Abstract
MicroRNAs (miRNAs) play an important role in fine-tuning host immune homeostasis and responses through the regulation of messenger RNA (mRNA) stability and translation. Studies have demonstrated that miRNA-mediated regulation of gene expression has a profound impact on immune cell development, function and response to invading pathogens. As we continue to examine the mechanisms by which miRNAs maintain the balance between robust protective host immune responses and dysregulated responses that promote immune pathology, careful consideration of the complexity of post-transcriptional immune regulation is needed. Distinct tissue- and stimulus-specific RNA-RNA and RNA-protein interactions can modulate the functions of a given miRNA. Thus, new challenges emerge in the identification of post-transcriptional co-regulatory modules and the genetic factors that impact miRNA function.
Keywords: microRNA, immunity, post-transcriptional regulation, RNA structure, lncRNAs, RNA-binding proteins
Emergence of miRNAs as immune regulators
It is widely established that non-coding RNAs (ncRNAs) play a critical role in the regulation of host innate and adaptive immune responses. Among these non-coding regulators, microRNAs (miRNAs) have been demonstrated to coordinate immune cell development and function [1–3]. MicroRNAs are small 19–22 nucleotide (nt), single-stranded RNAs transcribed by either RNA Pol II and RNA Pol III, as primary transcripts (pri-miRNAs). The RNase III endonuclease, Drosha, processes pri-miRNAs into precursor miRNAs (pre-miRNAs), which are shuttled to the cytosol by exportin 5 and RAN-GTP. Pre-miRNAs are then processed into mature miRNAs by the RNase III, Dicer. Mature miRNAs bind to complementary miRNA recognition elements (MRE) encoded within messenger RNAs (mRNA), leading to the recruitment and formation of the microRNA-induced silencing complex (miRISC). It is estimated that miRNAs regulate 30–80% of mammalian genes [4–6].
The first demonstration that miRNAs play a biological role in immune cells comes from the observation that certain miRNAs exhibit highly specific expression patterns in key organs of the immune system such as the thymus, bone marrow, and spleen [7]. Ectopic expression of specific miRNAs in primary hematopoietic progenitor cells was sufficient to alter lineage differentiation into either lymphoid or myeloid lineage both in vitro and in vivo, suggesting a functional role for miRNAs during immune cell development. Subsequent experiments with conditional deletion of key components of the miRNA biogenesis pathway (Drosha, Dicer, and Ago2) at different stages of hematopoiesis led to the same conclusion that additional layers of gene expression control at the post-transcriptional level are crucial for immune cell development and function [8, 9]. The natural transition from these discoveries was to focus on individual miRNAs and their biological effects were investigated using overexpression or deletion studies. These initial findings of the immunomodulatory function of key miRNAs have been extensively reviewed [1–3]. However, having a deeper understanding of the complexity of the post-transcriptional regulatory landscape, it is critical to consider the interactions among regulatory elements in our studies of miRNA-mediated gene regulation. Here, we highlight a few miRNAs that initially provided a conceptual framework of understanding how miRNAs exert their specific actions in the immune system and focus on pertinent approaches to determine the precise mechanism of action of miRNAs to overcome current challenges in accurately outlining miRNA function.
miRNAs in the immune system and challenges
The immune-related phenotypes arising from animal models either lacking or overexpressing certain miRNAs were not of surprise given the dramatic changes in specific miRNAs across cell types, observed in miRNA profiling studies of the hematopoietic system. Currently, a few models have been proposed based on these initial studies to account for miRNA-mediated immunological effects. One example comes from the study of miR-150, a miRNA shown to regulate B1 cell differentiation through the regulation of the transcription factor c-Myb [10]. As transcription factors possess the ability to affect gene expression of numerous genes, these findings led the authors to propose the ‘key target gene model,’ whereby changes in the concentration of a key cellular target (e.g. transcriptions factors) by a single miRNA could lead to profound immunological outcomes. A similar example is the observation that the miR-17-92 cluster targets RUNX1 (also known as AML1), a known transcription factor involved in monocytopoiesis. Ectopic expression of miR-17-92 is sufficient to suppress monocyte differentiation [11]. MicroRNAs have also been shown to be important in innate immunity by fine-tuning the threshold of innate immune sensors. NF-κB mediated induction of miR-146 has been demonstrated to target the TLR adaptors TRAF6 and IRAK1 mRNA, acting as a negative feedback system to prevent excessive inflammatory responses [12].
