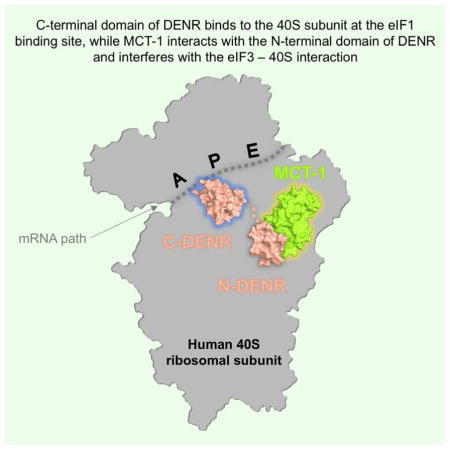
SUMMARY
The repertoire of the density regulated protein (DENR) and the malignant T-cell- amplified sequence 1 (MCT-1/MCTS1) oncoprotein was recently expanded to include translational control of a specific set of cancer-related mRNAs. DENR and MCT-1 form the heterodimer, which binds to the ribosome and operates at both translation initiation and reinitiation steps, though by a mechanism that is yet unclear. Here, we determined the first crystal structure of the human small ribosomal subunit in complex with DENR-MCT-1. The structure reveals the previously unknown location of the DENR-MCT-1 dimer bound to the small ribosomal subunit. The binding site of the C-terminal domain of DENR on the ribosome has a striking similarity with those of canonical initiation factor 1 (eIF1), which controls the fidelity of translation initiation and scanning. Our findings elucidate how the DENR-MCT-1 dimer interacts with the ribosome and have functional implications for the mechanism of unconventional translation initiation and reinitiation.
Graphical Abstract
INTRODUCTION
Eukaryotic translation initiation is a complex process that is coordinated by more than a dozen initiation factors (eIF), which facilitate the recruitment of mRNA and its scanning to locate the start codon (AUG), selection of initiator tRNA (tRNAi) and the joining of the small (40S) and large (60S) ribosomal subunits (Hinnebusch, 2017). After formation of the initiation complex, the 80S ribosome proceeds with protein synthesis until the stop codon on mRNA is placed in the ribosomal A site. Two eukaryotic release factors, eRF1 and eRF3, catalyze the recognition of the stop codon and hydrolysis of the peptidyl-tRNA, producing the post-termination complex (post-TC) (Dever and Green, 2012; Jackson et al., 2012). Post-TC is then recycled by being split into subunits by eRF1 and ABCE1 (Pisarev et al., 2010). After dissociation of the 60S subunit, the 40S subunit remains bound to mRNA along with a deacylated tRNA in the P site. Two alternatives are possible at this step. First, ejection of deacylated tRNA may cause dissociation of mRNA and free the 40S subunit for the next cycle of initiation. In vitro tRNA release from the P site can be stimulated either by a joint action of canonical initiation factors eIF1, eIF1A and eIF3 or by eIF2D or DENR-MCT-1 (Pisarev et al., 2007; Pisarev et al., 2010; Skabkin et al., 2010; Skabkin et al., 2013). Second, the 40S subunit remains bound to the mRNA, leading to reinitiation of protein synthesis from a nearest AUG (Jackson et al., 2012; Kozak, 2001).
Unlike in prokaryotes, reinitiation in eukaryotes was considered as an extremely rare event. For many mammalian mRNAs, the first AUG codon detected by the scanning complex (“first AUG rule”) is the initiation site of the protein-coding open reading frame (ORF), and the surrounding nucleotide sequence, together with eIF1 and eIF1A, modulate the efficiency of its recognition (Hinnebusch, 2017). However, recent advances in ribosome profiling reveal that many mRNAs have one or more functional start codons in their 5′ untranslated region (UTR), which are either bypassed (“leaky scanning”) or translated (Andreev et al., 2017; Archer et al., 2016). These may result in the production of functional peptides encoded by the upstream ORFs (uORFs) or different isoforms of the main protein. uORFs may inhibit translation of the main ORF or regulate it by reinitiation (Wethmar, 2014). Reinitiation is used for fine-tuning translation of some ORFs, and is widely used by viruses to express polycistronic RNAs (Hinnebusch et al., 2016; Ryabova et al., 2006). Though the mechanism of translation reinitiation is still an open question, experimental data suggest that reinitiation-competent 40S subunits are those that remain bound to mRNA after termination and to which some eIFs (eIF4F, eIF3) remain bound even after completing translation of the first uORF. According to recent data, among the initiation factors that promote reinitiation (eIF1, eIF1A, eIF2, eIF3), eIF3 plays a key role in the retention of mRNA on the post-TC 40S subunit and even alone is able to mediate reinitiation on some mRNAs (Jackson et al., 2012; Kozak, 1987; Mohammad et al., 2017; Poyry et al., 2004; Skabkin et al., 2013). In the case of bicistronic calicivirus mRNAs, a cis-acting mRNA element TURBS (“termination upstream ribosomal binding site”), located upstream of the restart AUG, interacts with the helix h26 of the 18S rRNA (Zinoviev et al., 2015). A detailed analysis in reconstituted in vitro system reveals that this interaction provides retention and restrains the 40S subunit on the RNA, eliminating eIF3 dependence. The TURBS-h26 interaction positions the restarting AUG codon directly in the P site of the 40S subunit. In vitro experiments suggest that the direct placement of AUG in the P site of the 40S subunit without scanning is required for translation reinitiation and initiation supported by eIF2D and DENR-MCT-1 (Dmitriev et al., 2010; Skabkin et al., 2010; Zinoviev et al., 2015). Recently DENR-MCT-1 was also implicated in the regulation of translation reinitiation after short uORFs of a specific set of mRNAs in flies (Schleich et al., 2014).
