Abstract
PHYTOCHROME INTERACTING FACTORs (PIFs) are members of the basic helix-loop-helix (bHLH) family of transcription factors in Arabidopsis. Since their discovery in phytochrome-mediated light signaling pathways, recent studies have unraveled new functions of PIFs in integrating multiple signaling pathways not only by their role as transcription factors directly targeting gene expression, but also by interacting with diverse groups of factors to optimize plant growth and development. These include endogenous (e.g., hormonal) as well as abiotic (light, circadian and elevated temperature) and biotic (defense responses) pathways. PIFs interact with key factors in each of these pathways, and tailor the outcome of the signal integration among these pathways. This review summarizes the role of PIFs as pivotal signal integrators in regulating plant growth and development.
Keywords: bHLH transcription factor, circadian clock, growth-defense tradeoff, hormone signaling, phytochrome-interacting factor, thermomorphogenesis, signal integration
INTRODUCTION
Plants continually adapt to natural light environments to optimize their growth and development. The information of surrounding light conditions is monitored and perceived by several classes of photoreceptors, and the light signals are eventually transduced to the transcriptional network that drives multiple facets of photomorphogenesis (Bae and Choi, 2008). Among the transcription factors that regulate light signaling pathways, phytochrome-interacting factors (PIFs) are well-characterized. PIFs are basic helix-loop-helix (bHLH) transcription factors, belonging to the fifteen-member of subgroup 15 among 162 members of the bHLH protein family in Arabidopsis thaliana (Bailey et al., 2003; Khanna et al., 2004; Toledo-Ortiz et al., 2003). To date, 7 of the 15 members in the subgroup have been shown to interact with at least one of the phytochromes (phy), known as PIFs including PIF1 (also referred to PIL5, At2g20180), PIF2 (PIL1, At2g46970), PIF3 (At1g09530), PIF4 (At2g43010), PIF5 (PIL6, At3g59060), PIF6 (PIL2, At3g62090), PIF7 (At5g61270), and PIF8 (At4g00050), whereas other members do not have any phytochrome-binding motif [see the references in a recent review (Lee and Choi, 2017)].
Since the discovery of PIF3, the founding member that negatively regulates phytochrome-mediated light signaling (Kim et al., 2003; Monte et al., 2004; Ni et al., 1998), PIFs have been described as central players in transducing light signals perceived by phytochromes (Castillon et al., 2007; Leivar and Quail, 2011). However, further studies have shown that PIFs are also involved in many other signaling pathways, such as thermal-induced responses, circadian clock or hormonal signaling, developmental and sugar-derived signaling, and responses to biotic and abiotic stresses. Tremendous progress has been made recently in dissecting phytochrome-PIF signaling interface by demonstrating new evidence that phytochrome acts as a kinase, identifying new kinases for PIFs, and understanding mechanistic details on PIF stability and their DNA binding ability. However, this review primarily focuses on the expanding roles of PIFs as signal integrators in plant growth and development by highlighting recent advances on the function of PIFs in regulating multiple processes. For details on PIFs role in phytochrome signaling, readers are directed to several recent research and review articles (Kim et al., 2016a; Kim et al., 2016b; Lee and Choi, 2017; Leivar and Monte, 2014; Leivar and Quail, 2011; Ni et al., 2017; Shin et al., 2016; Xu et al., 2015).
PIFs FUNCTION IN CIRCADIAN CLOCK INPUT AND OUTPUT PATHWAYS
Light is known as a crucial input to the circadian clock of living organisms. Light is perceived by a series of photoreceptors in plants, thereby making plants fit to changing environmental conditions. Since photoreceptors are responding at the forefront to the incoming light, it is reasonable that light input to the circadian clock is relayed by these photoreceptors. For example, phytochromes and cryptochromes (CRY) are responsible for the light entrainment of the circadian clock in A. thaliana, resulting in long period phenotypes in these photoreceptor mutants (Somers et al., 1998). As many studies continue to report the function of PIFs in photomorphogenesis and other signaling pathways, it is feasible that PIFs are also involved in regulation of the circadian clock. Central clock components CCA1/LHY, MYB transcription factors, contain G-box in their promoters which is known as the cis-element for binding of PIFs. Recent reports showed a direct in vivo and in vitro association of PIFs with the G-box element on CCA1/LHY promoters by chromatin immunoprecipitation (ChIP) and gel-shift assays (Martinez-Garcia et al., 2000; Oh et al., 2012). This indicates that PIFs might also have a role in the direct input to the clock. However, a role for PIFs in the Arabidopsis circadian clock has been elusive mainly because pif single and double mutants did not show any changes in the circadian clock (Nusinow et al., 2011; Viczian et al., 2005).
Strikingly, a recent study showed that pifQ (pif1, pif3, pif4, pif5 quadruple) mutant displays longer period compared to wild type in the presence of sucrose (Shor et al., 2017). This result appears to be inconsistent with the general roles of PIFs as negative regulators of the light signaling pathways, given that phytochrome mutants exhibit long period phenotype due to a lack of red light entrainment to the circadian clock (Somers et al., 1998). Thus, if PIFs are to relay photo-signal to the circadian clock, a short period phenotype is expected in the pifQ mutant. Therefore, the long period phenotype shown in pifQ suggests a lack of direct relationship between PIFs and the light input to the circadian clock. Instead, this study showed that metabolic input to the clock was impaired in the pifQ mutant and the longer period phenotype was suppressed by the inhibition of photosynthesis or lack of exogenously supplied sugar. More interestingly, the addition of sucrose significantly enhanced the binding of PIFs to the CCA1/LHY promoters (Figure 1). The photosynthesis-derived sugar can entrain the Arabidopsis circadian clock by suppressing a morning gene, PRR7 (Haydon et al., 2013). However, PIFs do not bind directly to the promoter of PRR7, but bind to the promoters of CCA1/LHY (Figure 1). These data suggest that PIFs mediate metabolic input signal to the clock independent of PRR7 expression, highlighting a unique role of PIFs in the sugar-mediated entrainment of the Arabidopsis circadian clock. However, the detailed mechanisms on how sugar enhances the binding of PIFs on the target promoters remain to be answered.
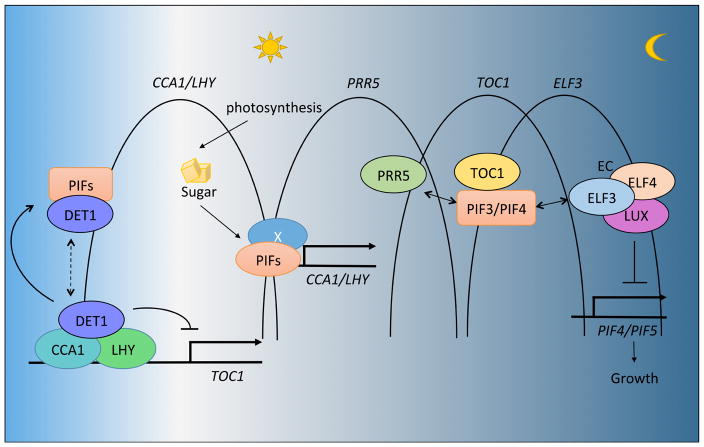
Figure 1.
Dynamic involvement of PIFs in regulating circadian clock and diurnal growth in Arabidopsis.
PIFs have been shown to participate in both the input and output pathways of circadian clock. (Left) DET1 forms a complex with CCA1/LHY to repress TOC1 expression in the morning. DET1 also directly interacts with and stabilizes PIFs in the dark. PIFs might form a complex with DET1-CCA1-LHY to form a transcription complex as indicated by a dotted line. (Middle) PIF1, PIF3, PIF4 and PIF5 are necessary to mediate metabolic signaling to the clock by directly binding to the CCA1/LHY promoters in response to sugar. X, indicates an unknown factor or sugar-induced modification necessary for enhanced PIF binding to the CCA1/LHY promoters. (Right) Sequential expression of PRR5 and TOC1 gates the growth by inhibiting PIF functions, while ELF3-ELF4-LUX forms an evening complex (EC) that represses PIF4/PIF5 expression to repress growth during early evening.
