Abstract
Electron cryotomography (ECT) enables intact cells to be visualized in 3D in an essentially native state to ‘macromolecular’ (~4 nm) resolution, revealing the basic architectures of complete nanomachines and their arrangements in situ. Since its inception, ECT has advanced our understanding of many aspects of prokaryotic cell biology, from morphogenesis to subcellular compartmentalization and from metabolism to complex interspecies interactions. In this Review, we highlight how ECT has provided structural and mechanistic insights into the physiology of bacteria and archaea and discuss prospects for the future.
Historically, bacteria and archaea were viewed mainly as undifferentiated sacs of jumbled enzymes (for example, see REFS 1,2). Technological advances, particularly in imaging, have given rise to a much more complicated and beautiful view of prokaryotic cells3. We now see these cells as organized assemblies of macromolecular machines4, optimized to travel through and interact with complex and dynamic environments. This increasing knowledge of finer and finer details of microbial cell biology has been enabled by astounding technological advances in imaging. In the seventeenth century, Antonie van Leeuwenhoek constructed microscopes with magnifying powers of several hundred times, enabling the first visualization of single-celled organisms. The development of electron microscopy in the first half of the twentieth century enabled magnification on the order of hundreds of thousands of times, bringing resolution from the level of microorganisms to that of atoms. This provided an unprecedented view inside cells, and much of what we know about cellular structure has come from studies using electron microscopy.
However, because of the vacuum that is necessary to operate an electron microscope, the traditional preparation of biological samples for electron microscopy involved fixation and dehydration, which can denature structures and introduce misleading artefacts5,6. This limitation was circumvented with the discovery that thin aqueous samples can be cooled so quickly that water molecules stop rearranging before they can crystallize, which results in the formation of a thin film of ‘vitreous ice’ (an amorphous solid that preserves native cellular structures)7. In electron cryotomography (ECT; also known as cryo-ET or CET), such plunge-frozen samples are imaged in an electron microscope at different angles as they are tilted, resulting in a series of projection images that can be computationally combined to produce a reconstruction of the specimen in 3D. Therefore, ECT enables entire cells or parts of cells to be viewed with macromolecular resolution (~4 nm) in 3D, in what is essentially a native state (BOX 1; FIG. 1) (Supplementary information S1 (movie)).
Box 1. Electron cryotomography.
In electron cryotomography (ECT), cells in standard aqueous media are plunge-frozen onto electron microscope grids using an efficient cryogen, such as a mixture of ethane and propane, which cools the sample at ~104 K s−1, leading to the formation of vitreous (non-crystalline) ice7,156. Grids are subsequently kept at the temperature of liquid nitrogen (~80 K). A series of images is acquired by transmission electron microscopy (TEM) as the grid is rotated, typically one or two degrees between images. Full 180° tilt is impossible owing to increasing sample thickness at high angles, as well as intrusion of the sample holder. In practice, 120° to 140° are usually collected. The resulting projection images are then digitally reconstructed into a 3D tomogram. Fiducial markers (usually electron-dense gold beads) can be added to the sample to assist in alignment during reconstruction. The lack of high-tilt-angle images introduces a ‘missing wedge’ artefact that decreases the resolution of the reconstruction along the z-axis (parallel to the beam).
The fundamental limitation to resolution in ECT is radiation damage; in imaging a unique object, the clarity of the final image is limited by how many electrons are scattered before the sample is destroyed. In addition, the high electron-scattering cross-section of biological material limits sample thickness to less than ~500 nm for reasonable image quality in currently available TEMs. Therefore, the small size of some prokaryotic cells makes them amenable to whole-cell ECT. Thicker cells can be prepared for ECT by sectioning, for example by vitreous cryosectioning157, although this technique can introduce artefacts, particularly compression artefacts from the pressure of the blade158,159. An alternative approach, focused ion beam (FIB) milling, uses an ion (typically gallium) source to precisely mill away material in a thick frozen sample, leaving a thin lamella, just a few hundred nanometres thick, that is suitable for imaging by ECT160–163. This expands the utility of ECT to larger prokaryotic and eukaryotic cells and tissues.
The typical resolution of whole-cell ECT is a few nanometres, enabling visualization of the shapes and organization of macromolecular complexes and bridging the gap between the near-atomic resolution of X-ray crystallography or NMR spectroscopy and the broader cellular-level resolution of light microscopy. However, the ongoing development of many technologies promises to increase resolution. For example, direct detectors are replacing phosphor charge-coupled devices. In a direct detector, electron hits are recorded directly (rather than through photon production) and can be counted individually, increasing resolution164. Resolution is also increasing through the adoption of energy filters, which block electrons that are inelastically scattered by thick biological samples and contribute to imaging noise. Zernike and Volta phase plates increase contrast in images, especially at low spatial frequencies, enabling the resolution of biological detail with lower electron doses, and therefore less damage to samples67,165–169.
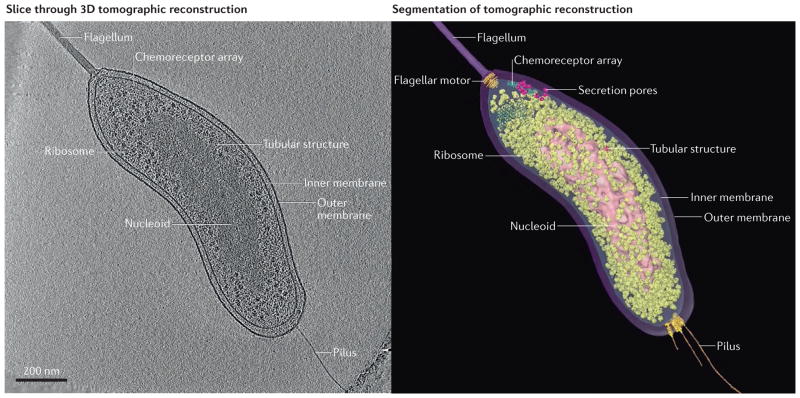
Figure 1. Example of electron cryotomography.
An intact Bdellovibrio bacteriovorus cell in standard media was plunge-frozen and imaged by electron cryotomography (ECT). The resulting tilt-series of images was reconstructed into a 3D tomogram. A slice through the reconstruction is shown (left panel), as well as a segmentation of visible cellular structures (right panel). To see the full reconstruction and segmentation, as well as fitting of crystal structures into electron microscopy densities, see Supplementary information S1 (movie).
A major limitation of ECT is the thickness of the sample, which should be less than ~500 nm for good resolution. However, the small size of many bacterial and archaeal cells makes them particularly amenable to ECT analysis. Indeed, ECT has enabled the observation of many components of these cells, including: the cell envelope and cytoskeleton; macromolecular machines involved in cell division, motility and navigation; and subcellular compartments. This has provided new insights into various aspects of prokaryotic physiology, including metabolism, interspecies cooperation and pathogenesis.
In this Review, we highlight the advances in our structural knowledge of prokaryotic cells that these efforts have produced (for more focused reviews of the technical aspects of ECT, see REFS 5,8,9; and for a review of eukaryotic applications of ECT, see REF. 10). In a previous review of this subject, in 2007, we discussed all of the relevant studies at that time, 15 in total11. Now that there is an order of magnitude more studies, we quickly summarize the existing published literature, highlighting what we feel are particularly interesting examples. As a narrative tool, we organize our discussion in terms of hypothetical challenges and opportunities that a single-celled organism might have encountered during the course of evolution.
Separation from the environment
One of the first requirements for the development of a cell is separation from the environment by a selectively porous envelope. As a first example of how ECT has provided new insights into cell biology, we discuss the mycobacterial cell envelope, an understanding of which is vital for the development of therapeutics.
Until fairly recently, the structure of pathogenic mycobacterial cell envelopes was debated, with various models proposing either a single membrane or a double membrane. Traditional electron microscopy of thin fixed sections could not resolve the structure of the mycobacterial cell envelope, as lipid structures are susceptible to rearrangements that are caused by dehydration and/or organic solvents during sample preparation. By contrast, ECT is particularly effective at preserving lipid structures, and studies using vitreous cryosectioning as well as whole cells have provided the first direct visualization of native envelopes in Mycobacterium bovis bacillus Calmette–Guérin (BCG) and Mycobacterium smegmatis, and the closely related bacterium Corynebacterium glutamicum. These studies revealed that the envelope of these bacteria is composed of an inner cytoplasmic membrane and a symmetrical outer membrane that is morphologically similar to that of Gram-negative bacteria12,13 (FIG. 2a), which rules out a fundamentally different envelope morphology for mycobacteria.
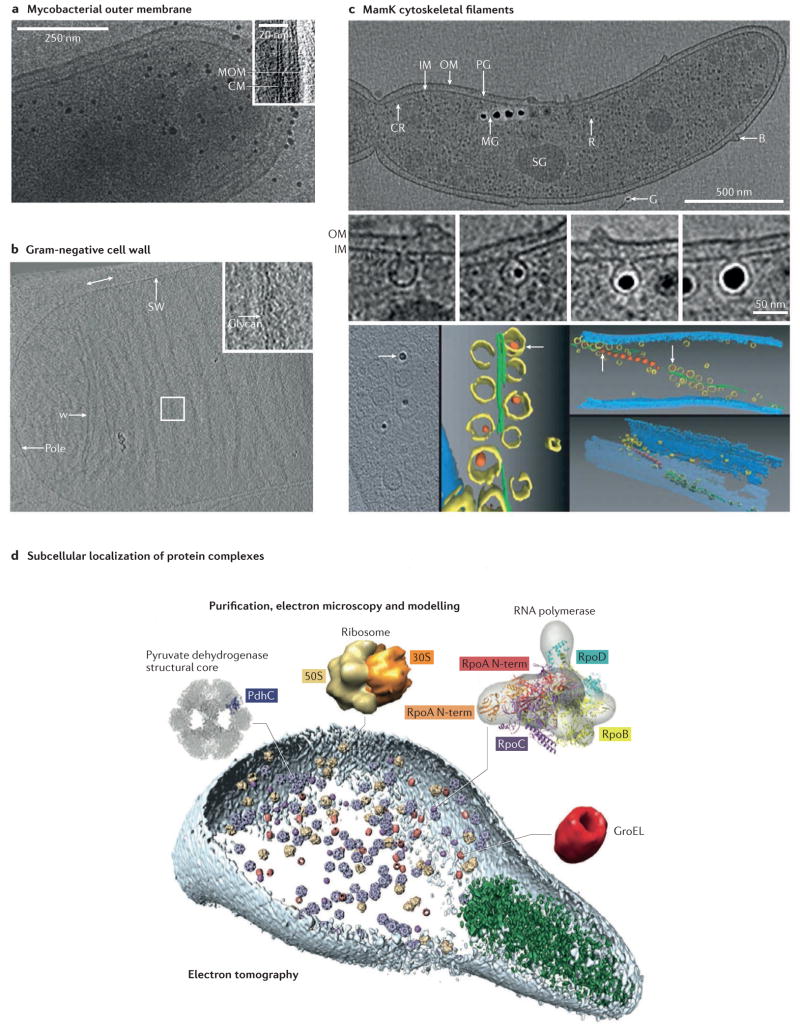
Figure 2. Cellular organization.
a | Electron cryotomography (ECT) provides excellent preservation of bacterial cell envelopes, enabling the direct visualization of the outer membrane bilayer of Mycobacterium bovis. b | ECT has also revealed the circumferential arrangement of glycan strands in the purified peptidoglycan sacculus of Escherichia coli. c | ECT has uncovered the existence and diversity of cytoskeletal filaments in bacteria, such as the filaments of MamK that organize the magnetosome chain in Magnetospirillum spp. In the segmentation, the inner membrane is shown in blue, magnetosomes are shown in yellow (with magnetite shown in orange) and MamK filaments are shown in green. d | ECT can be combined with knowledge about the structure of protein complexes to map their subcellular localization, a technique known as visual proteomics, as shown here in a Mycoplasma pneumoniae cell. The rod, a structural component of the specialized cell tip that is thought to be important for attachment to host cells, is shown in green. B, bleb; CM, cytoplasmic membrane; CR, chemoreceptor array; G, gold fiducial alignment marker; IM, inner membrane; MG, magnetosome chain; MOM, mycobacterial outer membrane; OM, outer membrane; PG, peptidoglycan; R, ribosome; SG, storage granule; SW, side wall; w, wrinkle. Part a is reproduced with permission from Hoffmann, C., Leis, A., Niederweis, M., Plitzko, J. M. & Engelhardt, H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl Acad. Sci. USA 105, 3963–3967 (2008). Copyright (2008) National Academy of Sciences, USA. Part b is reproduced with permission from Gan, L., Chen, S. & Jensen, G. J. Molecular organization of Gram-negative peptidoglycan. Proc. Natl Acad. Sci. USA 105, 18953–18957 (2008). Copyright (2008) National Academy of Sciences, USA. Top panel of part c is reproduced from Komeili, A., Li, Z., Newman, D. K. & Jensen, G. J. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311, 242–245 (2006). Reprinted with permission from AAAS. Middle and bottom panels of part c are from REF. 27, Nature Publishing Group. Part d is reproduced from Kuhner, S. et al. Proteome organization in a genome-reduced bacterium. Science 326, 1235–1240 (2009). Reprinted with permission from AAAS.
