Abstract
The hormone erythropoietin (Epo) is required for erythropoiesis, yet its molecular mechanism of action remains poorly understood, particularly with respect to chromatin dynamics. To investigate how Epo modulates the erythroid epigenome, we performed epigenetic profiling using an ex vivo murine cell system that undergoes synchronous erythroid maturation in response to Epo stimulation. Our findings define the repertoire of Epo-modulated enhancers, illuminating a new facet of Epo signaling. First, a large number of enhancers rapidly responded to Epo stimulation, revealing a cis-regulatory network of Epo-responsive enhancers. In contrast, most of the other identified enhancers remained in an active acetylated state during Epo signaling, suggesting that most erythroid enhancers are established at an earlier precursor stage. Second, we identified several hundred super-enhancers that were linked to key erythroid genes, such as Tal1, Bcl11a, and Mir144/451. Third, experimental and computational validation revealed that many predicted enhancer regions were occupied by TAL1 and enriched with DNA-binding motifs for GATA1, KLF1, TAL1/E-box, and STAT5. Additionally, many of these cis-regulatory regions were conserved evolutionarily and displayed correlated enhancer:promoter acetylation. Together, these findings define a cis-regulatory enhancer network for Epo signaling during erythropoiesis, and provide the framework for future studies involving the interplay of epigenetics and Epo signaling.
Erythropoiesis has long served as an experimental paradigm for understanding gene regulatory mechanisms governing cell identity and development. The hormone erythropoietin (Epo) is physiologically necessary and sufficient to trigger the initiation of terminal erythroid differentiation of pro-erythroblasts. Epo initiates erythroid differentiation and gene expression patterns by binding to its cognate receptor, which activates the Jak2–Stat5 signaling axis [1–3]. Erythroid expression patterns are highly dynamic and have been extensively studied by genome-wide expression profiling [4–9], providing numerous insights into the molecular pathways that control red blood cell development. Additionally, expression profiling of Epo starvation experiments in pro-erythroblasts derived from primary fetal liver revealed that Epo signaling modulates the expression of several hundred genes involved in cell survival signaling and cell identity [7].
Cell identity is established and propagated primarily through epigenetic mechanisms, including the cell type-specific repertoire of enhancers [10–12]. Enhancer segments typically are co-occupied by combinations of transcription factors (TFs) that influence gene expression programs, often integrating signals from multiple pathways. Collectively, TFs orchestrate cell type-specific transcription programs by binding to their cognate sequence motifs within specific enhancers and recruiting co-regulators and RNA polymerase II to promoters to initiate transcription [13–15]. Recent reports tracking histone modifications described the erythroid enhancer landscape in human and murine erythroid cells [16–18]. Interestingly, a previous study found that broad features of chromatin states remain largely unchanged during Gata1-induced differentiation in the murine G1E-ER4 cell line despite the dramatic transcriptional changes that accompany erythropoiesis [16]. The study suggested that erythroid enhancers are established in erythroid precursor cells, but precisely when this occurs remains unclear. Although the locations of erythroid enhancers have been determined in murine and human cell systems [16–18], the ways in which Epo influences the enhancer landscape are currently unknown. Defining the comprehensive set of Epo-modulated enhancers will better illuminate the molecular and epigenetic pathways controlled by Epo.
To investigate the interplay of Epo signaling and the chromatin landscape during erythroid development, we used highly purified pro-erythroblasts derived from mice injected with an anemia-inducing strain of the Friend virus (FVA), as previously described [19]. Physiologically, mice injected with the FVA strain rapidly accumulate immature pro-erythroblast precursor cells in the spleen, a condition termed erythroblastosis. FVA-derived pro-erythroblast proliferation depends on the truncated form of the stem cell-derived tyro-sine kinase receptor (sf-STK) but not JAK2-STAT5 activity [20,21]. Importantly, FVA pro-erythroblasts remain sensitive to physiological levels of Epo in an ex vivo culture system and, on Epo stimulation, will activate JAK2-STAT5 signaling and synchronously differentiate into fully mature, enucleated erythrocytes. Thus, FVA-derived pro-erythroblasts represent an ideal system for delineating the molecular action of Epo on a genomic scale and examining the epigenetic effects of Epo in a physiological context.
Here, we report the use of chromatin immunoprecipitation-exonuclease (ChIP-exo) analyses to profile the enhancer landscape during Epo-initiated erythropoiesis. Genome-wide locations for H3K4me1, H3K27ac, and H3K4me3 were examined in the FVA model system at baseline and 1 hour after Epo-initiated synchronous, terminal erythroid differentiation. This unique model cell system allowed us to uniquely define the immediate impact of Epo signaling on the epigenetic landscape during erythropoiesis.
Our work illustrates several new aspects of the role of Epo in erythroid chromatin biology. Remarkably, Epo stimulation alters the histone mark signatures across several thousand enhancer locations, highlighting Epo’s underap-preciated role in re-programming the epigenome. Enhancer marks were highly dynamic within the first hour of Epo stimulation, enabling classification of enhancer behavior based on the Epo-induced modulation of these histone marks. On Epo stimulation, a similar number of enhancers displayed increased (n = 1,589) and decreased (n = 1,529) acetylation. For example, loss of enhancer acetylation was linked to genes known to be downregulated during erythropoiesis, such as Cd44. Likewise, enhancers displaying increased acetylation levels in response to Epo were linked to genes involved in regulating the survival pathways, TFs, and chromatin regulators. Nevertheless, our finding that the vast majority of enhancers, including super-enhancers, were unaffected within the first hour of Epo stimulation fits with the notion that chromatin states remain largely unchanged during erythropoiesis [16].
Bioinformatic analyses of motif enrichment and H3K27ac enhancer:promoter correlation validated many of the enhancer locations identified in this study. Enhancer overlap with published data further validated enhancer locations, and revealed the location of evolutionarily conserved cis-regulatory modules across mouse and human erythroid cell genomes. Given that TAL1 is one of the master TF regulators of erythropoiesis, we also illustrate that TAL1 binds to thousands of enhancers, adding a layer of validation to our enhancer maps and suggesting a mechanistic link between Epo signaling, TAL1 occupancy, and chromatin dynamics. Together, these aspects of the presented study identify cis-regulatory enhancer circuits controlled by Epo signaling pathways. Importantly, the epigenetic profiles from the current study provide a framework for generating testable hypotheses about the ways in which Epo, enhancers, and TFs coordinately control erythroid expression programs.
Methods
Isolation of murine pro-erythroblasts
Highly purified pro-erythroblasts were obtained from spleens of mice infected with the Friend virus as previously described [22,23], with the following modifications. All animal procedures were performed in compliance with and approval from the Vanderbilt Division of Animal Care (DAC) and the Institutional Animal Care and Use Committee (IACUC). Female BALB/cJ mice (12 weeks old, Jackson Laboratories) were infected via intraperitoneal injection of ~ 104 spleen focus-forming units of FVA. At 13 to 15 days postinfection, the mice were sacrificed and their spleens removed. The spleens were homogenized to a single-cell suspension by mincing and passing through a sterile 100-μm nylon mesh filter into sterile solution of 0.2% bovine serum albumin (BSA) in 1 × phosphate-buffered saline (PBS). The filtrate was pi-petted repeatedly to ensure a single-cell suspension. The homogenized spleen cells were size-separated by gravity sedimentation for 4 hours at 4°C in a continuous gradient of 1% to 2% deionized BSA. The sedimentation apparatus consisted of a 25-cm-diameter sedimentation chamber containing a 2.4-L BSA gradient, two BSA gradient chambers containing 1.2-L 1% and 2% de-ionized BSA in 1 × PBS, and a cell-loading chamber (Pro-Science Inc.) containing the 50-mL cell suspension. After 4 hours of sedimentation, cells were collected in 50-mL fractions, with pro-erythroblasts typically enriched in fractions 5–20 of 24 total fractions. Typically, about 109 pro-erythroblasts were obtained from the separation of 1010 nucleated spleen cells (6–7 g spleen weight) across three 25-cm sedimentation chambers.
Cell culture
To study the effects of erythropoietin (Epo) on terminal erythroid differentiation, FVA-derived pro-erythroblasts were cultured at 106 cells/mL in Iscove’s modified Dulbecco’s medium (IMDM, Life Technologies No. 12440043), 30% heat-inactivated fetal bovine serum (Gibco, No. 26140-079), 1% penicillin–streptomycin (Gibco, No. 15140-122), 10% de-ionized BSA, and 100 μmol/L γ-thioglycerol (MP Biomedicals, No. 155723). Terminal erythroid differentiation of purified pro-erythroblasts was induced by the addition of 0.4 U/mL human recombinant Epo (10kU/ml Epogen by Amgen, NDC 55513-144-10) to medium. At the desired times after the addition of Epo, cells were crosslinked by the addition of 1% formaldehyde for 10 min. Crosslinking was quenched by adding 125 mmol/L glycine. Cross-linked cells were collected by centrifugation for 5 min at 1,000g at 4°C, washed once with 1 × PBS, flash frozen in liquid nitrogen, and stored at −80°C until used for ChIP analysis.