Recently, a number of reports suggested a role for PIFs in the diurnal regulation of growth in association with circadian clock factors. For example, the central circadian clock component TOC1 (TIMING OF CAB EXPRESSION 1) that suppresses the expression of CCA1 (CIRCADIAN CLOCK-ASSOCIATED 1) and LHY (LATE ELONGATED HYPOCOTYL) was shown to interact with PIF3 in yeast (Yamashino et al., 2003). This interaction has been recently confirmed and analyzed in planta (Soy et al., 2016). These authors showed that the physical interaction between PIFs and TOC1 results in ‘coincident co-binding’ to the promoters of dawn-phased genes under diurnal condition to optimize their expression (Figure 1). TOC1 inhibits the transcriptional activity of PIF3, thus TOC1-PIF3 interaction establishes a growth inhibition at early dusk and gates growth to predawn. Same group further analyzed genome wide gene expression under short day (SD) conditions (Martin et al., 2016). They identified a total of 349 PIFs- and SD-coregulated genes, among which 55% were induced and 42% were repressed by both SD and PIFs. These data suggest that PIF3 and possibly other PIFs are tightly interconnected with the circadian clock output through direct interaction with TOC1 to co-regulate the expression of target genes. In addition, a recent paper also reported direct interactions between TOC1/PRR5 (PSEUDO-RESPONSE REGULATOR 5) and PIF4, which results in TOC1/PRR5-mediated suppression of PIF4 transcription activity (Figure 1) (Zhu et al., 2016). These data again suggest a direct association of the circadian clock components with PIFs in Arabidopsis. Furthermore, the expression of PIF4 and PIF5 is regulated by circadian clock (Nozue et al., 2007; Yamashino et al., 2003). The transcription of PIF4 and PIF5 is repressed by the evening complex, ELF3 (EARLY FLOWERING 3)-ELF4 (EARLY FLOWERING 4)-LUX (LUX ARRHYTHMO, also known as PHYTOCLOCK 1), thus promoting cell elongation in a time-dependent manner at late night (Figure 1) (Nusinow et al., 2011).
It is notable that DET1 (DE-ETIOLOATED 1) possesses a transcriptional repression activity and directly associates with CCA1/LHY in vivo, thereby regulating TOC1 expression (Figure 1) (Lau et al., 2011). In a separate study, DET1 has been shown to directly interact with PIFs and stabilize them in the dark by unknown mechanisms (Dong et al., 2014). Thus, it is possible that PIFs are located in the same complex with CCA/LHY/DET1. However, further studies are necessary to examine any possible role of PIFs on the CCA1/LHY/DET1 transcriptional complex. Taken together, the circadian clock and PIFs have a mutual regulatory relationship to optimize growth and development of plants.
PIFs PLAY A CENTRAL ROLE IN THE THERMAL-INDUCED MORPHOGENESIS
Temperature is a major environmental cue that influences the distribution and seasonal responses of plants. Recent trend in increasing global temperature is likely to have an adverse effect on plant growth and development, resulting in decreased crop yields (Battisti and Naylor, 2009; Lobell and Gourdji, 2012; Quint et al., 2016). Thus, more attention has recently been given on thermal-induced morphogenesis, termed as thermomorphogenesis which is characterized by elongated hypocotyls and petioles, narrow leaves and accelerated flowering (Quint et al., 2016). Among PIFs, PIF4 has been reported as a key regulator of thermomorphogenesis in plants (Franklin et al., 2011; Koini et al., 2009; Sun et al., 2012). PIF4 was initially identified as a bHLH factor that negatively regulates phytochrome B (phyB) signaling (Huq and Quail, 2002). Later, it was found that PIF4 is also responsible for the hypocotyl elongation in response to high ambient temperature, in which high temperature regulates the transcription and posttranslational stabilization of PIF4. In addition, PIF4 promotes flowering at high temperature by directly binding to the promoter of the florigen, FT (Flowering Locus T) (Kumar et al., 2012).
The phenotypic similarities between thermomorphogenesis and shade avoidance response are striking. Both responses are characterized by a rapid and dramatic increase in the extension growth of stems and petioles at the expense of leaf growth, and reproductive development (Legris et al., 2017; Quint et al., 2016). Although, plants continue to monitor and respond to the changing environmental cues, especially light signals, light always coincides with the heat radiation emitted by the sun in nature. Therefore, it is not surprising that changes in plant morphology in response to high ambient temperature and by vegetation shade are very similar. Consistent with this, a light receptor, phyB has been identified as a plant thermosensor, as it undergoes thermal reversion, i.e., thermal reversion of the active Pfr form to an inactive Pr form (Figure 2) (Jung et al., 2016; Legris et al., 2016). This phenomenon is also more commonly referred to as “dark reversion”, although “thermal reversion” can occur under both dark and light conditions. As the active form of phyB inhibits PIF4 activity and also triggers rapid phosphorylation and degradation of PIF4 through ubiquitin-proteasome system (Leivar and Quail, 2011), it is possible that the high temperature-mediated thermal reversion of phyB results in enhanced PIF4 stability and activity to trigger thermomorphogenic responses. Similar to phyB, the blue light receptor CRY1 (Cryptochrome 1) has been shown to regulate PIF4 activity (Ma et al., 2016) (Figure 2). CRY1 directly interacts with PIF4 in a blue light dependent manner and inactivates PIF4 transcription activity. The blue light, therefore, is able to suppress thermomorphogenesis by suppressing PIF4 activity.
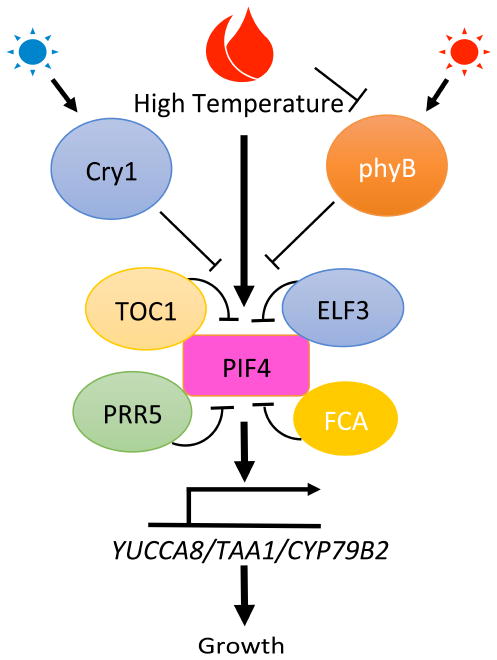
Figure 2.
PIF4 plays a central role in integrating light and high temperature signaling to promote growth.
High ambient temperature induces PIF4 expression and also stabilizes PIF4 protein. Multiple regulators have been shown to directly interact with PIF4 and regulate its function by inhibiting DNA binding, transcription activity and protein stability. While two photoreceptors, phyB and Cry1 function to inhibit PIF4 activity in response to red and blue light signals, respectively, only phyB has been shown to act as a thermosensor in Arabidopsis. PIF4 directly activates the expression of YUC/TAA1/CYP79B2 genes to promote growth.
Multiple factors have been identified that modulate the function of PIF4 in the context of thermomorphogenesis. For example, ELF3, as part of a component in the evening complex of circadian clock, has been shown to suppress the transcription of PIF4 and PIF5 (Nusinow et al., 2011). In addition, ELF3 also directly binds to PIF4 (Nieto et al., 2015), which prevents PIF4 from activating its transcriptional targets (Figure 2). Interestingly, ELF3 itself was shown to be necessary for temperature-mediated control of plant growth by targeting multiple loci in temperature-dependent manner (Box et al., 2015). FCA (FLOWERING TIME CONTROL PROTEIN) was found as another PIF4 regulator in thermomorphogenesis (Figure 2) (Lee et al., 2014). FCA is a RNA-binding protein that functions in autonomous flowering pathway to suppress FLC (FLOWERING LOCUS C) expression (Macknight et al., 1997). Notably, it was shown that FCA also mediates high ambient temperature-driven early flowering in Arabidopsis (Blázquez et al., 2003; Lee et al., 2014). In response to high temperature, FCA binds to PIF4, which dissociates PIF4 from its direct target gene YUC8 (YUCCA8) to suppress hypocotyl elongation. Moreover, a recent study has identified TOC1/PRR5 as PIF4 inhibitors (Figures 1 and 2) (Zhu et al., 2016). TOC1/PRR5 peaks in a sequential manner during light/dark cycle, so these authors hypothesize that the inhibition by TOC1/PRR5 controls circadian gating of PIF4 in thermomorphogenesis. It is notable that the circadian gating of thermomorphogenesis enables plants to respond to external temperatures differently at different times of the day. TOC1 peaks in the evening so that it can prevent overgrowth of plants by high temperature during evening. PRR5 peaks in the afternoon thus the sequential expression of PRR5/TOC1 ensures tight gating of plant growth in response to high ambient temperature (Figures 1 and 2). Taken together, PIF4 plays a central role as a key regulator in controlling thermomorphogenesis, and its activity is fine-tuned by multiple factors in a time-dependent manner.
