Abstract
MicroRNAs (miRNAs) are small non-coding RNAs that regulate gene expression. Compared to miRNA biogenesis, pathways that mediate mature miRNA decay are less-well understood. We report that biologically functional miRNAs are degraded in human cells by the endonuclease Tudor-SN (TSN). In vitro, recombinant TSN initiates the decay of both protein-free and AGO2-loaded miRNAs via endonucleolytic cleavage at CA and UA dinucleotides, preferentially at scissile bonds located > five nucleotides from miRNA ends. Consistent with this, cellular targets of TSN-mediated decay defined using miR-seq follow this rule. Inhibiting TSN-mediated miRNA decay by CRISPR-Cas9 knockout of TSN inhibits cell-cycle progression by upregulating a cohort of miRNAs that downregulates mRNAs encoding proteins such as CDK2, CCND1, E2F1 and E2F2, which are critical for G1-to-S phase transition.
Human TSN is an evolutionarily conserved nuclease that contains five staphylococcal/micrococcal-like nuclease (SN) domains and a Tudor domain. Bacterial homologs of TSN degrade RNA in vitro via endonucleolytic cleavage, which generates a 3′-phosphate, followed by 3′-to-5′ exonucleolytic cleavages, yielding mono- and di-nucleotides (1). In mammals, TSN is a multifunctional protein implicated in the degradation of A-to-I hyper-edited double-stranded RNAs (2) and inosine-containing primary (pri)-miRNAs (3). While TSN was the first RNA-induced silencing complex (RISC) subunit characterized to have endonuclease activity (4), subsequent reports demonstrating that AGO2 is the RISC catalytic component (5) spurred us to identify TSN function in RISC.
We first confirmed that cellular TSN co-immunoprecipitates with RISC components argonaute 2 (AGO2) and trinucleotide repeat-containing 6A (GW182) in a largely RNase I-resistant fashion (Fig. 1A and Table S1), which we corroborated by detecting cellular TSN in the reciprocal IP of FLAG-AGO2 (fig. S1).
Fig. 1. TSN mediates cellular degradation of mature and biologically active miRNAs.
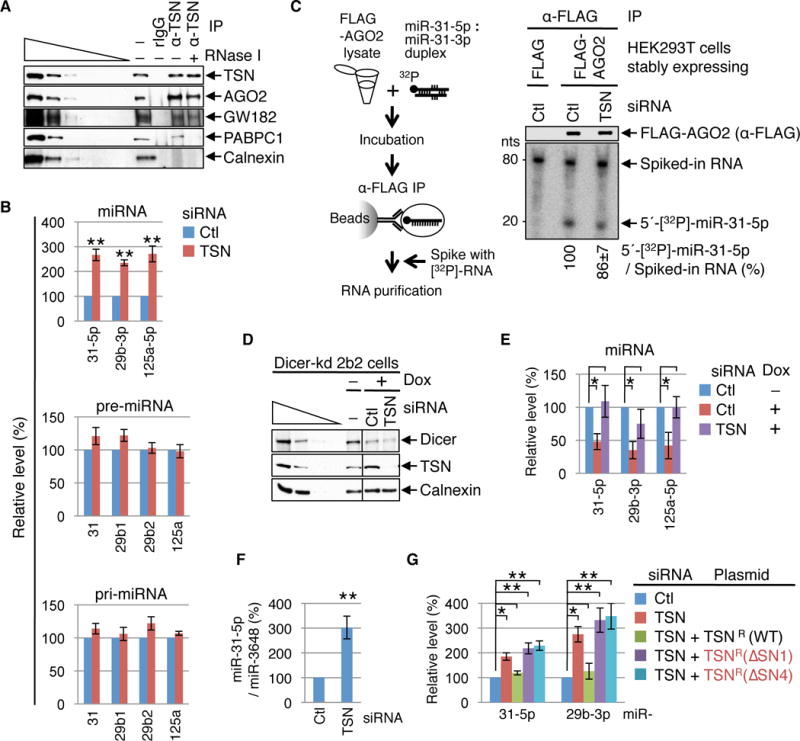
(A) Western blot (WB) demonstrating that HEK293T-cell TSN co-immunoprecipitates with RISC components in the presence (+) or absence (−) of RNase I. RNase I activity is confirmed by the loss of PABPC1 from the TSN IP, and IP specificity is evident by the absence of Calnexin. The wedge specifies three-fold dilutions of cell lysates. rIgG, control rabbit IgG; α-TSN, anti-TSN; − IP, prior to IP.