Protein sequences of eIF2D, DENR and MCT-1 share similar domains involved in RNA binding and start codon recognition (Figure 1A). In addition to recycling and reinitiation, they can stimulate eIF2- and GTP-independent recruitment of either aminoacyl- or deacylated tRNAi to the P-site of the 40S subunit bound to some specific mRNAs, including Hepatitis C virus (HCV)-like internal ribosome entry sites (IRESs) (Dmitriev et al., 2010; Skabkin et al., 2010; Zinoviev et al., 2015).
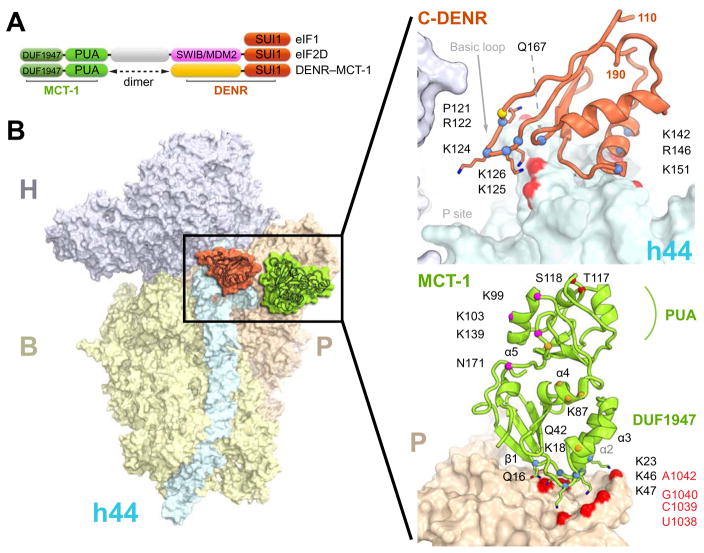
Figure 1. The crystal structure of human 40S-DENR-MCT-1 complex.
(A) Domain organization of human eIF1, eIF2D, MCT-1 and DENR. (B) H, head; B, body; P, platform; h44, helix 44 - domains of the 40S subunit. DENR (coral), MCT-1 (green) and its structural domains are shown. The basic loop of C-DENR is marked by an arrow; the N-terminus of C-DENR and MCT-1 are marked by N. Amino acid residues constituting ribosomal binding site and the basic loop of C-DENR, N-DENR binding site, phosphorylation site and suggested tRNA binding site of MCT-1 are marked in blue, gold, red, and magenta, respectively. Nucleotides of 18S rRNA interacting with C-DENR or MCT-1 are marked in red. See also Figures S1, S2, S4, S5.
MCT-1 is an oncoprotein originally identified in human lymphoma and it has been linked to increased cell proliferation and genome instability (Hsu et al., 2007; Prosniak et al., 1998). Its crystal structure revealed a highly conserved pseudouridine synthase and archaeosine transglycosylase (PUA) fold of the C-terminal domain, which is separated by two short linkers from the compact and globular N-terminal domain (Tempel et al., 2013). MCT-1 forms a heterodimer with DENR, both in vivo and in vitro (Haas et al., 2016; Reinert et al., 2006). MCT-1 sequence shares 47% identity with its yeast counterpart TMA20 and complements the translation defects observed in Δtma20 yeast strain (Fleischer et al., 2006). Like MCT-1, TMA20 forms a heterodimer with TMA22, the yeast homologue of DENR. These observations suggest that the DENR-MCT-1 binding interface is highly conserved between human and yeast.
DENR synthesis is upregulated with increasing cell density and it is also overexpressed in ovarian and breast cancers (Deyo et al., 1998; Oh et al., 1999). Sequence analysis revealed that the C-terminal part of DENR (C-DENR) comprises the SUI1 domain, which forms the core of the eIF1 structure (Figure 1A). It was shown that DENR in complex with MCT-1 alters the translational profile of a subset of cancer-related mRNAs, likely by interfering with the translation initiation complex (Mazan-Mamczarz et al., 2009; Reinert et al., 2006). Recently, DENR and MCT-1 were shown to be required for the efficient translation of a set of uORF-containing mRNAs, which are involved in cell proliferation and signaling in Drosophila and neuronal development in humans (Haas et al., 2016; Schleich et al., 2014). However, little is known about the interactions between DENR, MCT-1 and the ribosome. To obtain structural insights into the mechanism by which DENR-MCT-1 regulates translation initiation/reinitiation and recycling, we have determined the crystal structure of the human small ribosomal subunit in complex with DENR-MCT-1. The structure allows us to locate the position of DENR-MCT-1 on the 40S ribosomal subunit and map the binding interface between MCT-1 and DENR. This provides the basis for understanding the function of DENR-MCT-1 heterodimer during recycling, reinitiation by post-termination 40S ribosomal subunits and initiation on the HCV-like IRES RNAs.
RESULTS
Human 40S ribosomal subunit in complex with DENR-MCT-1 (40S-DENR-MCT-1) was crystallized and its structure was determined by X-ray crystallography (Figure 1B). The crystal belongs to the P21 monoclinic space group and contains six 40S ribosomal subunits per asymmetric unit (ASU). A complete data set was collected to 6.0 Å resolution (Table S1).
Strong electron density for ribosomal RNA (rRNA) and proteins is present in the map (Figure S1B). Core areas and main chains of the 18S rRNA and most ribosomal proteins are clearly seen in the electron density map, however, the exact protein side chain conformations could not be determined due to limitations of the current resolution. In this paper, we shall focus on the details related to interactions of MCT-1 and DENR with the 40S ribosomal subunit and their implication for the mechanistic understanding of translation initiation, recycling and reinitiation processes.