Despite these extensive reports, there are still many important questions yet to be answered. What is/are the upstream receptor(s) that perceive high ambient temperature and ultimately induce PIF4 expression? Recently, phyB has been shown to be one of the thermosensors. However, additional thermosensor(s) must be present, as phyB alone can’t explain all the phenotypic changes associated with thermomorphogenesis. In addition, what makes PIF4 to function uniquely in high temperature? What are the differences between PIF4 and other PIFs that uniquely position PIF4 as a central thermal transcription factor? Therefore, further studies are necessary to answer these questions.
PIFs INTEGRATE LIGHT AND HORMONE SIGNALING PATHWAYS
Plant hormones play pivotal roles in modulation of plant growth and development, as well as plant responses to external factors, including biotic and abiotic stresses. Plant hormones are essentially small chemical compounds derived from tightly regulated metabolic pathways. Several plant hormones have been identified and their biosynthetic and signaling pathways have been extensively characterized (Santner and Estelle, 2009). The linear signaling pathways for each of the hormones have been extensively characterized through forward genetics approaches. However, in recent years much of the research on hormone signaling has been focused on the interconnections that exist among different hormones, and also between hormones and external environmental signals, including light and temperature (Jaillais and Chory, 2010). These studies highlight the importance of signaling integrators that connect light signaling with those of hormonal pathways, although it is far from complete (de Wit et al., 2016; Lau and Deng, 2010). PIFs play key roles in signal integration between light and hormone signaling pathways (see below and Figure 3). In response to light quality and quantity, developmental or external cues, PIFs coordinate light and hormone signaling either by regulating the hormone biosynthesis and/or the expression of key components in hormone signal transduction, and/or by directly interacting with components of the hormone signaling pathways (de Wit et al., 2016; Lucas and Prat, 2014). PIFs role in signal integration for each of the hormones with light are briefly discussed below:
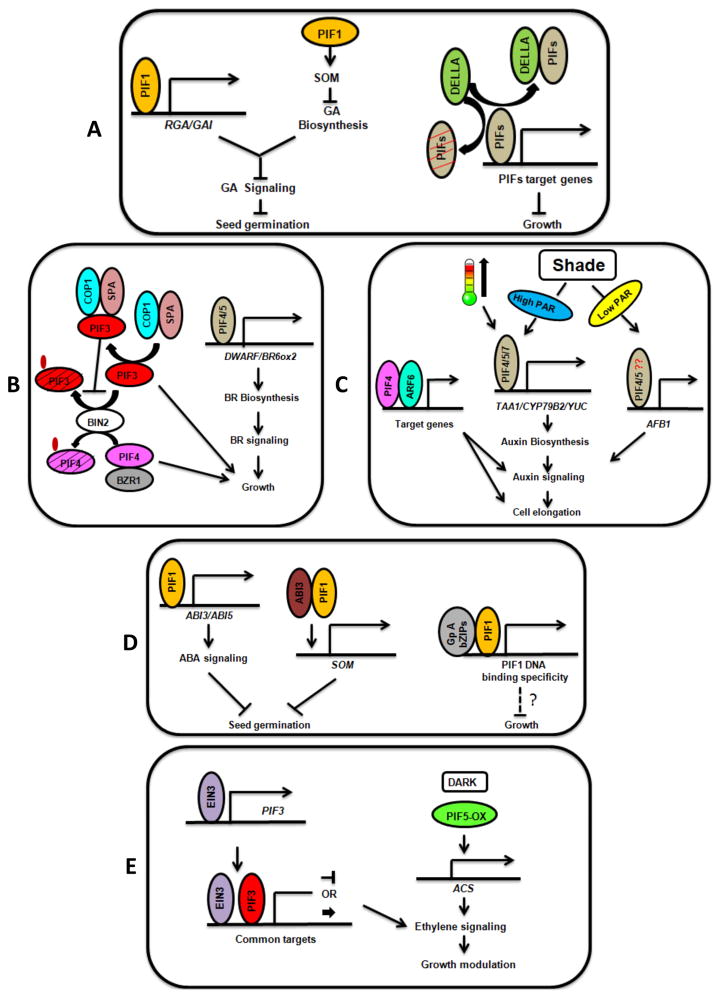
Figure 3.
PIFs integrate light and hormone signaling pathways to modulate growth in Arabidopsis.
(A, left) PIF1 has an exclusive role in regulating seed germination by directly activating RGA and GAI expression that inhibit GA signaling. PIF1 also inhibits GA biosynthesis indirectly by activating SOM expression, resulting in inhibition of seed germination in the dark. Light-induced degradation of PIF1 promotes seed germination. (Right) PIFs physically interact with DELLA proteins and this interaction results in inhibition of DNA binding activities of PIFs. DELLA proteins also induce degradation of PIFs in darkness and inhibit subsequent growth. (B, left) PIF4 interacts with BZR1 and regulates growth in response to BR and light signal. BIN2 phosphorylates PIF3 and PIF4 independent of light and promotes their degradation in darkness. However, COP1/SPA1 interact with PIF3 and prevent BIN2-mediated phosphorylation and degradation in the dark. (Right) PIF4 and PIF5 directly activate BR biosynthetic pathway genes to promote growth. (C, left) PIF4 forms a complex with ARF6 and promotes growth in response to light and auxin signaling. (Middle) In response to high ambient temperature and shade conditions, PIF4/PIF5/PIF7 promote auxin biosynthesis to promote cell elongation. (Right) PIF4/PIF5 and possibly other PIFs also activate the expression of auxin receptor AFB5 in response to low PAR shade conditions to promote auxin signaling and subsequent growth. (D, left) PIF1 directly activates the expression of the ABA signaling components (ABI3 and ABI5). PIF1 also interacts with ABI3 and the PIF1-ABI3 complex directly inhibits the expression of SOM, which in turn inhibits GA biosynthesis to suppress seed germination. (Right) PIF1 and possibly other PIFs directly interact with group A bZIP proteins (e.g., ABI5). This interaction regulates the DNA binding specificity and target gene selection of PIF1 and possibly other PIFs. (E, left) Ethylene signaling factor, EIN3 directly activates the expression of PIF3, which in turn binds to DNA along with EIN3 to regulate ethylene signaling as well as chlorophyll biosynthesis and growth. (Right) Overexpression of PIF5 activates the expression of ACC Synthase (ACS) genes, which results in increased ethylene biosynthesis and signaling to modulate growth.
PIFs mediate light and gibberellic acid (GA) pathways
PIFs and GA (a positive regulator of germination) regulate each other’s signaling pathways at multiple levels throughout the plant life cycle. PIF1, a strong inhibitor of seed germination in the dark, suppresses GA signaling both directly and indirectly. PIF1 directly binds to the promoters of two DELLA genes, RGA1 (REPRESSOR OF GA1-3) and GAI (GIBBERELLIC ACID INSENSITIVE), and activates the expression of these genes that act as suppressors of GA signaling (Figure 3A) (Oh et al., 2007). Moreover, PIF1 also regulates the biosynthesis of GA, but rather indirectly by activating the expression of a number of genes, including DAG1 (DOF AFFECTING GERMINATION 1), SOM (SOMNUS) and GA2ox2 (GIBBERELLIN 2-OXIDASE 2). DAG1 and SOM repress the expression of key GA biosynthetic genes GA3ox1 and GA3ox2, while activate the GA catabolic gene GA2ox2 (Gabriele et al., 2010; Kim et al., 2008). The SOM branch has been further elucidated by showing that SOM directly represses the expression of two Jumonji C (JmjC) domain-containing histone arginine demethylases encoded by JUMONJI 20 (JMJ20) and JMJ22 (Cho et al., 2012). The light-induced degradation of PIF1 results in decreased amount of SOM, which in turn increases the amount of JMJ20 and JMJ22. Removal of methyl groups from the arginine residues of histone H4 on the promoters of GA3ox1 and GA3ox2 results in higher expression of these biosynthetic enzymes promoting GA production and concomitant seed germination.