(B) RT-qPCR revealing that mature miRNA levels increased in cells transfected with TSN siRNA relative to Ctl siRNA, while the corresponding pre- or pri-miRNAs levels remained unchanged.
(C) Left: Lysates of HEK293T cells stably expressing FLAG or FLAG-AGO2 were incubated with 5′-[32P]-labeled miR-31-5p : miR-31-3p duplexes, and α-FLAG immunoprecipitates were spiked with in vitro-synthesized, internally [32P]-labeled RNA before RNA extraction. Right: WB (upper) and phosphorimage of RNAs extracted from α-FLAG IPs (lower) showing that TSN KD does not increase the efficiency of 5′-[32P]-miR-31-5p loading into FLAG-AGO2.
(D) WB showing that exposing Dicer-kd 2b2 cells (20) to doxycycline (Dox) to induce Dicer shRNA production inhibits Dicer expression. Cells were transfected with Ctl or TSN siRNA and exposed to Dox for 4 days prior to harvesting.
(E) RT-qPCR of RNA from (D) demonstrating that when doxycycline is used to inhibit miRNA biogenesis miRNAs are stabilized by TSN siRNA relative to Ctl siRNA.
(F) RT-qPCR showing that TSN siRNA upregulates the level of exogenously introduced miR-31-5p relative to the level of co-introduced miR-3648.
(G) RT-qPCR showing that TSNR(WT), but not the catalytically inactive variant TSNR(ΔSN1) or TSNR(ΔSN4), rescues the activity of cellular TSN.
Here and elsewhere, all results derive from ≥ 3 independent experiments. For RT-qPCR results, miRNA and pre-miRNA levels are relative to U6 snRNA, pri-miRNAs levels are relative to β-actin mRNA, and relative levels in the presence of Ctl siRNA are defined as 100. Histograms represent the average and SD. *P < 0.05, **P < 0.01.
To determine if RISC-associated TSN functions in miRNA metabolism, we quantitated the levels of select miRNAs and their precursor (pre)-miRNAs and pri-miRNAs in HEK293T cells transfected with TSN siRNA relative to Control (Ctl) siRNA (fig. S2A). RT-qPCR and Northern blotting demonstrated that TSN knockdown (KD) upregulates the abundance of mature miR-31-5p, miR-29b-3p and miR-125a-5p without significantly changing the levels of their pre-or pri-miRNAs (Fig. 1B and fig. S2B), providing the first indication that TSN decreases the abundance of mature miRNAs per se. TSN KD did not alter the abundance of mature miR-3648 and miR-128-3p (fig. S2C), suggesting that effects are not generalized to all miRNAs. Notably, these same miRNA-specific results also typified HeLa cells (fig. S2D).
TSN targets functional miRNAs as evidenced by the finding that TSN KD and the consequential accumulation of miR-31-5p and miR-29b-3p (Fig. 1B) are accompanied by decreased expression of integrin alpha 5 (ITGA5) and lysyl oxidase (LOX) (fig. S3A), which are their respective targets (6.7) (fig. S3, B–E). The decreased level of ITGA5 and LOX was restored by expressing a miR-31-5p inhibitor or a miR-29b-3p inhibitor, respectively (fig. S3A), confirming that the observed TSN regulation is miRNA-mediated.
The increased abundance of miRNAs and their mRNA-targeting activities upon TSN KD is not due to enhanced loading of miRNAs into AGO2. This was made clear by programming FLAG-AGO2 in lysates of HEK293T cells that stably express FLAG-AGO2 and had been transiently transfected with Ctl or TSN siRNA (fig. S3F) with 5′-[32P]-labeled miR-31-5p (Fig. 1C). IPs of FLAG-AGO2 revealed that TSN KD did not increase the level of 5′-[32P]-miR-31-5p in FLAG-AGO2 IPs (Fig. 1C).