Despite providing a plausible explanation of miRNA mediated biological phenotypes in certain immune-related contexts, it can be ambiguous as to what can be defined as a ‘key’ cellular target, especially in the absence of experiments involving transgenic animals or cell lines expressing mRNA targets lacking specific MREs (see below). It is also worth noting that systematic generation of numerous miRNA knockout mice, including the immune regulatory MyomiRs: miR208b and miR499, revealed little to no observable phenotype likely due to miRNA functional redundancies or the absence of a stress stimulus [13]. In fact, the potential key cellular target can also be entirely cell-type and context-dependent as the transgenic overexpression of miR-17-92 cluster in mice cause a severe form of lymphoproliferative autoimmune phenotype mediated mainly by T cells due to targeting of the pro-apoptotic protein BIM and the tumor suppressor PTEN [14].
These studies have yielded insights into the physiological and pathological roles of miRNAs while also highlighting the limitations of studying individual miRNAs and their function in selected cell types. To identify regulatory miRNAs in the immune system two main strategies have been implemented: a miRNA expression profiling approach and a host genetics approach. The first strategy combines a global survey of differential miRNAs expression profiles across tissue types, distinct developmental stages of immune cells, disease states, and in response to distinct immune stimuli. This is followed by statistical modeling and biochemical approaches to identify and validate miRNA target genes, and ultimately the generation of transgenic cell lines and mouse models to investigate the physiological consequences of modulating miRNA levels. The second approach favors the linkage of host genetic variance in or near miRNA genes or within regulatory elements encoded in their predicted target genes to study the association of specific miRNAs with immune disease.
The methods commonly used to validate miRNA-mediated action on target mRNAs in most studies up to date focus on the function of a single miRNA. First, a part of the RefSeq annotated target mRNA 3′UTR is cloned into a luciferase reporter with or without a mutated MRE site. Second, target miRNAs are either overexpressed or knocked down/out to recapitulate certain phenotypes exerted by the miRNA of interest. Third, endogenous miRNA binding site in target gene(s) is mutated to restore phenotypes [15]. Ideally, only the third type of approach allows us to dissect the effect of a single miRNA to its ‘key cellular target’ independent from all the other potential targets. With the advances in CRISPR/Cas9 mediated genome-editing technologies, we expect this approach to be more widely used in cultured cell lines. However, generation of miRNA binding site mutant animals for every single miRNA study may not be the most efficient nor cost-effective approach.
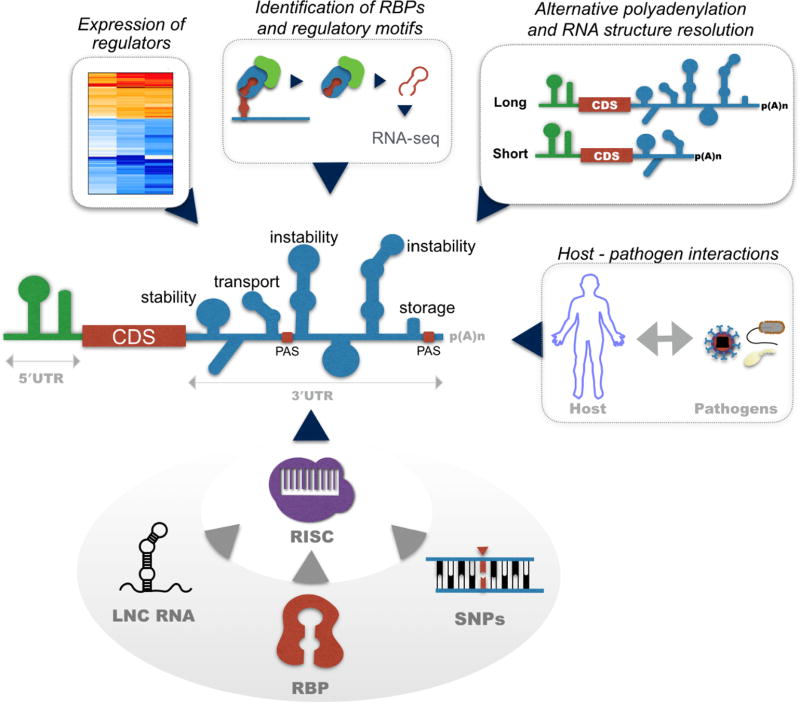
Another major challenge is determining miRNA-mediated target mRNA recognition and its silencing. This is exemplified by the fact that still up to this date, there is no clear distinction between miRNA-mediated mRNA stability control and translation control [16, 17]. Many earlier conclusions using genome-wide unbiased approaches to investigate the transcriptome (RNA-seq) and translatome (ribosome profiling) after transient miRNA transfection strategies should be interpreted cautiously due to the generation of high molecular weight RNA species that likely resulted in observed non-specific effects on gene expression [18]. It should also be noted that many groups have reported a poor overlap of expression changes in target genes, along with distinct biological phenotypes, even when the same miRNA is either overexpressed or depleted in the same system [19, 20]. These observations highlight the need to develop new molecular, statistical, and computational technologies to facilitate future studies of miRNA-mediated immune regulation. Moreover, defining key miRNAs that control immunological networks also necessitates a better understanding of the complex miRNA interactions with additional molecular regulators of post-transcriptional gene regulation (Figure 1).