Reciprocally, DELLAs also regulate the stability and activity of PIFs to regulate photomorphogenesis. Several independent studies demonstrated that the DELLA proteins physically interact with PIFs and sequester them into an inactive complex to restrain PIFs from binding to their targets (de Lucas et al., 2008; Feng et al., 2008; Gallego-Bartolomé et al., 2010). Recently it has been also shown that DELLAs not only sequester PIFs but also induce their degradation (Figure 3A), and more importantly, DELLAs-mediated degradation of PIFs functions independently of well-established phyB/LRBs pathways (Li et al., 2016). Thus, crosstalks between light and GA signaling pathways play an important role in fine-tuning germination and photomorphogenesis for optimal growth and development of plants.
PIFs role in light and brassinosteroids (BRs) pathways
BRs are steroid hormones that play diverse roles in plant growth and responses to external stresses, as well as act in a concert with light signaling pathway principally through PIFs (Lozano-Durán and Zipfel, 2015; Saini et al., 2015). In a study aimed at the identification of components of the light signaling pathway that interact with BZR1, PIF1 and PIF4 were identified as interacting partners of BZR1 (BRASSINAZOLE-RESISTANT 1) (Oh et al., 2012). The BZR1 is a transcription factor that selectively binds to BR response element (CGTG(T/C)G), and functions as both activator and repressor of distinct target genes. It was established that functional PIF4-BZR1 complex co-regulates the expression of both light and BR responsive genes, including PREs (PACLOBUTRAZOL RESISTANCE), which function in growth promotion over immunity (see below). In addition, BIN2 (BRASSINOSTEROID-INSENSITIVE 2), a glycogen synthase kinase 3 (GSK3) family of protein kinase that phosphorylates BZR1 in the BR signaling pathway, has been shown to phosphorylate PIF3 and PIF4. BIN2-mediated phosphorylation of PIF4 leads to the degradation of PIF4 through ubiquitin-proteasome system, and this process is required for diurnal growth of hypocotyls (Figure 3B) (Bernardo-García et al., 2014). Strikingly, COP1/SPA complex interacts with PIF3 and prevents BIN2-mediated phosphorylation of PIF3 in a non-proteolytic manner (Ling et al., 2017). In addition, very recently PIFs were also shown to modulate BR signaling by regulating BR biosynthesis (Wei et al., 2017). PIF4 and PIF5 bind to the promoter regions and induce the expression of DWF4 (DWARF4) and BR6ox2 (BRASSINOSTEROID-6-OXIDASE 2), two key enzymes involved in BR biosynthesis (Figure 3B). Thus, PIFs regulate both BR biosynthesis and signaling pathways to optimize plant growth in response to light and BR.
PIFs function in light and auxin signaling pathways
Auxin is a group of small organic molecules, which regulate almost every aspect of plant growth and development essentially by modulating the cell division and cell elongation (Santner and Estelle, 2009). To regulate growth, plants often modulate either the biosynthesis and/or the sensitivity of auxin signaling (de Wit et al., 2016). Arabidopsis seedlings grown in either shade or high temperature conditions display longer hypocotyls and petioles. However, mutants defective in auxin responses or in light signaling such as pif mutants do not show such phenotypes (Gray et al., 1998; Hornitschek et al., 2012; Tao et al., 2008). In accordance with the above phenotypic differences, pharmacological, molecular and biochemical assays have demonstrated that some of the PIFs (PIF4, 5, and 7) bind to promoter regions and induce the expression of TAA1 (TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS 1), CYP79B2 (CYTOCHROME P450, FAMILY 79, SUBFAMILY B, POLYPEPTIDE 2), and several YUC (YUCCA) genes selectively under shade and/or high temperature conditions (Figure 3C) (Franklin et al., 2011; Li et al., 2012; Sun et al., 2012). Similarly, several auxin signaling genes including AFB1 (AUXIN SIGNALING F-BOX1), IAA19 (INDOLE-3-ACETIC ACID INDUCIBLE 19) and IAA29 were also found to be either direct or indirect targets of PIF4 and PIF5 (Hersch et al., 2014; Hornitschek et al., 2012). Interestingly, alternating roles of PIF4 and PIF5 as modulators of auxin sensitivity or biosynthesis have been observed under low R:FR of different PAR (PHOTOSYNTHETICALLY ACTIVE RADIATION) levels (Hersch et al., 2014). In this study, it was shown that PIF4 and PIF5 under low R:FR of high PAR increase the auxin biosynthesis by directly up-regulating the expression of several auxin biosynthetic genes. However, under low R:FR and low PAR, PIF4 and PIF5 do not upregulate the auxin biosynthesis, rather they collectively enhance the auxin sensitivity, tentatively due to the increased expression of AFB1 auxin co-receptor (Figure 3C).
Adding to the complexity of already known interactions between light and auxin signaling, it was found that PIF4 strongly interacts with ARF6 (AUXIN RESPONSE FACTOR 6) to form a functional complex (similar to PIF4-BZR1) to cooperatively activate several genes in light and auxin signaling pathways (Figure 3C) (Oh et al., 2014). Thus, PIFs regulate both the biosynthesis and sensitivity of auxin signaling to optimize growth in response to light quality, quantity, and duration. However, it is not clear how the PIF4-ARF6 transcription complex selectively binds to target promoters and regulates their expression.
During seedling de-etiolation, light signal inhibits hypocotyl elongation while it promotes the cotyledon expansion. One of the long-standing questions in the field is how the light signaling achieves this contrasting growth patterns in these two organs. In a recent study, it was shown that light oppositely regulates the organ-specific expression of a group of SAUR (Small Auxin Upregulated RNA) genes, named lirSAUR (light-induced in cotyledons and/or repressed in hypocotyls SAURs) (Sun et al., 2016). These genes are direct targets of PIFs: PIFs activate the expression of these genes in the hypocotyls to promote hypocotyl elongation, while inhibit their expression in the cotyledons to repress cotyledon expansion. Although PIFs are equally degraded in both hypocotyls and cotyledons, light-induced degradation of PIFs results in reduced expression of SAURs in the hypocotyls but increased expression in the cotyledons driving the contrasting growth pattern in an organ-specific manner. However, it is still unknown how PIFs activate gene expression in one organ while they inhibit the gene expression in another organ. As the authors suggested, the promoters of these genes might be epigenetically regulated in different organs. Alternatively or in addition to epigenetic changes, organ-specific specificity factors might be necessary to establish this contrasting expression patterns as recently shown for PIF1 (see below) (Kim et al., 2016a).
PIFs mediate light and abscisic acid (ABA) signaling pathways
Similar to GA, BRs, and auxin, the light-signaling pathway is also closely associated with ABA pathway. Unlike complex interactions between light signaling and GA, BR, and auxin signaling pathways, molecular interactions with ABA are relatively less complex (de Wit et al., 2016; Yu and Huang, 2017). In imbibed seeds incubated in darkness, PIF1 binds to the promoter regions and activates the transcription of ABA signaling factors, including ABI3 (ABA INSENSITIVE 3) and ABI5 (Figure 3D). These transcription factors not only enhance the ABA biosynthesis and signaling, but also repress GA signaling to inhibit the germination (Oh et al., 2009; Park et al., 2011). In addition, PIF1 physically interacts with ABI3 in imbibed seeds to cooperatively regulate the expression of SOM, a negative regulator of seed germination (Park et al., 2011). Moreover, the group-A bZIP proteins including ABI5 directly interact with PIF1 and modulate the DNA binding specificity of PIF1 (Kim et al., 2016a). However, this study does not demonstrate the biological significance of the modulation of DNA binding specificity of PIF1. Thus, although PIF1 in cooperation with components of ABA signaling controls the developmentally crucial phase of seed germination in response to endogenous and external cues, further studies are necessary to understand how the group-A bZIP transcription factors including the ABA signaling factors modulate the DNA binding specificity of PIFs to regulate seed germination and seedling establishment.
PIFs function in light and ethylene signal integration
The interactions between light and ethylene pathways has been extensively reviewed recently (Yu and Huang, 2017). Similar to other hormones, PIFs mediate interactions between light and ethylene pathway by regulating both biosynthesis and signaling of ethylene responses. For ethylene biosynthesis, overexpression of PIF5, but not other PIFs, has been shown to upregulate the expression of members of the ACS (1-aminocyclopropane-1-carboxylic acid (ACC) SYNTHASE) genes (Figure 3E), resulting in an increased production of ethylene in the dark (Khanna et al., 2007). Therefore, PIF5 overexpression lines display triple responses, characteristic of either ethylene overproduction or constitutively active ethylene signaling. However, the study could not confirm whether the endogenous PIF5 regulates ethylene biosynthesis as pif5 mutant neither displayed defects in ACS gene expression nor ethylene-related phenotypes. This is a typical complicacy with overexpression studies for transcription factors that are able to homo- and hetero-dimerize. It is still possible that PIF5 might regulate ethylene biosynthesis in a tissue or cell-type specific manner, which might have been masked in this study due to the use of whole seedlings. Alternatively, PIF5 might interact with other bHLH proteins that regulate ethylene biosynthesis. Thus, overexpression of PIF5 may simply show a dominant negative phenotype due to the titration of other bHLH proteins.