Additional support for TSN promoting miRNA turnover derives from our finding that TSN KD increased the abundance of miR-31-5p, miR-29b-3p and miR-125a-5p when miRNA biogenesis was inhibited by conditional Dicer KD (Fig. 1, D and E and fig. S4, A–D, which shows no effect on the corresponding two miR* that were quantifiable). Furthermore, TSN KD increased the abundance of miR-31-5p that derived from an exogenously introduced miR-31-5p mimic (Fig. 1F), indicating that TSN-mediated miRNA turnover does not require Drosha- or Dicer-dependent processing steps that precede miR : miR* duplex formation. We call this Tudor SN-mediated miRNA decay pathway “TumiD”. TumiD adds a new dimension to reports that miRNA stability can be regulated by exonucleases and complementarity to target mRNAs (8).
TumiD requires TSN nuclease activity as evidenced by the ability of wild-type TSN (i.e. TSNR(WT)) but not catalytically inactive TSNR(∆SN1) or TSNR(ΔSN4) (fig. S4, E–I) to rescue the TumiD activity of cellular TSN (Fig. 1G). Moreover, incubating HEK293T cells with 2′-deoxythymidine 5′,3′-bisphosphate (pdTp), a general inhibitor of staphylococcal nucleases (9) (fig. S4J), at a concentration (200 μM) that inhibits TSN nuclease activity in human cells (10) increased the abundance of TumiD targets (fig. S4K).
Two independent miR-seq experiments (Run #1 and Run #2), each in biological triplicate, expanded the number of cellular TumiD targets. Upon TSN KD, the levels of 49 and 88 miRNAs were significantly upregulated (adjusted P-value “Padj” < 0.05) in Run #1 and #2, respectively, of which 35 were in common to both experiments (Fig. 2A and figs. S5–S10; Tables S2–S4).
Fig. 2. miR-seq, combined with RT-qPCR, defines HEK293T-cell TumiD targets.
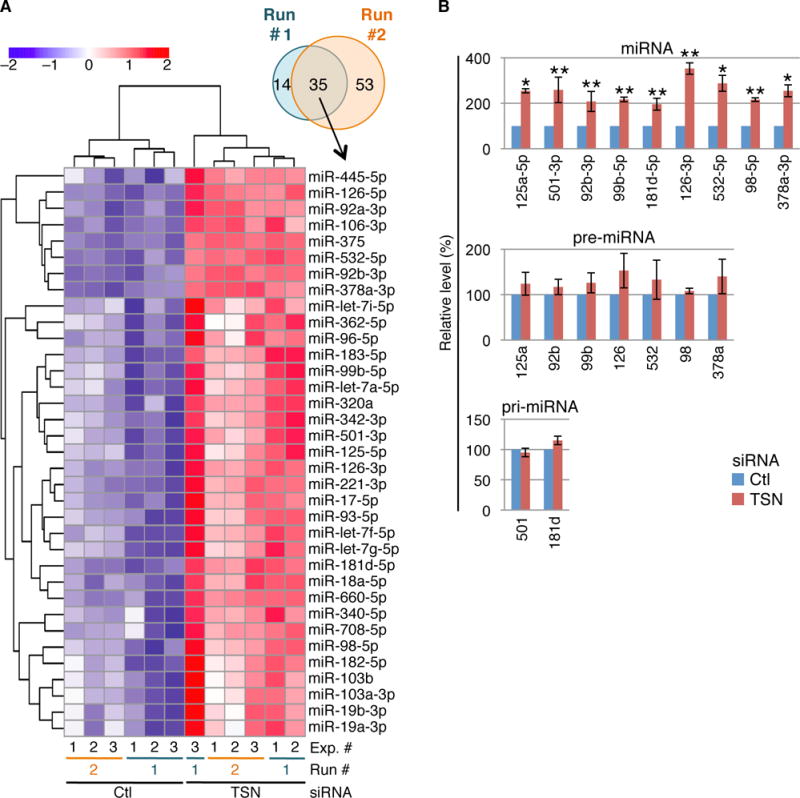
(A) Venn diagram and heat map of miRNA expression from miR-seq Runs #1 and #2 (Tables S3 and S4), identifying miRNAs exhibiting significant upregulation upon TSN KD. Color key represents row-scaling of the batch-corrected DESeq rLog-transformed expression values.