ChIP-exo and antibodies
With the following modifications, ChIP-exo was performed as previously described [24] with chromatin extracted from 50 million cells, ProteinG MagSepharose resin (GE Healthcare), and 5 μg of antibody directed against the H3K4me1, H3K4me3, H3K27ac, or Tal1 (Abcam ab8895, ab8580, ab4729, and Santa Cruz Biotech sc-12984, respectively). First, formaldehyde cross-linked cells were lysed with buffer 1 (50 mmol/L HEPES–KOH, pH 7.5; 140 mmol/L NaCl; 1 mmol/L EDTA; 10% glycerol; 0.5% Nonidet P-40; 0.25% Triton X-100) and washed once with buffer 2 (10 mmol/L Tris–HCL, pH 8; 200 mmol/L NaCl; 1 mmol/L EDTA; 0.5 mmol/L EGTA), and the nuclei were lysed with buffer 3 (10 mmol/L Tris–HCl, pH 8; 100 mmol/L NaCl; 1 mmol/L EDTA; 0.5 mmol/L EGTA; 0.1% sodium deoxycholate; 0.5% N-lauroylsarcosine). All cell lysis buffers were supplemented with fresh EDTA-free complete protease inhibitor cocktail (CPI, Roche, No. 11836153001). Purified chromatin was sonicated with a Bioruptor (Diagenode) to obtain fragments ranging from 100 bp to 500 bp. Triton X-100 was added to extract at 1% to neutralize sarcosine. Insoluble chromatin debris was removed by centrifugation, and sonication extracts were stored at −80°C until used for ChIP analysis.
Illumina sequencing and data pre-processing
Libraries were sequenced using an Illumina NextSeq500 sequencer as single-end reads 75 nucleotides in length on high output mode (Supplementary Table E1, online only, available at www.exphem.org). The sequence reads were aligned to the mouse mm10 reference genome using BWA-MEM algorithm with default parameters [25]. Because the patterns described here were evident among individual biological replicates and replicates were well correlated (Supplementary Figure E1, online only, available at www.exphem.org), we merged all tags from biological replicate data sets for final analyses.
Data visualization
Raw sequencing tags were smoothed (20bp bin, 100bp sliding window) and normalized to reads per kilobase per million (RPKM) using deepTOOLS [26] and visualized with an Integrative Genomics Viewer (IGV) [27]. The ChAsE (Chromatin Analysis and Exploration) visualization suite [28] was used to display the distribution of histone marks relative to the transcription start site (TSS). RPKM normalized density plots were generated using Java TreeView [29].
Chromatin state mapping
To identify genomic intervals whose patterns of histone marks were consistent with enhancers, we applied a hidden Markov modeling algorithm using ChromHMM [30]. To obtain a unified set of enhancer intervals, the data for each histone mark were merged across both time points. Briefly, the merged files were binarized using the BinarizeBed function with the signal threshold option set to 100. The minimum number of nonredundant chromatin states was heuristically determined to be six using the LearnModel function with default parameters.
Quantification and annotation of ChromHMM enhancer intervals
Chromatin states 1 and 2 (Supplementary Table E2, online only, available at www.exphem.org), whose chromatin patterns were consistent with enhancers, were annotated and quantified using the Hypergeometric Optimization of Motif EnRichment (HOMER) suite [31]. Briefly, bam files were converted to tag directories using the makeTagDirectory function with the –genome, –checkGC, and –format options. To quantify and normalize tags within enhancer regions to RPKM, the analyzeRepeats function was used with the –rpkm and –d options. The log2 fold change (from 0 to 1 hour after Epo) was then computed for each histone mark. Finally, enhancer–gene associations were inferred based on the nearest gene paradigm [32–34], and each enhancer interval was linked to a gene using the annotatePeaks function with the –noadj and –d options.
Super-enhancer analysis
Super-enhancers were identified based on the algorithm originally described by Whyte et al. [35], and implemented by HOMER using the findPeaks function with the –style option set to super. For this analysis, the minimum distance to stitch peaks together (-minDist) and the local fold change thresholds (-L) were set to 6,000 and 1, respectively. Data for H3K4me1 and H3K27acmarks were combined to generate a single “enhancer” tag directory for 0 and 1 hour, individually. Enhancer intervals whose midpoints were within 1 kb of a TSS were filtered. The remaining enhancer boundaries were trimmed 3 kb distal to a TSS if their interval edges overlapped with a TSS. As with the ChromHMM enhancer analysis described previously, the super-enhancer regions were quantified and annotated using the analyzeRepeats and annotatePeaks functions.
Classification of enhancers
Similar to the criteria detailed in Ostuni et al. [36], enhancer intervals were assigned to seven enhancer classes based on H3K27ac and H3K4me1 RPKM values and H3K27ac fold-change for signals in response to Epo stimulation (Supplementary Tables E3 and E4, online only, available at www.exphem.org). Briefly, RPKM values within the lowest quartile and the upper three quartiles were designated as “low” and “high, ” respectively. Occupancy changes in response to Epo were designated as “up” or “down” based on whether RPKM fold-changes increased or decreased by more than twofold, respectively. RPKM fold changes that were between these values were considered as no change. The criteria for each enhancer class are summarized below: (1) Constitutive activated enhancers displayed “high” H3K27ac and “up” in H3K27ac. (2) Constitutive not-activated enhancers displayed “high” H3K27ac and no change in H3K27ac. (3) Poised activated enhancers displayed “high” H3K4me1 levels, “low” H3K27ac, and “up” in H3K27ac. (4) Poised not-activated enhancers displayed “high” H3K4me1 levels, “low” H3K27ac, and no change in H3K27ac. (5) Latent enhancers displayed “low” H3K4me1 and H3K27ac levels and “up” in H3K4me1 and H3K27ac levels. (6) Latent to poised enhancers displayed “low” H3K4me1 and H3K27ac levels, “up” in H3K4me1, and no change or “down” in H3K27ac levels. (7) Repressed enhancers displayed “high” H3K27ac and “down” in H3K27ac. Lastly, 3,279 intervals were excluded from further analysis because they displayed biologically irrelevant characteristics, such as low H3K4me1 and/or H3K27ac levels that further decreased on Epo stimulation.
Motif discovery
De novo motif discovery for all enhancer regions was conducted using the findMotifsGenome function in the HOMER suite. To comprehensively identify locations for motifs related to erythroid cell function that were enriched in the de novo search, we conducted a directed search for the following consensus motifs [37] with zero mismatches allowed: GATA1 (WGATAR), KLF1 (YMCDCCCW), TAL1-Ebox (CANNTG), ETS (YWTCCK), and STAT5 (TTCYHDGAA). The scanMotifGenomeWide function in HOMER and intersectBED function in BEDtools [38] were used to find all instances of listed motifs residing within enhancer intervals (Supplementary Tables E3 and E5, online only, available at www.exphem.org).
Data access
Processed data from data analyses are available in the Supplementary Material (online only, available at www.exphem.org). Raw sequencing data are available at NCBI Sequence Read Archive (SRP082181).
Results
Experimental overview and distribution of histone marks at the β-globin locus control region
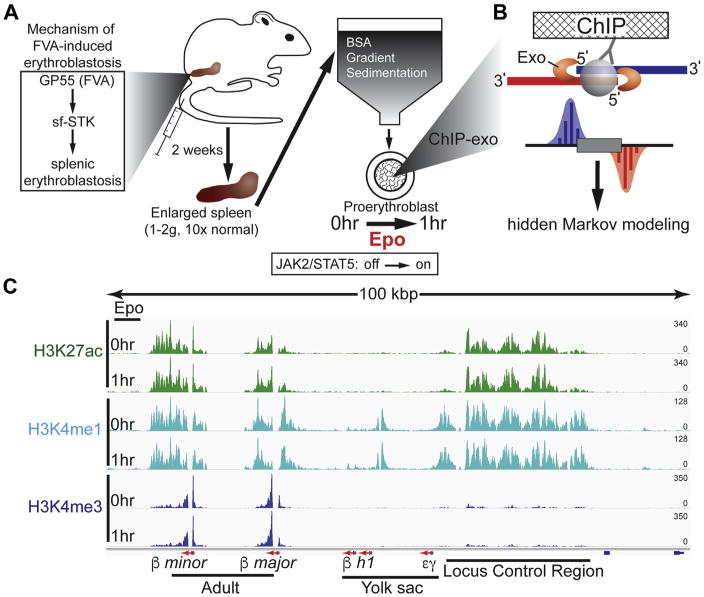
To study the molecular action of Epo, we leveraged the anemia-inducing strain of the Friend virus (FVA) murine model system that has been reported to recapitulate normal erythropoiesis, as evidenced by JAK2-STAT5 signaling, globin expression kinetics, cell morphology, cell surface marker kinetics, and cellular enucleation [19,21,39–43]. Indeed, the FVA-derived pro-erythroblasts were recently used as a standard for developing an improved flow cytometry sorting scheme for bone marrow-derived erythroblasts [39]. This system enables facile large-scale procurement of highly purified murine pro-erythroblast cell populations that synchronously respond to Epo (Fig. 1A) [22].
Figure 1.
Experimental overview and distribution of histone marks at the β-globin locus control region. (A) Shown is the workflow for generating and isolating highly purified Epo-responsive pro-erythroblasts from a mouse injected with FVA. (B) ChIP-exo cartoon illustrates how the 5′ to 3′ lambda exonuclease trims DNA to the crosslink point, effectively footprinting the protein–DNA interaction. Mapping the 5′ ends of the sequence tags to the reference genome demarcates the exonuclease barrier and, thus, the precise site of protein-DNA crosslinking. (C) Genome browser view of H3K4me1, H3K27ac, and H3K4me3 ChIP-exo signal in murine pro-erythroblasts shown at the β-globin (Hbb) LCR. Tag distributions were smoothed and RPKM was normalized using deepTools [88].