Structural support
Cellular membranes must be supported against turgor and environmental pressures. The solution most bacteria use is the peptidoglycan cell wall, which is created by crosslinking long stiff glycan strands into a mesh-like network with short peptide crosslinks. Despite the ubiquity of peptidoglycan in microorganisms and decades of study of its structure, details of the arrangement of the glycan strands remained unclear until fairly recently. Competing models proposed either a circumferential arrangement of the glycan strands parallel to the cell surface, a scaffold-like arrangement perpendicular to the surface, or coiled cables. ECT of purified peptidoglycan sacculi revealed that strands are aligned circumferentially around the cell in both Gram-negative bacteria (such as Caulobacter crescentus and Escherichia coli)14 (FIG. 2b) and Gram-positive bacteria (such as Bacillus subtilis)15, with the main difference between these sacculi simply being the presence of multiple layers in the thicker Gram-positive cell wall16. This insight eroded the notion that Gram-negative and Gram-positive phyla are only distantly related (although some debate continues regarding the Gram-positive architecture)17.
ECT also helped to clarify the related ‘chlamydial anomaly’: the long-standing debate over whether Chlamydiae have a peptidoglycan cell wall. This debate arose from the observation that Chlamydiae are susceptible to peptidoglycan-targeting antibiotics, but attempts to purify sacculi from these bacteria have been unsuccessful. This contradiction was resolved when ECT revealed a peptidoglycan cell wall in two Chlamydiae, ‘Candidatus Protochlamydia amoebophila’ and Simkania negevensis18.
Cytoskeleton
A reinforced cell envelope enables cells to adopt different shapes. Most cells maintain specialized shapes by building an internal scaffold of filaments, or cytoskeleton. The cytoskeleton, which is well understood in eukaryotes, has been the subject of much debate in prokaryotes. Homologues of all three main classes of eukaryotic cytoskeletal elements (actin microfilaments, microtubules and intermediate filaments) have been found in bacterial genomes, but traditional electron microscopy methods largely failed to identify any of these filaments in bacterial cells. However, by providing better structural preservation than traditional electron microscopy methods, ECT has now revealed an impressive diversity of cytoskeletal filaments in bacterial cells that mediate a range of processes.
The actin homologue MreB has a role in shape determination in many rod-shaped bacteria, although its exact function remains unclear. Based on light microscopy of fluorescently tagged MreB, it was thought that long helical filaments of MreB wrapped around the cell, perhaps globally coordinating cell wall synthesis. However, long filaments were not seen in the cryotomograms of six bacterial species (E. coli, C. crescentus, B. subtilis, Vibrio cholerae, Borrelia burgdorferi and Acetonema longum)19, and such filaments were later determined to be artefacts, at least in the case of one MreB fluorescent protein fusion20. These studies highlight the power of ECT to elucidate what is not present, in addition to what is. Subsequent fluorescence light microscopy studies showed small patches of MreB dynamically travelling around the cell21,22. These data support the idea that MreB does not form long helical filaments around the cell but also highlight a major limitation of ECT: it can only provide snapshots of cell structures, missing the dynamics that are inherent in many cellular processes.
An intermediate filament homologue, crescentin, is known to have an important role in defining the curved cell shape of C. crescentus. ECT helped to identify novel filaments of a metabolic enzyme, CTP synthase, that seem to negatively regulate crescentin to define the correct curvature of the cell23,24. Interestingly, the assembly of CTP synthase into filaments seems to inhibit its activity, reflecting a more general mechanism of regulating enzymatic activity by polymerization. Notably, this regulatory mechanism seems to be conserved across prokaryotes and eukaryotes, and may in fact have given rise to the bacterial cytoskeleton25.
ECT has also helped to identify some cytoskeletal filaments that are unique to bacteria, such as a unique class of polymeric proteins called bactofilins, which are nearly universally conserved in bacteria. ECT of C. crescentus revealed that two bactofilins form a scaffold to recruit cell wall biosynthetic enzymes to the stalked cell pole, probably supporting the formation of this thin adhesion structure26. The functions of bactofilin in other organisms are still being discovered, but seem to reflect an intermediate-filament-like role in the bacterial cytoskeleton.
Other cytoskeletal elements function in processes other than cell shape determination. MamK, another protein related to MreB (and actin), is found in magnetotactic bacteria and, as shown by ECT, forms filaments that help to align magnetosomes into a compass that orients the bacterium in a magnetic field27–31 (FIG. 2c). Two tubulin homologues, bacterial tubulin A (BtubA) and BtubB, were identified in Prosthecobacter spp.32, but were not seen to form microtubule-like filament bundles in vivo32 or in vitro33 by traditional electron microscopy. However, ECT revealed that BtubA and BtubB form filament tubes that are very similar to eukaryotic microtubules, although they comprise only five protofilaments rather than the 13 usually found in eukaryotes34. These findings challenge the idea that microtubules are a uniquely eukaryotic structure. ECT has also been helpful in characterizing the structure and function of several other filaments that are involved in cell division (see below).
Subcellular organization
In addition to having a cytoskeleton and a defined shape, cells benefit from organizing their interiors in other ways. An early goal of ECT was to address the question of whether protein components localize to specialized locations in bacterial cells. Visual proteomics, whereby protein complexes are surveyed in individual cells using template matching algorithms that are based on avail able structures35,36, has been used to count proteins in the cell37,38 and to determine their subcellular localization. For example, an estimated 15% of the 70S ribosomes in Spiroplasma melliferum were found to be associated with the membrane in a preferred orientation39, and ribosomes were seen to be similarly peripherally localized in ultra-small archaeal Richmond Mine acidophilic nanoorganism (ARMAN) cells40. In Leptospira interrogans41 and Mycoplasma pneumoniae42, the cellular distribution of several molecular complexes was mapped by visual proteomics, which showed that large complexes can be located within the cell and paved the way for future work to reveal the finer details of proteome organization (FIG. 2d).
One way that prokaryotic cells organize their interior is by clustering functionally related enzymes into specialized compartments, which are functional analogues of eukaryotic organelles. For example, green photosynthetic bacteria use compartments called chlorosomes to aggregate light-harvesting pigments, with each chlorosome containing up to 250,000 bacterio chlorophyll molecules43. ECT was used to reveal the structure and distribution of chlorosomes (covering roughly 70% of the cytoplasmic membrane) and their association with reaction centres in the membrane44,45. This enabled, for the first time, the mapping of reaction centres, which were found to be irregularly clustered throughout the membrane45. Similarly, cyanobacteria concentrate photosynthetic enzymes in the inner membrane, expanding their energy-harvesting capabilities by elaborating invaginations of the membrane. The extensive lamellar structure46,47, and assembly in response to light48, of these intracytoplasmic membranes were characterized by ECT. The additional finding that these membranes can bud into fully detached vesicular structures challenges the assumption that membrane-enclosed organelles that are not attached to the plasma membrane are found only in eukaryotic cells49.
In addition to photosynthetic compartments, ECT has also elucidated the structure of carboxysomes, which are used by cyanobacteria and chemo autotrophic bacteria to concentrate ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO), probably the most abundant protein on earth and a crucial catalyst in carbon fixation50. ECT revealed that carboxysomes have irregular proteinaceous icosahedral shells and that these shells and the internal RuBisCO lattice probably form simultaneously51–53. Interestingly, carboxysomes were seen to associate with other subcellular structures, polyphosphate storage granules (discussed below), which suggests a functional relationship between these two compartments53. ECT also revealed the fairly heterogeneous polyhedral structure of another bacterial microcompartment that is responsible for the utilization of propanediol. This microcompartment also has clustered enzymes surrounded by a thin protein shell, which indicates that this organization may represent a general structural feature of bacterial microcompartments54.
ECT has even been able to reveal some information about how DNA is organized within living cells. Similarly to eukaryotes, bacterial cells have subcellular organization of their genetic material, packing their chromosomes into the nucleoid (the genome-containing region of the cell). ECT of Bdellovibrio bacteriovorus, a small predatory bacterium, revealed a twisted spiral compaction of the nucleoid with ribosomes located along the periphery. Different levels of compaction were observed in different cells, including an MreB mutant, which suggests that DNA packing may regulate transcription, similarly to what is observed for eukaryotic chromatin55. ECT has shown that Leptospira spp. organize their DNA into bundles of parallel filaments56 and that ultra-small bacteria, which were recently discovered with the help of a portable field system to plunge-freeze samples from diverse natural environments for ECT57, have similar dense packing of putative DNA filaments58. Further study is required to examine how microorganisms may use DNA packing to regulate transcription.
Another kind of subcellular differentiation occurs in C. crescentus, which attaches to a surface by means of a long thin stalk. Correlated light microscopy and ECT helped to identify four proteins (StpABCD) that form bands in the stalk, which act as general barriers to diffusion of periplasmic, inner membrane and outer membrane proteins, compartmentalizing the cell and further blurring the boundary between eukaryotic and prokaryotic features59.
Finally, some extracellular appendages also act to spatially differentiate cellular processes. For example, ECT revealed the stalk structures that iron-oxidizing bacteria (Mariprofundus ferrooxydans and Gallionella-like bacteria) use to concentrate iron precipitation away from their interior or overall surface, where it would be detrimental60. Similarly, ECT has elucidated the structure of the ferric nano-aggregates that decorate the surface of iron-reducing planktonic bacteria61. Gram-negative bacteria in aqueous environments frequently secrete outer membrane vesicles for metabolic purposes, and ECT revealed a clever adaptation of this system for the partially hydrated environment of the soil, in which a Delftia sp. Cs1-4 forms ‘nanopods’ that are composed of extended tubes contiguous with the surface layer (S-layer) and that deliver vesicles some distance from the cell62.
Motility
Prokaryotic cells have evolved complicated mechanisms that enable them to move to new and possibly more beneficial environments. Perhaps because motility is of such fundamental importance, or because of the diversity of environments that microorganisms inhabit, there is a surprising variety of ways by which these cells move.
In many cases, motility is mediated by cellular nanomachines (large, nanometre-scale multi-protein complexes) the overall architectures of which have been solved by ECT. Nanomachines that are involved in motility frequently span the cell envelope and contain extra cellular, periplasmic and cytoplasmic components. The structures of the individual pieces of nano machines can be solved by X-ray crystallography and/or NMR spectroscopy, but it often remains unclear how the pieces fit together, because either the complex cannot be purified intact or it cannot be assembled in vitro. Therefore, ECT is invaluable in revealing the structure of the complete complex in its intact state in vivo. The location of specific components can then be determined by comparing wild-type ECT structures to those of mutants in which individual proteins are either knocked out or tagged. This localization map can then be used to inform electron microscopy docking of atomic models into the ECT density, ultimately enabling the construction of pseudo-atomic models of the complete system.
One of the most common nanomachines involved in motility is the flagellum. ECT studies, and in particular subtomogram averaging, have shown the detailed location of most of the components of the flagellar motor63–65 and its architecture across many diverse bacterial species66 (FIG. 3a). This has revealed a surprising number of species-specific peripheral adaptations around a common structural core64–67. For example, motors in some organisms, such as Borrelia burgdorferi, contain collars that may function to stabilize the stator at higher torque. ECT was used successfully to visualize key structures that were lost in previous purified preparations, including the stator and the export apparatus, which is responsible for self-assembly of the flagellum63,68. Furthermore, the ability of ECT to resolve the complete macro molecular structure of the entire machine, as opposed to individual parts, has provided insights into its mechanical properties. For instance, in species that use periplasmic flagella to drive the rotation of the cell body (rather than an oar-like extracellular flagellum), ECT revealed larger rings that may produce the increased torque that is required to drive whole-cell rotation63. Finally, ECT also uncovered the modular sequence of assembly of the flagellum in B. burgdorferi: opening of the central channel and assembly of the rod, assembly of the hook and finally formation of the filament68.