(B) RT-qPCR corroboration of miR-seq data defines miRNAs that are upregulated upon TSN KD without significant changes in the levels of their pre-miRNAs or, when pre-miRNAs were undetectable, pri-miRNAs.
Nine of 10 chosen miRNAs that were significantly upregulated in both Runs (Fig. 2A and Table S4) proved to be bona fide TumiD targets (Fig. 2B and fig. S11A). Evaluation of the tenth, miR-221-3p, was obscured by upregulation of its pri-mRNA upon TSN KD (fig. S11B). miR-seq and RT-qPCR demonstrating that TSN KD did not alter the abundance of the miR* for miR-125a-5p, miR-99b-5p or miR-98-5p (fig. S11C and Table S3) provides additional evidence that TumiD degrades mature miRNAs. It is noteworthy that the miR* of some TumiD targets is itself a TumiD target, as exemplified by miR-126-5p (Figure 2, A and B; Tables S3 and S4).
TumiD also contributes to the differential regulation of the six miR-17-92 cluster-derived miRNAs (fig. S11D and E; Table S3), and to miR-15a/16-1 and miR-23a/27a/24-2 cluster-derived miRNAs (fig. S12, A–C; Table S3). Notably, miRNA members of each cluster have distinct functions in development and disease.
TSN preferentially cleaves at CA and UA dinucleotides, as revealed when 5′-[32P]-labeled miR-31-5p, miR-29b-3p or miR-126-3p were incubated with recombinant HIS-TSN and reaction products were analyzed in sequencing gels (Fig. 3A and fig. S13A). Endonucleolytic cleavage is followed by 3′-to-5′ exonucleolytic degradation (Fig. 3A and fig. S13A) as typifies the mode of the bacterial homologue of TSN, micrococcal nuclease (1). miR-3648, which lacks CA and UA, is resistant to HIS-TSN-mediated degradation (fig. S13B), as is a hybrid miRNA generated by swapping a cleavable CA-containing region of miR-31-5p with a region from miR-3648 (fig. S13C, compare to Fig. 3A). HIS-TSN-mediated degradation depends on Ca2+ (fig. S13B) and is inhibited by converting the phosphodiester bond of a susceptible dinucleotide to a phosphorothioate bond (fig. S14), which has lower affinity for Ca2+ (9). Inserting a CA dinucleotide into HIS-TSN-resistant miR-3648 induced cleavage at the inserted CA (fig. S15), and cleavage was least optimal at CA dinucleotides situated within the five 5′-most or five 3′-most nucleotides of a miRNA (fig. S16, A and B). Consistent with this, swapping the eleven 3′-most nucleotides of miR-29b-3p with the corresponding region of miR-150-3p, which contains a CA in the five 3′-most nucleotides, abolished cleavage in the swapped region (fig. S16, C and D, compare to Fig. 3A).
Fig. 3. TSN degrades naked and AGO2-loaded miRNAs at CA and UA dinucleotides in vitro.
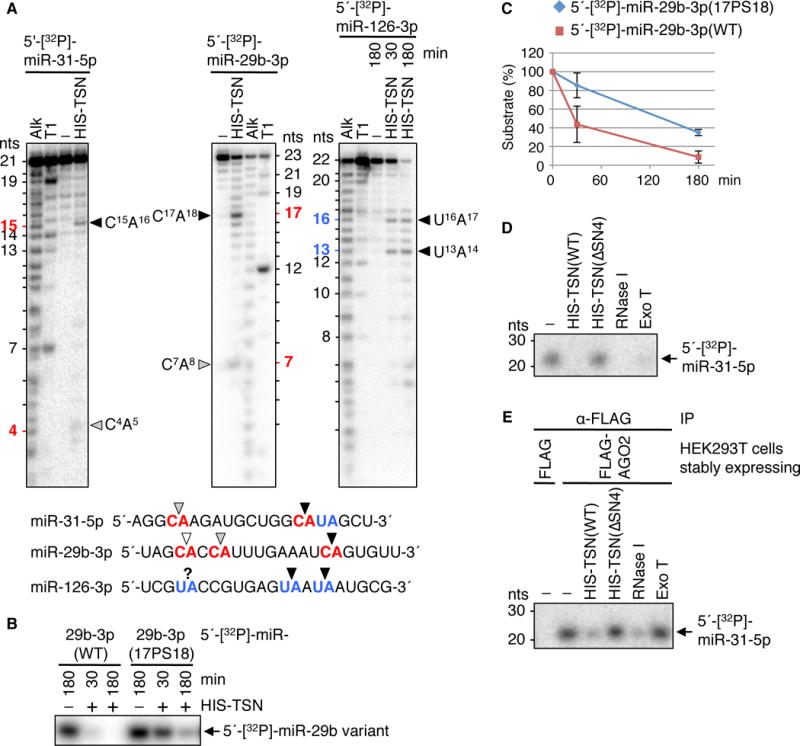
(A) Phosphorimages of sequencing gels reveal that recombinant TSN cleaves protein-free 5′-[32P]-labeled miRNAs at CA and UA. Alk, alkaline hydrolysis to generate a ladder of nucleotides (nts); T1, cleavage after guanine by RNase T1; black, gray and white arrowheads, respectively, strong, medium and weak cleavage sites. Unlabeled bands are decay intermediates resulting from TSN exonuclease activity since they were not detectable upon 3′-[32P]Cp-labeled miRNA incubation with HIS-TSN (fig. S13A).