For interaction with ethylene signaling, PIFs have been shown to genetically interact with ethylene signaling factors. For example, ethylene signaling stabilizes the downstream transcription factors EIN3/EIL1 (ETHYLENE INSENSITIVE 3 and EIN3-like 1) that induce the expression of PIF3 in the cotyledon of growing seedlings (Figure 3E) (Zhong et al., 2014). EIN3/EIL1 also activates ERF1 (ETHYLENE RESPONSE FACTOR 1) in the hypocotyls to inhibit hypocotyl elongation. Thus, EIN3/EIL1 coordinately regulate the expression of PIF3 in the cotyledons and ERF1 in the hypocotyls to prevent photo-oxidative damage of seedlings emerging from subterranean darkness. Recently, PIFs and EIN3 have been shown to mediate transcriptional co-regulation of common target genes. In this study, PIFs and EIN3 did not physically interact with each other. However, they bind to several common target promoters and regulate their expression in the same direction either interdependently or additively (Figure 3E) (Jeong et al., 2016). The authors performed ChIP assays to examine interdependency between PIF4 and EIN3. However, instead of using the mutant backgrounds, the authors used dark vs. light (assuming PIFs will be degraded under light) or presence of exogenously added Ag+ or ACC (mimicking ⩲ ethylene). Under these conditions, they did not observe any interdependent DNA binding in vivo. Thus, these data are inconclusive whether PIFs and EIN3 can bind to DNA interdependently in vivo. Further studies are necessary to understand how PIF3 and EIN3 target downstream common targets.
PIFs INVOLVE IN GROWTH-DEFENSE TRADEOFFS
Plant growth is plastic throughout its life cycle and displays remarkable adaptation in response to biotic and abiotic threats. Plants growing under less external stress show a robust growth with relatively high accumulation of biomass before they enter into the reproductive stage. On the contrary, under sub-optimal conditions, plants not only slow down the rate of growth but also accelerates the developmental transition from vegetative to reproductive growth resulting in significantly lower biomass accumulation and fewer seed production (Ballaré, 2014; Zust and Agrawal, 2017).
In recent years, PIFs have been shown to govern both growth and defense responses in plants (Leivar and Monte, 2014). Recent studies have shown that the loss-of-function mutation or ectopic expression of one or more of the eight closely related PIFs severely alters the fine balance between growth-defense tradeoffs in plants (Campos et al., 2016; Gangappa et al., 2017; Yang et al., 2012). Therefore, plants carefully regulate the level of active PIFs through multiple mechanisms including transcriptional regulation and post-transcriptional regulation via ubiquitin-proteasome-mediated degradation, heterodimerization-mediated protein sequestration, and probably other yet unknown mechanisms (Lucas and Prat, 2014; Xu et al., 2017; Xu et al., 2015; Yu and Huang, 2017; Zhu et al., 2015). One of the classical examples of growth-defense tradeoffs in plants occurs during the pathogen attack. To spread the disease rapidly, pathogens typically promote the plant growth by altering their signaling networks, while plants tend to reduce growth by repressing several growth promoting genes including, PIFs to limit the spread the disease (Figure 4A) (Windram et al., 2012).
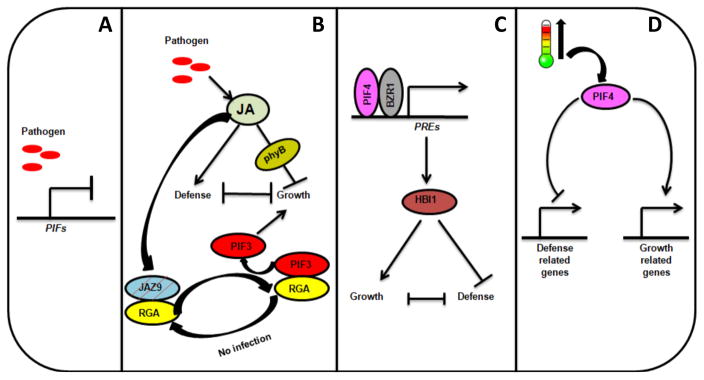
Figure 4.
PIFs regulate growth-defense trade-offs in Arabidopsis.
Plants use multiple mechanisms involving PIFs as core factors to fine-tune growth in response to pathogen attacks. (A) In response to pathogen attack, plants down-regulate the expression of PIFs to suppress growth. (B) To counter the pathogen attacks, plants activate the JA signaling which promotes defense responses, while suppressing growth by modulating the functions of light signaling components. Activated JA signaling promotes the degradation of JAZ9, releasing RGA from JAZ9-RGA complex, which in-turn forms another complex with PIF3 to inactivate PIF3 activity. Secondly, activated JA signaling genetically interacts with phyB to repress plant growth. (C) PIF4 interacts with BZR1 to form a complex and collectively activates the expression of PREs. PREs in turn indirectly activate HBI1. HBI1 promotes growth, while suppresses defense signaling in the absence of pathogen challenge. (D) High temperature-activated PIF4 actively promotes growth by upregulating the growth-related genes and simultaneously represses the defense responses by suppressing the expression of genes involved in plant defense.
Jasmonic acid (JA) regulates plant growth and development as well as plant responses to biotic and abiotic stresses. JA elicits the defense signaling by promoting the degradation of JAZ (JASMONATE-ZIM DOMAIN) proteins, i.e., repressors of JA-responsive genes involved in defense signaling (Campos et al., 2016). A recent study demonstrated the molecular link through which JA participates in plant growth-defense tradeoff (Yang et al., 2012). JAZ9, one of the JAZ proteins, inhibits the interaction between RGA (a DELLA repressor protein) and PIF3. This study suggests that, in plants growing under near-optimal growth conditions, JAZ9 levels are high due to low amounts of JA, which releases PIF3 from RGA-PIF3 complex, resulting in growth promotion. On the contrary, in response to pathogen attacks, plants activate JA signaling, leading to enhanced degradation of JAZ9, which in-turn stabilizes RGA-PIF3 interaction and sequesters PIF3 away from target genes (Figure 4B). Consistently, JA treatment activates defense responses and represses the growth, while overexpression of PIF3 partially suppresses JA-induced inhibition of growth. Therefore, JA signaling indirectly regulates the growth-defense tradeoff via PIF3.
It is interesting to note that PIF3’s primary interacting partner phyB is also involved in the growth-defense tradeoff (Figure 4B). It is widely accepted that the defense responses against pathogen attacks and plant growth are coupled processes, occurring primarily due to the partitioning of photoassimilates (Zust and Agrawal, 2017). Accordingly, healthy plants are believed to proportionate nutrients and energy requirements to balance the active growth and basal level of immunity for possible pathogen attacks. Nevertheless, under the pathogen attacks, plants divert most of the nutrients and energy from growth towards the defense, leading to relatively stunted growth (Huot et al., 2014). However, the above assumption was shown to be untrue, at least partially, in a recent study. It has been demonstrated that the JA-mediated growth-defense tradeoff is rather uncoupled in jazQ phyB (jaz1/3/4/9/10 phyB) mutant (Campos et al., 2016), as it shows physiological traits of constitutively heightened defense responses as well as an active growth. Such an enhanced defense and an active growth was absent from either jazQ (enhanced resistance but stunted growth) or phyB (active growth but diminished resistance), concluding that at least the JA-mediated signaling network, which is activated in response to pathogen attack, attenuates the growth signaling. It is possible that the elevated level of PIFs in the jazQ phyB mutant might have contributed to the uncoupling of the growth-defense tradeoff in this background.
Seedlings growing under darkness exhibit rapid growth and low level of immunity against plant pathogens (Roden and Ingle, 2009). PIFs, which are relatively stable in the dark, activate the transcription of growth-promoting genes. Nevertheless, experimental evidences gathered in recent studies suggest that PIF4 regulates growth-defense tradeoff in the dark indirectly by increasing the activity of HBI1 (HOMOLOG OF BEE2 INTERACTING WITH IBH1) (Lozano-Durán and Zipfel, 2015). HBI1 is a bHLH protein that suppresses the defense signaling, while promoting an active growth (Fan et al., 2014). In darkness, PIF4 interacts with BZR1 and the PIF4-BZR1 complex cooperatively activates the expression of PREs (PACLOBUTRAZOL RESISTANCE) (Oh et al., 2012; Zhang et al., 2009). PREs interact with IBH1 (ILI1 BINDING BHLH 1; a protein that inactivates HBI1 upon interaction) and inhibit the activity of IBH1, thus promoting the activity of HBI1 resulting in reduced defense response (Figure 4C).