MCT-1 binding to the 40S ribosomal subunit
The recently published crystal structure of human MCT-1 perfectly fits the unbiased additional density in the Fo-Fc electron density map of the 40S-DENR-MCT-1 complex (Figure S1A) (Tempel et al., 2013). Strong electron density is observed on the right side of the platform domain at the interface of the 40S subunit where the N-terminal domain (residues 1-92) of MCT-1 interacts with helix h24 of the 18S rRNA. Conserved amino acid residues of the β1 strand and helix α2 of MCT-1 (Gln16, Ser20, Lys18 and 23) form the binding interface with the ribosome. Positively charged Lys46 and Lys47 of the α3-β2 loop are within the interacting distance to the phosphate backbone of U1038 and C1039 of h24 (Figures 1B, S4). The abundance of positively charged residues at the binding interface suggests that there are electrostatic interactions with the phosphate backbone of h24 of the 18S rRNA, which supports the observation that MCT-1 alone binds the 40S subunit in vitro (Skabkin et al., 2010).
The interactions with the ribosome are similar for all six copies of MCT-1 in the ASU, barring minor differences. In addition to the main binding site on the 40S subunit described above, five MCT-1 molecules in ASU have contacts with the neighboring ribosome. Ser12 and/or His56 of MCT-1 may interact with the tip of the helical terminus of the ribosomal protein eS8, which belongs to the neighboring molecule. Two from these five MCT-1 molecules contact the neighboring ribosome through interactions between the α5-α6 loop and the base of U368 of the extension segment ES13S of h11 of the 18S rRNA (Figure S4). Because we did not observe these interactions for all molecules in ASU, these relatively weak interactions are likely a product of crystal packing and are not relevant to MCT-1 functions.
MCT-1 interferes with the eIF3 interaction with the 40S subunit
The position of MCT-1 on the 40S subunit is incompatible with the locations of the C-terminal domain of eIF3a and the N-terminus of eIF3c subunits of the initiation factor eIF3. Human eIF3 is comprised of 13 subunits and it is the largest initiation factor with the mass of about 800 kDa. eIF3a and eIF3c are core subunits, which are present in mammals and yeast (Hershey, 2015). The structure of the yeast translation initiation complex (py48S-closed IC) revealed that the C-terminal part of eIF3a and the N-terminus of eIF3c bind at the interface of the 40S subunit (Llacer et al., 2015). However, location of these binding sites remains ambiguous, because corresponding electron density can also be attributed to the RRM domain of eIF3b (Simonetti et al., 2016). Superposition of the structure of py48S-closed IC with the structure of 40S-DENR-MCT-1 revealed potential steric clashes between MCT-1 and the C-terminus of eIF3a and the N-terminus of eIF3c (or eIF3b RRM) (Figure 2). This suggests that the DENR-MCT-1 activity in translation may not require eIF3. Indeed, this correlates well with the ability of DENR-MCT-1 to mediate reinitiation on the rabbit hemorrhagic disease virus (RHDV) RNA without eIF3 (Zinoviev et al., 2015). In addition, competition for the binding with the 40S subunit between MCT-1 and eIF3 may explain how DENR-MCT-1 can facilitate translation initiation on HCV-like IRESs. It was shown in vitro that eIF2D, which shares homologous domains with DENR-MCT-1 (Figure 1A), stimulates formation of the 48S complex on HCV-like IRESs (HCV, CSFV) in eIF2 and eIF3 independent mode (Skabkin et al., 2010). Binding of the HCV-like IRES to the ribosomal preinitiation complex induces conformational changes and subsequent dissociation of eIF3 from the interface of the 40S subunit (Hashem et al., 2013). Our structure explains these effects and also suggests two independent stages of the reaction. At the first stage MCT-1 competes with the eIF3a and eIF3c (or eIF3b RRM) for the 40S subunit, facilitating their release and promoting correct interaction between IRES RNA and the 40S subunit. At the second stage, DENR-MCT-1 facilitates recruitment of the tRNAi to the P site of the 40S subunit (see below), which has the initiation codon, placed directly by viral RNA.
Figure 2. Binding site of the human MCT-1 on the 40S subunit overlaps with the binding site of the eIF3a and eIF3c (or eIF3b RRM) subunits of eIF3.
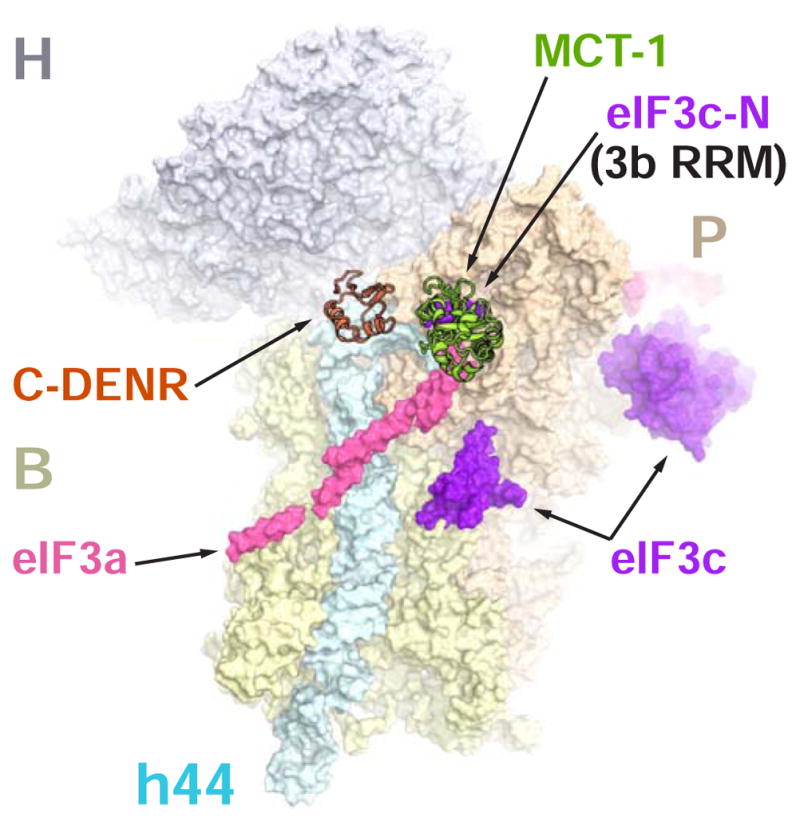
The crystal structure of the human 40S-DENR-MCT-1 complex was superposed with the structure of the partial yeast 48S complex in closed conformation (PDB ID 3JAP). Only eIF3a (pink) and eIF3c (purple) subunits from the yeast structure are shown. Electron density corresponding to eIF3a and eIF3c-N can also be attributed to eIF3b RRM (PDB ID 5K1H). C-DENR and MCT-1are shown in coral and green, respectively. H, head; B, body; P, platform; h44, helix 44 - domains of the 40S subunit.