Figure 1. Schematic of microRNA interactions with post-transcriptional regulators.
Recruitment of the RNA-induced silencing complex (RISC) to miRNA recognition elements (MRE) leads to ribonuclease-mediated mRNA decay and alterations in gene expression. The co-expression of post-transcriptional modulators such as RNA-binding proteins (RBP) and long non-coding RNAs (lncRNAs) can impact miRNA biogenesis and function. RBPs can inhibit miRNA expression and prevent the miRNA recruitment to target mRNAs through competitive binding or mRNA compartmentalization. On the other hand, RBPs can cooperatively enhance miRNA-mediated mRNA decay. LncRNAs can interact with DNA, RNA and RBPs, suppressing both miRNA and RBP activity through competitive binding. Additionally, the function of miRNAs can be impacted by changes in the sequence and structure of the 3′ UTR. Single nucleotide polymorphisms (SNPs) can disrupt or create novel MREs. SNPs can also change the structure of the 3′ UTR, hindering the accessibility of miRNA and RBPs to their target elements. Finally, dynamic changes in the length of the 3′ UTR through usage of alternative polyadenylation sites (pA) results in changes in the number of potential interaction sites/motifs for the previously mentioned post-transcriptional modulators, impacting mRNA stability. The development of novel molecular and computational tools provide means to understand global changes in post-transcriptional regulator expression, the identification of mRNA regulatory motifs, and changes in the sequence and structure of the 3′ UTR. Building a comprehensive post-transcriptional regulome will allow us to contextualize the function of miRNAs in the regulation of host gene expression during immune development, infection, and immunological disease.
miRNA function in the context of other post-transcriptional regulators
Along with miRNAs, the post-transcriptional effectors involved in regulation of gene expression include other noncoding RNAs (ncRNAs) and RNA binding proteins (RBPs) that can influence the potency of miRNA-mediated gene regulation [21]. To understand the effect of a given miRNA in pathogen clearance and predisposition to immune disease, knowledge of the regulatory modules that are involved in orchestrating immunological processes must be considered. Thus, we favor studying a framework that considers the integrative effects of miRNAs alongside other post-transcriptional regulators in conferring protection or susceptibility to immune pathology.
Long non-coding RNAs affecting miRNA function
Increasing evidence shows that MREs are present on other ncRNAs, such as pseudogenes and long non-coding RNAs (lncRNAs) [22]. LncRNAs, are transcripts longer than 200 nt that control gene expression through their ability to interact with DNA, RNA, and proteins [23]. When localized in the cytosol, lncRNAs can compete with miRNA binding to mRNAs that share common MREs, acting as miRNA sponges [22]. One such interaction has been shown to impact monocyte/macrophage differentiation. miR-199a-5p inhibits monocyte/macrophage differentiation through targeting of activin A receptor type 1B (ACVR1B) [24]. Sequestration of miR-199a-5p by a long non-coding monocytic RNA (lnc-MC), rescues miRNA-mediated ACVR1B repression and promotes monocyte/macrophage differentiation [25]. As lncRNAs continue to emerge as important regulators of host immunity [23, 26, 27], both molecular [28] and computational tools [29] are being used to derive a comprehensive lncRNA-miRNA interactome. Public repositories that document lncRNA expression profiles [30–32] and miRNA-ncRNA interactions [33] provide a powerful tool to guide future studies in understanding the functional relationship between co-expressed ncRNAs.