Plants growing at higher than ambient temperature display rapid growth of multiple organs, however, at the cost of relatively diminished immunity against pathogens (Gangappa et al., 2017). As discussed above, a number of studies in the past decade have demonstrated the involvement of PIF4 as one of the crucial components in high temperature-mediated growth (Franklin et al., 2011; Koini et al., 2009; Sun et al., 2012). However, only very recently it was shown that PIF4 signaling not only modulates growth, but also controls the plant defense responses under elevated temperature (Gangappa et al., 2017). Nucleotide-binding and leucine-rich repeat (NB-LRR) proteins like SNC1 (suppressor of npr1-1, constitutive 1) are important modulators of temperature-sensitive plant defense responses. The snc1-1 mutant displays constitutive activation of immune responses and severe growth defects under ambient temperature, but not at higher temperature (Zhu et al., 2010). The recent study showed that the temperature-induced suppression of defense responses in snc1-1 is nullified in snc1-1 pif4 double mutant background (Gangappa et al., 2017). They also showed that the expression of a number of defense-related genes is significantly increased in the pif4 mutant, while expression of growth-related genes decreased significantly. On the contrary, overexpression of PIF4 behaves oppositely, indicating that PIF4 indeed modulates growth-defense tradeoff under higher temperature (Figure 4D). Interestingly, they also found that overexpression of PIF4 lacking the basic domain (PIF4Δb) functions as a dominant negative, as it significantly downregulates the expression of growth-associated genes, while upregulating defense-responsive genes. However, the major drawback of the study is the use of PIF4Δb itself. It is a well-known that PIF4 forms heterodimers with other PIFs and HLH proteins through its HLH domain. Hence, overexpression of PIF4Δb might result in excessive sequestration of other PIFs and HLH proteins through heterodimerization, which in-turn might have caused some of the genetic and physiological responses they observed in PIF4Δb transgenic plants. Therefore, further studies are necessary to understand how PIF4 and/or other PIFs regulate these responses.
CONCLUSIONS AND FUTURE PERSPECTIVES
PIFs have expanded their horizon from central phytochrome signaling components to crucial signal integrators of multiple signaling pathways regulating plant growth and development. They do so by expanding their repertoire of genetic and physical interactions with components of multiple pathways. Although a considerable amount of information about signal integration by PIFs has been learned in recent years, we are still at an early stage of our understanding of the molecular details. A comprehensive analysis of PIF-interactome might uncover new roles of PIFs in signal integration from other pathways. Our knowledge about the known physical interactions of PIFs is also rudimentary. Apart from DELLA-mediated inhibition of PIF-DNA binding, the significance of PIFs interactions with ARF6 and BZR1 is only at a genetic level. How these transcription factors directly interact with DNA? The fact that PIFs DNA binding ability is facilitated by the bZIP and other transcription factor families, suggests a cooperative DNA binding. Thus, PIF4 and BZR1 might form a complex and bind to DNA as heterotetramer. But PIF4-BZR1-ARF6 might form a complex on the DNA with neighboring DNA binding sites facilitating each other’s binding. PIFs also function as an activator in one organ while as a repressor in another organ. It is still unknown whether PIFs achieve this bifunctional transcriptional regulation by interacting with other transcriptional coregulators and/or by interacting with chromatin modifying enzyme complexes to open up chromatin for activation, while close chromatin for repression.
In recent years, PIFs have been discovered from Physcomitrella to higher plants including a variety of crop plants. A major future challenge will be to transfer the knowledge gained from model to crop plants to produce genetically tailored plants that can grow under adverse climate (e.g., elevated temperature) with increased biomass and yield. In summary, due to their extensive networking capabilities, PIFs hold a great potential for future biotechnological applications.
Acknowledgments
FUNDING
We acknowledge support by grants from the National Institute of Health (NIH) (1R01 GM-114297), National Science Foundation (MCB-1543813), U.S.-Israel Binational Science Foundation (BSF#2015316) to E.H, and Rural Development Administration, Republic of Korea (PJ01104001) to J.-I.K.
We thank members of the Huq laboratory for critical reading of this manuscript. Due to space constraints, we would like to apologize for the work from other colleagues that could not be discussed. No conflict of interest declared.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bae G, Choi G. Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol. 2008;59:281–311. doi: 10.1146/annurev.arplant.59.032607.092859. [DOI] [PubMed] [Google Scholar]
- Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim M, Jakoby M, Werber M, Weisshaar B. Update on the Basic/Helix-Loop-Helix Transcription Factor Gene Family in Arabidopsis thaliana. The Plant Cell. 2003;15:2497–2501. doi: 10.1105/tpc.151140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL. Light regulation of plant defense. Annual Review of Plant Biology. 2014;65:335–363. doi: 10.1146/annurev-arplant-050213-040145. [DOI] [PubMed] [Google Scholar]
- Battisti DS, Naylor RL. Historical warnings of future food insecurity with unprecedented seasonal heat. Science. 2009;323:240–244. doi: 10.1126/science.1164363. [DOI] [PubMed] [Google Scholar]
- Bernardo-García S, de Lucas M, Martínez C, Espinosa-Ruiz A, Daviere JM, Prat S. BR-dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes & Development. 2014;28:1681–1694. doi: 10.1101/gad.243675.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blázquez MA, Ahn JH, Weigel D. A thermosensory pathway controlling flowering time in Arabidopsis thaliana. Nature genetics. 2003;33:168–171. doi: 10.1038/ng1085. [DOI] [PubMed] [Google Scholar]
- Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, Webb AA. ELF3 controls thermoresponsive growth in Arabidopsis. Current biology. 2015;25:194–199. doi: 10.1016/j.cub.2014.10.076. [DOI] [PubMed] [Google Scholar]
- Campos ML, Yoshida Y, Major IT, de Oliveira Ferreira D, Weraduwage SM, Froehlich JE, Johnson BF, Kramer DM, Jander G, Sharkey TD. Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nature Communications. 2016;7:12570. doi: 10.1038/ncomms12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillon A, Shen H, Huq E. Phytochrome Interacting Factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007;12:514–521. doi: 10.1016/j.tplants.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Cho JN, Ryu JY, Jeong YM, Park J, Song JJ, Amasino RM, Noh B, Noh YS. Control of seed germination by light-induced histone arginine demethylation activity. Developmental cell. 2012;22:736–748. doi: 10.1016/j.devcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- de Lucas M, Davière JM, Rodríguez-Falcón M, Pontin M, Iglesias-Pedraz JM, Lorrain S, Fankhauser C, Blaezquez MA, Titarenko E, Prat S. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- de Wit M, Galvão VC, Fankhauser C. Light-mediated hormonal regulation of plant growth and development. Annual review of plant biology. 2016;67:513–537. doi: 10.1146/annurev-arplant-043015-112252. [DOI] [PubMed] [Google Scholar]
- Dong J, Tang D, Gao Z, Yu R, Li K, He H, Terzaghi W, Deng XW, Chen H. Arabidopsis DE-ETIOLATED1 Represses Photomorphogenesis by Positively Regulating Phytochrome-Interacting Factors in the Dark. Plant Cell. 2014;26:3630–3645. doi: 10.1105/tpc.114.130666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Bai MY, Kim JG, Wang T, Oh E, Chen L, Park CH, Son SH, Kim SK, Mudgett MB. The bHLH transcription factor HBI1 mediates the trade-off between growth and pathogen-associated molecular pattern–triggered immunity in Arabidopsis. The Plant Cell. 2014;26:828–841. doi: 10.1105/tpc.113.121111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng SH, Martinez C, Gusmaroli G, Wang Y, Zhou JL, Wang F, Chen LY, Yu L, Iglesias-Pedraz JM, Kircher S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–U479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, et al. PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) regulates auxin biosynthesis at high temperature. Proceedings of the National Academy of Sciences. 2011;108:20231–20235. doi: 10.1073/pnas.1110682108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriele S, Rizza A, Martone J, Circelli P, Costantino P, Vittorioso P. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. The Plant Journal. 2010;61:312–323. doi: 10.1111/j.1365-313X.2009.04055.x. [DOI] [PubMed] [Google Scholar]