DENR interacts with MCT-1 through its N-terminal domain, providing an interface to stabilize initiator tRNA
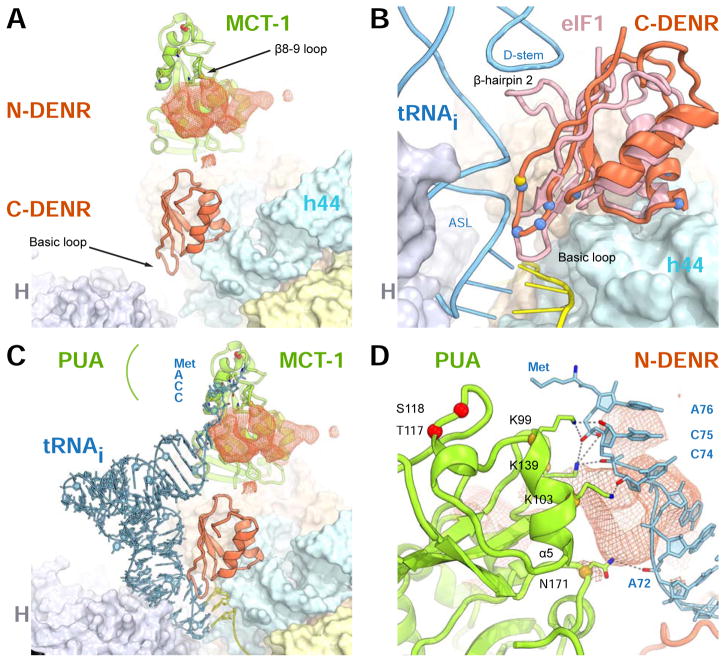
We attributed the strong unbiased electron density connected to the PUA domain of MCT-1 to the N-terminal part of DENR (N-DENR) (Figures 3, S1A, S3A). This electron density extends toward the N-terminal domain of MCT-1 revealing also interactions between DENR and helices α3, α4 and α5 of MCT-1 (Figures 1B, S3A). To confirm that the MCT-1 binding site is located on the N-terminal part of DENR, we performed the pull-down experiment using T7-tagged N- and C-DENR (amino acid residues 11-98 and 110-198, respectively, Figure S3B). It shows that MCT-1 indeed binds to the N-terminal but not C-terminal part of DENR. However, we did not build the model for the N-DENR due to limitations imposed by resolution. We do not see any electron density connecting the N-DENR with C-DENR, which suggests that this connection is flexible or unstructured.
Figure 3. Binding of N-DENR to MCT-1 provides interface to interact with the P-site tRNA.
(A) Unbiased additional electron density (Fo-Fc map contoured at σ=2.0, coral mesh) connected to the PUA domain of MCT-1 is attributed to N-DENR. (B) Superposition of the human 40S-DENR-MCT-1 complex and rabbit PICs (PDB ID 4KZX, 4KZZ), only the P site region of the 40S subunit is shown, eIF1A is omitted. Blue spheres show conserved amino acid residues of the basic loop and the ribosome binding site, gold sphere shows P121 of C-DENR. (C) same as b, eIF1 is omitted. To show the location of the Met-moiety of tRNAi, aminoacyl- tRNA (fMet-tRNAi Met) from the structure of bacterial 70S complex (PDB ID 1VY4, blue) was superposed on the tRNAi in the 48S PIC. (D) a close-up view of c, showing proposed interaction of the PUA domain of MCT-1 with the acceptor stem of the tRNAi. Dashed lines show possible interactions between MCT-1 and tRNA. Red spheres represent phosphorylation site; yellow spheres show amino acid residues interacting with the tRNA. H, head; h44, helix 44 - domains of the 40S subunit. See also Figure S3.
The PUA domain is present in many RNA modification enzymes (Cerrudo et al., 2014). It binds tRNA or rRNA through a structurally conserved motif, which comprises α5-β7 loop and β10 strand in the human MCT-1 (Figure 1B). However, the amino acid sequence in these regions of the PUA domain is not conserved, which reflects different RNA-binding specificity (Perez-Arellano et al., 2007). The loop β8-β9 (Gly138-His141) of the MCT-1 PUA domain interacts with N-DENR (Figures 3A, D, S3). The large DENR-MCT-1binding interface formed by the PUA domain is in agreement with biochemical data showing that ΔPUA MCT-1 was not able to pulldown DENR in the cap-binding assay (Reinert et al., 2006). The DENR-MCT-1 binding interface is also formed in part by helices α3 (Ile36-Gln42) and α4 (Tyr87-His88) of the MCT-1 N-terminal domain (Figures 1B, S3A, S4). It was shown that the C37Y mutation impairs DENR function in the radial migration of neurons within the murine embryonic cerebral cortex and disrupts positioning of cortical neurons and their morphology (Haas et al., 2016). C37Y may affect MCT-1-DENR interaction, which provides additional evidence that correct binding between DENR and MCT-1 is important for their function in vivo.