RNA-binding proteins affecting miRNA function
A number of RNA-binding proteins (RBPs) have been implicated in controlling the efficacy of both miRNA biogenesis and their regulatory functions [34]. The overall mechanisms by which RBPs can directly or indirectly affect miRNA functions include: the regulation of miRNA expression, inhibition of miRNA recruitment to target mRNAs by competitive binding and mRNA compartmentalization, and enhanced mRNA decay through miRNA and RBP cooperation. The repressive functions of RBPs is exemplified by the negative regulation of let-7 miRNAs expression by the RBP, LIN28, through direct binding to the pri- and pre-let-7 and blocking of mature miRNA processing [35–37]. Ectopic expression of Lin28b is sufficient for the reprogramming of adult hematopoetic stem cells (HSCs) into fetal-like HSCs and the regulation of fetal lymphopoiesis [38] as well as fetal T cell differentiation [39] in mice. RBPs can also diminish gene repression by either preventing their recruitment to MREs through competitive binding or by inducing structural change in the target mRNA. Under conditions of normoxia, miR-297 or miR-299 can target the 3′UTR of VEGFA in myeloid cells. Following hypoxic stress, the heterogenous nuclear ribonucleoprotein L (hnRNP L) binds to the CA-rich element targeted by miR-297 and miR-299, inhibiting miRNA-mediated decay of VEGFA [40]. Lastly, RBP-mRNA interactions can lead to the subcellular compartmentalization of mRNAs protecting them from miRNA targeting [41].
Support for cooperation between miRNA and RBPs in the control of inflammatory responses was first described in studies of tumor necrosis factor-α (TNFA) and COX2 gene regulation. These observations suggested that interactions between tristetraprolin (TTP, ZFP36) and the miRISC subunit Ago2 enable miR-16 enhancement of AU-rich element mediated decay (AMD) of these two genes [42]. Other studies implicated miR-221 and TTP interactions in the enhanced degradation of TNFA mRNA during LPS tolerance [43]. Interestingly, RBPs may also potentiate the activity of miRNAs indirectly by altering mRNA accessibility. The RBP Pumilio can mediate structural changes that relieve target site hindrance, indirectly allowing for the recruitment of miR-221 and miR-222 to the 3′UTR of the cell cycle regulator p27 [44].
Despite evidence of complex post-transcriptional regulation by miRNAs and RBPs, studies focusing on the combinatorial effects of miRNA and RBPs on immune outcomes are limited. Mapping of RBP and miRNA transcript specificity using high-throughput approaches like crosslinking and immunoprecipitation (CLIP) technologies [45, 46] will allow us to resolve the cell type and stimulus-specific RBP-miRNA interactions that shape host immunity.
Defining the 3′ untranslated region in host immunoregulatory genes
Another critical limitation to the study of miRNA-mediated immune regulation is the lack of an accurate annotation of the 3′UTRs sequence and structure of miRNA-target genes. For example, due to the use of partial interferon gamma (IFNG) 3′UTRs, conflicting results have been derived from previous studies of miR-29-mediated control of IFNG expression. miR-29 has been proposed to inhibit IFNG expression, while others show that the impact of miR-29 control is minimal relative to IFNG transcript instability promoted by AMD [47, 48]. This warrants accurate annotation of the sequences and structural topography of the 3′UTR of genes involved in innate and adaptive immunity.
Alternative polyadenylation impacting miRNA-mediated regulation
Early studies demonstrated that immune activation of lymphocytes and myeloid cells triggers the alternative polyadenylation (APA) of transcripts that harbor distinct polyadenylation sites (PAS) in their 3′UTR [49, 50]. Alternative polyadenylation is a means by which the length of the 3′UTR can be dynamically altered through differential usage of PAS. About 50% of genes contain several PAS [51, 52], resulting in changes to the length and sequence of the 3′UTR of a given gene. The regulatory mechanisms that control APA and the potential consequences to gene expression have been previously reviewed [53]. Acute viral infection of macrophages [54] and T cell activation [55] result in the global shortening in the 3′UTR of immune genes, resulting in the exclusion of MREs found the distal 3′UTR. Thus, in this context, miRNA-regulated gene expression is dependent on both changes in miRNA concentration and PAS utilization.
The importance of 3′UTR reprogramming and its positive and negative effects on the strength of miRNA-dependent repression [56, 57] in immunological outcomes is beginning to be recognized. The development of high-throughput mRNA 3′end mapping methodologies [58–60] has demonstrated that PAS utilization is dynamic. These technologies are amenable for detection of global in vivo dynamic changes in 3′UTR definitions and have identified novel functional miRNA binding sites in alternatively polyadenylated transcripts [61]. Utilizing such tools in the context of immune cell development or immune stimulus could further refine our understanding of both the importance of previously observed APA events (e.g. during immune cell development, differentiation, and activation [49, 50, 54, 62, 63]) and also allow us explore the involvement of miRNA-dependent regulation in these processes.