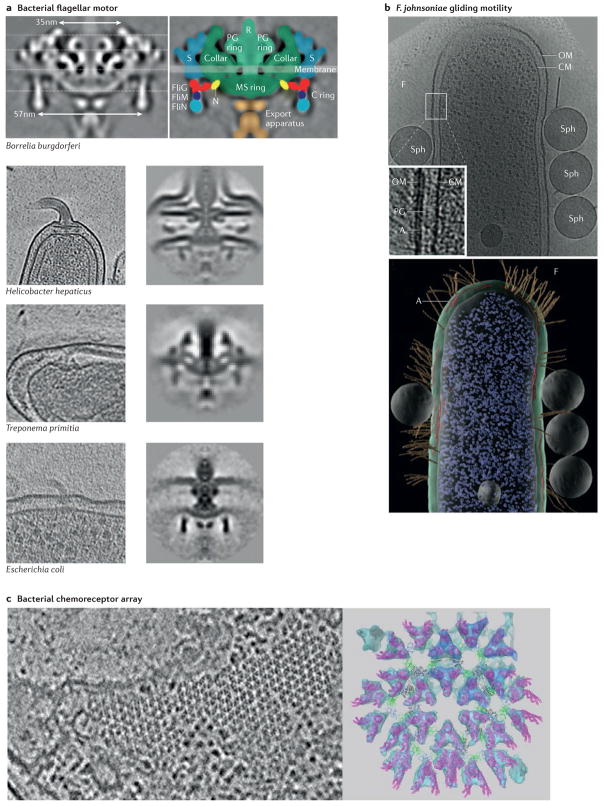
Figure 3. Motility and navigation.
a | Electron cryotomography (ECT) and subtomogram averaging have revealed the high-resolution structure of the bacterial flagellar motor (top panel), and imaging different species has uncovered species-specific structural adaptations (bottom panels). Top left and bottom right columns show subtomogram averages; bottom left column shows slices through tomographic reconstructions. b | ECT has also helped to identify the different motility mechanisms used by other bacteria, such as the filament-mediated gliding motility of Flavobacterium johnsoniae. The top panel shows a slice through a tomographic reconstruction of the bacterium, and the bottom panel a 3D segmentation. The electron-dense patches underlying filaments (A) and the latex spheres used to identify regions of motility (Sph) are indicated. c | High-resolution ECT and subtomogram averaging revealed the hexagonal lattice structure of the chemoreceptor arrays that bacteria and archaea use to direct motility. The image on the left is a slice through a tomographic reconstruction of an Escherichia coli cell, and the image on the right is a top view of a pseudo-atomic model of crystal structures docked into a subtomogram average (shown in cyan). Chemoreceptors are shown in magenta, the coupling protein CheW is shown in green and two domains of the kinase CheA are shown in black and blue. CM, cytoplasmic membrane; F, filament; MS ring, membrane and supramembrane ring; N, amino terminus; OM, outer membrane; PG, peptidoglycan; R, rod; S, stator. Top panel of part a is from J. Bacteriol., 2009, 191, 5026–5036 http://dx.doi.org/10.1128/JB.00340-09 and reproduced with permission from American Society for Microbiology. Bottom panel of part a is modified with permission from REF. 66, EMBO press. Part b is from J. Bacteriol., 2007, 189, 7503–7506 http://dx.doi.org/10.1128/JB.00957-07 and reproduced with permission from American Society for Microbiology. Left panel of part c is modified with permission from REF. 99, Wiley. The right panel of part c is modified with permission from REF. 97, National Academy of Sciences.
In addition to the structure of flagella, ECT has helped to identify details of their function in different motility systems. For example, in pathogenic spirochetes, such as Treponema denticola, Treponema pallidum and B. burgdorferi, periplasmic flagella enable the cells to penetrate tissue64,65,69–71. Whereas traditional electron microscopy preparations of B. burgdorferi showed a bundle of flagella, similar to that found in other species72, ECT revealed that in this case the flagella form a flat ribbon. This structure is indicative of a mechanism to exert force on the cell cylinder and propagate a wave along the long axis of the cell, which propels the cell forward73. Other ECT studies have identified the roles of individual proteins in B. burgdorferi flagellar assembly74–76 and have shown that motor rotation is required for the formation of the flagellar ribbon77. ECT of a mutualistic spirochete, Treponema primitia, lent support to a model of motility in which the rigid helical cell cylinder rolls within the sheath of the outer membrane78. ECT of the magnetotactic bacterium MO-1 identified a sheathed bundle of extracellular flagella that, along with associated thin filaments, may rotate in an interlocking-gear configuration that can generate tremendous power and speed79.
Other bacteria use non-flagellar systems for motility. For example, Flavobacterium johnsoniae exhibits gliding motility but does not contain genes that encode known motility structures. The mechanism of motility of this bacterium remained a mystery until ECT revealed tufts of thin filaments attached to the outer membrane. These filaments were shown to have a role in adherence-based motility80 (FIG. 3b). Furthermore, ECT revealed long cytoskeletal ribbons that are used by helical Mollicutes to propel themselves across surfaces; changes in the relative length of these ribbons are thought to turn the whole cell into a helical propeller81. ECT also identified that M. pneumoniae uses an elaborate attachment structure that consists of a large, jointed core in the cytoplasm and associated adhesion proteins on the surface to mediate what is thought to be inchworm-like movement37,82. In yet another mechanism, the iron-oxidizing bacteria discussed above are thought to propel themselves, at least in part, by the extrusion of their mineralizing stalks60. Finally, Listeria monocytogenes can remodel the actin cytoskeleton of its host into ‘comet tails’, which ECT has shown to comprise bundles of parallel, hexagonally packed filaments that power movement, perhaps by pushing or squeezing the bacterium forward, highlighting the remarkable ability of pathogens to remodel host cells to suit their needs83.
Navigation
With motility comes the problem of deciding where to go. We have already mentioned one potential orienting mechanism — magnetotaxis. ECT has revealed the structure and formation of magnetosome chains in several species27–30,84,85, including Magnetospirillum magneticum sp. AMB-1, which have vesicles loaded with ferrous crystals that are attached to the cytoplasmic membrane28 (FIG. 2c), and Magnetovibrio blakemorei, which have magnetosomes that are fully detached from the cytoplasmic membrane85. Interestingly, ECT of Desulfovibrio magneticus RS-1 led to the suggestion that magnetosomes in this bacterium are not surrounded by membranes84.
More widespread than magnetotaxis is chemotaxis, which integrates cues about the chemical environment into a signal transduction cascade that controls flagellar rotation or pilus extension. Much of our knowledge about the macromolecular structure of the chemotaxis system comes from a series of ECT studies showing that chemoreceptors are coupled together into a hexagonal lattice of interconnected rings71,86–93 (FIG. 3c). ECT revealed that this architecture is conserved across bacteria94 and archaea95, as well as between membrane-bound and cytoplasmic arrays96, suggestive of the fundamental utility of this architecture for chemosensory transduction. In another example of the power of subtomogram averaging and electron microscopy docking, ECT was used to generate a pseudo-atomic model of this extensive subcellular structure, revealing the interconnections between chemosensory components and supporting the idea that conformational changes in one hexagon of receptors could be transmitted to adjacent hexagons, thus amplifying the signal and explaining the remarkable sensitivity of the system97,98 (FIG. 3c). Other ECT studies have provided insights into the mechanism of activation of the system (in which substrate binding at the proximal end of the chemoreceptor triggers a conformational change in the kinase bound at its distal tip)99 and into the assembly of arrays from subunits of trimers-of-receptor-dimers100.
Division
Successful cells propagate through division, a process that presents many challenges. Below, we discuss how ECT has elucidated the molecular mechanisms of two of these processes — DNA segregation and cytokinesis.
First, to ensure that both daughter cells are functional copies, the cell must evenly distribute low-copy-number structures. The most obvious low-copy-number structure in the cell is the chromosome. ECT has helped to identify a ribosome exclusion zone at the poles of C. crescentus cells where PopZ tethers replicated chromosomes at either end of the cell to facilitate segregation101,102. In addition to the chromosome, different extrachromosomal plasmids are present at different copy numbers, sometimes as few as one per cell. ECT of E. coli revealed one solution to ensure that these plasmids are equally segregated: the actin homologue ParM, present as double helical filaments, positions the plasmids at the ends of the nucleoid for division103, and each replicated plasmid pair is probably pushed apart by a dedicated pair of antiparallel ParM doublets104 (FIG. 4a). Other bacteria, including Bacillus thuringiensis, use a tubulin homologue, TubZ, to segregate plasmids. TubZ also forms a double helical superstructure, which suggests the convergent evolution of bacterial filaments for this role of plasmid segregation105.
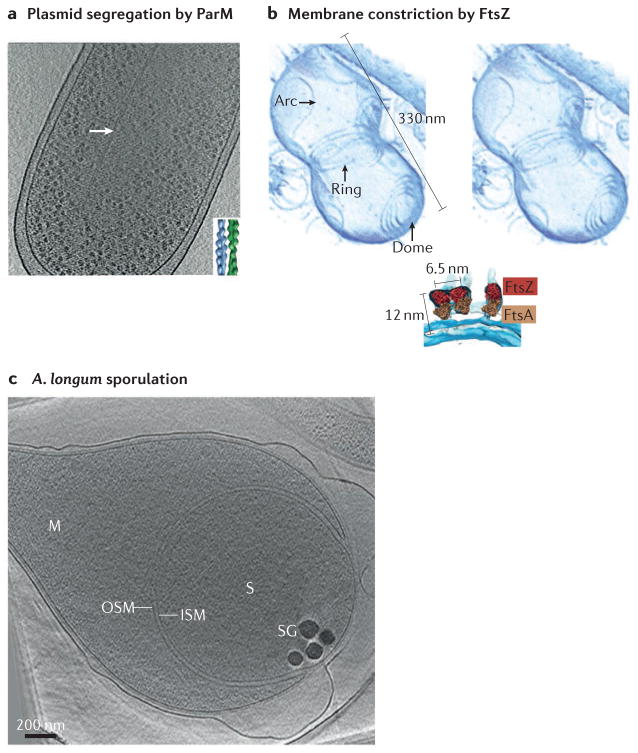
Figure 4. Division and differentiation.
Electron cryotomography (ECT) has provided mechanistic details of bacterial division, including the antiparallel doublets of ParM filaments that segregate plasmids (part a), and the copolymers of FtsZ and FtsA that can constrict membranes (part b). ECT has also revealed details of how bacterial cells differentiate into resistant spores (part c). Part a shows a slice through a tomographic reconstruction of an Escherichia coli cell expressing a high-copy-number plasmid. Inset shows a model of a ParM doublet based on a class average of 2D images of filaments formed in vitro. The top of part b is a stereoscopic image of a tomographic reconstruction of an in vitro system consisting of liposomes and FtsZ and FtsA purified from Thermotoga maritima. Below is a fit of the crystal structures of FtsZ from Staphylococcus aureus and FtsA from T. maritima into the ECT density. Part c shows a tomographic slice through an Acetonema longum cell. ISM, inner spore membrane; M, mother cell; OSM, outer spore membrane; S, spore; SG, storage granule. Part a is modified from REF. 104, Nature Publishing Group. Part b is modified from REF. 110. Part c is reproduced with permission from REF. 120, Cell Press.
Once contents are segregated, the cell must physically divide. Visualization of this process by ECT has provided mechanistic insights for both bacteria and archaea. Most bacteria use FtsZ, a GTPase that is homologous to eukaryotic tubulin, and FtsA, an actin homologue, to constrict the cell. ECT has shown that FtsA forms protofilaments that tether FtsZ filaments to the membrane106. However, details of the mechanism of action of FtsZ remained unclear until the filaments and division process were visualized directly by ECT in C. crescentus. In this organism, division is asymmetrical, and an early ECT study showed that the inner membrane completes constriction before the outer membrane107. Later tomograms revealed relatively few arcing FtsZ filaments positioned around the division site108, in contrast to previous models of thick, complete FtsZ rings based on early fluorescence microscopy. These observations support a model of division109 in which the FtsZ filaments themselves provide the force of constriction through iterative rounds of polymerization, attachment, conformational changes gathering in the membrane and depolymerization108,109. By contrast, later ECT work provided evidence for bundles of FtsZ filaments forming complete rings that encircle cells during the late stages of division and proposed a different model for constriction that was based on filament sliding110 (FIG. 4b). The discrepancies between these observations highlight two drawbacks of ECT imaging: the ‘missing wedge’ effect (see BOX 1) means that features, such as filaments, cannot be tracked all the way around the circumference of the cell; and static images, no matter how high their resolution, can only provide hints to the mechanisms of dynamic processes. FtsZ filaments are indeed highly dynamic and additional work is needed to resolve their constrictive mechanism.
ECT has also recently helped to show that two proteins, MinC and MinD, form copolymers that interact with the membrane and position FtsZ filaments at the division plane111. The use of ECT also led to the description of a C. crescentus protein, DipM, that coordinates cell wall remodelling for constriction during cell division112,113.
Instead of using FtsZ, some archaea use endosomalsorting complexes required for transport (ESCRT) proteins to divide. ESCRT mediates membrane scission events across many archaea and eukaryotes, but the exact mechanism has been the subject of much debate. However, ECT of dividing Sulfolobus acidocaldarius revealed a belt of filaments at the constriction zone, supporting a model in which spiralling filaments constrict the cell membrane114, although more work is required to test this.
Cytokinesis itself helps to divide some cellular features, such as the magnetosome chain in magnetotactic bacteria mentioned above. ECT identified a mechanism in which MamK localizes the magnetosome chain to the mid-cell, which ensures proper segregation of the chain to both daughter cells by cytokinesis115.