The DENR-MCT-1 binding interface forms a surface with protruding positively charged amino acid residues. In addition to the helix α5 of MCT-1, the middle of the N-DENR also contains a conserved cluster of positively charged residues, which is possibly located in this area judging from the size of the electron density and the distance from C-DENR. We speculate that this surface made by the N-DENR/MCT-1 interface may serve as a binding site for the CCA end of the P-site tRNA. Indeed, superposition of our structure with the structure of the 48S PIC containing mRNA and P-site tRNAi shows that the acceptor stem of the tRNAi is oriented toward DENR-MCT-1 binding interface and that the CCA end is perfectly positioned for interaction with MCT-1 and, possibly, N-DENR (Figure 3C, D). Though there are other structures of 48S-like complexes published, we used our structure of rabbit 48S PIC because this is the only available complex structure containing in the P site codon-anticodon base-paired tRNA, which is not bound to eIF2. eIF2-free tRNA, base-paired with mRNA in the P site, resembles not only intermediate state during translation initiation but also the state of the post-TC after eRF1 and ABCE1 dissociation. Therefore, this 48S PIC is a suitable model for a substrate for DENR-MCT-1. Superposition suggests that Lys99, Lys103 and Lys139 of MCT-1 may stabilize the CCA end of tRNA and Asn171 may interact with the phosphate backbone of A72 (Figure 3D). Amino acid residues Lys99, Lys103 and Lys139 are absolutely conserved in metazoan MCT-1 and in their yeast counterpart TMA20 (Figure S5). These interactions will facilitate binding of the tRNAi to the P site of the 40S subunit, which is consistent with biochemical data, demonstrating that DENR-MCT-1 promotes eIF2-independent recruitment of tRNAi to the 40S subunit if the start codon of mRNA is placed directly in the P site (Skabkin et al., 2010; Zinoviev et al., 2015).
It was reported that phosphorylation of Thr117 and/or Ser118 of the MCT-1, which are positioned in the vicinity to the aminoacyl moiety of the tRNAi in our model (Figure 3D), is important for reinitiation activity of DENR-MCT-1 in vivo (Nandi et al., 2007; Schleich et al., 2014). Indeed, the additional negative charge provided by phosphorylation may be important for stabilization of the methionine moiety and protection of the ester bond from non-enzymatic hydrolysis. This hypothesis agrees with the observation that reinitiation activity of MCT-1 is abolished in vivo by mutations blocking these phosphorylation sites (Schleich et al., 2014).
The C-terminal domain of DENR binds near the P site of the 40S subunit
Electron density for C-DENR is observed at the top of h44 adjacent to h45 and h24 of the 18S rRNA in the vicinity of the P site (Figures 1B, S1D). The binding site of C-DENR on the 18S rRNA resembles that of eIF1 (Lomakin and Steitz, 2013). eIF1 allows proofreading and sensing a non-optimal context of an AUG codon, dissociation of aberrantly assembled complexes during scanning and promotes ejection of deacylated tRNA during recycling (Hinnebusch, 2017; Jackson et al., 2012). Analysis of the amino acid sequence of DENR predicted that its C-terminal part forms a domain with a fold similar to that of eIF1 (SUI1 in yeast) (Reinert et al., 2006). Based on sequence and structural homology we built a model of C-DENR and manually fitted it in the additional unbiased electron density (Figure S1).
As expected, the overall structure of C-DENR resembles that of eIF1 (Figure 4). Conserved basic loop (122Arg-Lys126) protrudes towards the mRNA binding channel in the P site between nucleotides 1827-1829 of h44 and 1058 of h24 of the 18S rRNA, as seen previously for eIF1 (Figures 1B, 3B) (Hussain et al., 2014; Lomakin and Steitz, 2013; Rabl et al., 2011; Weisser et al., 2013). The basic surface of this loop and conserved basic residues Lys142, Arg146 and Lys151 of the helix of C-DENR contact the phosphate backbone of the h24 and h44 in the mRNA binding channel and h44 and h45 in the platform of the 18S rRNA, respectively (Figure 1B). The basic loop of eIF1 plays the main role in eIF1 interaction with the tRNA. During scanning, the basic loop displaces the anticodon stem loop (ASL) of tRNAi and prevents the locking of the tRNAi in the P site. The AUG base pairing with the anticodon locks tRNAi in the Pin state, moving the basic loop of eIF1 out of the P site, which subsequently results in dissociation of eIF1 from the ribosome (Hinnebusch, 2017; Llacer et al., 2015; Lomakin and Steitz, 2013). The ASL of only initiator tRNA allows displacing of the basic loop from the P site. eIF1 discriminates between initiator and other tRNAs base pared in the P site by sensing the presence of the three universally conserved G-C base pairs in the ASL (Lomakin et al., 2006). This allows eIF1 to facilitate ejection of deacylated tRNA from the P site during recycling and be ejected itself from the P site at the end of the initiation stage. Superposition of the rabbit PIC1, PIC2 and 48S PIC on the structure of 40S-DENR-MCT-1 shows that the basic loop of C-DENR is positioned closer to the ASL of the tRNAi compared to that of eIF1 in the rabbit complexes (Figure 3B) (Lomakin and Steitz, 2013). The potential clash of the C-DENR’s basic loop with the ASL of tRNAi is more severe than in the case of eIF1. In all DENR from vertebrates Pro (Pro121 in human) is the amino acid residue before the first Arg of the basic loop (Figures 1B, 3B). Pro121 provides a structural kink and may impose local rigidity for the basic loop, which may enhance its activity in the dissociation of the P-site tRNA. It is not clear if Pro121 is important for scanning, since currently there are no data available implicating DENR-MCT-1 in the scanning. Experiments in vivo demonstrated that P121L mutation in human DENR is associated with Asperger syndrome and impairs positioning, terminal arborization, and dendritic spine densities of cerebral cortical neurons (Haas et al., 2016). In human eIF1, Gln37, which corresponds to P121 of DENR, may provide an additional interaction with the ASL of the tRNAi in transient conformations during scanning, as seen in the structure of the yeast py48S-closed complex. In such conformations, both eIF1 and tRNAi are shuttling in and out of the P site in accordance with our current model of scanning (Llacer et al., 2015; Lomakin and Steitz, 2013). Recently, it was demonstrated that DENR-MCT-1 is required for translation of the specific set of mRNAs in flies by the reinitiation pathway, however whether or not other protein factors and scanning are necessary for reinitiation on these mRNAs remains to be elucidated (Schleich et al., 2014).