Host genetic variance impacts miRNA function
Host genetic variance can have a significant impact on the expression of miRNA and their function. On the other hand, variations within the 3′UTR of miRNA targets can also result in the disruption or creation of MRE sites [64]. A wealth of information of genetic variation and gene expression is available through the Encyclopedia of DNA Elements (ENCODE; www.encodeproject.org), 1000 genomes (www.internationalgenome.org), and the Genotype-Tissue Expression (GTEx) project (www.gtexportal.org) data repositories. Additionally, a comprehensive summary of genome-wide association studies (GWAS) is available through the NHGRI-EBI Catalog of published GWAS [65]. Leveraging these data to identify novel and disease relevant miRNA-mRNA interactions can facilitate the implementation of genetic approaches to identify the mutational effects that SNPs have on the miRNA potency in the regulation of genes linked to disease phenotypes.
One can prioritize the study of miRNAs with significant potential impact on disease outcome by coupling GWAS to detect disease-associated SNPs and analysis of differential miRNA expression. Indeed, we have demonstrated that polymorphisms within MRE encoded in the 3′UTR of immune genes alter the predisposition to viral susceptibility and persistence [66–68]. A single nucleotide polymorphism (SNP) in the 3′UTR of HLA-C regulates binding of miR-148 to its target site, associating strongly with HIV control [67]. A SNP (rs4803217) in the 3′UTR of IFNL3 can partly alter transcript stability through disruption of the MRE sites targeted by the virus-induced miRNAs, miR-208b and miR-499-5p. The disruption of this site results in enhanced IFNL3 mRNA stability and promotes clearance of HCV [68].
mRNA structure influencing miRNA-mediated control
A key feature in ncRNA function is its ability to form secondary and tertiary structures [69]. Structural variation of the 3′UTR dictates RNA- and RBP-mRNA interactions and needs to be considered in the design and interpretation of studies on miRNA-mediated immune regulation [70]. In addition to the potent effect target structure has on miRNA recognition, it is important to acknowledge the relationship between the structure of the 3′UTR and the accessibility of other post-transcriptional effectors to their regulatory elements [71, 72]. AGO-CLIP studies show that genes with greater structural accessibility within the 3′UTR are targeted by miR-142 and miR-148 more effectively [73]. Therefore, understanding RNA structure can separate true target genes from genes that contain “appropriate” but poorly accessible regulatory sequences.
In addition to the impact that host genetic variance has on RNA structure, post-transcriptional RNA modifications can also alter its structure [69]. A riboSNitch is an RNA element that undergoes structural changes in the presence of single nucleotide variants (SNV). The ability to predict the extent to which a SNV disrupts RNA function/structure is paramount to our understanding of how SNPs found in intergenic and non-coding regions of the genome contribute to disease phenotypes. The combination of genetic variance data and next-generation sequencing approaches with structure-dependent chemical modifications in the RNA (SHAPE-seq [74] and DMS-seq [75, 76], icSHAPE[77]), the use of structure-specific RNAses (PARS [73] and FragSeq [78]), or RNA proximity-ligation assays (RPL [79]) provide a powerful toolkit to map changes in RNA topography. Altogether, this structural information provides critical insights towards the understanding of how a given sequence change can affect the regulation of a gene.
Concluding Remarks
Extensive studies in immunology over decades have primarily focused on major transcriptional programs that dictate immunological outcomes such as immune cell subset identity and function. Post-transcriptional control through miRNAs provide a new layer of gene expression control through either fine-tuning or rapid mRNA turnover, an essential feature to achieve immune homeostasis. Understanding miRNA-regulated immunological networks and the specific molecular mechanisms by which miRNAs shape the immune system is therefore an outstanding area of research. Conventional approaches have focused primarily on uncovering the relationship between the inhibition or overexpression of a single miRNA and the expression of a handful of target genes placing several miRNAs at the center of immune development, cell function, and resistance/susceptibility to infectious and immune disease. However, as we expand our understanding of the dynamic post-transcriptional regulatory landscape in the regulation of immunity, several questions remain (see Outstanding Questions).