Surviving lean times
Nutrient sources in the environment are by no means guaranteed, and cells frequently face shortages. To prepare for such lean times, both bacteria and archaea store essential nutrients in storage granules during times of sufficiency, and details of these granules have been revealed by ECT. In both Gram-positive and Gram-negative bacteria, including C. crescentus, polyphosphate is stored in uniform, spherical structures that lack any membrane boundary, indicating that they form through a mechanism of local aggregation116,117. C. crescentus also stores carbon in slightly larger, less regular structures116. D. magneticus has been shown to concentrate iron and phosphorus in a membrane-bound compartment, although it is not clear whether this is for purposes of sequestration or storage84. Cupriavidus necator stores polyhydroxybutyrate in compartments that are not membrane bound but instead are coated by a protein shell and form as small granules that then coalesce118. ECT also revealed that the archaeon Methanospirillum hungatei contains several amorphous polyphosphate bodies that are located at the ends of the cell119.
To deal with lean times, some bacterial species form metabolically inert and environmentally protective spores. ECT studies of a rare Gram-negative endospore-forming member of the Firmicutes phylum revealed that the inner membrane of the mother cell surrounding the spore is inverted and eventually becomes the outer membrane of the germinating Gram-negative cell120 (FIG. 4c). This unexpected finding suggests that the sporulation process might have given rise to the diderm cell plan. ECT has also helped to identify polyphosphate storage granules in Gram-negative but not Gram-positive spores, which suggests that these granules may help to maintain the outer membrane during spore outgrowth16. Furthermore, ECT has also shown that membrane vesicles that are secreted by sporulating colonies of Streptomyces coelicolor are densely packed with proteins (including metabolic enzymes, antioxidants and resistance factors) that may aid survival121.
Cooperation and competition
In nature, prokaryotic cells are surrounded by other organisms, both single-celled and multicellular; these include conspecifics, symbionts, competitors, predators and prey. To adapt to such an environment, cells may coat their envelopes with proteins, creating a layer that provides protection against predation or mediates biofilm attachment. The S-layer can represent 10–15% of the total protein in the cell122, but its structure was difficult to resolve using traditional methods; although the S-layer is a crystalline lattice, it makes a poor sample for methods such as X-ray crystallography, at least in part because of its natural curvature around the cell. One of the first applications of ECT was to visualize the S-layer of intact Pyrobaculum aerophilum archaeal cells123, and ECT was later applied to the Gram-negative bacterium C. crescentus124. Surprisingly, these studies revealed that the S-layer is not a uniform lattice; instead, it has significant heterogeneity across the cell and includes regions of double-stacked layers, suggesting assembly from multiple nucleation points.
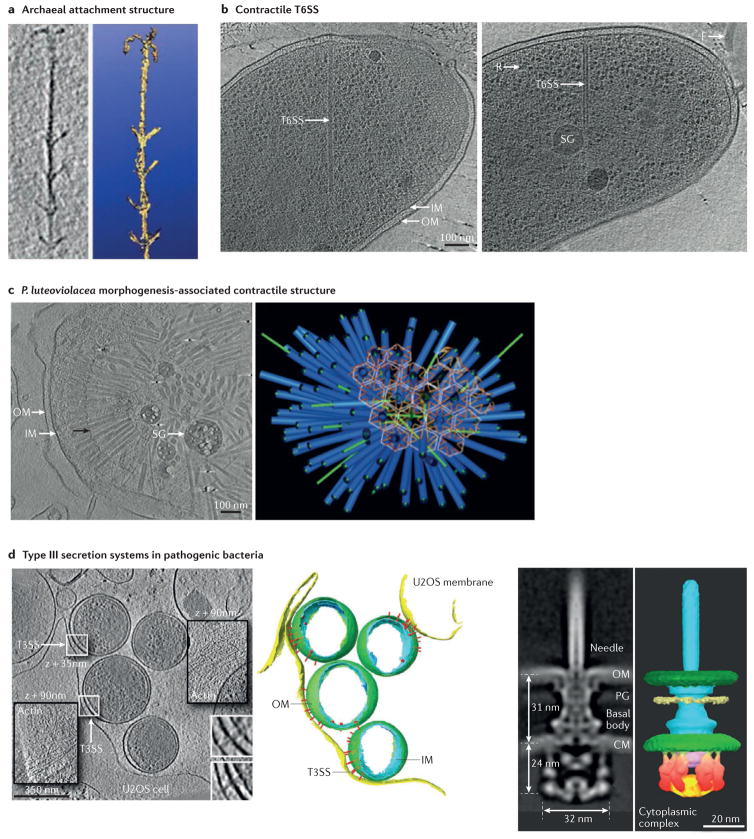
Cooperation can occur within or between species and even kingdoms, and it often involves specialized structures that enable physical contact between organisms. For example, studies have used ECT to obtain images of: the vast extracellular matrix of hollow tubules (known as cannulae) that anchors a community of Pyrodictium abyssi to one another125; interspecies cell–cell contacts of various archaeal communities126,127; and the grappling hook structures known as hami that the SM1 euryarchaeon uses to anchor itself in symbiotic communities with bacteria128 (FIG. 5a).
Figure 5. Interaction with other cells.
Electron cryotomography (ECT) has revealed some of the mechanisms prokaryotic cells use to interact with other prokaryotic cells. a | One example is the grappling-hook-like extracellular appendages, known as hami, that archaea use to anchor themselves in communities with bacteria. The image on the left is a slice through a tomographic reconstruction of a hamus from an SM1 euryarchaeal cell plunge-frozen in the marsh water from which it was isolated, and the image on the right is a 3D surface rendering. The hamus is approximately 60 nm across. b | Bacteria also use spring-loaded ‘molecular daggers’ to kill neighbouring competitors. The image shows slices through tomograms of the extended (left) and contracted (right) conformations of the type VI secretion system (T6SS) in intact Vibrio cholerae cells. c | ECT has also revealed the structures prokaryotic organisms use to interact with eukaryotic organisms. These include a T6SS-related structure that triggers differentiation in a marine tubeworm. The image on the left shows a slice through a tomographic reconstruction of a Pseudoalteromonas luteoviolacea cell in the process of lysis to produce an extracellular metamorphosis-associated contractile structure aggregate, shown as a 3D segmentation on the right. Sheaths are shown in blue, cores are shown in green, tail fibres are shown in orange and filamentous connections are shown in white. d | The oriented arrays of type III secretion systems (T3SS) that Chlamydia spp. use to attack human cells are another example of structures that are used to interact with eukaryotes. The image on the left is a slice through a tomographic reconstruction of Chlamydia trachomatis elementary bodies infecting U2OS cells, and in the middle is a 3D segmentation. The image on the right shows a slice through (left) and 3D surface rendering (right) of a subtomogram average of T3SSs in intact Shigella flexneri minicells. Insets show slices at different z-heights in the tomographic volume (35 or 90 nm above the background slice, as indicated) to best display features of interest. White boxes are magnified at bottom right. CM, cytoplasmic membrane; F, flagellum; IM, inner membrane; OM, outer membrane; PG, peptidoglycan; R, ribosome; SG storage granule. Part a is reproduced with permission from REF. 128, Blackwell. Part b is reproduced from REF. 129, Nature Publishing Group. Part c is reproduced from Shikuma, N. J. et al. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science 343, 529–533 (2014). Reprinted with permission from AAAS. Left and middle panels of part d are reproduced from REF. 140. The right panel of part d is reproduced with permission from REF. 141, National Academy of Sciences.
Instead of cooperating, prokaryotic cells might benefit more by killing their neighbours. For example, ECT revealed that the type VI secretion system (T6SS), a cellular nanomachine that was first characterized in V. cholerae and is used by many Gram-negative bacteria to combat bacterial and eukaryotic cells, is a spring-loaded ‘molecular dagger’ that is structurally homologous to a contractile phage tail (BOX 2). ECT visualized the dynamic conformations of T6SSs, including extended filled (loaded) and contracted empty (fired) tubes, thereby uncovering a mechanism by which the force of contraction drives the translocation of the T6SS needle into the target cell129 (FIG. 5b). In addition, correlated cryo-PALM and ECT enabled the identification of transient assembly and disassembly intermediates, highlighting the utility of this technique for identifying previously uncharacterized structures and conformations130. A T6SS-like structure is also responsible for a remarkable interaction between bacteria and multicellular eukaryotes. Larvae of a marine tubeworm select surfaces covered by bacterial biofilms of Pseudoalteromonas luteoviolacea to settle and differentiate into sessile adults. ECT revealed that the signals that the tubeworm recognizes are metamorphosis-associated contractile structures that are formed by an interconnected web of T6SS-like pyocins that develop inside P. luteoviolacea cells and are released by lysis to form a micrometre-scale, well-organized extracellular spike ball consisting of roughly 100 pyocins and associated proteins131 (FIG. 5c). The function of this structure for the bacterium remains unknown.
Box 2. Prokaryotic viruses.
Another important aspect of prokaryotic cell biology is the interaction of prokaryotes with viruses or phage. Electron cryotomography (ECT) studies have elucidated the structural features of viral capsids170,171 and detailed phage attachment to bacterial cells172–175, including a particularly elegant mechanism by which phage wrap a filament around the flagellum of Caulobacter crescentus, taking advantage of flagellar rotation to move toward the cell pole where they are concentrated around their pilus entry point, increasing the chance of infection in a dilute environment176 (see the figure, part a). Another study of a virus that attacks the conjugative F-pilus used subtomogram averaging to reveal asymmetries in the capsid that may lead to viral entry177. An early application of ECT was to show the contractile mechanism of the phage tail that injects the viral genome into the host cell178, which was further elucidated in later studies172,174,179–181. Further work probed the mechanism of phage assembly within bacterial cells182,183, including outward expansion of the nascent capsid due to pressure from RNA genome packaging184. Another study imaged host lysis, completing the infection cycle185.
Archaea are also prey to specific viruses, notably archaeal pyramidal viruses. ECT was used to elucidate the entry mechanism of these viruses, which involves tracking along pilus-like filaments186, as well as the striking release mechanism, in which a seven-faced pyramidal structure penetrates the membrane and surface layer (S-layer) of the host cell and opens to release mature virions187,188 (see the figure, part b). Archaea can also be infected by lemon-shaped viruses, the pleomorphic structure of which was recently resolved by ECT189.
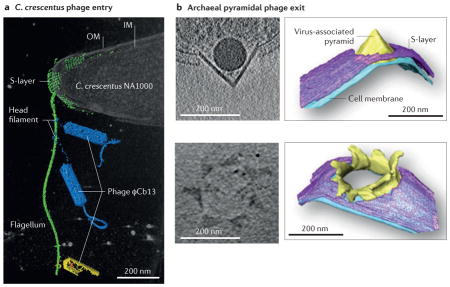
IM, inner membrane; OM, outer membrane. Figure part a shows a 3D segmentation overlaid on a volume rendering of a tomographic reconstruction of phage interacting with an intact C. crescentus cell. Figure part b shows slices (left) and 3D segmentations (right) of tomographic reconstructions of Sulfolobus islandicus cells infected with S. islandicus rod-shaped virus 2. Part a is reproduced with permission from REF. 176, National Academy of Sciences. Part b is modified with permission from REF. 188, National Academy of Sciences.
Alternatively, prokaryotic cells might eat their neighbours, as exemplified by B. bacteriovorus, which pierces the periplasm of its prey and eats their cytoplasmic contents. ECT provided new insights into this process by revealing an unusual flexibility of the cell wall that probably underlies the ability of B. bacteriovorus to squeeze into its prey132.
Pathogenicity
One particularly important form of interaction between prokaryotes and their neighbouring cells is pathogenicity to human hosts. Many of the adaptations that turn a bacterium into an efficient pathogen involve structural changes, and ECT has provided insights into the mechanisms of pathogenesis of several bacteria.
First, pathogens must sense and enter their host. In addition to pathogenic adaptations to the flagellar motor that are discussed above, many structural adaptations occur in the cell tip, which ECT has shown can be complex in pathogenic bacteria. For example, ECT of Campylobacter jejuni revealed an elaborate cell tip with storage granules, extensive chemoreceptor arrays and a complex flagellar motor, all of which are thought to help mediate invasion of host cells133. Similarly, ECT of Leptospira spp. revealed extensive chemoreceptor arrays that are probably important for sensing host cells56. ECT of T. pallidum and T. denticola identified complex conelike periplasmic structures at the cell tip, although the function of these structures remains unknown65,134.
Once a cell enters its host, it must evade the host immune system, and studies using ECT have provided key insights into how pathogens adapt their cell surface for this purpose. For example, T. pallidum exhibits a complex and labile outer membrane that is not tethered to the peptidoglycan layer as in most Gram-negative cells, enabling the bacterium to shed the outer membrane (which contains multiple components that are recognized by the immune system) and thereby evade the host immune response65. Similarly, ECT of B. burgdorferi revealed a labile outer membrane sheath70. Interestingly, ECT also observed cell fusion events in Borrelia spp., as well as two cells sharing a single outer membrane, suggesting that these bacteria use this as a strategy to diversify surface antigens to evade host immunity135.