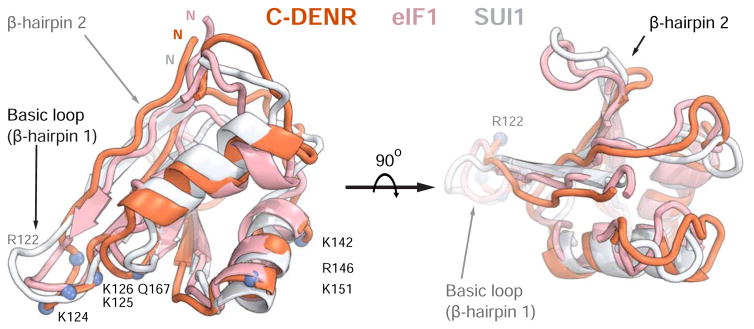
Figure 4. The structure of the C-terminal domain of human DENR.
The crystal structure of C-DENR (coral) from the 40S-DENR-MCT-1 complex was superposed with the NMR structures of the human eIF1 (pink, PDB ID 2IF1, amino acid residues 28-108) and yeast eIF1/SUI1 (white, PDB ID 2OGH, amino acid residues 24-100). The basic loop and β-hairpin 2 loop is marked by an arrow; the N-terminus of C-DENR is marked by N. Amino acid residues of C-DENR constituting the basic loop and binding site on the 18S rRNA are marked in blue.
Biochemical analysis in the reconstituted system showed that DENR-MCT-1 mediates eIF2-independent initiation on HCV-like IRESs and reinitiation on RHDV mRNA. In both cases RNA attachment to the 40S subunit places the initiation codon directly in the P site without scanning. This suggests that like eIF1, C-DENR should be displaced from its binding site when the AUG codon is base-paired with the anticodon of the tRNAi and rotation of the head domain locked tRNAi in the Pin state (Lomakin and Steitz, 2013). In addition to the basic loop clash, displacement of eIF1 is promoted by repulsion between the negatively charged β-hairpin loop 2 and the backbone of the D stem of the tRNAi (Llacer et al., 2015; Lomakin and Steitz, 2013). The β-hairpin loop 2 of C-DENR is smaller and probably does not interact with the tRNA (Figures 3B, 4). This suggests that the nature of the ASL of the tRNA in the P site may determine the fate of C-DENR during interaction with the tRNA. Unlike other tRNAs, including methionine elongator tRNA, ASL of the tRNAi has three G:C base pairs at positions 29–31:39–41 in all three domains of life (Kolitz and Lorsch, 2010). It has been shown in vitro, that eIF1 and its functional homologue bacterial IF3 discriminate against tRNAs with mutated conserved G:C base pairs (Lomakin et al., 2006). We suggest that C-DENR has similar discriminating activity, which is consistent with its ability to facilitate ejection of tRNA from the P site during recycling and promote fidelity of the P-site tRNAi during initiation and reinitiation (Skabkin et al., 2010; Zinoviev et al., 2015).
DENR-MCT-1 prevents subunits joining
The positions of DENR and MCT-1 on the 40S subunit are incompatible with the binding of the large 60S ribosomal subunit to the 40S subunit. The region of interaction between DENR and 18S rRNA comprises the intersubunit bridge B2a and part of bridge B2b. C-DENR will sterically clash with H69 of the 23S rRNA. MCT-1 binds to the 18S rRNA in the location of the intersubunit bridges B7b-B7c and it is very close to the intersubunit bridge B7a. MCT-1 will also sterically clash with H68 and H75 of the 23S rRNA. These observations suggest that during recycling both DENR and MCT-1 should bind the 40S subunit at a stage after ABCE1 action and 60S subunit dissociation, and that during initiation/reinitiation they should dissociate from the 40S subunit before or during formation of the 80S complex.
Conformation of the 40S subunit
All six 40S subunits observed in the ASU of the crystal structure of the 40S-DENR-MCT-1 complex resemble a conformation that is similar to the rabbit 48S PIC, with only minor rotations of the head of about 1–3 degrees. An important structural feature of the mRNA binding channel, the latch, is closed as it has been closed in almost all PICs (Hinnebusch, 2017). The closed conformation of the latch ensures that mRNA remains in the binding channel during translation initiation or reinitiation.
DISCUSSION
Mechanism of DENR-MCT-1 dependent translation initiation/reinitiation and recycling
Our structure of the human 40S-DENR-MCT-1 complex illuminates important details of interactions between DENR, MCT-1, and the ribosome. It reveals that the C-terminal domain of DENR bears a strong structural resemblance to eIF1 and occupies the same binding site on the 40S subunit, suggesting that they use similar mechanisms in their function in discriminating initiator tRNA in the P site of the 40S subunit. The DENR-MCT-1 binding interface constitutes the surface that may bind/stabilize the acceptor stem of the P-site tRNA, facilitating translation initiation/reinitiation on the mRNAs with AUG positioned directly in the P site of the 40S subunit. Interestingly, superposition of the mammalian 48S initiation complex with 40S-DENR-MCT-1 did not reveal steric clashes between DENR-MCT-1 and eIF2-tRNAi (Simonetti et al., 2016). These raise the question of whether DENR-MCT-1 can replace eIF1 in scanning during translation initiation, and whether it can be done without eIF3 or eIF2. The binding site of MCT-1 on the 40S subunit overlaps with binding sites of eIF3a and eIF3c (or eIF3b RRM). This probably facilitates dissociation of eIF3 from the interface of the 40S subunit induced by the HCV-like IRESs and promotes their translation initiation. Importantly, DENR-MCT-1 could sense the presence of eIF3 on the ribosome during recycling and translation reinitiation. If no eIF3 is present on post-TC ribosome, DENR-MCT-1 binds to it and ejects deacylated tRNA from the P site, promoting recycling. If eIF3 is present, DENR-MCT-1 cannot bind, which leads to reinitiation. These observations suggest that DENR-MCT-1 could function as a switch directing post-TCs either to the recycling or to the reinitiation pathway. Alternatively, DENR-MCT-1 and eIF3 may sense and respond to different features of 5′ UTR of the upstream ORFs and thus promote reinitiation on different classes of mRNAs in mutually independent manner. Future high-resolution structures of the ribosomal complexes with DENR-MCT-1, eIFs, mRNA and tRNA coupled with biochemical studies are needed to refine and complete the model presented here.