- Gallego-Bartolomé J, Minguet EG, Marín JA, Prat S, Blázquez MA, Alabadí D. Transcriptional diversification and functional conservation between DELLA proteins in Arabidopsis. Molecular Biology and Evolution. 2010;27:1247–1256. doi: 10.1093/molbev/msq012. [DOI] [PubMed] [Google Scholar]
- Gangappa SN, Berriri S, Kumar SV. PIF4 Coordinates Thermosensory Growth and Immunity in <em>Arabidopsis</em>. Current Biology. 2017;27:243–249. doi: 10.1016/j.cub.2016.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Östin A, Sandberg G, Romano CP, Estelle M. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proceedings of the National Academy of Sciences. 1998;95:7197–7202. doi: 10.1073/pnas.95.12.7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon MJ, Mielczarek O, Robertson FC, Hubbard KE, Webb AA. Photosynthetic entrainment of the Arabidopsis thaliana circadian clock. Nature. 2013;502:689–692. doi: 10.1038/nature12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersch M, Lorrain S, de Wit M, Trevisan M, Ljung K, Bergmann S, Fankhauser C. Light intensity modulates the regulatory network of the shade avoidance response in Arabidopsis. Proceedings of the National Academy of Sciences. 2014;111:6515–6520. doi: 10.1073/pnas.1320355111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012;71:699–711. doi: 10.1111/j.1365-313X.2012.05033.x. [DOI] [PubMed] [Google Scholar]
- Huot B, Yao J, Montgomery BL, He SY. Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Molecular plant. 2014;7:1267–1287. doi: 10.1093/mp/ssu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Quail PH. PIF4, a phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. Embo J. 2002;21:2441–2450. doi: 10.1093/emboj/21.10.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y, Chory J. Unraveling the paradoxes of plant hormone signaling integration. Nature structural & molecular biology. 2010;17:642–645. doi: 10.1038/nsmb0610-642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong J, Kim K, Kim ME, Kim HG, Heo GS, Park OK, Park Y-I, Choi G, Oh E. Phytochrome and Ethylene Signaling Integration in Arabidopsis Occurs via the Transcriptional Regulation of Genes Co-targeted by PIFs and EIN3. Frontiers in Plant Science. 2016:7. doi: 10.3389/fpls.2016.01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung JH, Domijan M, Klose C, Biswas S, Ezer D, Gao M, Khattak AK, Box MS, Charoensawan V, Cortijo S, et al. Phytochromes function as thermosensors in <em>Arabidopsis</em>. Science. 2016;354:886–889. doi: 10.1126/science.aaf6005. [DOI] [PubMed] [Google Scholar]
- Khanna R, Huq E, Kikis EA, Al-Sady B, Lanzatella C, Quail PH. A novel molecular recognition motif necessary for targeting photoactivated phytochrome signaling to specific basic helix-loop-helix transcription factors. Plant Cell. 2004;16:3033–3044. doi: 10.1105/tpc.104.025643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R, Shen Y, Marion CM, Tsuchisaka A, Theologis A, Schäfer E, Quail PH. The Basic Helix-Loop-Helix Transcription Factor PIF5 Acts on Ethylene Biosynthesis and Phytochrome Signaling by Distinct Mechanisms. Plant Cell. 2007;19:3915–3929. doi: 10.1105/tpc.107.051508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Yamaguchi S, Lim S, Oh E, Park J, Hanada A, Kamiya Y, Choi G. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell. 2008;20:1260–1277. doi: 10.1105/tpc.108.058859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kang H, Park J, Kim W, Yoo J, Lee N, Kim J, Yoon T-y, Choi G. PIF1-interacting transcription factors and their binding sequence elements determine the in vivo targeting sites of PIF1. Plant Cell. 2016a;28:1388–1405. doi: 10.1105/tpc.16.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Song K, Park E, Kim K, Bae G, Choi G. Epidermal Phytochrome B Inhibits Hypocotyl Negative Gravitropism Non-Cell-Autonomously. The Plant Cell. 2016b;28:2770–2785. doi: 10.1105/tpc.16.00487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yi H, Choi G, Shin B, Song PS, Choi G. Functional characterization of phytochrome interacting factor 3 in phytochrome-mediated light signal transduction. Plant Cell. 2003;15:2399–2407. doi: 10.1105/tpc.014498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Current Biology. 2009;19:408–413. doi: 10.1016/j.cub.2009.01.046. [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, Harberd NP, Wigge PA. Transcription factor PIF4 controls the thermosensory activation of flowering. Nature. 2012;484:242–245. doi: 10.1038/nature10928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau OS, Deng XW. Plant hormone signaling lightens up: integrators of light and hormones. Current Opinion in Plant Biology. 2010;13:571–577. doi: 10.1016/j.pbi.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Lau OS, Huang X, Charron JB, Lee JH, Li G, Deng XW. Interaction of Arabidopsis DET1 with CCA1 and LHY in Mediating Transcriptional Repression in the Plant Circadian Clock. Molecular Cell. 2011;43:703–712. doi: 10.1016/j.molcel.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-J, Jung J-H, Llorca LC, Kim S-G, Lee S, Baldwin IT, Park C-M. FCA mediates thermal adaptation of stem growth by attenuating auxin action in Arabidopsis. Nature communications. 2014:5. doi: 10.1038/ncomms6473. [DOI] [PubMed] [Google Scholar]
- Lee N, Choi G. Phytochrome-interacting factor from Arabidopsis to liverwort. Current Opinion in Plant Biology. 2017;35:54–60. doi: 10.1016/j.pbi.2016.11.004. [DOI] [PubMed] [Google Scholar]
- Legris M, Klose C, Burgie ES, Costigliolo C, Neme M, Hiltbrunner A, Wigge PA, Schäfer E, Vierstra RD, Casal JJ. Phytochrome B integrates light and temperature signals in Arabidopsis. Science. 2016;354:897–900. doi: 10.1126/science.aaf5656. [DOI] [PubMed] [Google Scholar]
- Legris M, Nieto C, Sellaro R, Prat S, Casal JJ. Perception and signalling of light and temperature cues in plants. The Plant Journal. 2017;90:683–697. doi: 10.1111/tpj.13467. [DOI] [PubMed] [Google Scholar]
- Leivar P, Monte E. PIFs: Systems Integrators in Plant Development. Plant Cell. 2014;26:56–78. doi: 10.1105/tpc.113.120857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH. PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci. 2011;16:19–28. doi: 10.1016/j.tplants.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Yu R, Fan LM, Wei N, Chen H, Deng XW. DELLA-mediated PIF degradation contributes to coordination of light and gibberellin signalling in Arabidopsis. Nat Commun. 2016;7:11868. doi: 10.1038/ncomms11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ljung K, Breton G, Schmitz RJ, Pruneda-Paz J, Cowing-Zitron C, Cole BJ, Ivans LJ, Pedmale UV, Jung HS, et al. Linking photoreceptor excitation to changes in plant architecture. Genes & Development. 2012;26:785–790. doi: 10.1101/gad.187849.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling JJ, Li J, Zhu D, Deng XW. Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proceedings of the National Academy of Sciences. 2017;114:3539–3544. doi: 10.1073/pnas.1700850114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell DB, Gourdji SM. The influence of climate change on global crop productivity. Plant Physiology. 2012;160:1686–1697. doi: 10.1104/pp.112.208298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R, Zipfel C. Trade-off between growth and immunity: role of brassinosteroids. Trends in plant science. 2015;20:12–19. doi: 10.1016/j.tplants.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Lucas M, Prat S. PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytologist. 2014;202:1126–1141. doi: 10.1111/nph.12725. [DOI] [PubMed] [Google Scholar]
- Ma D, Li X, Guo Y, Chu J, Fang S, Yan C, Noel JP, Liu H. Cryptochrome 1 interacts with PIF4 to regulate high temperature-mediated hypocotyl elongation in response to blue light. Proceedings of the National Academy of Sciences. 2016;113:224–229. doi: 10.1073/pnas.1511437113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macknight R, Bancroft I, Page T, Lister C, Schmidt R, Love K, Westphal L, Murphy G, Sherson S, Cobbett C. FCA, a gene controlling flowering time in Arabidopsis, encodes a protein containing RNA-binding domains. Cell. 1997;89:737–745. doi: 10.1016/s0092-8674(00)80256-1. [DOI] [PubMed] [Google Scholar]