Pathogenic bacteria use another nanomachine that is related to the flagellar motor to facilitate infection. ECT of Shigella flexneri, Salmonella enterica and Yersinia entero colitica identified the complete structure of the type III secretion system (T3SS) injectisome. This structure consists of a basal body and a secretion channel that delivers effector proteins directly into a target cell. The basal body is structurally similar to the flagellar motor, although it exhibits key differences, including a wider channel that may allow the secretion of at least partially folded substrates (rather than unfolded flagellin sub units) and greater elasticity136–138. Furthermore, pathogenic T3SSs in Y. enterocolitica were seen to cluster, perhaps to enhance secretion into a target cell139, and Chlamydia trachomatis was also seen to orient an array of T3SSs to contact the target cell140 (FIG. 5d). Later, higher-resolution ECT and subtomogram averaging of S. flexneri minicells revealed, for the first time, how the flagellar C-ring has been adapted into the sorting platform of the injectisome, a cytoplasmic complex that selects specific effector proteins for secretion141 (FIG. 5d).
Once in a host cell, some pathogenic bacteria, such as Chlamydia spp., undergo differentiation from infectious elementary bodies to metabolically active reticulate bodies. During this process, many pathogenic adaptations that are no longer required for survival within the host are lost. For example, ECT of C. trachomatis showed that T3SS-mediated cell contact induces bacterial internalization into early vacuoles, where polarization and T3SSs are lost140. Conversely, ECT also revealed the adaptations that accompany the differentiation of C. trachomatis from reticulate bodies into elementary bodies, such as a near doubling in the thickness of the outer membrane142. ECT of another pathogen, Chlamydophila abortus, showed that it has a similar T3SS structure and differentiation strategy to C. trachomatis143. ECT also identified that a previously identified Chlamydial development stage, the crescent-shaped stage, was actually an artefact of the chemical fixation and dehydration used in traditional electron microscopy preparations6, highlighting the power of ECT to image native structures.
ECT can be useful to characterize therapeutic agents that target pathogenic bacteria. For example, ECT was used to see how an antibody against B. burgdorferi lyses the cells by inducing outer membrane projections that probably increase permeability144. ECT may also be useful in identifying future therapeutic targets. For example, ECT has shown that a known Mycobacterium marinum virulence factor, SecA2, helps to maintain cell wall integrity145, which suggests that targeting this protein could be an effective strategy to combat mycobacterial infections. Similarly, ECT of Acinetobacter baumannii identified a pathogenic mechanism that is based on outer membrane vesicle delivery of effector molecules. Interestingly, these vesicles were seen to form in areas with irregular peptidoglycan, and sub-lethal concentrations of a peptidoglycan-targeting antibiotic enhanced vesicle formation146.
Finally, ECT may also help to design strategies against antibiotic-resistant pathogens. Many bacterial cells have evolved defence mechanisms against anti biotics, including the multidrug efflux pump, a cellular nanomachine that spans the inner and outer membranes and selectively exports antibiotics from the cytoplasm. The structure is too unstable to be purified, precluding most structure determination methods. ECT and subtomogram averaging revealed part of the structure and possible assembly of a reconstituted Pseudomonas aeruginosa multidrug efflux pump147, which offers promise for solving the full structure. This in situ structure would probably elucidate the mechanism of the efflux pump, which could inform new therapeutic strategies to combat antibiotic resistance.
Outlook
Compared with the scarcity of information available before 2002, we are witnessing an explosion of new, high-resolution information about molecular arrangements inside prokaryotic cells. This Review highlights the impressive diversity of structural insights into bacterial and archaeal physiology that have already arisen from ECT of whole cells in a native state. For example, for the first time, the arrangement of polymers in the cell wall can be directly visualized, which has enabled the observation of previously unappreciated structural similarities between Gram-positive and Gram-negative bacteria. Similarly, ECT revealed the interconversion of outer and inner membranes during sporulation, suggesting a simple new hypothesis for the evolution of the diderm cell plan. Also for the first time, ECT has directly proven that bacteria not only have a cytoskeleton, but that this cytoskeleton is rich and varied in function. Similarly, cell compartmentalization is no longer a feature restricted to eukaryotes; membrane-bound, even budded, compartments have been observed, and many structures have restricted subcellular localizations. ECT has also enabled the visualization of intact nanomachines, the macro molecular complexes that carry out diverse functions, from motility to interspecies interactions and pathogenicity. The complexity of these machines (which can comprise many copies of dozens of distinct proteins) and their locations spanning cellular compartments (cytoplasm, periplasm and extracellular space) mean that many cannot be purified or reconstituted intact. Now, by visualizing them in situ and creating pseudoatomic models, we can finally begin to understand them in context.
Much work remains to be done, and continuing technological developments (BOX 1) should expand the power of ECT in the near future. A key difficulty in ECT is identifying macromolecules of interest; the structures of many protein complexes remain undetermined because they cannot be unambiguously identified in the crowded cellular environment. For that reason, many ECT studies to date have focused on structures for which the morphology or localization was already known. To identify new structures, we need improved technologies to locate them. Correlated light microscopy and ECT has been successfully applied to identify several structures in vivo, and the recent development of correlated cryo-PALM and ECT has increased the precision of this technique, enabling the localization of tagged proteins to within a few hundred nanometres in a cell130. Anticipated technological improvements could further enhance localization to within tens of nanometres, in 3D. However, adding a bulky fluorophore to a protein of interest can introduce structural and localization artefacts, so the potential application of fluorescent small molecules148 to in vivo protein labelling, perhaps through the use of unnatural amino acids149, is intriguing. Another approach is to locate proteins of interest directly in tomograms, potentially with the help of electron-microscopy-specific tags, such as genetically encoded metallothionein-based or ferritin-based tags with a density that would stand out in electron microscopy images150–153. Other complex structures (for example, REFS 40,65,134,154,155) have been identified visually using ECT, but their functions remain a mystery. And even when ECT successfully identifies a protein complex, it can miss the dynamics of its localization.
Ultimately, we would want to map the structure and distribution of every protein in a prokaryotic cell. By combining ECT with other structure determination methods (such as X-ray crystallography, NMR spectroscopy and single-particle reconstruction), as well as with information about location and dynamics gained from studies using fluorescence light microscopy, we are getting ever closer to the goal of fully resolving the structure of prokaryotic cells.
Supplementary Material
Acknowledgments
The authors apologize that they could not discuss all of the work in this burgeoning field. The authors thank members of the Jensen laboratory for helpful comments on the manuscript, and J. Ding and Y.-W. Chang for producing the accompanying movie. The authors also thank L. Sockett (University of Nottingham) for the gift of the Bdellovibrio bacteriovorus strain imaged in figure 1 and shown in the accompanying movie. Microbial electron cryotomography (ECT) in the Jensen laboatory is supported, in part, by the Howard Hughes Medical Institute, the US National Institutes of Health (grants RO1 GM101425 and RO1 GM094800), the Beckman Institute at Caltech, Caltech’s Center for Environmental Microbial Interactions, and gifts to Caltech from the Gordon and Betty Moore Foundation and the Agouron Institute.
Glossary
- Phase plates
Electron microscope components that shift the phases of the scattered beam with respect to the unscattered beam to boost contrast.
- Vitreous cryosectioning
A sample preparation technique in which thick samples are frozen rapidly to prevent ice formation and then cut with a diamond blade into thin (50–400 nm) slices that can be imaged by electron cryotomography.
- Peptidoglycan
A polymer crosslinked into a mesh-like network that forms the cell walls of most bacteria.
- Sacculi
Sac-like exoskeletons (peptidoglycan cell walls) of bacterial cells.
- Stalked cell
In the context of Caulobacter crescentus, one of the two cells produced by asymmetric division. Unlike the motile swarmer cell, the stalked cell is attached to a surface and is capable of replication and division. Swarmer cells later differentiate into stalked cells.
- Magnetosomes
Compartments containing ferrous microcrystals that are used by magnetotactic bacteria to orient the cell in a magnetic field.
- Template matching
A digital image processing technique to search a 3D tomogram for an object of interest (the template). The template is typically a single-particle reconstruction or X-ray crystallographical structure of a macromolecule or complex of interest.
- Archaeal Richmond Mine acidophilic nanoorganisms
(ARMAN). A highly divergent group of archaea isolated from an extremely acidic environment in California.
- Reaction centres
Complexes of enzymes, pigments and cofactors that convert solar energy that is captured by light-harvesting antennae into chemical energy during photosynthesis.
- Correlated light microscopy and ECT
A specialization of correlated light microscopy and electron microscopy (CLEM) that is used to locate structures of interest within visually crowded tomograms. Cells containing a fluorescently labelled target protein are rapidly frozen on electron microscope grids that contain landmarks for correlation, and first imaged by light microscopy to identify the location of fluorescent signals. Grids are then transferred to the electron microscope and the same locations are imaged at high resolution by electron cryotomography (ECT).
- Electron microscopy docking
A method for modelling the structure of macromolecular complexes by fitting high-resolution models (like X-ray crystallographical structures) of components into electron microscopy density maps, often informed by biochemical studies detailing component interactions.
- Subtomogram averaging
An image processing technique to increase the clarity of structures that are present in multiple copies and/or in multiple tomograms. Averaging particles increases the signal-to-noise ratio and can yield reliable high-resolution (< 1 nm) detail, including secondary protein structure.
- Endosomal-sorting complexes required for transport
(ESCRT). Protein complexes that remodel cellular membranes to carry out various processes, including cell division and viral budding.
- Diderm cell plan
Under a classification system not based on the Gram stain, diderm bacteria are surrounded by two lipid bilayer membranes, as opposed to monoderms, which have only one.
- Correlated cryo-PALM and ECT
A specialized application of correlated light microscopy and electron cryotomography (ECT) using a super-resolution microscopy technique — photoactivated localization microscopy (PALM) — to increase the localization precision of a structure of interest in fluorescence images for correlation with high-resolution ECT images.
- Pyocins
Type VI secretion system (T6SS)-like contractile nanomachines, related to phage tails, that bacteria use to kill other bacteria.
- Elementary bodies
The non-replicating forms of Chlamydia spp., which are responsible for cellular infection.
- Reticulate bodies
The metabolically active forms of Chlamydia spp., which are found in the cytoplasm of infected cells.
- Single-particle reconstruction
A transmission electron microscopy technique in which many identical copies of a purified macromolecule or complex are imaged, providing different projection views of the particle that can then be computationally combined into a 3D reconstruction.
Footnotes
Competing interests statement
The authors declare no competing interests.