EXPERIMENTAL PROCEDURES
Detailed methods are provided in the Supplemental Experimental Procedures.
Sample Preparation
Human 40S ribosomal subunits were purified from HEK293 cells. Recombinant human DENR and MCT-1 were overexpressed in E. coli BL21 (DE3) strain.
Structure determination
The structure of the 40S subunit, taken from the cryo-EM reconstruction of the human 80S ribosome was used as a search model to determine phases by molecular replacement for calculating the electron density map (Khatter et al., 2015; Quade et al., 2015). The structure of the 40S subunit was separated into 8 domains, which were treated as rigid bodies during refinement (Tables S1,2). The presence of unbiased additional electron densities in the map allows us to unambiguously identify the locations of MCT-1 and DENR (Figure S1A). At a later stage, the structures of MCT-1 and C-DENR were added to the refinement. The high-resolution structure of human MCT-1 was used as a model (Tempel et al., 2013), while the structure of C-DENR was modeled based on sequence homology using the I-TASSER server and then manually fitted in the electron density map (Yang and Zhang, 2015). Six 40S-DENR-MCT-1 monomers in the ASU are arranged as two trimers related by a noncrystallographic twofold axis of symmetry (Figure S2).
Supplementary Material
Acknowledgments
We thank the members of the Steitz lab for useful suggestions, the staff of the Advanced Photon Source beamline 24-ID, the Richards Center facility at Yale University and the Yale Liver Center for support. Special thanks to J. Wang and Y. Xiong for advice with crystallographic software. This work was supported by the National Institutes of Health (NIH) grant GM022778 (to T.A.S.), NIH NIDDK grant P30KD034989 (to I.B.L.) and the Russian Science Foundation (RSF) grant 14-50-00060 (to S.E.D.).
Footnotes
ACCESSION NUMBERS
Coordinates and structure factors have been deposited in the Protein Data Bank under accession code 5VYC.
Supplemental Information includes Supplemental Experimental Procedures, five figures and two tables and can be found with this article online at XXXX.
AUTHOR CONTRIBUTIONS
I.B.L. designed and performed experiments, analyzed data, wrote the paper and directed research; E.A.S, C.Z., A.V. performed experiments, analyzed data; M.B.G. analyzed data and wrote the paper, S.E.D designed experiments, analyzed data and wrote the paper; T.A.S. analyzed data, wrote the paper and directed research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andreev DE, O’Connor PB, Loughran G, Dmitriev SE, Baranov PV, Shatsky IN. Insights into the mechanisms of eukaryotic translation gained with ribosome profiling. Nucleic Acids Res. 2017;45:513–526. doi: 10.1093/nar/gkw1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Beilharz TH, Preiss T. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature. 2016;535:570–574. doi: 10.1038/nature18647. [DOI] [PubMed] [Google Scholar]
- Cerrudo CS, Ghiringhelli PD, Gomez DE. Protein universe containing a PUA RNA-binding domain. FEBS J. 2014;281:74–87. doi: 10.1111/febs.12602. [DOI] [PubMed] [Google Scholar]
- Dever TE, Green R. The elongation, termination, and recycling phases of translation in eukaryotes. Cold Spring Harb Perspect Biol. 2012;4:a013706. doi: 10.1101/cshperspect.a013706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deyo JE, Chiao PJ, Tainsky MA. drp, a novel protein expressed at high cell density but not during growth arrest. DNA Cell Biol. 1998;17:437–447. doi: 10.1089/dna.1998.17.437. [DOI] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischer TC, Weaver CM, McAfee KJ, Jennings JL, Link AJ. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas MA, Ngo L, Li SS, Schleich S, Qu Z, Vanyai HK, Cullen HD, Cardona-Alberich A, Gladwyn-Ng IE, Pagnamenta AT, et al. De Novo Mutations in DENR Disrupt Neuronal Development and Link Congenital Neurological Disorders to Faulty mRNA Translation Re-initiation. Cell Rep. 2016;15:2251–2265. doi: 10.1016/j.celrep.2016.04.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature. 2013;503:539–543. doi: 10.1038/nature12658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey JW. The role of eIF3 and its individual subunits in cancer. Biochim Biophys Acta. 2015;1849:792–800. doi: 10.1016/j.bbagrm.2014.10.005. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science. 2016;352:1413–1416. doi: 10.1126/science.aad9868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. Structural Insights into the Mechanism of Scanning and Start Codon Recognition in Eukaryotic Translation Initiation. Trends Biochem Sci. 2017 doi: 10.1016/j.tibs.2017.03.004. [DOI] [PubMed] [Google Scholar]
- Hsu HL, Choy CO, Kasiappan R, Shih HJ, Sawyer JR, Shu CL, Chu KL, Chen YR, Hsu HF, Gartenhaus RB. MCT-1 oncogene downregulates p53 and destabilizes genome structure in the response to DNA double-strand damage. DNA repair. 2007;6:1319–1332. doi: 10.1016/j.dnarep.2007.02.028. [DOI] [PubMed] [Google Scholar]
- Hussain T, Llacer JL, Fernandez IS, Munoz A, Martin-Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell. 2014;159:597–607. doi: 10.1016/j.cell.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. Termination and post-termination events in eukaryotic translation. Adv Protein Chem Struct Biol. 2012;86:45–93. doi: 10.1016/B978-0-12-386497-0.00002-5. [DOI] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. Structure of the human 80S ribosome. Nature. 2015;520:640–645. doi: 10.1038/nature14427. [DOI] [PubMed] [Google Scholar]