The rapid advances and implementation of novel genetic, biochemical, computational and statistical approaches to map the post-transcriptional landscape can facilitate the studies on the immune regulatory roles of miRNAs. These tools have provided a wealth of information available through various public repositories of differential miRNA expression across tissue types and immune stimuli, statistical predictions of miRNA targets, information of non-coding RNA interactions, and RBP expression data. However, due to differences in biological models and the choices of immune stimuli, the integration of this information is cumbersome. As has been achieved with Immgen [80], a concerted effort to generate the post-transcriptional regulome of immune genes would be beneficial to extend our understanding of the impact of miRNA in immunity and immune disease.
Acknowledgments
This work was supported in part by R01AI108765 (R.S.) and 5T32HL007312-37 (A.F.). The views and conclusions in this report are those of the authors and do not necessarily reflect the views of the funding agency.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.O'Connell RM, et al. Physiological and pathological roles for microRNAs in the immune system. Nat Rev Immunol. 2010;10(2):111–22. doi: 10.1038/nri2708. [DOI] [PubMed] [Google Scholar]
- 2.Xiao C, Rajewsky K. MicroRNA control in the immune system: basic principles. Cell. 2009;136(1):26–36. doi: 10.1016/j.cell.2008.12.027. [DOI] [PubMed] [Google Scholar]
- 3.Josefowicz SZ, et al. Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol. 2012;30:531–64. doi: 10.1146/annurev.immunol.25.022106.141623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis BP, et al. Prediction of mammalian microRNA targets. Cell. 2003;115(7):787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 5.Brennecke J, et al. Principles of microRNA-target recognition. PLoS Biol. 2005;3(3):e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.John B, et al. Human MicroRNA targets. PLoS Biol. 2004;2(11):e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen CZ, et al. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303(5654):83–6. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 8.Cobb BS, et al. A role for Dicer in immune regulation. J Exp Med. 2006;203(11):2519–27. doi: 10.1084/jem.20061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muljo SA, et al. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202(2):261–9. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131(1):146–59. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 11.Fontana L, et al. MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat Cell Biol. 2007;9(7):775–87. doi: 10.1038/ncb1613. [DOI] [PubMed] [Google Scholar]
- 12.Taganov KD, et al. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103(33):12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park CY, et al. Analysis of microRNA knockouts in mice. Hum Mol Genet. 2010;19(R2):R169–75. doi: 10.1093/hmg/ddq367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat Immunol. 2008;9(4):405–14. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu LF, et al. A Single miRNA-mRNA Interaction Affects the Immune Response in a Context- and Cell-Type-Specific Manner. Immunity. 2015;43(1):52–64. doi: 10.1016/j.immuni.2015.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwakawa HO, Tomari Y. The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol. 2015;25(11):651–65. doi: 10.1016/j.tcb.2015.07.011. [DOI] [PubMed] [Google Scholar]
- 17.Filipowicz W, et al. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–14. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 18.Jin HY, et al. Transfection of microRNA Mimics Should Be Used with Caution. Front Genet. 2015;6:340. doi: 10.3389/fgene.2015.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–4. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin HY, et al. Differential Sensitivity of Target Genes to Translational Repression by miR-17~92. PLoS Genet. 2017;13(2):e1006623. doi: 10.1371/journal.pgen.1006623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwerk J, Savan R. Translating the Untranslated Region. J Immunol. 2015;195(7):2963–71. doi: 10.4049/jimmunol.1500756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poliseno L, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465(7301):1033–8. doi: 10.1038/nature09144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atianand MK, Fitzgerald KA. Long non-coding RNAs and control of gene expression in the immune system. Trends Mol Med. 2014;20(11):623–31. doi: 10.1016/j.molmed.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin HS, et al. miR-199a-5p inhibits monocyte/macrophage differentiation by targeting the activin A type 1B receptor gene and finally reducing C/EBPalpha expression. J Leukoc Biol. 2014;96(6):1023–35. doi: 10.1189/jlb.1A0514-240R. [DOI] [PubMed] [Google Scholar]
- 25.Chen MT, et al. PU.1-Regulated Long Noncoding RNA lnc-MC Controls Human Monocyte/Macrophage Differentiation through Interaction with MicroRNA 199a-5p. Mol Cell Biol. 2015;35(18):3212–24. doi: 10.1128/MCB.00429-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heward JA, Lindsay MA. Long non-coding RNAs in the regulation of the immune response. Trends Immunol. 2014;35(9):408–19. doi: 10.1016/j.it.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Turner M, et al. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat Immunol. 2014;15(6):484–91. doi: 10.1038/ni.2887. [DOI] [PubMed] [Google Scholar]