(B) Replacing a susceptible CA phosphodiester bond in protein-free 5′-[32P]-labeled miR-29b-3p with a CA phosphorothioate bond inhibits HIS-TSN-mediated degradation. WT, wild-type; 17PS18, phosphorothioate bond between nucleotides 17 and 18.
(C) Plot of results shown in (B). The amount of each 5′-[32P]-labeled miRNA in the absence of HIS-TSN is defined as 100.
(D) Protein-free 5′-[32P]-miR-31-5p (−) is degraded by HIS-TSN(WT), endonuclease RNase I, or 3′-to-5′ exonuclease Exo T but not catalytically inactive (fig. S4G) HIS-TSN(ΔSN4).
(E) FLAG-AGO2-loaded 5′-[32P]-miR-31-5p is protected from degradation by Exo T, and susceptible to degradation by HIS-TSN(WT) and RNase I but not HIS-TSN(ΔSN4).
Investigating cellular TumiD revealed that artificially inserting CA dinucleotides into TumiD-resistant miR-3648 at two positions favorable for TSN cleavage in vitro (fig. S17A) converted the miRNA to a TumiD target (fig. S17B). Of the 35 miRNAs that were significantly upregulated upon TSN KD in both miR-seq Runs (Table S4), the two lacking CA or UA were upregulated prior to miRNA generation (fig. S17C) and the remaining 33 contain CA and/or UA at favorable positions for TSN cleavage. Notably, 31 mouse miRNAs that we identify as orthologs of putative or proven human TumiD targets that were upregulated upon TSN KD in both Runs are themselves predicted to be TumiD targets and have CA and/or UA dinucleotides at the same position(s) (Table S5).
We are necessarily underestimating the number of cellular TumiD targets since miRNAs with half-lives sufficiently long to preclude a detectable change during the period of TSN KD will escape definition. We limited our analyses to ≤ 4 days after transient transfection using TSN siRNA because longer transient KD or constitutive knockout (KO) of TSN inhibits cell-cycle progression (see below). Moreover, unknown factors must affect the specificity of cellular TumiD given that short-lived miR-503-5p (12) is not a TumiD target despite harboring an internal CA (fig. S17D).
AGO2-loaded miRNAs are protected from 3′-to-5′ exonucleolytic degradation by bacterial exonuclease T (Exo T) but susceptible to endonucleolytic degradation mediated by bacterial RNase I, which cleaves between any two nucleotides, or HIS-TSN (Fig. 3, D and E; fig. S4G and fig. S18).
