., Law M. F., Martin M. A. A rapid method for detecting and mapping homology between heterologous DNAs. Evaluation of polyomavirus genomes. J Biol Chem. 1979 Jun 10;254(11):4876–4883. [PubMed] [Google Scholar]
- Huang L. H., Farnet C. M., Ehrlich K. C., Ehrlich M. Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res. 1982 Mar 11;10(5):1579–1591. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JERNE N. K., AVEGNO P. The development of the phage-inactivating properties of serum during the course of specific immunization of an animal: reversible and irreversible inactivation. J Immunol. 1956 Mar;76(3):200–208. [PubMed] [Google Scholar]
- KAY D., FILDES P. Hydroxymethylcytosine-containing and tryptophan-dependent bacteriophages isolated from city effluents. J Gen Microbiol. 1962 Jan;27:143–146. doi: 10.1099/00221287-27-1-143. [DOI] [PubMed] [Google Scholar]
- Kim J. S., Davidson N. Electron microscope heteroduplex study of sequence relations of T2, T4, and T6 bacteriophage DNAs. Virology. 1974 Jan;57(1):93–111. doi: 10.1016/0042-6822(74)90111-1. [DOI] [PubMed] [Google Scholar]
- LANNI F., LANNI Y. T. Antigenic structure of bacteriophage. Cold Spring Harb Symp Quant Biol. 1953;18:159–168. doi: 10.1101/sqb.1953.018.01.026. [DOI] [PubMed] [Google Scholar]
- Manning P. A., Puspurs A., Reeves P. Outer membrane of Escherichia coli K-12: isolation of mutants with altered protein 3A by using host range mutants of bacteriophage K3. J Bacteriol. 1976 Sep;127(3):1080–1084. doi: 10.1128/jb.127.3.1080-1084.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConaughy B. L., Laird C. D., McCarthy B. J. Nucleic acid reassociation in formamide. Biochemistry. 1969 Aug;8(8):3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- Moreno F., Wandersman C. OmpC and LamB proteins can serve as substitute receptors for host range mutants of coliphage TuIa. J Bacteriol. 1980 Dec;144(3):1182–1185. doi: 10.1128/jb.144.3.1182-1185.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. B., Crowther R. A. DNA sequence of the tail fibre genes 36 and 37 of bacteriophage T4. J Mol Biol. 1981 Dec 15;153(3):545–568. doi: 10.1016/0022-2836(81)90407-1. [DOI] [PubMed] [Google Scholar]
- PORTER R. R. The hydrolysis of rabbit y-globulin and antibodies with crystalline papain. Biochem J. 1959 Sep;73:119–126. doi: 10.1042/bj0730119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel H. R. Molecular cloning of the T4 genome: organization and expression of the tail fiber gene cluster 34--38. Mol Gen Genet. 1981;182(3):445–455. doi: 10.1007/BF00293934. [DOI] [PubMed] [Google Scholar]
- Russell R. L. Comparative genetics of the T-even bacteriophages. Genetics. 1974 Dec;78(4):967–988. doi: 10.1093/genetics/78.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüther U., Koenen M., Otto K., Müller-Hill B. pUR222, a vector for cloning and rapid chemical sequencing of DNA. Nucleic Acids Res. 1981 Aug 25;9(16):4087–4098. doi: 10.1093/nar/9.16.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Hutchinson C. A., 3rd, Harris J. I. A thermostable sequence-specific endonuclease from Thermus aquaticus. Proc Natl Acad Sci U S A. 1977 Feb;74(2):542–546. doi: 10.1073/pnas.74.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer M., Hindennach I., Garten W., Henning U. Major proteins of the Escherichia coli outer cell envelope membrane. Interaction of protein II with lipopolysaccharide. Eur J Biochem. 1978 Jan 2;82(1):211–217. doi: 10.1111/j.1432-1033.1978.tb12013.x. [DOI] [PubMed] [Google Scholar]
- Skurray R. A., Hancock R. E., Reeves P. Con--mutants: class of mutants in Escherichia coli K-12 lacking a major cell wall protein and defective in conjugation and adsorption of a bacteriophage. J Bacteriol. 1974 Sep;119(3):726–735. doi: 10.1128/jb.119.3.726-735.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Wahl G. M., Stern M., Stark G. R. Efficient transfer of large DNA fragments from agarose gels to diazobenzyloxymethyl-paper and rapid hybridization by using dextran sulfate. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3683–3687. doi: 10.1073/pnas.76.8.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandersman C., Schwartz M. Protein Ia and the lamB protein can replace each other in the constitution of an active receptor for the same coliphage. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5636–5639. doi: 10.1073/pnas.75.11.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G. G., Tanyashin V. I., Murray N. E. Molecular cloning of fragments of bacteriophage T4 DNA. Mol Gen Genet. 1977 Nov 14;156(2):203–214. doi: 10.1007/BF00283493. [DOI] [PubMed] [Google Scholar]
- Wilson J. H., Luftig R. B., Wood W. B. Interaction of bacteriophage T4 tail fiber components with a lipopolysaccharide fraction from Escherichia coli. J Mol Biol. 1970 Jul 28;51(2):423–434. doi: 10.1016/0022-2836(70)90152-x. [DOI] [PubMed] [Google Scholar]
- Yamamoto K. R., Alberts B. M., Benzinger R., Lawhorne L., Treiber G. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology. 1970 Mar;40(3):734–744. doi: 10.1016/0042-6822(70)90218-7. [DOI] [PubMed] [Google Scholar]