- 28.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol. 2016;1402:271–86. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 29.Furio-Tari P, et al. spongeScan: A web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res. 2016;44(W1):W176–80. doi: 10.1093/nar/gkw443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quek XC, et al. lncRNAdb v2.0: expanding the reference database for functional long noncoding RNAs. Nucleic Acids Res. 2015;43(Database issue):D168–73. doi: 10.1093/nar/gku988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Josset L, et al. Annotation of long non-coding RNAs expressed in collaborative cross founder mice in response to respiratory virus infection reveals a new class of interferon-stimulated transcripts. RNA Biol. 2014;11(7):875–90. doi: 10.4161/rna.29442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou KR, et al. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 2017;45(D1):D43–D50. doi: 10.1093/nar/gkw965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li JH, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(Database issue):D92–7. doi: 10.1093/nar/gkt1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Kouwenhove M, et al. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11(9):644–56. doi: 10.1038/nrc3107. [DOI] [PubMed] [Google Scholar]
- 35.Piskounova E, et al. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147(5):1066–79. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Newman MA, et al. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14(8):1539–49. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viswanathan SR, et al. Selective blockade of microRNA processing by Lin28. Science. 2008;320(5872):97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan J, et al. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science. 2012;335(6073):1195–200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bronevetsky Y, et al. Lin28b Regulates Fetal Regulatory T Cell Differentiation through Modulation of TGF-beta Signaling. J Immunol. 2016;197(11):4344–4350. doi: 10.4049/jimmunol.1601070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jafarifar F, et al. Repression of VEGFA by CA-rich element-binding microRNAs is modulated by hnRNP L. EMBO J. 2011;30(7):1324–34. doi: 10.1038/emboj.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136(4):719–30. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jing Q, et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell. 2005;120(5):623–34. doi: 10.1016/j.cell.2004.12.038. [DOI] [PubMed] [Google Scholar]
- 43.El Gazzar M, McCall CE. MicroRNAs distinguish translational from transcriptional silencing during endotoxin tolerance. J Biol Chem. 2010;285(27):20940–51. doi: 10.1074/jbc.M110.115063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kedde M, et al. A Pumilio-induced RNA structure switch in p27-3' UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12(10):1014–20. doi: 10.1038/ncb2105. [DOI] [PubMed] [Google Scholar]
- 45.Bottini S, et al. From benchmarking HITS-CLIP peak detection programs to a new method for identification of miRNA-binding sites from Ago2-CLIP data. Nucleic Acids Res. 2017;45(9):e71. doi: 10.1093/nar/gkx007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ennajdaoui H, et al. IGF2BP3 Modulates the Interaction of Invasion-Associated Transcripts with RISC. Cell Rep. 2016;15(9):1876–83. doi: 10.1016/j.celrep.2016.04.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma F, et al. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-gamma. Nat Immunol. 2011;12(9):861–9. doi: 10.1038/ni.2073. [DOI] [PubMed] [Google Scholar]
- 48.Smith KM, et al. miR-29ab1 deficiency identifies a negative feedback loop controlling Th1 bias that is dysregulated in multiple sclerosis. J Immunol. 2012;189(4):1567–76. doi: 10.4049/jimmunol.1103171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shell SA, et al. Elevated levels of the 64-kDa cleavage stimulatory factor (CstF-64) in lipopolysaccharide-stimulated macrophages influence gene expression and induce alternative poly(A) site selection. J Biol Chem. 2005;280(48):39950–61. doi: 10.1074/jbc.M508848200. [DOI] [PubMed] [Google Scholar]