Friend virus infection in susceptible mice leads to a multistage disease, characterized initially by erythroblastosis and then later by erythroleukemia [44]. FVA-derived pro-erythroblasts used in this study were harvested during the initial cell expansion stage of erythroblastosis and were not erythroleukemic. The two strains of the Friend virus result in distinct phenotypic outcomes caused by amino acid differences in the Friend virus glycoprotein, GP55. The polycythemia-inducing GP55P variant requires both EpoR receptor and sf-STK, thereby constitutively activating JAK2-STAT5 signaling and Epo-independent differentiation [45]. Mechanistically, the anemia-inducing GP55A variant directly interacts with the sf-STK receptor to enable erythroid precursor proliferation in the absence of Epo (Fig. 1A). Indeed, a previous report indicated that neither EPOR nor STAT5 is required for FVA-induced erythroblastosis [21].
To assess the extent to which Epo influences the epigenetic landscape, we performed ChIP-exo on three key histone H3 modifications (H3K4me1, H3K27ac, and H3K4me3) before and after cells were exposed to Epo (0 and 1 hour Epo, Fig. 1B). For each antibody, ChIP-exo signals across biological replicates were highly correlated (R = 0.96–0.99), indicating high reproducibility (Supplementary Figure E1, online only, available at www.exphem.org). Importantly, these three histone marks enable complex pattern recognition algorithms, such as hidden Markov modeling, to identify candidate enhancers genome-wide and to distinguish them from promoter-specific patterns [30]. The putative functional nature of enhancers in the present study should be noted, because enhancer predictions are based on structural histone modification patterns. In Figure 1C, the histone modification patterns are illustrated at the well-characterized β-globin locus and nearby locus control region (LCR), a cluster of enhancers that control β-globin transcription [46]. Consistent with the marks found at enhancers, H3K4me1 and H3K27ac, but not H3K4me3, were enriched at the LCR. H3K4me3 was enriched at the promoters of the β-major and -minor globin genes that are expressed in adult erythroid cells. In contrast, the embryonically expressed βh1 and γε loci lacked both H3K27ac and H3K4me3 associated with active promoters.
ChIP-exo analysis reveals histone modification patterns at gene promoters
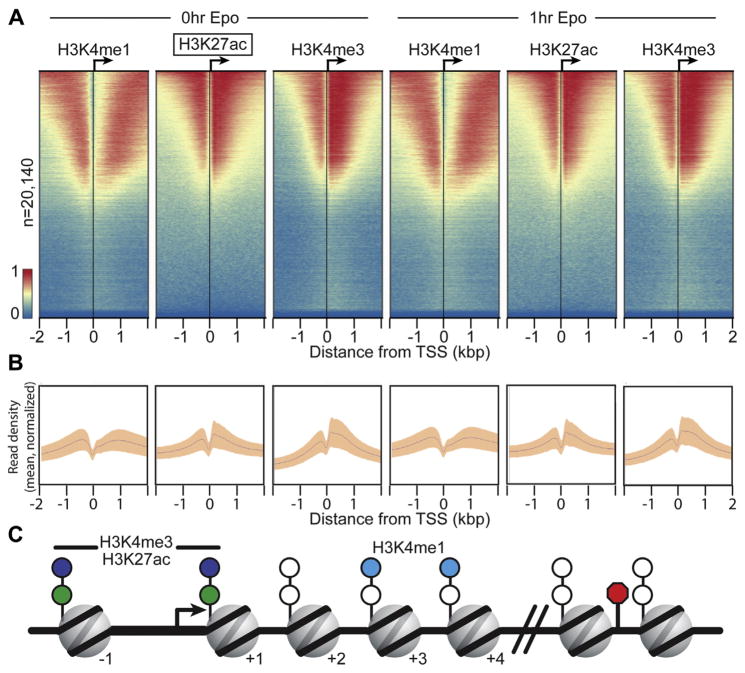
Next, we examined the global ChIP-exo patterns for H3K4me1, H3K27ac, and H3K4me3 at promoters of protein-coding genes. The ChIP-exo signal for each mark before and after Epo treatment (0 and 1 hour, respectively) was aligned to the coding TSS and displayed as a heatmap (Fig. 2A; sorted by maximal peak position in Supplementary Figure E2, online only, available at www.exphem.org). Each histone mark was detected at nearly one half of all protein-coding genes (Supplementary Table E6, online only, available at www.exphem.org), consistent with previous reports indicating that these marks are not found at all genes, but rather, are enriched in the subset of genes actively transcribed in erythroid cells [4–6].
Figure 2.
ChIP-exo reveals histone modification patterns at gene promoters. (A) ChIP-exo libraries were prepared for H3K4me1, H3K27Ac, and H3K4me3 and visualized using ChAsE [89]. Aligned heatmaps indicate RPKM normalized number of reads across a 4-kb genomic interval in 20-bp bins relative to the TSS before and after Epo stimulation. Regions are sorted in descending order based on average row tag density for 0-hour H3K27ac. Each row represents one protein-coding gene, with 20,140 lines/genes present. Red and blue reflect high and low read densities, respectively. (B) Composite plots below each heatmap quantify the normalized tag density. The central trace denotes the average tag density for each 20-bp bin, and the orange fill reflects the standard deviation. (C) Model illustrating the nucleosome position for enriched histone marks, which are derived from the tag enrichment patterns in (A) and Supplementary Figure E2 (online only, available at www.exphem.org).
It is well established that the genome-wide distribution of histone marks at promoters is not random, but exhibits distinct preferences to specific nucleosomes within promoter regions [47]. Given the mapping precision afforded by ChIP-exo analysis, we examined the spatial distribution of histone marks relative to each other and nucleosome positions relative to the TSS. As expected, we found that H3K4me1 and H3K4me3 tend to be mutually exclusive in their preferred nucleosomes, with H3K4me3 most enriched at the −1 and +1 nucleosomes. In contrast, H3K4me1 marks were enriched at nucleosomes distal to the TSS, particularly in the +3 and +4 positions, but decayed rapidly thereafter. Notably, the H3K27ac ChIP-exo signal was enriched in a pattern similar to that of H3K4me3 at the −1 and +1 nucleosomes, reflecting the expected signature pattern at the nucleosomes flanking core promoter regions [11]. Composite plot distribution peaks (in Fig. 2B and C) further highlight the nucleosomal preferences of specific histone marks at promoters of protein-coding genes in pro-erythroblasts.
Hidden Markov modeling of chromatin states reveals unique enhancer signatures
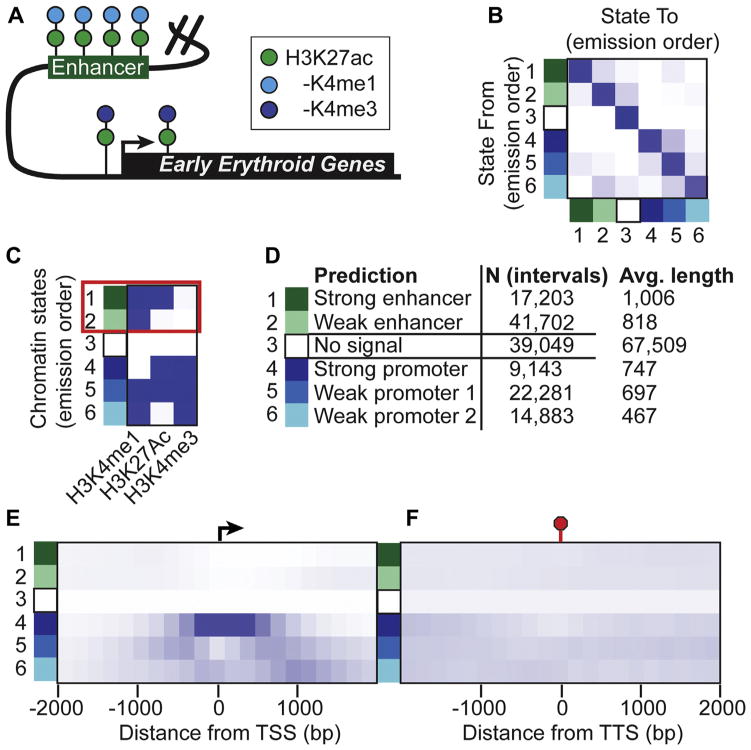
Pioneering studies by the ENCODE consortium suggest that the mammalian genome may harbor hundreds of thousands of enhancer regions, with each of the approximately 200 different cell types containing distinct repertoires of enhancer elements [48–51]. Given this complexity and despite recent advances in high-throughput technologies, it remains a challenge to identify the complete set of bona fide functional enhancers in a given cell type. Nevertheless, this large-scale project and other studies have found that enhancers and promoters display distinct epigenetic signatures (Fig. 3A), which can be used to predict enhancer locations in differing cell types with a relatively high success rate [52]. In particular, enhancer elements are demarcated by histone 3 lysine 4 monomethylation (H3K4me1) and histone 3 lysine 27 acetylation (H3K27ac) [47,53,54]. This signature distinguishes enhancers from promoters, which are marked by histone 3 lysine 4 trimethylation (H3K4me3).
Figure 3.
Hidden Markov modeling of chromatin states identify unique enhancer signatures. (A) The cartoon illustrates distinct chromatin signatures associated with enhancers and promoters. (B) The transition matrix illustrates the probability that the state in the row (State From) will transition to the state in the column (State To) using hidden Markov modeling of chromatin states [33]. (C) Heatmap representation of chromatin state emissions from hidden Markov modeling that predicts enhancer intervals from H3K4me1, H3K27ac, and H3K4me3. The red box highlights states 1 and 2, which display chromatin state patterns consistent with enhancers that enrich H3K4me1 and/or H3K27ac but not H3K4me3. (D) Functional annotation inferences for each chromatin state are listed next to the number of intervals comprising the state and the average length. (E, F) Heatmaps illustrating the enrichment of each learned chromatin state ± 2000 bp from the TSS and transcription termination site (TTS), respectively.