To assess if TSN facilitates the G1/S-phase transition in human cells as is does in mouse embryonic fibroblasts (13), we established two independent stable TSN KO HEK293T cell lines using CRISPR, the D10A nickase variant of Cas9 (14) and, to control for off-target effects of CRISPR/Cas9, two different pairs of single guide RNAs (fig. S19). Synchronizing WT and TSN KO cells at G2/M using nocodazole or at G1/S using a double-thymidine block followed by flow cytometry analyses after release from synchronization revealed a prolonged cell cycle in both TSN KO #1 or #2 cell lines due to slow progression through G1/S (Fig. 4A and fig. S20). Stably expressing FLAG-TSN in each TSN KO cell line rescued the pace of G1/S progression (fig. S21, A and B). Consistent with results from transient TSN KDs (Fig. 1B and fig. S11E), the levels of the TumiD targets miR-31-5p, miR-17-5p and miR-20a-5p were significantly upregulated in G1/S in both TSN KO cell lines compared to WT cells (Fig. 4B and fig. S22A).
Fig. 4. TumiD regulates proteins critical for HEK293T-cell G1-to-S phase transition.
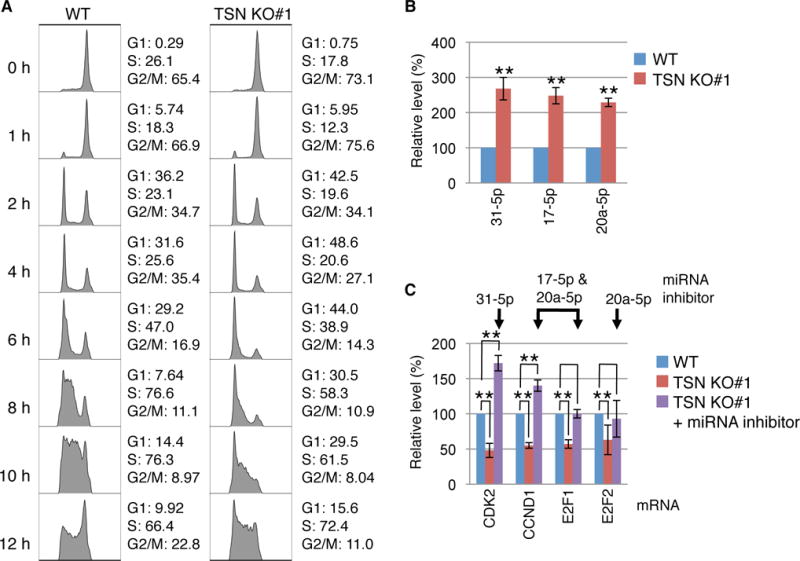
(A) Flow cytometry analyses of the cell cycle demonstrating delayed G1-to-S phase progression in TSN KO#1 cells compared to WT cells. Cells were synchronized in G2/M using nocodazole, released from synchronization, and analyzed at the designated times. The percent (%) of cells in each cell-cycle phase is shown.
(B) RT-qPCR showing upregulation of TumiD targets in TSN KO#1 cells compared to WT cells. Cells were synchronized at G1/S using a double-thymidine block.
(C) RT-qPCR demonstrating that the levels of CDK2, CCND1, E2F1 and E2F2 mRNAs are downregulated in TSN KO#1 cells compared to WT cells when synchronized using a double-thymidine block, and that transfecting TSN KO#1 cells with miRNA inhibitors rescues the levels of CDK2, CCND1, E2F1 and E2F2 mRNAs.
Using miRNA mimics, we confirmed previous reports (15–18) that these three TumiD targets downregulate the expression of genes critical for G1/S transition: cyclin dependent kinase 2 (CDK2) mRNA was downregulated by a miR-31-5p mimic, both cyclin D1 (CCND1) and E2F transcription factor 1 (E2F1) mRNAs were downregulated by a miR-17-5p mimic and a miR-20a-5p mimic, and E2F2 mRNA was downregulated by a miR-20a-5p mimic (fig. S22B). Consistent with these results, increased G1/S levels of the three miRNAs in TSN KO cells were accompanied by decreased levels of their target CDK2, CCND1, E2F1 and E2F2 mRNAs (Fig. 4C and fig. S22C) and target-encoded proteins (fig. S22D). Furthermore, transfecting TSN KO cells with miRNA inhibitors rescued the levels of CDK2, CCND1, E2F1 and E2F2 mRNAs (Fig. 4C and fig. S22C) and proteins (fig. S22D), indicating that TSN regulates the expression of these cell-cycle regulatory proteins via TumiD. Notably, while the level of E2F3 mRNA was also downregulated in TSN KO#1 cells, miRNA inhibitors did not restore the level of E2F3 mRNA (fig. S22E), indicating that its downregulation by TSN is not mediated by miR-17-5p or miR-20a-5p. Stably expressing FLAG-TSN in TSN KO cells rescued the expression levels of the three miRNAs and their target CDK2, CCND1, E2F1 and E2F2 mRNAs (fig. S22, F and G).