- 50.Beisang D, et al. Alternative polyadenylation regulates CELF1/CUGBP1 target transcripts following T cell activation. Gene. 2014;550(1):93–100. doi: 10.1016/j.gene.2014.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tian B, et al. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 2005;33(1):201–12. doi: 10.1093/nar/gki158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beaudoing E, et al. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 2000;10(7):1001–10. doi: 10.1101/gr.10.7.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Di Giammartino DC, et al. Mechanisms and consequences of alternative polyadenylation. Mol Cell. 2011;43(6):853–66. doi: 10.1016/j.molcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jia X, et al. The role of alternative polyadenylation in the antiviral innate immune response. Nat Commun. 2017;8:14605. doi: 10.1038/ncomms14605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sandberg R, et al. Proliferating cells express mRNAs with shortened 3' untranslated regions and fewer microRNA target sites. Science. 2008;320(5883):1643–7. doi: 10.1126/science.1155390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liaw HH, et al. Differential microRNA regulation correlates with alternative polyadenylation pattern between breast cancer and normal cells. PLoS One. 2013;8(2):e56958. doi: 10.1371/journal.pone.0056958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas LF, Saetrom P. Single nucleotide polymorphisms can create alternative polyadenylation signals and affect gene expression through loss of microRNA-regulation. PLoS Comput Biol. 2012;8(8):e1002621. doi: 10.1371/journal.pcbi.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Derti A, et al. A quantitative atlas of polyadenylation in five mammals. Genome Res. 2012;22(6):1173–83. doi: 10.1101/gr.132563.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shepard PJ, et al. Complex and dynamic landscape of RNA polyadenylation revealed by PAS-Seq. RNA. 2011;17(4):761–72. doi: 10.1261/rna.2581711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin G, et al. Genome-wide analysis of pre-mRNA 3' end processing reveals a decisive role of human cleavage factor I in the regulation of 3' UTR length. Cell Rep. 2012;1(6):753–63. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 61.Hwang HW, et al. PAPERCLIP Identifies MicroRNA Targets and a Role of CstF64/64tau in Promoting Non-canonical poly(A) Site Usage. Cell Rep. 2016;15(2):423–35. doi: 10.1016/j.celrep.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Edwalds-Gilbert G, Milcarek C. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol Cell Biol. 1995;15(11):6420–9. doi: 10.1128/mcb.15.11.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lassman CR, et al. Plasma cell-regulated polyadenylation at the Ig gamma 2b secretion-specific poly(A) site. J Immunol. 1992;148(4):1251–60. [PubMed] [Google Scholar]
- 64.Saunders MA, et al. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007;104(9):3300–5. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.MacArthur J, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45(D1):D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jarret A, et al. Hepatitis-C-virus-induced microRNAs dampen interferon-mediated antiviral signaling. Nat Med. 2016;22(12):1475–1481. doi: 10.1038/nm.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kulkarni S, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472(7344):495–8. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McFarland AP, et al. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs. Nat Immunol. 2014;15(1):72–9. doi: 10.1038/ni.2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Incarnato D, Oliviero S. The RNA Epistructurome: Uncovering RNA Function by Studying Structure and Post-Transcriptional Modifications. Trends Biotechnol. 2017;35(4):318–333. doi: 10.1016/j.tibtech.2016.11.002. [DOI] [PubMed] [Google Scholar]
- 70.Long D, et al. Potent effect of target structure on microRNA function. Nat Struct Mol Biol. 2007;14(4):287–94. doi: 10.1038/nsmb1226. [DOI] [PubMed] [Google Scholar]
- 71.Meisner NC, et al. mRNA openers and closers: modulating AU-rich element-controlled mRNA stability by a molecular switch in mRNA secondary structure. Chembiochem. 2004;5(10):1432–47. doi: 10.1002/cbic.200400219. [DOI] [PubMed] [Google Scholar]
- 72.Chen JM, et al. A systematic analysis of disease-associated variants in the 3' regulatory regions of human protein-coding genes II: the importance of mRNA secondary structure in assessing the functionality of 3' UTR variants. Hum Genet. 2006;120(3):301–33. doi: 10.1007/s00439-006-0218-x. [DOI] [PubMed] [Google Scholar]
- 73.Wan Y, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505(7485):706–9. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lucks JB, et al. Multiplexed RNA structure characterization with selective 2'-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc Natl Acad Sci U S A. 2011;108(27):11063–8. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505(7485):696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 76.Rouskin S, et al. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature. 2014;505(7485):701–5. doi: 10.1038/nature12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Flynn RA, et al. Transcriptome-wide interrogation of RNA secondary structure in living cells with icSHAPE. Nat Protoc. 2016;11(2):273–90. doi: 10.1038/nprot.2016.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Underwood JG, et al. FragSeq: transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods. 2010;7(12):995–1001. doi: 10.1038/nmeth.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramani V, et al. High-throughput determination of RNA structure by proximity ligation. Nat Biotechnol. 2015;33(9):980–4. doi: 10.1038/nbt.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heng TS, et al. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol. 2008;9(10):1091–4. doi: 10.1038/ni1008-1091. [DOI] [PubMed] [Google Scholar]