- Martin G, Soy J, Monte E. Genomic Analysis Reveals Contrasting PIFq Contribution to Diurnal Rhythmic Gene Expression in PIF-Induced and-Repressed Genes. Frontiers in Plant Science. 2016:7. doi: 10.3389/fpls.2016.00962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia JF, Huq E, Quail PH. Direct targeting of light signals to a promoter element-bound transcription factor. Science. 2000;288:859–863. doi: 10.1126/science.288.5467.859. [DOI] [PubMed] [Google Scholar]
- Monte E, Tepperman JM, Al-Sady B, Kaczorowski KA, Alonso JM, Ecker JR, Li X, Zhang Y, Quail PH. The phytochrome-interacting transcription factor, PIF3, acts early, selectively, and positively in light-induced chloroplast development. Proc Natl Acad Sci U S A. 2004;101:16091–16098. doi: 10.1073/pnas.0407107101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Ni W, Xu SL, González-Grandío E, Chalkley RJ, Huhmer AFR, Burlingame AL, Wang ZY, Quail PH. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nature Communications. 2017;8:15236. doi: 10.1038/ncomms15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C, López-Salmerón V, Davière JM, Prat S. ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Current Biology. 2015;25:187–193. doi: 10.1016/j.cub.2014.10.070. [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain AA, Fankhauser C, Harmer SL, Maloof JN. Rhythmic growth explained by coincidence between internal and external cues. Nature. 2007;448:358–361. doi: 10.1038/nature05946. [DOI] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farre EM, Kay SA. The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature. 2011;475:398–U161. doi: 10.1038/nature10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G. Genome-Wide Analysis of Genes Targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during Seed Germination in Arabidopsis. Plant Cell. 2009;21:403–419. doi: 10.1105/tpc.108.064691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Yamaguchi S, Huc J, Yusukeb J, Jung B, Paik I, Leed HS, Sun TP, Kamiya Y, Choi G. PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by directly binding to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell. 2007;19:1192–1208. doi: 10.1105/tpc.107.050153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Bai MY, Arenhart RA, Sun Y, Wang ZY. Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife. 2014;3:e03031. doi: 10.7554/eLife.03031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu J-Y, Wang Z-Y. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nature Cell Biology. 2012:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lee N, Kim W, Lim S, Choi G. ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. The Plant Cell. 2011;23:1404–1415. doi: 10.1105/tpc.110.080721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M. Molecular and genetic control of plant thermomorphogenesis. Nature plants. 2016;2:15190. doi: 10.1038/nplants.2015.190. [DOI] [PubMed] [Google Scholar]
- Roden LC, Ingle RA. Lights, rhythms, infection: the role of light and the circadian clock in determining the outcome of plant–pathogen interactions. The Plant Cell. 2009;21:2546–2552. doi: 10.1105/tpc.109.069922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S, Sharma I, Pati PK. Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Frontiers in plant science. 2015:6. doi: 10.3389/fpls.2015.00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santner A, Estelle M. Recent advances and emerging trends in plant hormone signalling. Nature. 2009;459:1071–1078. doi: 10.1038/nature08122. [DOI] [PubMed] [Google Scholar]
- Shin AY, Han YJ, Baek A, Ahn T, Kim SY, Nguyen TS, Son M, Lee KW, Shen Y, Song PS, et al. Evidence that phytochrome functions as a protein kinase in plant light signalling. Nature Communications. 2016;7:11545. doi: 10.1038/ncomms11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shor E, Paik I, Kangisser S, Green R, Huq E. PHYTOCHROME INTERACTING FACTORS mediate metabolic control of the circadian system in Arabidopsis. New Phytologist. 2017;215:217–228. doi: 10.1111/nph.14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Devlin PF, Kay SA. Phytochromes and cryptochromes in the entrainment of the Arabidopsis circadian clock. Science. 1998;282:1488–1490. doi: 10.1126/science.282.5393.1488. [DOI] [PubMed] [Google Scholar]
- Soy J, Leivar P, Gonzalez-Schain N, Martin G, Diaz C, Sentandreu M, Al-Sady B, Quail PH, Monte E. Molecular convergence of clock and photosensory pathways through PIF3-TOC1 interaction and co-occupancy of target promoters. Proc Natl Acad Sci U S A. 2016;113:4870–4875. doi: 10.1073/pnas.1603745113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C. PIF4–mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genet. 2012;8:e1002594. doi: 10.1371/journal.pgen.1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Wang J, Gao Z, Dong J, He H, Terzaghi W, Wei N, Deng XW, Chen H. Arabidopsis SAURs are critical for differential light regulation of the development of various organs. Proc Natl Acad Sci U S A. 2016;113:6071–6076. doi: 10.1073/pnas.1604782113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viczian A, Kircher S, Fejes E, Millar AJ, Schafer E, Kozma-Bognar L, Nagy F. Functional characterization of phytochrome interacting factor 3 for the Arabidopsis thaliana circadian clockwork. Plant Cell Physiol. 2005;46:1591–1602. doi: 10.1093/pcp/pci175. [DOI] [PubMed] [Google Scholar]
- Wei Z, Yuan T, Tarkowská D, Kim J, Nam HG, Novák O, He K, Gou X, Li J. Brassinosteroid Biosynthesis Is Modulated via a Transcription Factor Cascade of COG1, PIF4, and PIF5. Plant Physiology. 2017;174:1260–1273. doi: 10.1104/pp.16.01778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windram O, Madhou P, McHattie S, Hill C, Hickman R, Cooke E, Jenkins DJ, Penfold CA, Baxter L, Breeze E. Arabidopsis defense against Botrytis cinerea: chronology and regulation deciphered by high-resolution temporal transcriptomic analysis. The Plant Cell. 2012;24:3530–3557. doi: 10.1105/tpc.112.102046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Kathare PK, Pham VN, Bu Q, Nguyen A, Huq E. Reciprocal proteasome-mediated degradation of PIFs and HFR1 underlies photomorphogenic development in <em>Arabidopsis</em>. Development. 2017;144:1831–1840. doi: 10.1242/dev.146936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Paik I, Zhu L, Huq E. Illuminating Progress in Phytochrome-Mediated Light Signaling Pathways. Trends in plant science. 2015;20:641–650. doi: 10.1016/j.tplants.2015.06.010. [DOI] [PubMed] [Google Scholar]
- Yamashino T, Matsushika A, Fujimori T, Sato S, Kato T, Tabata S, Mizuno T. A Link between circadian-controlled bHLH factors and the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol. 2003;44:619–629. doi: 10.1093/pcp/pcg078. [DOI] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, Tong XH, Zeng LJ, Li Q, Xiao LT, Sun T-p, Li J, Deng XW. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proceedings of the National Academy of Sciences. 2012;109:E1192–E1200. doi: 10.1073/pnas.1201616109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Huang R. Integration of ethylene and light signaling affects hypocotyl growth in Arabidopsis. Frontiers in plant science. 2017:8. doi: 10.3389/fpls.2017.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang LY, Bai MY, Wu J, Zhu JY, Wang H, Zhang Z, Wang W, Sun Y, Zhao J, Sun X. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. The Plant Cell. 2009;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Shi H, Xue C, Wei N, Guo H, Deng XW. Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proceedings of the National Academy of Sciences. 2014;111:3913–3920. doi: 10.1073/pnas.1402491111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JY, Oh E, Wang T, Wang ZY. TOC1–PIF4 interaction mediates the circadian gating of thermoresponsive growth in Arabidopsis. Nature Communications. 2016;7:13692. doi: 10.1038/ncomms13692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L, Bu Q, Xu X, Paik I, Huang X, Hoecker U, Deng XW, Huq E. CUL4 forms an E3 ligase with COP1 and SPA to promote light-induced degradation of PIF1. Nature commun. 2015;6:7245. doi: 10.1038/ncomms8245. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Xu F, Zhang Y, Cheng YT, Wiermer M, Li X, Zhang Y. Arabidopsis resistance protein SNC1 activates immune responses through association with a transcriptional corepressor. Proceedings of the National Academy of Sciences. 2010;107:13960–13965. doi: 10.1073/pnas.1002828107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zust T, Agrawal AA. Trade-Offs Between Plant Growth and Defense Against Insect Herbivory: An Emerging Mechanistic Synthesis. Annu Rev Plant Biol. 2017;68:10.11–10.22. doi: 10.1146/annurev-arplant-042916-040856. [DOI] [PubMed] [Google Scholar]