Interestingly, E2F proteins promote transcription of the miR-17-92 cluster (11). Consistent with this, E2F1-3 KD in TSN KO#1 cells compared to WT cells was accompanied by an ~30% reduced level of pri-miR-17-92 but increased levels of miR-17-5p and miR-20a-5p in TSN KO cells because TumiD was inhibited (fig. S22H). Thus, TSN KD disrupts the tight control of mRNAs encoding cell-cycle regulatory proteins via TumiD, offering an explanation for the elevated levels of TSN that can typify rapidly proliferating cells (19).
Our studies provide the first demonstration of endonuclease-mediated miRNA decay that degrades specific miRNAs and properties of TSN that may be useful as a new biochemical tool.
Supplementary Material
One Sentence Summary.
Tudor-SN mediates endonucleolytic decay of CA- and UA-containing microRNAs to promote the cell cycle during G1/S.
Acknowledgments
Raw data are deposited in the Gene Expression Omnibus (accession no. GSE77484). We thank F. Lopez for plasmid preparations; H. Siomi, K. Nishikura, and H. Yuan for plasmids; P. Svoboda for Dicer-kd 2b2 cells; M. Gleghorn for protein-structure advice; A. Rosenberg for statistical analyses; and M. Popp for comments on the manuscript. This work was supported by NIH R37 GM74593 to L.E.M. and R01 GM084089 to B.T. Partial salary support derived from NIH P30 AR061307 (R.A.E.), the Uehara Memorial Foundation (K.M.) and the Mochida Memorial Foundation (K.M.).
REFERENCES AND NOTES
- 1.Reddi KK. Mode of action of micrococcal phosphodiesterase. Nature. 1960;187:74–75. doi: 10.1038/187074a0. [DOI] [PubMed] [Google Scholar]
- 2.Scadden AD. The RISC subunit Tudor-SN binds to hyper-edited double-stranded RNA and promotes its cleavage. Nat Struct Mol Biol. 2005;12:489–496. doi: 10.1038/nsmb936. [DOI] [PubMed] [Google Scholar]
- 3.Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caudy AA, Ketting RF, Hammond SM, Denli AM, Bathoorn AM, Tops BBJ, Silva JM, Myers MM, Hannon GJ, Plasterk RHA. A micrococcal nuclease homologue in RNAi effector complexes. Nature. 2003;425:411–414. doi: 10.1038/nature01956. [DOI] [PubMed] [Google Scholar]
- 5.Hammond SM. Dicing and slicing: the core machinery of the RNA interference pathway. FEBS Lett. 2005;579:5822–5829. doi: 10.1016/j.febslet.2005.08.079. [DOI] [PubMed] [Google Scholar]
- 6.Augoff K, Das M, Bialkowska K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 is a broad regulator of beta1-integrin expression and function in cancer cells. Mol Cancer Res. 2011;9:1500–1508. doi: 10.1158/1541-7786.MCR-11-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou J, Lin JH, Brenot A, Kim J, Provot S, Werb Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol. 2013;15:201–213. doi: 10.1038/ncb2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15:509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 9.Schwarz DS, Tomari Y, Zamore PD. The RNA-induced silencing complex is a Mg2+-dependent endonuclease. Curr Biol. 2004;14:787–791. doi: 10.1016/j.cub.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 10.Yoo BK, Santhekadur PK, Gredler R, Chen D, Emdad L, Bhutia S, Pannell L, Fisher PB, Sarkar D. Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma. Hepatology. 2011;53:1538–1548. doi: 10.1002/hep.24216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mogilyansky E, Rigoutsos I. The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603–1614. doi: 10.1038/cdd.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rissland OS, Hong SJ, Bartel DP. MicroRNA destabilization enables dynamic regulation of the miR-16 family in response to cell-cycle changes. Mol Cell. 2011;43:993–1004. doi: 10.1016/j.molcel.2011.