References
- 1.Loewy AG, Siekevitz P. Cell Structure and Function. Brooks/Cole; 1991. [Google Scholar]
- 2.Alberts B, et al. Molecular Biology of the Cell. Garland Publishing; 1983. [Google Scholar]
- 3.Goodsell DS. Escherichia coli Biochem Mol Biol Educ. 2009;37:325–332. doi: 10.1002/bmb.20345. [DOI] [PubMed] [Google Scholar]
- 4.Alberts B. The cell as a collection of protein machines: preparing the next generation of molecular biologists. Cell. 1998;92:291–294. doi: 10.1016/s0092-8674(00)80922-8. [DOI] [PubMed] [Google Scholar]
- 5.Pilhofer M, Ladinsky MS, McDowall AW, Jensen GJ. Bacterial TEM: new insights from cryo-microscopy. Methods Cell Biol. 2010;96:21–45. doi: 10.1016/S0091-679X(10)96002-0. [DOI] [PubMed] [Google Scholar]
- 6.Pilhofer M, et al. Architecture and host interface of environmental chlamydiae revealed by electron cryotomography. Environ Microbiol. 2014;16:417–429. doi: 10.1111/1462-2920.12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dubochet J, et al. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 8.Gan L, Jensen GJ. Electron tomography of cells. Q Rev Biophys. 2012;45:27–56. doi: 10.1017/S0033583511000102. [DOI] [PubMed] [Google Scholar]
- 9.Lucic V, Rigort A, Baumeister W. Cryo-electron tomography: the challenge of doing structural biology in situ. J Cell Biol. 2013;202:407–419. doi: 10.1083/jcb.201304193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fridman K, Mader A, Zwerger M, Elia N, Medalia O. Advances in tomography: probing the molecular architecture of cells. Nat Rev Mol Cell Biol. 2012;13:736–742. doi: 10.1038/nrm3453. [DOI] [PubMed] [Google Scholar]
- 11.Jensen GJ, Briegel A. How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr Opin Struct Biol. 2007;17:260–267. doi: 10.1016/j.sbi.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Zuber B, et al. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol. 2008;190:5672–5680. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H. Disclosure of the mycobacterial outer membrane: cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci USA. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci USA. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beeby M, Gumbart JC, Roux B, Jensen GJ. Architecture and assembly of the Gram-positive cell wall. Mol Microbiol. 2013;88:664–672. doi: 10.1111/mmi.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tocheva EI, et al. Peptidoglycan transformations during Bacillus subtilis sporulation. Mol Microbiol. 2013;88:673–686. doi: 10.1111/mmi.12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner RD, Vollmer W, Foster SJ. Different walls for rods and balls: the diversity of peptidoglycan. Mol Microbiol. 2014;91:862–874. doi: 10.1111/mmi.12513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pilhofer M, et al. Discovery of chlamydial peptidoglycan reveals bacteria with murein sacculi but without FtsZ. Nat Commun. 2013;4:2856. doi: 10.1038/ncomms3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swulius MT, et al. Long helical filaments are not seen encircling cells in electron cryotomograms of rodshaped bacteria. Biochem Biophys Res Commun. 2011;407:650–655. doi: 10.1016/j.bbrc.2011.03.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swulius MT, Jensen GJ. The helical MreB cytoskeleton in Escherichia coli MC1000/pLE7 is an artifact of the N-terminal yellow fluorescent protein tag. J Bacteriol. 2012;194:6382–6386. doi: 10.1128/JB.00505-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dominguez-Escobar J, et al. Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science. 2011;333:225–228. doi: 10.1126/science.1203466. [DOI] [PubMed] [Google Scholar]
- 22.Garner EC, et al. Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science. 2011;333:222–225. doi: 10.1126/science.1203285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briegel A, et al. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol. 2006;62:5–14. doi: 10.1111/j.1365-2958.2006.05355.x. [DOI] [PubMed] [Google Scholar]
- 24.Ingerson-Mahar M, Briegel A, Werner JN, Jensen GJ, Gitai Z. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat Cell Biol. 2010;12:739–746. doi: 10.1038/ncb2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barry RM, Gitai Z. Self-assembling enzymes and the origins of the cytoskeleton. Curr Opin Microbiol. 2011;14:704–711. doi: 10.1016/j.mib.2011.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuhn J, et al. Bactofilins, a ubiquitous class of cytoskeletal proteins mediating polar localization of a cell wall synthase in Caulobacter crescentus. EMBO J. 2010;29:327–339. doi: 10.1038/emboj.2009.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scheffel A, et al. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature. 2006;440:110–114. doi: 10.1038/nature04382. [DOI] [PubMed] [Google Scholar]
- 28.Komeili A, Li Z, Newman DK, Jensen GJ. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science. 2006;311:242–245. doi: 10.1126/science.1123231. Together with REF. 27, these papers describe the cytoskeletal filaments of MamK that align magnetosomes into chains. [DOI] [PubMed] [Google Scholar]
- 29.Scheffel A, Schuler D. The acidic repetitive domain of the Magnetospirillum gryphiswaldense MamJ protein displays hypervariability but is not required for magnetosome chain assembly. J Bacteriol. 2007;189:6437–6446. doi: 10.1128/JB.00421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katzmann E, Scheffel A, Gruska M, Plitzko JM, Schuler D. Loss of the actin-like protein MamK has pleiotropic effects on magnetosome formation and chain assembly in Magnetospirillum gryphiswaldense. Mol Microbiol. 2010;77:208–224. doi: 10.1111/j.1365-2958.2010.07202.x. [DOI] [PubMed] [Google Scholar]
- 31.Draper O, et al. MamK, a bacterial actin, forms dynamic filaments in vivo that are regulated by the acidic proteins MamJ and LimJ. Mol Microbiol. 2011;82:342–354. doi: 10.1111/j.1365-2958.2011.07815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jenkins C, et al. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc Natl Acad Sci USA. 2002;99:17049–17054. doi: 10.1073/pnas.012516899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sontag CA, Staley JT, Erickson HP. In vitro assembly and GTP hydrolysis by bacterial tubulins BtubA and BtubB. J Cell Biol. 2005;169:233–238. doi: 10.1083/jcb.200410027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. Microtubules in bacteria: Ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol. 2011;9:e1001213. doi: 10.1371/journal.pbio.1001213. This work describes bacterial microtubules containing five protofilaments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohm J, et al. Toward detecting and identifying macromolecules in a cellular context: template matching applied to electron tomograms. Proc Natl Acad Sci USA. 2000;97:14245–14250. doi: 10.1073/pnas.230282097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frangakis AS, et al. Identification of macromolecular complexes in cryoelectron tomograms of phantom cells. Proc Natl Acad Sci USA. 2002;99:14153–14158. doi: 10.1073/pnas.172520299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seybert A, Herrmann R, Frangakis AS. Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J Struct Biol. 2006;156:342–354. doi: 10.1016/j.jsb.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 38.Malmstrom J, et al. Proteome-wide cellular protein concentrations of the human pathogen Leptospira interrogans. Nature. 2009;460:762–765. doi: 10.1038/nature08184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortiz JO, Forster F, Kurner J, Linaroudis AA, Baumeister W. Mapping 70S ribosomes in intact cells by cryoelectron tomography and pattern recognition. J Struct Biol. 2006;156:334–341. doi: 10.1016/j.jsb.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 40.Comolli LR, Baker BJ, Downing KH, Siegerist CE, Banfield JF. Three-dimensional analysis of the structure and ecology of a novel, ultra-small archaeon. ISME J. 2009;3:159–167. doi: 10.1038/ismej.2008.99. [DOI] [PubMed] [Google Scholar]
- 41.Beck M, et al. Visual proteomics of the human pathogen Leptospira interrogans. Nat Methods. 2009;6:817–823. doi: 10.1038/nmeth.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuhner S, et al. Proteome organization in a genome-reduced bacterium. Science. 2009;326:1235–1240. doi: 10.1126/science.1176343. Together with REF. 41, this study uses visual proteomics to survey the in situ locations of protein complexes in intact cells. [DOI] [PubMed] [Google Scholar]
- 43.Martinez-Planells A, et al. Determination of the topography and biometry of chlorosomes by atomic force microscopy. Photosynth Res. 2002;71:83–90. doi: 10.1023/A:1014955614757. [DOI] [PubMed] [Google Scholar]
- 44.Psencik J, et al. Structure of chlorosomes from the green filamentous bacterium Chloroflexus aurantiacus. J Bacteriol. 2009;191:6701–6708. doi: 10.1128/JB.00690-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kudryashev M, Aktoudianaki A, Dedoglou D, Stahlberg H, Tsiotis G. The ultrastructure of Chlorobaculum tepidum revealed by cryo-electron tomography. Biochim Biophys Acta. 2014;1837:1635–1642. doi: 10.1016/j.bbabio.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 46.Ting CS, Hsieh C, Sundararaman S, Mannella C, Marko M. Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium. J Bacteriol. 2007;189:4485–4493. doi: 10.1128/JB.01948-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konorty M, Kahana N, Linaroudis A, Minsky A, Medalia O. Structural analysis of photosynthetic membranes by cryo-electron tomography of intact Rhodopseudomonas viridis cells. J Struct Biol. 2008;161:393–400. doi: 10.1016/j.jsb.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 48.Konorty M, et al. Photosynthetic system in Blastochloris viridis revisited. Biochemistry. 2009;48:4753–4761. doi: 10.1021/bi900267r. [DOI] [PubMed] [Google Scholar]
- 49.Tucker JD, et al. Membrane invagination in Rhodobacter sphaeroides is initiated at curved regions of the cytoplasmic membrane, then forms both budded and fully detached spherical vesicles. Mol Microbiol. 2010;76:833–847. doi: 10.1111/j.1365-2958.2010.07153.x. [DOI] [PubMed] [Google Scholar]
- 50.Ellis RJ. The most abundant protein in the world. Trends Biochem Sci. 1979;4:241–244. [Google Scholar]
- 51.Schmid MF, et al. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol. 2006;364:526–535. doi: 10.1016/j.jmb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iancu CV, et al. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J Mol Biol. 2007;372:764–773. doi: 10.1016/j.jmb.2007.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Iancu CV, et al. Organization, structure, and assembly of α-carboxysomes determined by electron cryotomography of intact cells. J Mol Biol. 2010;396:105–117. doi: 10.1016/j.jmb.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tocheva EI, et al. Structure and expression of propanediol utilization microcompartments in Acetonema longum. J Bacteriol. 2014;196:1651–1658. doi: 10.1128/JB.00049-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Butan C, et al. Spiral architecture of the nucleoid in Bdellovibrio bacteriovorus. J Bacteriol. 2011;193:1341–1350. doi: 10.1128/JB.01061-10. This paper describes the organization of the nucleoid in an intact cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raddi G, et al. Three-dimensional structures of pathogenic and saprophytic Leptospira species revealed by cryo-electron tomography. J Bacteriol. 2012;194:1299–1306. doi: 10.1128/JB.06474-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Comolli LR, et al. A portable cryo-plunger for on-site intact cryogenic microscopy sample preparation in natural environments. Microsc Res Tech. 2012;75:829–836. doi: 10.1002/jemt.22001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Luef B, et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun. 2015;6:6372. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 59.Schlimpert S, et al. General protein diffusion barriers create compartments within bacterial cells. Cell. 2012;151:1270–1282. doi: 10.1016/j.cell.2012.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Comolli LR, Luef B, Chan CS. High-resolution 2D and 3D cryo-TEM reveals structural adaptations of two stalk-forming bacteria to an Fe-oxidizing lifestyle. Environ Microbiol. 2011;13:2915–2929. doi: 10.1111/j.1462-2920.2011.02567.x. [DOI] [PubMed] [Google Scholar]
- 61.Luef B, et al. Iron-reducing bacteria accumulate ferric oxyhydroxide nanoparticle aggregates that may support planktonic growth. ISME J. 2013;7:338–350. doi: 10.1038/ismej.2012.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shetty A, Chen S, Tocheva EI, Jensen GJ, Hickey WJ. Nanopods: a new bacterial structure and mechanism for deployment of outer membrane vesicles. PLoS ONE. 2011;6:e20725. doi: 10.1371/journal.pone.0020725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Murphy GE, Leadbetter JR, Jensen GJ. In situ structure of the complete Treponema primitia flagellar motor. Nature. 2006;442:1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- 64.Liu J, et al. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191:5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu J, et al. Cellular architecture of Treponema pallidum: novel flagellum, periplasmic cone, and cell envelope as revealed by cryo electron tomography. J Mol Biol. 2010;403:546–561. doi: 10.1016/j.jmb.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen S, et al. Structural diversity of bacterial flagellar motors. EMBO J. 2011;30:2972–2981. doi: 10.1038/emboj.2011.186. This paper describes the structural diversity of flagellar motors in 11 bacterial species. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hosogi N, Shigematsu H, Terashima H, Homma M, Nagayama K. Zernike phase contrast cryo-electron tomography of sodium-driven flagellar hook-basal bodies from Vibrio alginolyticus. J Struct Biol. 2011;173:67–76. doi: 10.1016/j.jsb.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 68.Zhao X, et al. Cryoelectron tomography reveals the sequential assembly of bacterial flagella in Borrelia burgdorferi. Proc Natl Acad Sci USA. 2013;110:14390–14395. doi: 10.1073/pnas.1308306110. This work uses high-resolution subtomogram averaging and mutant analysis to construct a detailed structural picture of flagellar assembly in vivo in B. burgdorferi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Izard J, Hsieh CE, Limberger RJ, Mannella CA, Marko M. Native cellular architecture of Treponema denticola revealed by cryo-electron tomography. J Struct Biol. 2008;163:10–17. doi: 10.1016/j.jsb.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kudryashev M, et al. Comparative cryo-electron tomography of pathogenic Lyme disease spirochetes. Mol Microbiol. 2009;71:1415–1434. doi: 10.1111/j.1365-2958.2009.06613.x. [DOI] [PubMed] [Google Scholar]