Thus, to systematically identify the genomic locations of chromatin signatures consistent with enhancers in FVA-derived pro-erythroblasts, we applied hidden Markov modeling (HMM) to ChIP-exo data (Fig. 3). ChromHMM employs a powerful algorithm for finding unique patterns in complex data [30]. HMM analysis of the ChIP-exo data for H3K4me1, H3K27ac, and H3K4me3 at 0- and 1-hour Epo time points revealed six non-redundant chromatin states (Fig. 3B, C). Chromatin states 1 and 2 represent nearly 60,000 candidate enhancers with an average length of ~900 bp and signatures consistent with strong and weak enhancers, respectively—that is, having H3K4me1 and/or H3K27ac but lacking H4K4me3, respectively. As expected, these candidate enhancer regions tended to be distal to transcription start and end sites (Fig. 3E, F; quantified in Supplementary Figure E3, online only, available at www.exphem.org). In contrast, chromatin states 4–6 were strongly associated with promoter regions (Fig. 3E) and were each strongly enriched with H3K4me3 relative to states 1 and 2. Lastly, chromatin state 3 was depleted of ChIP signal for the three histone marks in this study and encompassed the majority of the genome, suggesting low representation of these marks and/or the possible presence of other histone modifications in the intervals of this state.
Epo-induced remodeling of the erythroid enhancer landscape
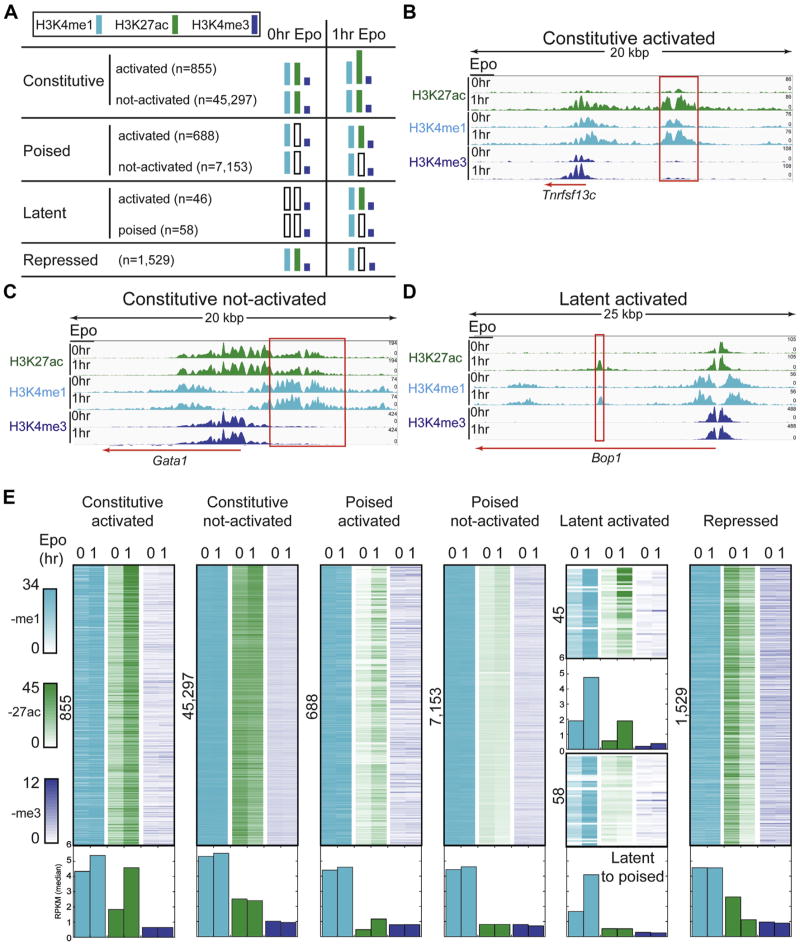
Tens of thousands of enhancers are scattered across mammalian genomes in a cell type-specific manner and are dynamically shaped in response to environmental stimuli [11]. Although a handful of studies have focused on erythroid enhancers [16–18], the ways in which Epo influences erythroid enhancers remain unclear. We hypothesized that Epo signaling reshapes the epigenetic landscape of a specific set of erythroid enhancers and drives the erythroid transcriptional program. To test this hypothesis, we systematically classified the nearly 60,000 candidate enhancer regions based on their response to Epo stimulation, in a manner similar to that previously described [40]. The presence or absence of H3K27ac is associated with enhancer activation or repression, respectively [54]. Poised enhancers initially lack H3K27ac but can acquire the acetyl mark with cellular stimulation from environmental cues, such as cytokines and hormones [33,36,55,56]. Latent enhancers were recently described as regions of the genome that lack enhancer histone marks, but acquire these marks on stimulation [36].
Overall, response to Epo stimulation was highly dynamic, enabling stratification of the ~60,000 genomic intervals into four major classes (Fig. 4A). Candidate enhancers across each class shared several features, including commonly residing within intergenic and intronic regions of the genome, averaging about 1 kb in length, and varying quite dramatically in their distances to the nearest gene (Supplementary Figure E4, online only, available at www.exphem.org). First, the class of constitutive enhancers was associated with both H3K4me1 and H3K27ac in unstimulated pro-erythroblasts (0-hour Epo). Although most (45,297) were unaffected after 1 hour of Epo stimulation (constitutive not-activated, Fig.4E), 855 displayed increased acetylation within the first hour of Epo stimulation (constitutive activated, Fig. 4E). Figure 4B illustrates the Epo-induced dynamics of a constitutive activated enhancer at the Tnfrsf13c gene, which interestingly, encodes a receptor known to promote survival in B cells [57]. Indeed, transcriptional profiling in murine pro-erythroblasts revealed that Tnfrsf13c was strongly induced in response to Epo treatment [7]. This observation supports the notion that the enhancer we identified upstream of the Tnfrsf13c locus may be mediating its transcriptional response to Epo to further promote pro-survival pathways that Epo is known to regulate [19]. As illustrated in Figure 4C, the Gata1 locus, which encodes one of the three master regulators of the erythroid expression program, exemplifies the constitutive not-activated class of enhancers. Consistent with this observation, previous studies have indicated that maximal Gata1 gene expression, and presumably increased enhancer acetylation, occurs in basophilic and orthochromatic erythroblasts [4–6], which are not present in the model system until 12–36 hours after Epo stimulation.
Figure 4.
Epo-induced remodeling of the erythroid enhancer landscape. (A) Classification of erythroid enhancers based on their response to Epo. (B–D) Representative genome browser views of Epo-responsive enhancers (B and D: constitutive activated and latent activated, respectively) and an Epo non-responsive enhancer (C, constitutive not activated). Red boxes denote genomic intervals identified as enhancers by hidden Markov modeling. (E) Density plots illustrating the dynamic behavior of histone marks for each enhancer interval induced by a 1-hour Epo treatment. The number of enhancer intervals shown is indicated to the left of each plot. The normalized RPKM median value for each column is represented as a bar graph below the density plots.
Next, the class of poised enhancers was characterized by H3K4me1, but not H3K27ac, in unstimulated pro-erythroblasts. Of the 7,841 poised enhancers we detected, 688 were Epo-responsive and displayed increased acetylation after 1 hour of Epo stimulation (poised activated, Fig. 4E). Gene ontology analysis (Supplementary Table E7, online only, available at www.exphem.org) revealed that the set of genes proximal to poised activated enhancers was most associated with positive regulation of TF activity (p = .001), including Kit, the gene encoding the receptor tyrosine kinase that activates a number of signaling pathways critical to cell survival during erythropoiesis. Also included in the group of poised activated enhancers were genes encoding Wnt signaling proteins (WNT3A and WNT10B), critical regulators that balance self-renewal, proliferation, and differentiation in hematopoietic cells [58].
We also identified a class of more than 100 latent enhancers, which gained H3K4me1 and/or H3K27ac on Epo stimulation (Fig. 4E). Remarkably, nearly one half of these latent enhancers (45) were activated within 1 hour of Epo stimulation. Figure 4D illustrates the Epo-induced dynamics of the class of latent activated enhancers at the Bop1 (Block of proliferation 1) gene locus. The precise timing of the Bop1 latent enhancer activation with Epo-stimulated erythropoiesis is consistent with the cellular transition to the inherently non-proliferating mode of terminal differentiation. Interestingly, as a protein involved in rRNA processing, Bop1 has been implicated in ribosomopathies [59,60], such as Diamond–Blackfan anemia.
Finally, the class of enhancers consistent with repression displayed both H3K4me1 and H3K27ac in untreated pro-erythroblasts, but lost or exhibited diminished H3K27ac within 1 hour of Epo stimulation (Fig. 4E). This class of enhancers was enriched near numerous genes encoding TF regulators (p = 10−7), including MAX. MAX is the primary partner for MYC, conferring the ability of MYC/MAX dimers to bind E-boxes. Together, MYC/MAX dimers control the opposing cell fate decisions of cell cycle progression and apoptosis [61], suggesting that Epo-induced enhancer remodeling may contribute to cell fate decisions during erythropoiesis via MAX. Consistent with the observation that Epo simulation leads to repression of an enhancer linked to the binding partner for MYC, a previous report indicated that downregulation of MYC is important during erythropoiesis [62].