- Kolitz SE, Lorsch JR. Eukaryotic initiator tRNA: finely tuned and ready for action. FEBS Lett. 2010;584:396–404. doi: 10.1016/j.febslet.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Effects of intercistronic length on the efficiency of reinitiation by eucaryotic ribosomes. Mol Cell Biol. 1987;7:3438–3445. doi: 10.1128/mcb.7.10.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llacer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG, Ramakrishnan V. Conformational Differences between Open and Closed States of the Eukaryotic Translation Initiation Complex. Mol Cell. 2015;59:399–412. doi: 10.1016/j.molcel.2015.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Shirokikh NE, Yusupov MM, Hellen CU, Pestova TV. The fidelity of translation initiation: reciprocal activities of eIF1, IF3 and YciH. The EMBO journal. 2006;25:196–210. doi: 10.1038/sj.emboj.7600904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Steitz TA. The initiation of mammalian protein synthesis and mRNA scanning mechanism. Nature. 2013;500:307–311. doi: 10.1038/nature12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazan-Mamczarz K, Hagner P, Dai B, Corl S, Liu Z, Gartenhaus RB. Targeted suppression of MCT-1 attenuates the malignant phenotype through a translational mechanism. Leuk Res. 2009;33:474–482. doi: 10.1016/j.leukres.2008.08.012. [DOI] [PubMed] [Google Scholar]
- Mohammad MP, Munzarova Pondelickova V, Zeman J, Gunisova S, Valasek LS. In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S, Reinert LS, Hachem A, Mazan-Mamczarz K, Hagner P, He H, Gartenhaus RB. Phosphorylation of MCT-1 by p44/42 MAPK is required for its stabilization in response to DNA damage. Oncogene. 2007;26:2283–2289. doi: 10.1038/sj.onc.1210030. [DOI] [PubMed] [Google Scholar]
- Oh JJ, Grosshans DR, Wong SG, Slamon DJ. Identification of differentially expressed genes associated with HER-2/neu overexpression in human breast cancer cells. Nucleic Acids Res. 1999;27:4008–4017. doi: 10.1093/nar/27.20.4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Arellano I, Gallego J, Cervera J. The PUA domain - a structural and functional overview. FEBS J. 2007;274:4972–4984. doi: 10.1111/j.1742-4658.2007.06031.x. [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Skabkin MA, Pisareva VP, Skabkina OV, Rakotondrafara AM, Hentze MW, Hellen CU, Pestova TV. The role of ABCE1 in eukaryotic posttermination ribosomal recycling. Mol Cell. 2010;37:196–210. doi: 10.1016/j.molcel.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyry TA, Kaminski A, Jackson RJ. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosniak M, Dierov J, Okami K, Tilton B, Jameson B, Sawaya BE, Gartenhaus RB. A novel candidate oncogene, MCT-1, is involved in cell cycle progression. Cancer Res. 1998;58:4233–4237. [PubMed] [Google Scholar]
- Quade N, Boehringer D, Leibundgut M, van den Heuvel J, Ban N. Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-A resolution. Nat Commun. 2015;6:7646. doi: 10.1038/ncomms8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- Reinert LS, Shi B, Nandi S, Mazan-Mamczarz K, Vitolo M, Bachman KE, He H, Gartenhaus RB. MCT-1 protein interacts with the cap complex and modulates messenger RNA translational profiles. Cancer Res. 2006;66:8994–9001. doi: 10.1158/0008-5472.CAN-06-1999. [DOI] [PubMed] [Google Scholar]
- Ryabova LA, Pooggin MM, Hohn T. Translation reinitiation and leaky scanning in plant viruses. Virus Res. 2006;119:52–62. doi: 10.1016/j.virusres.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Schleich S, Strassburger K, Janiesch PC, Koledachkina T, Miller KK, Haneke K, Cheng YS, Kuchler K, Stoecklin G, Duncan KE, et al. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature. 2014;512:208–212. doi: 10.1038/nature13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A, Brito Querido J, Myasnikov AG, Mancera-Martinez E, Renaud A, Kuhn L, Hashem Y. eIF3 Peripheral Subunits Rearrangement after mRNA Binding and Start-Codon Recognition. Mol Cell. 2016;63:206–217. doi: 10.1016/j.molcel.2016.05.033. [DOI] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Hellen CU, Pestova TV. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol Cell. 2013;51:249–264. doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tempel W, Dimov S, Tong Y, Park HW, Hong BS. Crystal structure of human multiple copies in T-cell lymphoma-1 oncoprotein. Proteins. 2013;81:519–525. doi: 10.1002/prot.24198. [DOI] [PubMed] [Google Scholar]
- Weisser M, Voigts-Hoffmann F, Rabl J, Leibundgut M, Ban N. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nature structural & molecular biology. 2013;20:1015–1017. doi: 10.1038/nsmb.2622. [DOI] [PubMed] [Google Scholar]
- Wethmar K. The regulatory potential of upstream open reading frames in eukaryotic gene expression. Wiley Interdiscip Rev RNA. 2014;5:765–778. doi: 10.1002/wrna.1245. [DOI] [PubMed] [Google Scholar]
- Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43:W174–181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinoviev A, Hellen CU, Pestova TV. Multiple mechanisms of reinitiation on bicistronic calicivirus mRNAs. Mol Cell. 2015;57:1059–1073. doi: 10.1016/j.molcel.2015.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.