- 71.Xu H, Raddi G, Liu J, Charon NW, Li C. Chemoreceptors and flagellar motors are subterminally located in close proximity at the two cell poles in spirochetes. J Bacteriol. 2011;193:2652–2656. doi: 10.1128/JB.01530-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Motaleb MA, et al. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci USA. 2000;97:10899–10904. doi: 10.1073/pnas.200221797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Charon NW, et al. The flat-ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J Bacteriol. 2009;191:600–607. doi: 10.1128/JB.01288-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sze CW, et al. Carbon storage regulator A (CsrA(Bb)) is a repressor of Borrelia burgdorferi flagellin protein FlaB. Mol Microbiol. 2011;82:851–864. doi: 10.1111/j.1365-2958.2011.07853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang K, Tong BA, Liu J, Li C. A single-domain FlgJ contributes to flagellar hook and filament formation in the Lyme disease spirochete Borrelia burgdorferi. J Bacteriol. 2012;194:866–874. doi: 10.1128/JB.06341-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Motaleb MA, Pitzer JE, Sultan SZ, Liu J. A novel gene inactivation system reveals altered periplasmic flagellar orientation in a Borrelia burgdorferi fliL mutant. J Bacteriol. 2011;193:3324–3331. doi: 10.1128/JB.00202-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sultan SZ, et al. Motor rotation is essential for the formation of the periplasmic flagellar ribbon, cellular morphology, and Borrelia burgdorferi persistence within Ixodes scapularis tick and murine hosts. Infect Immun. 2015;83:1765–1777. doi: 10.1128/IAI.03097-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murphy GE, Matson EG, Leadbetter JR, Berg HC, Jensen GJ. Novel ultrastructures of Treponema primitia and their implications for motility. Mol Microbiol. 2008;67:1184–1195. doi: 10.1111/j.1365-2958.2008.06120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ruan J, et al. Architecture of a flagellar apparatus in the fast-swimming magnetotactic bacterium MO-1. Proc Natl Acad Sci USA. 2012;109:20643–20648. doi: 10.1073/pnas.1215274109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu J, McBride MJ, Subramaniam S. Cell surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol. 2007;189:7503–7506. doi: 10.1128/JB.00957-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kurner J, Frangakis AS, Baumeister W. Cryoelectron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science. 2005;307:436–438. doi: 10.1126/science.1104031. This early description of bacterial cytoskeletal filaments proposed a model for motility of helical Mollicutes based on cytoskeletal ribbons. [DOI] [PubMed] [Google Scholar]
- 82.Henderson GP, Jensen GJ. Three-dimensional structure of Mycoplasma pneumoniae’s attachment organelle and a model for its role in gliding motility. Mol Microbiol. 2006;60:376–385. doi: 10.1111/j.1365-2958.2006.05113.x. [DOI] [PubMed] [Google Scholar]
- 83.Jasnin M, et al. Three-dimensional architecture of actin filaments in Listeria monocytogenes comet tails. Proc Natl Acad Sci USA. 2013;110:20521–20526. doi: 10.1073/pnas.1320155110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Byrne ME, et al. Desulfovibrio magneticus RS-1 contains an iron- and phosphorus-rich organelle distinct from its bullet-shaped magnetosomes. Proc Natl Acad Sci USA. 2010;107:12263–12268. doi: 10.1073/pnas.1001290107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Abreu F, et al. Cryo-electron tomography of the magnetotactic vibrio Magnetovibrio blakemorei: insights into the biomineralization of prismatic magnetosomes. J Struct Biol. 2013;181:162–168. doi: 10.1016/j.jsb.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang P, Khursigara CM, Hartnell LM, Subramaniam S. Direct visualization of Escherichia coli chemotaxis receptor arrays using cryo-electron microscopy. Proc Natl Acad Sci USA. 2007;104:3777–3781. doi: 10.1073/pnas.0610106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Khursigara CM, Wu X, Zhang P, Lefman J, Subramaniam S. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc Natl Acad Sci USA. 2008;105:16555–16560. doi: 10.1073/pnas.0806401105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Briegel A, et al. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol. 2008;69:30–41. doi: 10.1111/j.1365-2958.2008.06219.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Khursigara CM, Wu X, Subramaniam S. Chemoreceptors in Caulobacter crescentus: trimers of receptor dimers in a partially ordered hexagonally packed array. J Bacteriol. 2008;190:6805–6810. doi: 10.1128/JB.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Briegel A, Beeby M, Thanbichler M, Jensen GJ. Activated chemoreceptor arrays remain intact and hexagonally packed. Mol Microbiol. 2011;82:748–757. doi: 10.1111/j.1365-2958.2011.07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Khursigara CM, et al. Lateral density of receptor arrays in the membrane plane influences sensitivity of the E coli chemotaxis response. EMBO J. 2011;30:1719–1729. doi: 10.1038/emboj.2011.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang K, et al. Two CheW coupling proteins are essential in a chemosensory pathway of Borrelia burgdorferi. Mol Microbiol. 2012;85:782–794. doi: 10.1111/j.1365-2958.2012.08139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamaichi Y, et al. A multidomain hub anchors the chromosome segregation and chemotactic machinery to the bacterial pole. Genes Dev. 2012;26:2348–2360. doi: 10.1101/gad.199869.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Briegel A, et al. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci USA. 2009;106:17181–17186. doi: 10.1073/pnas.0905181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Briegel A, et al. Structural conservation of chemotaxis machinery across Archaea and Bacteria. Environ Microbiol Rep. 2015;7:414–419. doi: 10.1111/1758-2229.12265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Briegel A, et al. Structure of bacterial cytoplasmic chemoreceptor arrays and implications for chemotactic signaling. eLife. 2014;3:e02151. doi: 10.7554/eLife.02151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Briegel A, et al. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci USA. 2012;109:3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu J, et al. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci USA. 2012;109:E1481–E1488. doi: 10.1073/pnas.1200781109. Together with REF. 97, this study provides a detailed structural model of the bacterial chemoreceptor array. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Briegel A, et al. The mobility of two kinase domains in the Escherichia coli chemoreceptor array varies with signalling state. Mol Microbiol. 2013;89:831–841. doi: 10.1111/mmi.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Briegel A, et al. New insights into bacterial chemoreceptor array structure and assembly from electron cryotomography. Biochemistry. 2014;53:1575–1585. doi: 10.1021/bi5000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ebersbach G, Briegel A, Jensen GJ, Jacobs-Wagner C. A self-associating protein critical for chromosome attachment, division, and polar organization in. Caulobacter Cell. 2008;134:956–968. doi: 10.1016/j.cell.2008.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bowman GR, et al. Caulobacter PopZ forms a polar subdomain dictating sequential changes in pole composition and function. Mol Microbiol. 2010;76:173–189. doi: 10.1111/j.1365-2958.2010.07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Salje J, Zuber B, Lowe J. Electron cryomicroscopy of E coli reveals filament bundles involved in plasmid DNA segregation. Science. 2009;323:509–512. doi: 10.1126/science.1164346. [DOI] [PubMed] [Google Scholar]
- 104.Bharat TA, Murshudov GN, Sachse C, Lowe J. Structures of actin-like ParM filaments show architecture of plasmid-segregating spindles. Nature. 2015;523:106–110. doi: 10.1038/nature14356. This study reveals plasmid segregation by antiparallel doublets of ParM filaments. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aylett CH, Wang Q, Michie KA, Amos LA, Lowe J. Filament structure of bacterial tubulin homologue TubZ. Proc Natl Acad Sci USA. 2010;107:19766–19771. doi: 10.1073/pnas.1010176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Szwedziak P, Wang Q, Freund SM, Lowe J. FtsA forms actin-like protofilaments. EMBO J. 2012;31:2249–2260. doi: 10.1038/emboj.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Judd EM, et al. Distinct constrictive processes, separated in time and space, divide Caulobacter inner and outer membranes. J Bacteriol. 2005;187:6874–6882. doi: 10.1128/JB.187.20.6874-6882.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li Z, Trimble MJ, Brun YV, Jensen GJ. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J. 2007;26:4694–4708. doi: 10.1038/sj.emboj.7601895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lu C, Reedy M, Erickson HP. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol. 2000;182:164–170. doi: 10.1128/jb.182.1.164-170.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Szwedziak P, Wang Q, Bharat TA, Tsim M, Lowe J. Architecture of the ring formed by the tubulin homologue FtsZ in bacterial cell division. eLife. 2014;3:e04601. doi: 10.7554/eLife.04601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ghosal D, Trambaiolo D, Amos LA, Lowe J. MinCD cell division proteins form alternating copolymeric cytomotive filaments. Nat Commun. 2014;5:5341. doi: 10.1038/ncomms6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moll A, Schlimpert S, Briegel A, Jensen GJ, Thanbichler M. DipM, a new factor required for peptidoglycan remodelling during cell division in Caulobacter crescentus. Mol Microbiol. 2010;77:90–107. doi: 10.1111/j.1365-2958.2010.07224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Goley ED, Comolli LR, Fero KE, Downing KH, Shapiro L. DipM links peptidoglycan remodelling to outer membrane organization in Caulobacter. Mol Microbiol. 2010;77:56–73. doi: 10.1111/j.1365-2958.2010.07222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dobro MJ, et al. Electron cryotomography of ESCRT assemblies and dividing Sulfolobus cells suggests that spiraling filaments are involved in membrane scission. Mol Biol Cell. 2013;24:2319–2327. doi: 10.1091/mbc.E12-11-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Katzmann E, et al. Magnetosome chains are recruited to cellular division sites and split by asymmetric septation. Mol Microbiol. 2011;82:1316–1329. doi: 10.1111/j.1365-2958.2011.07874.x. [DOI] [PubMed] [Google Scholar]
- 116.Comolli LR, Kundmann M, Downing KH. Characterization of intact subcellular bodies in whole bacteria by cryo-electron tomography and spectroscopic imaging. J Microsc. 2006;223:40–52. doi: 10.1111/j.1365-2818.2006.01597.x. [DOI] [PubMed] [Google Scholar]
- 117.Tocheva EI, et al. Polyphosphate storage during sporulation in the Gram-negative bacterium Acetonema longum. J Bacteriol. 2013;195:3940–3946. doi: 10.1128/JB.00712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Beeby M, Cho M, Stubbe J, Jensen GJ. Growth and localization of polyhydroxybutyrate granules in Ralstonia eutropha. J Bacteriol. 2012;194:1092–1099. doi: 10.1128/JB.06125-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Toso DB, Henstra AM, Gunsalus RP, Zhou ZH. Structural mass and elemental analyses of storage granules in methanogenic archaeal cells. Environ Microbiol. 2011;13:2587–2599. doi: 10.1111/j.1462-2920.2011.02531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tocheva EI, et al. Peptidoglycan remodeling and conversion of an inner membrane into an outer membrane during sporulation. Cell. 2011;146:799–812. doi: 10.1016/j.cell.2011.07.029. This work reveals the interconversion of inner and outer membranes during Gram-negative sporulation, suggesting a potential evolutionary source of the outer membrane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schrempf H, Koebsch I, Walter S, Engelhardt H, Meschke H. Extracellular Streptomyces vesicles: amphorae for survival and defence. Microb Biotechnol. 2011;4:286–299. doi: 10.1111/j.1751-7915.2011.00251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sleytr UB, Messner P, Pum D, Sara M. Crystalline bacterial cell surface layers. Mol Microbiol. 1993;10:911–916. doi: 10.1111/j.1365-2958.1993.tb00962.x. [DOI] [PubMed] [Google Scholar]
- 123.Grimm R, et al. Electron tomography of iceembedded prokaryotic cells. Biophys J. 1998;74:1031–1042. doi: 10.1016/S0006-3495(98)74028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Amat F, et al. Analysis of the intact surface layer of Caulobacter crescentus by cryo-electron tomography. J Bacteriol. 2010;192:5855–5865. doi: 10.1128/JB.00747-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Nickell S, Hegerl R, Baumeister W, Rachel R. Pyrodictium cannulae enter the periplasmic space but do not enter the cytoplasm, as revealed by cryoelectron tomography. J Struct Biol. 2003;141:34–42. doi: 10.1016/s1047-8477(02)00581-6. [DOI] [PubMed] [Google Scholar]
- 126.Junglas B, et al. Ignicoccus hospitalis and Nanoarchaeum equitans: ultrastructure, cell-cell interaction, and 3D reconstruction from serial sections of freeze-substituted cells and by electron cryotomography. Arch Microbiol. 2008;190:395–408. doi: 10.1007/s00203-008-0402-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Comolli LR, Banfield JF. Inter-species interconnections in acid mine drainage microbial communities. Front Microbiol. 2014;5:367. doi: 10.3389/fmicb.2014.00367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moissl C, Rachel R, Briegel A, Engelhardt H, Huber R. The unique structure of archaeal ‘hami’, highly complex cell appendages with nano-grappling hooks. Mol Microbiol. 2005;56:361–370. doi: 10.1111/j.1365-2958.2005.04294.x. [DOI] [PubMed] [Google Scholar]