In summary, we identified more than 3,000 Epo-responsive enhancers, of which 1,588 and 1,529 were activated and repressed, respectively, within the first hour of terminal erythroid differentiation. This suggests that Epo rapidly remodels a subset of the epigenetic landscape of pro-erythroblasts undergoing erythropoiesis. Consistent with a previous report [16] indicating that chromatin states remain largely unchanged during erythroid differentiation, the vast majority of the candidate enhancer regions identified in this study (n = 52,508) were static during the first hour of Epo exposure. This indicates that they are established at a prior precursor stage and are not Epo-responsive or that they respond to Epo at some point after 1 hour of treatment.
Validation and evolutionary conservation of candidate enhancers
The possibility of false positives is a caveat to algorithms that predict enhancer regions based on functional genomic data. To provide corroborative evidence for the validity of candidate enhancers, we applied four complementary computational and experimental approaches: (1) DNA motif analysis, (2) correlation to promoter acetylation, (3) overlap analysis with orthogonal published data sets, and (4) TF binding via ChIP-exo analysis. Transcription factors regulate cell type-specific gene expression patterns by binding to their cognate motifs within enhancer regions and recruiting the transcription machinery to target genes [13]. In erythroid cells, GATA1, KLF1, and TAL1 are master TF regulators and play a major role in coordinating the precise timing of gene expression patterns during erythropoiesis [63]. GATA1, the most well studied of the trio of master regulators, binds to the WGATAR consensus motif [64,65], which can have a positive or negative influence on transcription, depending on the composition of the LDB1-nucleated complex [16,66,67].
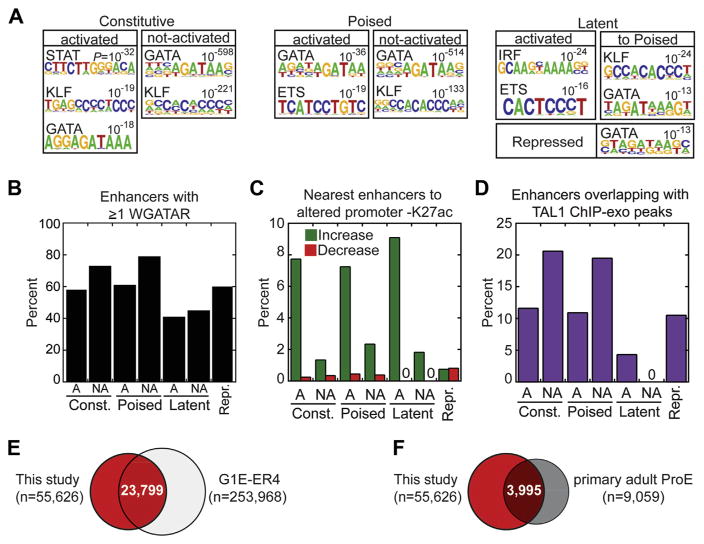
Therefore, we hypothesized that HMM-identified candidate enhancers that are physiologically relevant in erythroid cells would be enriched for motifs corresponding to the master regulator TFs of erythropoiesis. To test this hypothesis, we conducted de novo motif discovery analysis of all ~60,000 enhancer regions from Figure 4. Strikingly, the Gata motif was the most commonly enriched (p = 10−13 to 10−598) across the enhancer classes (Fig. 5A). Other erythroid-related motifs found in the de novo search included the KLF1, ETS, and STAT5 consensus motifs.
Figure 5.
Validation of candidate enhancers. (A) TF binding motifs over-represented in each enhancer class are shown as motif logos based on their respective position weight matrices (PWMs). De novo discovery of PWMs, p values, and best Jaspar motif match were generated using HOMER. (B) Bar graph illustrating percentage of each enhancer class containing at least one GATA1 consensus motif (WGATAR) with zero mismatches. Activated and not-activated subclasses are denoted by “A” and “NA, ” respectively. (C) Bar graph of the percentage of enhancers, separated by class, that are nearest to genes with greater than twofold increase (green) or decrease (red) in H3K27ac at promoters (±1 kb). Activated and not-activated subclasses are denoted by “A” and “NA, ” respectively. (D) Percentage of enhancer intervals occupied by ≥1 ChIP-exo Tal1 peaks and broken out by enhancer class. Activated and not-activated subclasses are denoted by “A” and “NA, ” respectively. (E) Venn overlap between enhancer intervals in this study (red fill) and mouse G1E-ER4 chromatin states 2 and 3 (H3Kme1 and H3K4me1/H3K27me3, respectively) from Wu et al. [16]. (F) Venn overlap between enhancer intervals in this study (red fill) and enhancer regions in human primary adult pro-erythroblasts from Huang et al. [18]
Because de novo motif analysis provides initial clues to over-represented motifs in the genomic regions, we next conducted a more sensitive, direct motif search. Given the connection between Epo signaling and GATA1 phosphorylation [68], we focused on identifying all occurrences of the GATA consensus motif within enhancers (Fig. 5B, Supplementary Figure E5, online only, available at www.exphem.org). Indeed, for each class of enhancers, the observed frequency of the GATA motif was significantly greater than an equivalent number of randomly sampled genomic regions (Fig. 5B, empirical p value <.001). Altogether, our motif analysis provides corroborative evidence for validating candidate enhancers.
Previous studies indicated that enhancer:promoter interactions may be inferred by correlating H3K27ac patterns [69]. Therefore, we examined the relationship between Epo-stimulated changes in H3K27ac at enhancers and those of the nearest gene promoter. In general, genes that increased in promoter acetylation were linked to nearby enhancers that also acquired H3K27ac in response to Epo (e.g., activated enhancer subclasses denoted by ‘A’) (Fig. 5C), with the repressed enhancer class least represented. Overall, the correlation between decreased promoter and enhancer acetylation was far weaker, although the repressed class of enhancers was most associated with a decrease in promoter acetylation. Not only do these results support the enhancer classification methodology (Fig. 4), they provide further corroborative evidence for the candidate enhancers in this study.
Given the previous finding that TF binding is an accurate predictor of enhancer activity [70] and Epo stimulation rapidly activates TAL1 in FVA cells [71], we performed TAL1 ChIP-exo analysis in FVA-derived pro-erythroblasts and identified 19,025 and 13,913 enriched peaks for the 0- and 1-hour Epo treatments, respectively. Strikingly, nearly 60% (20,289 of 32,938) of all TAL1 peaks resided within candidate enhancer regions identified in this study, which are separated by class in Figure 5D. In other words, TAL1 alone occupied 33% of all candidate enhancers (18,370 of 55,626 enhancers with one or more TAL1 peaks), suggesting that these enhancer regions are valid.
Lastly, we compared our set of enhancer locations to published orthogonal data sets from mouse and human studies [16,18]. We found that nearly 24,000 enhancers identified in this study also were present in G1E-ER4 cells [16] (Fig. 5E). In addition, 44% (3,995 of 9,059) of enhancers previously identified in primary human pro-erythroblasts [18] were shared with the current study (Fig. 5F), indicating substantial evolutionary conservation of erythroid cis-regulatory modules. In summary, based on evidence from the four aforementioned complementary approaches used to validate enhancers, we conclude that candidate enhancer regions identified in this study generally represent bona fide enhancers and are likely to be physiologically and functionally relevant to terminal erythroid differentiation.
Identification of super-enhancers at erythroid genes involved in cell fate decisions
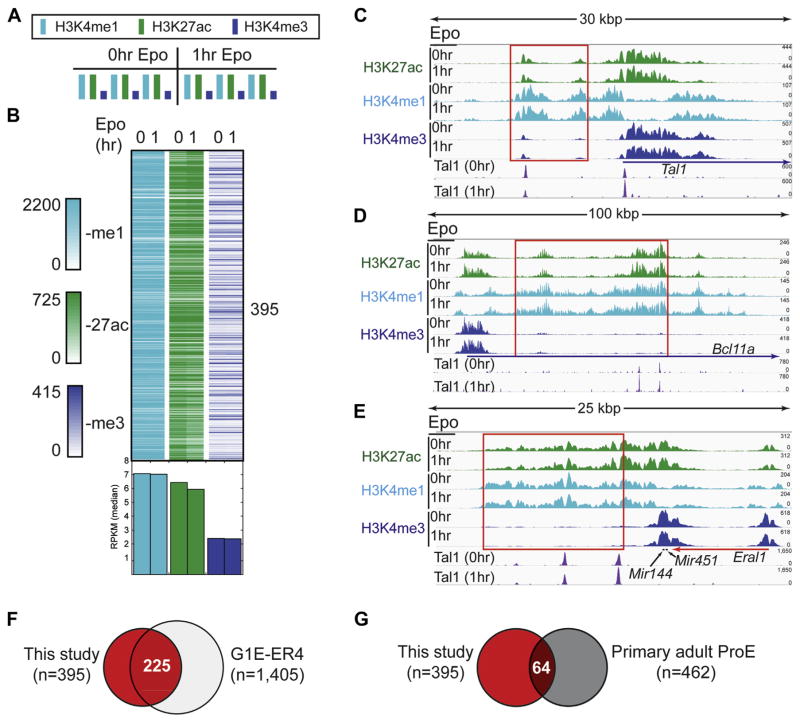
Super-enhancers are an emerging class of long, clustered enhancers important for establishing cell fate and identity [35]. A recent study examined super-enhancers in CD34+ derived pro-erythroblasts and murine G1E cell lines [18], but whether Epo stimulation affects super-enhancers remains unclear. Therefore, to investigate the extent to which Epo signaling influences super-enhancer dynamics in pro-erythroblasts, we applied the algorithm previously described for identifying super-enhancers [60] to our H3K4me1 and H3K27ac ChIP-exo data sets. We identified 395 super-enhancers dispersed throughout the mouse genome that were characterized by their length (21,192 bp on average; Supplementary Figure E4) and exceptionally high H3K4me1 and H3K27ac levels (Fig. 6A,B). We found that the super-enhancers were generally nonresponsive to Epo with respect to H3K27ac (Fig. 6B). Consistent with the notion that super-enhancers tend to be associated with genes controlling cell fate and identity, they were discovered near a number of genes known to play roles in erythroid cell physiology, such as Tal1, Bcl11a, and Mir144/451 (Fig. 6C–E, Supplementary Figure E6 for additional screen shots, and a full list of super-enhancer locations and nearby genes may be found in Supplementary Table E3, online only, available at www.exphem.org). Tal1 and Bcl11a encode TFs that are known to regulate erythroid gene expression patterns [71–73]. The Mir144/451 polycistronic gene produces microRNAs that promote erythroid maturation and are strongly induced by GATA1 during erythropoiesis [74,75]. Interestingly, beyond the representative genes in Figure 6C–E, TAL1 bound to 92% of all super-enhancer regions (365 of 395 enhancers; Supplementary Figure E7, online only, available at www.exphem.org). In addition, GATA motifs were found in nearly all super-enhancer regions (99.7%, 394 of 395 super-enhancers; Supplementary Figure E5, online only, available at www.exphem.org). Indeed, the GATA motif overlapped with 95.6% of TAL1 peaks within super-enhancers (345 of 361 Tal1 peaks for 0-hour Epo; Supplementary Figure E8, online only, available at www.exphem.org). Overlap analysis between super-enhancer locations revealed that 57% (225 of 395; Fig. 6F) of super-enhancer locations from this study were shared with a previous report in mouse G1E-ER4 cells [18], thereby corroborating our super-enhancer analysis. In contrast, super-enhancer locations were poorly conserved evolutionarily when compared to super-enhancer locations from CD34+ derived pro-erythroblasts (16%, 64 of 395; Fig. 6G).