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su C, Zhang C, Tecle A, Fu X, He J, Song J, Zhang W, Sun X, Ren Y, Silvennoinen O, Yao Z, Yang X, Wei M, Yang J. Tudor staphylococcal nuclease (Tudor-SN), a novel regulator facilitating G1/S phase transition, acting as a co-activator of E2F-1 in cell cycle regulation. J Biol Chem. 2015;290:7208–7220. doi: 10.1074/jbc.M114.625046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino A, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.SKim H, Lee KS, Bae HJ, Eun JW, Shen Q, Park SJ, Shin WC, Yang HD, Park M, Park WS, Kang Y-K, Nam SW. MicroRNA-31 functions as a tumor suppressor by regulating cell cycle and epithelial-mesenchymal transition regulatory proteins in liver cancer. Oncotarget. 2015;6:8089–8102. doi: 10.18632/oncotarget.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu Z, Wang C, Wang M, Li Z, Casimiro MC, Liu M, Wu K, Whittle J, Ju X, Hyslop T, McCue P, Pestell RG. A cyclin D1/microRNA 17/20 regulatory feedback loop in control of breast cancer cell proliferation. J Cell Biol. 2008;182:509–517. doi: 10.1083/jcb.200801079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 18.Sylvestre Y, Guire VD, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, Ferbeyre G, Chartrand P. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 19.Jariwala N, Rajasekaran D, Srivastava J, Gredler R, Akiel MA, Robertson CL, Emdad L, Fisher PB, Sarkar D. Role of the staphylococcal nuclease and tudor domain containing 1 in oncogenesis. Int J Oncol. 2015;46:465–473. doi: 10.3892/ijo.2014.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmitter D, Filkowski J, Sewer A, Pillai RS, Oakeley EJ, Zavolan M, Svoboda P, Filipowicz W. Effects of Dicer and Argonaute down-regulation on mRNA levels in human HEK293 cells. Nucleic Acids Res. 2006;34:4801–4815. doi: 10.1093/nar/gkl646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li CL, Yang WZ, Chen YP, Yuan HS. Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic Acids Res. 2008;36:3579–3589. doi: 10.1093/nar/gkn236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Popp MW, Maquat LE. Attenuation of nonsense-mediated mRNA decay facilitates the response to chemotherapeutics. Nat Commun. 2015;6:6632. doi: 10.1038/ncomms7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gong C, Kim YK, Woeller CF, Tang Y, Maquat LE. SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs. Genes Dev. 2009;23:54–66. doi: 10.1101/gad.1717309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singh G, Kucukural A, Cenik C, Leszyk JD, Shaffer SA, Weng Z, Moore MJ. The cellular EJC interactome reveals higher-order mRNP structure and an EJC-SR protein nexus. Cell. 2012;151:750–764. doi: 10.1016/j.cell.2012.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ishizuka A, Siomi MC, Siomi H. A Drosophila fragile X protein interacts with components of RNAi and ribosomal proteins. Genes Dev. 2002;16:2497–2508. doi: 10.1101/gad.1022002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gleghorn ML, Gong C, Kielkopf CL, Maquat LE. Staufen1 dimerizes through a conserved motif and a degenerate dsRNA-binding domain to promote mRNA decay. Nat Struct Mol Biol. 2013;20:515–524. doi: 10.1038/nsmb.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park E, Gleghorn ML, Maquat LE. Staufen2 functions in Staufen1-mediated mRNA decay by binding to itself and its paralog and promoting UPF1 helicase but not ATPase activity. Proc Natl Acad Sci U S A. 2013;110:405–412. doi: 10.1073/pnas.1213508110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolde R. pheatmap: Pretty Heatmaps. R package version 1.0.8. 2015 [Google Scholar]
- 32.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Iimma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. https://www.R-project.org/ [Google Scholar]
- 34.Wei T, Simko V. corrplot: Visualization of a Correlation Matrix. R package version 0.77. 2016 https://CRAN.R-project.org/package=corrplot.
- 35.Ichim G, Mola M, Finkbeiner MG, Cros MP, Herceg Z, Hernandez-Vargas H. The histone acetyltransferase component TRRAP is targeted for destruction during the cell cycle. Oncogene. 2014;33:181–192. doi: 10.1038/onc.2012.570. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