- 129.Basler M, Pilhofer M, Henderson GP, Jensen GJ, Mekalanos JJ. Type VI secretion requires a dynamic contractile phage tail-like structure. Nature. 2012;483:182–186. doi: 10.1038/nature10846. This study reveals the phage tail-like contractile mechanism of the T6SS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chang YW, et al. Correlated cryogenic photoactivated localization microscopy and cryoelectron tomography. Nat Methods. 2014;11:737–739. doi: 10.1038/nmeth.2961. This paper describes the technique of correlated cryo-PALM and ECT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Shikuma NJ, et al. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science. 2014;343:529–533. doi: 10.1126/science.1246794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Borgnia MJ, Subramaniam S, Milne JL. Three-dimensional imaging of the highly bent architecture of Bdellovibrio bacteriovorus by using cryo-electron tomography. J Bacteriol. 2008;190:2588–2596. doi: 10.1128/JB.01538-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Muller A, et al. Ultrastructure and complex polar architecture of the human pathogen Campylobacter jejuni. Microbiologyopen. 2014;3:702–710. doi: 10.1002/mbo3.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Izard J, et al. Cryo-electron tomography elucidates the molecular architecture of Treponema pallidum, the syphilis spirochete. J Bacteriol. 2009;191:7566–7580. doi: 10.1128/JB.01031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kudryashev M, et al. Evidence of direct cell-cell fusion in Borrelia by cryogenic electron tomography. Cell Microbiol. 2011;13:731–741. doi: 10.1111/j.1462-5822.2011.01571.x. [DOI] [PubMed] [Google Scholar]
- 136.Abrusci P, et al. Architecture of the major component of the type III secretion system export apparatus. Nat Struct Mol Biol. 2013;20:99–104. doi: 10.1038/nsmb.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kawamoto A, et al. Common and distinct structural features of Salmonella injectisome and flagellar basal body. Sci Rep. 2013;3:3369. doi: 10.1038/srep03369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kudryashev M, et al. In situ structural analysis of the Yersinia enterocolitica injectisome. eLife. 2013;2:e00792. doi: 10.7554/eLife.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kudryashev M, et al. Yersinia enterocolitica type III secretion injectisomes form regularly spaced clusters, which incorporate new machines upon activation. Mol Microbiol. 2014;95:875–884. doi: 10.1111/mmi.12908. [DOI] [PubMed] [Google Scholar]
- 140.Nans A, Saibil HR, Hayward RD. Pathogen–host reorganization during Chlamydia invasion revealed by cryo-electron tomography. Cell Microbiol. 2014;16:1457–1472. doi: 10.1111/cmi.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hu B, et al. Visualization of the type III secretion sorting platform of Shigella flexneri. Proc Natl Acad Sci USA. 2015;112:1047–1052. doi: 10.1073/pnas.1411610112. This work provides a pseudo-atomic model of the pathogenic T3SS, including the substrate sorting platform. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Huang Z, et al. Cryo-electron tomography of Chlamydia trachomatis gives a clue to the mechanism of outer membrane changes. J Electron Microsc (Tokyo) 2010;59:237–241. doi: 10.1093/jmicro/dfp057. [DOI] [PubMed] [Google Scholar]
- 143.Wilkat M, Herdoiza E, Forsbach-Birk V, Walther P, Essig A. Electron tomography and cryo-SEM characterization reveals novel ultrastructural features of host-parasite interaction during Chlamydia abortus infection. Histochem Cell Biol. 2014;142:171–184. doi: 10.1007/s00418-014-1189-y. [DOI] [PubMed] [Google Scholar]
- 144.LaRocca TJ, et al. The bactericidal effect of a complement-independent antibody is osmolytic and specific to Borrelia. Proc Natl Acad Sci USA. 2009;106:10752–10757. doi: 10.1073/pnas.0901858106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Watkins BY, et al. Mycobacterium marinum SecA2 promotes stable granulomas and induces tumor necrosis factor-α in vivo. Infect Immun. 2012;80:3512–3520. doi: 10.1128/IAI.00686-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Koning RI, et al. Cryo-electron tomography analysis of membrane vesicles from Acinetobacter baumannii ATCC19606 T. Res Microbiol. 2013;164:397–405. doi: 10.1016/j.resmic.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 147.Trepout S, et al. Structure of reconstituted bacterial membrane efflux pump by cryo-electron tomography. Biochim Biophys Acta. 2010;1798:1953–1960. doi: 10.1016/j.bbamem.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 148.Wysocki LM, Lavis LD. Advances in the chemistry of small molecule fluorescent probes. Curr Opin Chem Biol. 2011;15:752–759. doi: 10.1016/j.cbpa.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 149.Zhang WH, Otting G, Jackson CJ. Protein engineering with unnatural amino acids. Curr Opin Struct Biol. 2013;23:581–587. doi: 10.1016/j.sbi.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 150.Mercogliano CP, DeRosier DJ. Concatenated metallothionein as a clonable gold label for electron microscopy. J Struct Biol. 2007;160:70–82. doi: 10.1016/j.jsb.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Wang Q, Mercogliano CP, Lowe J. A ferritin-based label for cellular electron cryotomography. Structure. 2011;19:147–154. doi: 10.1016/j.str.2010.12.002. [DOI] [PubMed] [Google Scholar]
- 152.Diestra E, Fontana J, Guichard P, Marco S, Risco C. Visualization of proteins in intact cells with a clonable tag for electron microscopy. J Struct Biol. 2009;165:157–168. doi: 10.1016/j.jsb.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 153.Bouchet-Marquis C, Pagratis M, Kirmse R, Hoenger A. Metallothionein as a clonable high-density marker for cryo-electron microscopy. J Struct Biol. 2012;177:119–127. doi: 10.1016/j.jsb.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Delgado L, Martinez G, Lopez-Iglesias C, Mercade E. Cryo-electron tomography of plunge-frozen whole bacteria and vitreous sections to analyze the recently described bacterial cytoplasmic structure, the Stack. J Struct Biol. 2015;189:220–229. doi: 10.1016/j.jsb.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 155.Bernadac A, et al. Structural properties of the tubular appendage spinae from marine bacterium Roseobacter sp strain YSCB. Sci Rep. 2012;2:950. doi: 10.1038/srep00950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tivol WF, Briegel A, Jensen GJ. An improved cryogen for plunge freezing. Microsc Microanal. 2008;14:375–379. doi: 10.1017/S1431927608080781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Al-Amoudi A, Norlen LP, Dubochet J. Cryo-electron microscopy of vitreous sections of native biological cells and tissues. J Struct Biol. 2004;148:131–135. doi: 10.1016/j.jsb.2004.03.010. This paper describes the technique of vitreous cryosectioning to expand the power of ECT to thicker biological samples. [DOI] [PubMed] [Google Scholar]
- 158.Al-Amoudi A, Studer D, Dubochet J. Cutting artefacts and cutting process in vitreous sections for cryo-electron microscopy. J Struct Biol. 2005;150:109–121. doi: 10.1016/j.jsb.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 159.Dubochet J, et al. How to “read” a vitreous section. Methods Cell Biol. 2007;79:385–406. doi: 10.1016/S0091-679X(06)79015-X. [DOI] [PubMed] [Google Scholar]
- 160.Marko M, Hsieh C, Schalek R, Frank J, Mannella C. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat Methods. 2007;4:215–217. doi: 10.1038/nmeth1014. [DOI] [PubMed] [Google Scholar]
- 161.Rigort A, et al. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc Natl Acad Sci USA. 2012;109:4449–4454. doi: 10.1073/pnas.1201333109. This paper illustrates the power of FIB milling to enable high-resolution ECT of thick cells without the artefacts introduced by vitreous cryosectioning. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rigort A, Villa E, Bauerlein FJ, Engel BD, Plitzko JM. Integrative approaches for cellular cryo-electron tomography: correlative imaging and focused ion beam micromachining. Methods Cell Biol. 2012;111:259–281. doi: 10.1016/B978-0-12-416026-2.00014-5. [DOI] [PubMed] [Google Scholar]
- 163.Strunk KM, Wang K, Ke D, Gray JL, Zhang P. Thinning of large mammalian cells for cryo-TEM characterization by cryo-FIB milling. J Microsc. 2012;247:220–227. doi: 10.1111/j.1365-2818.2012.03635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.McMullan G, Clark AT, Turchetta R, Faruqi AR. Enhanced imaging in low dose electron microscopy using electron counting. Ultramicroscopy. 2009;109:1411–1416. doi: 10.1016/j.ultramic.2009.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Murata K, et al. Zernike phase contrast cryo-electron microscopy and tomography for structure determination at nanometer and subnanometer resolutions. Structure. 2010;18:903–912. doi: 10.1016/j.str.2010.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Dai W, et al. Zernike phase-contrast electron cryotomography applied to marine cyanobacteria infected with cyanophages. Nat Protoc. 2014;9:2630–2642. doi: 10.1038/nprot.2014.176. This study uses Zernike phase contrast ECT to describe the cyanophage maturation process inside cyanobacterial cells. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fukuda Y, Laugks U, Lucic V, Baumeister W, Danev R. Electron cryotomography of vitrified cells with a Volta phase plate. J Struct Biol. 2015;190:143–154. doi: 10.1016/j.jsb.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 168.Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc Natl Acad Sci USA. 2014;111:15635–15640. doi: 10.1073/pnas.1418377111. This paper describes the Volta phase plate for improving contrast in transmission electron microscopy imaging. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Asano S, et al. Proteasomes. A molecular census of 26S proteasomes in intact neurons. Science. 2015;347:439–442. doi: 10.1126/science.1261197. This paper illustrates the power of Volta phase plate ECT, distinguishing conformational states of a protein complex in an intact eukaryotic cell. [DOI] [PubMed] [Google Scholar]
- 170.Hu GB, Wei H, Rice WJ, Stokes DL, Gottlieb P. Electron cryo-tomographic structure of cystovirus Φ12. Virology. 2008;372:1–9. doi: 10.1016/j.virol.2007.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Leo-Macias A, et al. Toroidal surface complexes of bacteriophage Φ12 are responsible for host-cell attachment. Virology. 2011;414:103–109. doi: 10.1016/j.virol.2011.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chang JT, et al. Visualizing the structural changes of bacteriophage ε15 and its Salmonella host during infection. J Mol Biol. 2010;402:731–740. doi: 10.1016/j.jmb.2010.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Dai W, et al. Three-dimensional structure of tropism-switching Bordetella bacteriophage. Proc Natl Acad Sci USA. 2010;107:4347–4352. doi: 10.1073/pnas.0915008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage T7 virion undergoes extensive structural remodeling during infection. Science. 2013;339:576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Parent KN, et al. OmpA and OmpC are critical host factors for bacteriophage Sf6 entry in Shigella. Mol Microbiol. 2014;92:47–60. doi: 10.1111/mmi.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Guerrero-Ferreira RC, et al. Alternative mechanism for bacteriophage adsorption to the motile bacterium Caulobacter crescentus. Proc Natl Acad Sci USA. 2011;108:9963–9968. doi: 10.1073/pnas.1012388108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dent KC, et al. The asymmetric structure of an icosahedral virus bound to its receptor suggests a mechanism for genome release. Structure. 2013;21:1225–1234. doi: 10.1016/j.str.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bohm J, et al. FhuA-mediated phage genome transfer into liposomes: a cryo-electron tomography study. Curr Biol. 2001;11:1168–1175. doi: 10.1016/s0960-9822(01)00349-9. This early application of ECT reveals the contractile injection mechanism of the phage tail. [DOI] [PubMed] [Google Scholar]
- 179.Liu X, et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17:830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Fu X, Walter MH, Paredes A, Morais MC, Liu J. The mechanism of DNA ejection in the Bacillus anthracis spore-binding phage 8a revealed by cryoelectron tomography. Virology. 2011;421:141–148. doi: 10.1016/j.virol.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Liu J, Chen CY, Shiomi D, Niki H, Margolin W. Visualization of bacteriophage P1 infection by cryoelectron tomography of tiny Escherichia coli. Virology. 2011;417:304–311. doi: 10.1016/j.virol.2011.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Nemecek D, Heymann JB, Qiao J, Mindich L, Steven AC. Cryo-electron tomography of bacteriophage Φ6 procapsids shows random occupancy of the binding sites for RNA polymerase and packaging NTPase. J Struct Biol. 2010;171:389–396. doi: 10.1016/j.jsb.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Dai W, et al. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature. 2013;502:707–710. doi: 10.1038/nature12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Nemecek D, et al. Stepwise expansion of the bacteriophage Φ6 procapsid: possible packaging intermediates. J Mol Biol. 2011;414:260–271. doi: 10.1016/j.jmb.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Dewey JS, et al. Micron-scale holes terminate the phage infection cycle. Proc Natl Acad Sci USA. 2010;107:2219–2223. doi: 10.1073/pnas.0914030107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Quemin ER, et al. First insights into the entry process of hyperthermophilic archaeal viruses. J Virol. 2013;87:13379–13385. doi: 10.1128/JVI.02742-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Fu CY, et al. In vivo assembly of an archaeal virus studied with whole-cell electron cryotomography. Structure. 2010;18:1579–1586. doi: 10.1016/j.str.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Daum B, et al. Self-assembly of the general membrane-remodeling protein PVAP into sevenfold virus-associated pyramids. Proc Natl Acad Sci USA. 2014;111:3829–3834. This study reveals the lytic mechanism of pyramidal archaeal viruses. [Google Scholar]
- 189.Hong C, et al. Lemon-shaped halo archaeal virus His1 with uniform tail but variable capsid structure. Proc Natl Acad Sci USA. 2015;112:2449–2454. doi: 10.1073/pnas.1425008112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.