Figure 6.
Identification of super-enhancers at erythroid genes involved in cell fate decisions. (A) Histone modification enrichment across super-enhancers in erythroid cells before and after Epo stimulation. (B) Density plots illustrating the H3K4me1, H3K27ac, and H3K4me3 ChIP-exo signal (RPKM normalized) across super-enhancer intervals before and after Epo treatment. The normalized RPKM median value for each column is represented as a bar graph below the density plots. (C–E) Genome browser views of super-enhancers before and after Epo stimulation at the Tal1, Bcl11a, and Mir144/Mir451 genes, respectively. Red boxes denote genomic intervals identified as super-enhancers by the Whyte et al. algorithm employed by HOMER. (F,G) Venn overlap of super-enhancer intervals identified in this study (red fill) compared with super-enhancer regions from Huang et al. [18] in mouse (panel F, G1E-ER4) and human (panel G, primary adult ProE) erythroid cells, respectively.
Epo-modulated enhancers integrate erythroid signaling pathways
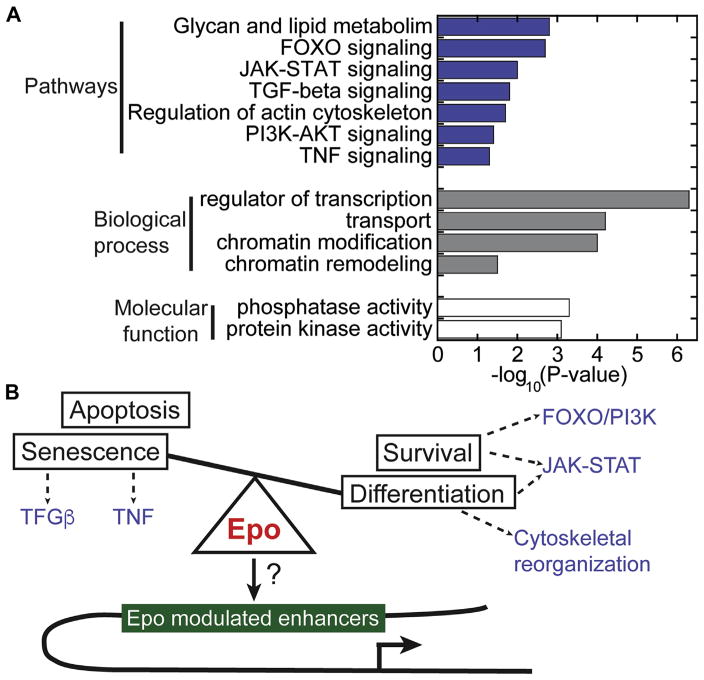
To investigate what types of genes are associated with Epo-modulated enhancers, we conducted gene ontology and pathway enrichment analysis (Fig. 7A). We found that many enhancers modulated by Epo stimulation were linked to genes involved in cell-signaling pathways, such as JAK-STAT (n = 24), PI3K (n = 45), and FOXO (n = 25) signaling. Although these pathways are known to regulate erythroid differentiation [76], it is surprising that enhancers linked to these pathways are remodeled in response to Epo. Nevertheless, this observation is consistent with the notion that Epo-driven epigenetic changes may form feed-forward loops that serve to strengthen the kinetics and robustness of Epo-responsive pathways. During terminal erythroid differentiation, the cytoskeletal structure undergoes a dramatic transformation, which involves actin and the glycosylation of a number of erythroid transmembrane proteins, such as Band 3 and Glycophorin A [77]. Indeed, within the first hour of exposure to Epo, the epigenetic landscape is altered near genes involved in regulating the actin cytoskeleton (n = 31), glycosylation (n = 153), and transport (n = 221). This suggests that cytoskeletal reorganization may be regulated in part at the epigenetic level.
Figure 7.
Epigenetic integration of signaling pathways by Epo-modulated enhancers. (A) Gene ontology and pathway enrichment analysis for genes linked to Epo-modulated enhancers. Top enriched pathways, biological processes, and molecular functions are represented by blue, gray, and white fill bars, respectively. (B) Model illustrating the interplay of pathways enriched in the set of genes linked to Epo-modulated enhancers.
Collectively, TFs orchestrate cell type-specific transcription programs by binding to their cognate sequence motifs within specific enhancers. TFs then recruit co-regulators and RNA polymerase II to promoters to initiate erythroid transcription programs. Strikingly, analysis of genes linked to Epo-modulated enhancers revealed an enrichment of numerous regulators of transcription (n = 284), chromatin regulators (remodeling and modification, n = 60), and phosphorylation regulators (kinase and phosphatase activity, n = 97). In particular, Epo simulation altered enhancers linked to Bach1, Bcl11a, Elf1, Klf10, Sox6, Tcf3, and Tif1γ, each of which has previously been reported to be involved in controlling erythroid expression programs [78–83]. Together, these observations suggest that Epo stimulation influences a network of erythroid-relevant TFs at the epigenetic level, and may serve to coordinately mobilize TF activity to orchestrate erythroid expression programs.
Discussion
How hormone signaling is manifested in chromatin dynamics remains an important, unanswered question. Here, we have illustrated that the hormone Epo reprograms a repertoire of enhancers in murine pro-erythroblasts that are linked to integrated networks of erythroid transcriptional regulators and signaling pathways. Identifying where and how these histone marks are altered in response to stimuli reveals the plasticity of erythroid enhancers and provides new insights into the molecular action of Epo.
Our experimental murine model uniquely allowed us to investigate how the enhancer landscape changes on Epo stimulation. We identified ~60,000 enhancers in pro-erythroblasts, and in response to Epo stimulation, their histone mark dynamics enabled stratification of enhancers into four major classes in addition to the super-enhancer class. Consistent with the findings of a previous study [16], we found that the enhancer landscape remains largely unchanged before and after Epo stimulation of pro-erythroblasts. However, nearly 3,000 enhancers were remodeled within the first hour of Epo treatment, suggesting that Epo initiates terminal erythroid differentiation in part by influencing a portion of the erythroid enhancer repertoire. Whether Epo continues to influence and reshape the epigenetic landscape throughout erythropoiesis remains unclear.
Two common themes emerged among the genes linked to Epo-responsive enhancers: (1) over-representation of pathways involved in coordinating the transition from a proliferative state to a non-proliferative differentiating state (Fig. 7B), and (2) enrichment of genes encoding TFs. The transition to a non-proliferative differentiating state is exemplified by Epo-responsive enhancers linked to Tnfrsf13c and Bop1 (Fig. 4B, D) and is consistent with the physiological transitions that accompany the cell fate decision to undergo terminal erythroid differentiation. In particular, once Epo triggers the cellular commitment to undergo erythropoiesis, the cell must halt proliferative signaling pathways and commence differentiation pathways, while blocking the cellular inclination toward apoptosis. Interestingly, Epo starvation and add-back experiments resulted in strongly upregulated Tnfrsf13c expression in murine fetal pro-erythroblasts [7]. This finding is in agreement with our discovery of a constitutive enhancer upstream of Tnfrsf13c that is further activated within the first hour of Epo stimulation.
Mechanistically, this study provides insight into how Epo stimulation alters the enhancer landscape in erythroid cells, which are presumably bound by TFs to orchestrate the dynamic gene expression programs during erythropoi-esis. Indeed, the top Gene Ontology category for genes linked to the Epo-responsive class of constitutive activated enhancers is the positive regulation of transcription from Pol II promoters (Supplementary Table E7, online only, available at www.exphem.org; p = .0001), which included the genes for TFs Lmo2, Klf5, and Klf13 and the Ep300 gene, which encodes an acetyltransferase. Furthermore, we found super-enhancers near a number of important genes encoding TFs that regulate erythropoiesis, such as Bcl11a and Tal1. It is interesting to note that a recent study reported a Tal1 super-enhancer specific to T-cell acute lymphoblastic leukemia cells, leading the study’s authors to suggest that the Tal1 super-enhancer is oncogenic [84]. However, in the context of normal erythropoiesis, we propose that the Tal1 super-enhancer identified in the present study serves a physiological function controlling early Tal1 expression kinetics in murine pro-erythroblasts. With respect to super-enhancers, a recent report described a super-enhancer at the α-globin locus and elegantly dissected the cis-regulatory requirements controlling α-globin expression [85]. This super-enhancer also was found in the present study (Supplementary Figure E6D; online only; www.exphem.org), further supporting the validity of the enhancers presented.
Several previous studies have used epigenetic profiling to identify candidate enhancers in human and mouse erythroid cells [16–18]. Although these studies provided detailed insights into erythroid enhancers during erythroid differentiation, it is important to note that the inherent limitations of these model systems precluded an assessment of Epo-dependent enhancer dynamics. Furthermore, putative enhancers were identified without considering H3K4me1 [17,18] or H3K27ac [16], which would miss the dynamic behavior of activated, repressed, poised, and latent enhancer states.
Specifically, our analysis of both H3K4me1 and H3K27ac histone modifications by ChIP-exo analysis in the FVA system circumvents these limitations and provides new insights into erythroid biology by tying dynamic enhancer states directly to Epo stimulation. In addition, our finding that Epo uncovers nearly 100 latent enhancers in erythroid cells further expands the repertoire of environmental stimuli that reveal cryptic enhancer locations [36,55].
The primary strength of using chromatin state mapping to identify enhancers is that it provides insights into the plasticity of the enhancer landscape in an unbiased and genome-wide manner, particularly in response to stimuli. We acknowledge the limitations of the present study for experimental models and approaches. First, enhancer predictions based on epigenetic profiling alone do not address functional significance. Corroborative evidence presented in Figure 5 sought to mitigate this limitation, particularly with functional genomic assays, such as TAL1 ChIP-exo analysis (Fig. 5F). Still, establishing a causal link between enhancer activation or repression and a corresponding gene activity is a challenging task, particularly on a genome-wide scale. Further confounding this pairwise enhancer–promoter association is the observation made in previous studies that, together, multiple enhancers can fine-tune the expression of a single gene [86,87]. In agreement, the present study found that on average, a given gene was linked to approximately five discrete enhancer regions. However, whether multiple nonclustered enhancers (i.e., not super-enhancers) synergistically or antagonistically regulate a given gene locus requires a gene-by-gene analysis. Lastly, our study along with previous erythroid enhancer reports [16–18] inferred enhancer–gene linkages using the nearest gene paradigm. A previous report using human cancer cell lines suggested that as many as 40% of enhancers can skip the nearest gene to loop to a more distant gene [88], but the extent to which this gene skipping occurs in a normal physiological context remains unclear. Although limited by resolution, long-range chromatin interaction assays performed on a genome-wide scale would extend previous locus-specific looping studies in erythroid cells [89] and would complement the current study by furthering our understanding of the dynamics of enhancer repertoire usage during terminal erythroid differentiation.
Here, we used the FVA murine model system of erythro-poiesis that uniquely allowed us to investigate how the hormone Epo shapes the enhancer landscape. This study provides novel insights into how hormone signaling pathways are able to control transcriptional programs during cellular differentiation by altering enhancer-associated marks. Together, these findings define a cis-regulatory enhancer network for Epo signaling during erythropoiesis and provide the framework for future studies involving the interplay of epigenetics and Epo signaling.
Supplementary Material
Acknowledgments
We thank the Vanderbilt Technologies for Advanced Genomics (VANTAGE) Core, Olivia Koues, and Ivo Violovich for technical and computational support with Illumina sequencing. Special thanks also to Scott Beeler for additional computational assistance.
AAP was supported by Vanderbilt Molecular Endocrinology Training Program Grant 5T32 DK07563.
References
- 1.Wojchowski DM, Sathyanarayana P, Dev A. Erythropoietin receptor response circuits. Curr Opin Hematol. 2010;17:169–176. doi: 10.1097/MOH.0b013e328338008b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koulnis M, Porpiglia E, Hidalgo D, Socolovsky M. Erythropoiesis: From molecular pathways to system properties. Adv Exp Med Biol. 2014;844:37–58. doi: 10.1007/978-1-4939-2095-2_3. [DOI] [PubMed] [Google Scholar]
- 3.Richmond TD, Chohan M, Barber DL. Turning cells red: Signal transduction mediated by erythropoietin. Trends Cell Biol. 2005;15:146–155. doi: 10.1016/j.tcb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 4.An X, Schulz VP, Li J, et al. Global transcriptome analyses of human and murine terminal erythroid differentiation. Blood. 2014;123:3466–3477. doi: 10.1182/blood-2014-01-548305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kingsley PD, Greenfest-Allen E, Frame JM, et al. Ontogeny of erythroid gene expression. Blood. 2013;121:e5–e13. doi: 10.1182/blood-2012-04-422394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pishesha N, Thiru P, Shi J, Eng JC, Sankaran VG, Lodish HF. Transcriptional divergence and conservation of human and mouse erythropoiesis. Proc Natl Acad Sci USA. 2014;111:4103–4108. doi: 10.1073/pnas.1401598111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh S, Dev A, Verma R, et al. Defining an EPOR-regulated transcriptome for primary progenitors, including Tnfr-sf13c as a novel mediator of EPO- dependent erythroblast formation. PloS One. 2012;7:e38530. doi: 10.1371/journal.pone.0038530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tallack MR, Magor GW, Dartigues B, et al. Novel roles for KLF1 in erythropoiesis revealed by mRNA-seq. Genome Res. 2012;22:2385–2398. doi: 10.1101/gr.135707.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng Y, Wu W, Kumar SA, et al. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 2009;19:2172–2184. doi: 10.1101/gr.098921.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li W, Notani D, Rosenfeld MG. Enhancers as non-coding RNA transcription units: Recent insights and future perspectives. Nat Rev Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- 11.Rivera CM, Ren B. Mapping human epigenomes. Cell. 2013;155:39–55. doi: 10.1016/j.cell.2013.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shlyueva D, Stampfel G, Stark A. Transcriptional enhancers: From properties to genome-wide predictions. Nat Rev Genet. 2014;15:272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 13.Lee TI, Young RA. Transcriptional regulation and its misregulation in disease. Cell. 2013;152:1237–1251. doi: 10.1016/j.cell.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol. 2009;44:117–141. doi: 10.1080/10409230902858785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu W, Cheng Y, Keller CA, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 2011;21:1659–1671. doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su MY, Steiner LA, Bogardus H, et al. Identification of biologically relevant enhancers in human erythroid cells. J Biol Chem. 2013;288:8433–8444. doi: 10.1074/jbc.M112.413260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Liu X, Li D, et al. Dynamic control of enhancer repertoires drives lineage and stage-specific transcription during hematopoiesis. Dev Cell. 2016;36:9–23. doi: 10.1016/j.devcel.2015.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koury MJ, Bondurant MC. Erythropoietin retards DNA breakdown and prevents programmed death in erythroid progenitor cells. Science. 1990;248:378–381. doi: 10.1126/science.2326648. [DOI] [PubMed] [Google Scholar]
- 20.Finkelstein LD, Ney PA, Liu QP, Paulson RF, Correll PH. Sf-Stk kinase activity and the Grb2 binding site are required for Epo-independent growth of primary erythroblasts infected with Friend virus. Oncogene. 2002;21:3562–3570. doi: 10.1038/sj.onc.1205442. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Randall MS, Loyd MR, et al. Role of erythropoietin receptor signaling in Friend virus-induced erythroblastosis and polycythemia. Blood. 2006;107:73–78. doi: 10.1182/blood-2005-05-1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sawyer ST, Koury MJ, Bondurant MC. Large-scale procurement of erythropoietin-responsive erythroid cells: Assay for biological activity of erythropoietin. Methods Enzymol. 1987;147:340–352. doi: 10.1016/0076-6879(87)47123-1. [DOI] [PubMed] [Google Scholar]
- 23.Koury MJ, Sawyer ST, Bondurant MC. Splenic erythroblasts in anemia-inducing Friend disease: A source of cells for studies of erythropoietin-mediated differentiation. J Cell Physiol. 1984;121:526–532. doi: 10.1002/jcp.1041210311. [DOI] [PubMed] [Google Scholar]
- 24.Pugh BF, Venters BJ. Genomic organization of human transcription initiation complexes. PLoS One. 2016;11:e0149339. doi: 10.1371/journal.pone.0149339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernstein BE, Stamatoyannopoulos JA, Costello JF, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Younesy H, Nielsen CB, Moller T, et al. An interactive analysis and exploration tool for epigenomic data. Computer Graphics Forum. 2013;32:91–100. [Google Scholar]
- 29.Saldanha AJ. Java Treeview–extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 30.Ernst J, Kellis M. ChromHMM: Automating chromatin-state discovery and characterization. Nat Methods. 2012;9:215–216. doi: 10.1038/nmeth.1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heinz S, Benner C, Spann N, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Visel A, Blow MJ, Li Z, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heintzman ND, Hon GC, Hawkins RD, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J, Shao Z, Glass K, et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev Cell. 2012;23:796–811. doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Whyte WA, Orlando DA, Hnisz D, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ostuni R, Piccolo V, Barozzi I, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 37.Mathelier A, Zhao X, Zhang AW, et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quinlan AR. BEDTools: The Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:11.12.11-11.1–34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen K, Liu J, Heck S, Chasis JA, An X, Mohandas N. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci USA. 2009;106:17413–17418. doi: 10.1073/pnas.0909296106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bondurant MC, Lind RN, Koury MJ, Ferguson ME. Control of globin gene transcription by erythropoietin in erythroblasts from friend virus-infected mice. Mol Cell Biol. 1985;5:675–683. doi: 10.1128/mcb.5.4.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koury MJ, Bondurant MC. Maintenance by erythropoietin of viability and maturation of murine erythroid precursor cells. J Cell Physiol. 1988;137:65–74. doi: 10.1002/jcp.1041370108. [DOI] [PubMed] [Google Scholar]
- 42.Rhodes MM, Kopsombut P, Bondurant MC, Price JO, Koury MJ. Bcl-x(L) prevents apoptosis of late-stage erythroblasts but does not mediate the antiapoptotic effect of erythropoietin. Blood. 2005;106:1857–1863. doi: 10.1182/blood-2004-11-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Koury MJ. Abnormal erythropoiesis and the pathophysiology of chronic anemia. Blood Rev. 2014;28:49–66. doi: 10.1016/j.blre.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 44.Friend C. Cell-free transmission in adult Swiss mice of a disease having the character of a leukemia. J Exp Med. 1957;105:307–318. doi: 10.1084/jem.105.4.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li JP, D’Andrea AD, Lodish HF, Baltimore D. Activation of cell growth by binding of Friend spleen focus-forming virus gp55 glycoprotein to the erythropoietin receptor. Nature. 1990;343:762–764. doi: 10.1038/343762a0. [DOI] [PubMed] [Google Scholar]
- 46.Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calo E, Wysocka J. Modification of enhancer chromatin: What, how, and why? Mol Cell. 2013;49:825–837. doi: 10.1016/j.molcel.2013.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu J, Adli M, Zou JY, et al. Genome-wide chromatin state transitions associated with developmental and environmental cues. Cell. 2013;152:642–654. doi: 10.1016/j.cell.2012.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yue F, Cheng Y, Breschi A, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515:355–364. doi: 10.1038/nature13992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat Biotechnol. 2010;28:817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 54.Creyghton MP, Cheng AW, Welstead GG, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vahedi G, Takahashi H, Nakayamada S, et al. STATs shape the active enhancer landscape of T cell populations. Cell. 2012;151:981–993. doi: 10.1016/j.cell.2012.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brown JD, Lin CY, Duan Q, et al. NF-kappaB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol Cell. 2014;56:21–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rauch M, Tussiwand R, Bosco N, Rolink AG. Crucial role for BAFF-BAFF-R signaling in the survival and maintenance of mature B cells. PLoS One. 2009;4:e5456. doi: 10.1371/journal.pone.0005456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lento W, Congdon K, Voermans C, Kritzik M, Reya T. Wnt signaling in normal and malignant hematopoiesis. Cold Spring Harb Perspect Biol. 2013;5:a008011. doi: 10.1101/cshperspect.a008011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chiabrando D, Tolosano E. Diamond Blackfan anemia at the crossroad between ribosome biogenesis and heme metabolism. Adv Hematol. 2010;2010:790632. doi: 10.1155/2010/790632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pestov DG, Strezoska Z, Lau LF. Evidence of p53-dependent crosstalk between ribosome biogenesis and the cell cycle: Effects of nucleolar protein Bop1 on G(1)/S transition. Mol Cell Biol. 2001;21:4246–4255. doi: 10.1128/MCB.21.13.4246-4255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amati B, Littlewood TD, Evan GI, Land H. The c-Myc protein induces cell cycle progression and apoptosis through dimerization with Max. EMBO J. 1993;12:5083–5087. doi: 10.1002/j.1460-2075.1993.tb06202.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jayapal SR, Lee KL, Ji P, Kaldis P, Lim B, Lodish HF. Down-regulation of Myc is essential for terminal erythroid maturation. J Biol Chem. 2010;285:40252–40265. doi: 10.1074/jbc.M110.181073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Love PE, Warzecha C, Li L. Ldb1 complexes: The new master regulators of erythroid gene transcription. Trends Genet. 2014;30:1–9. doi: 10.1016/j.tig.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Orkin SH. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- 65.Evans T, Reitman M, Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci USA. 1988;85:5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wadman IA, Osada H, Grutz GG, et al. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Soler E, Andrieu-Soler C, de Boer E, et al. The genome-wide dynamics of the binding of Ldb1 complexes during erythroid differentiation. Genes Dev. 2010;24:277–289. doi: 10.1101/gad.551810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhao W, Kitidis C, Fleming MD, Lodish HF, Ghaffari S. Erythropoietin stimulates phosphorylation and activation of GATA-1 via the PI3-kinase/AKT signaling pathway. Blood. 2006;107:907–915. doi: 10.1182/blood-2005-06-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dogan N, Wu W, Morrissey CS, et al. Occupancy by key transcription factors is a more accurate predictor of enhancer activity than his-tone modifications or chromatin accessibility. Epigenetics Chromatin. 2015;8:16. doi: 10.1186/s13072-015-0009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Prasad KS, Jordan JE, Koury MJ, Bondurant MC, Brandt SJ. Erythropoietin stimulates transcription of the TAL1/SCL gene and phosphorylation of its protein products. J Biol Chem. 1995;270:11603–11611. doi: 10.1074/jbc.270.19.11603. [DOI] [PubMed] [Google Scholar]
- 72.Sankaran VG, Xu J, Ragoczy T, et al. Developmental and species-divergent globin switching are driven by BCL11A. Nature. 2009;460:1093–1097. doi: 10.1038/nature08243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu W, Morrissey CS, Keller CA, et al. Dynamic shifts in occupancy by TAL1 are guided by GATA factors and drive large-scale reprogramming of gene expression during hematopoiesis. Genome Res. 2014;24:1945–1962. doi: 10.1101/gr.164830.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dore LC, Amigo JD, Dos Santos CO, et al. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc Natl Acad Sci USA. 2008;105:3333–3338. doi: 10.1073/pnas.0712312105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yu D, dos Santos CO, Zhao G, et al. miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 2010;24:1620–1633. doi: 10.1101/gad.1942110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kuhrt D, Wojchowski DM. Emerging EPO and EPO receptor regulators and signal transducers. Blood. 2015;125:3536–3541. doi: 10.1182/blood-2014-11-575357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.SEt Lux. Anatomy of the red cell membrane skeleton: Unanswered questions. Blood. 2016;127:187–199. doi: 10.1182/blood-2014-12-512772. [DOI] [PubMed] [Google Scholar]
- 78.Perkins A, Xu X, Higgs DR, et al. Kruppeling erythropoiesis: An unexpected broad spectrum of human red blood cell disorders due to KLF1 variants. Blood. 2016;127:1856–1862. doi: 10.1182/blood-2016-01-694331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bai X, Kim J, Yang Z, et al. TIF1gamma controls erythroid cell fate by regulating transcription elongation. Cell. 2010;142:133–143. doi: 10.1016/j.cell.2010.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bauer DE, Orkin SH. Hemoglobin switching’s surprise: The versatile transcription factor BCL11A is a master repressor of fetal hemoglobin. Curr Opin Genet Dev. 2015;33:62–70. doi: 10.1016/j.gde.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tanimura N, Miller E, Igarashi K, et al. Mechanism governing heme synthesis reveals a GATA factor/heme circuit that controls differentiation. EMBO Rep. 2016;17:249–265. doi: 10.15252/embr.201541465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu J, Sankaran VG, Ni M, et al. Transcriptional silencing of {gamma}-globin by BCL11A involves long-range interactions and cooperation with SOX6. Genes Dev. 2010;24:783–798. doi: 10.1101/gad.1897310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Calero-Nieto FJ, Wood AD, Wilson NK, Kinston S, Landry JR, Gottgens B. Transcriptional regulation of Elf-1: Locus-wide analysis reveals four distinct promoters, a tissue-specific enhancer, control by PU.1 and the importance of Elf-1 downregulation for erythroid maturation. Nucleic Acids Res. 2010;38:6363–6374. doi: 10.1093/nar/gkq490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mansour MR, Abraham BJ, Anders L, et al. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hay D, Hughes JR, Babbs C, et al. Genetic dissection of the alpha-globin super-enhancer in vivo. Nat Genet. 2016;48:895–903. doi: 10.1038/ng.3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guerrero L, Marco-Ferreres R, Serrano AL, Arredondo JJ, Cervera M. Secondary enhancers synergise with primary enhancers to guarantee fine-tuned muscle gene expression. Dev Biol. 2010;337:16–28. doi: 10.1016/j.ydbio.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 87.Hong JW, Hendrix DA, Levine MS. Shadow enhancers as a source of evolutionary novelty. Science. 2008;321:1314. doi: 10.1126/science.1160631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Li G, Ruan X, Auerbach RK, et al. Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell. 2012;148:84–98. doi: 10.1016/j.cell.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lee K, Hsiung CC, Huang P, Raj A, Blobel GA. Dynamic enhancer-gene body contacts during transcription elongation. Genes Dev. 2015;29:1992–1997. doi: 10.1101/gad.255265.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.