Abstract
We have determined the nucleotide sequences of the two internal transcribed spacers, the adjacent ribosomal coding sequences and the boundary between the external transcribed spacer and the 18S coding sequence in a cloned ribosomal transcription unit from Xenopus borealis. The transcribed spacers differ very extensively from those of X. laevis. Nevertheless, embedded in the internal transcribed spacers are several short sequence elements which are identical between the two species. These conserved elements are laterally displaced by substantial distances in the X. borealis sequence with respect to that of X. laevis. These relative displacements imply that insertions and deletions have played a major role in transcribed spacer divergence in Xenopus. This in turn implies that large regions of the transcribed spacers do not play a sequence-specific role in ribosome maturation. In contrast, the sequenced parts of the ribosomal coding regions, which encompass 670 nucleotides, differ at only three points from the corresponding sequences in X. laevis, each by a single substitution. These substitutions are readily accommodated by current models for rRNA higher order structure.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Dawid I. B., Reeder R. H. Xenopus borealis misidentified as Xenopus mulleri. Dev Biol. 1977 Sep;59(2):266–267. doi: 10.1016/0012-1606(77)90263-9. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Wensink P. C., Jordan E. A comparison of the ribosomal DNA's of Xenopus laevis and Xenopus mulleri: the evolution of tandem genes. J Mol Biol. 1972 Jan 14;63(1):57–73. doi: 10.1016/0022-2836(72)90521-9. [DOI] [PubMed] [Google Scholar]
- Ford P. J., Mathieson T. The nucleotide sequences of 5.8-S ribosomal RNA from Xenopus laevis and Xenopus borealis. Eur J Biochem. 1978 Jun 1;87(1):199–214. doi: 10.1111/j.1432-1033.1978.tb12367.x. [DOI] [PubMed] [Google Scholar]
- Forsheit A. B., Davidson N., Brown D. D. An electron microscope heteroduplex study of the ribosomal DNAs of Xenopus laevis and Xenopus mulleri. J Mol Biol. 1974 Dec 5;90(2):301–314. doi: 10.1016/0022-2836(74)90375-1. [DOI] [PubMed] [Google Scholar]
- Hall L. M., Maden B. E. Nucleotide sequence through the 18S-28S intergene region of a vertebrate ribosomal transcription unit. Nucleic Acids Res. 1980 Dec 20;8(24):5993–6005. doi: 10.1093/nar/8.24.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan B. R., Latil-Damotte M., Jourdan R. Sequence of the 3'-terminal portion of Drosophila melanogaster 18 S rRNA and of the adjoining spacer: comparison with corresponding prokaryotic and eukaryotic sequences. FEBS Lett. 1980 Aug 11;117(1):227–231. doi: 10.1016/0014-5793(80)80951-3. [DOI] [PubMed] [Google Scholar]
- Khan M. S., Maden B. E. Nucleotide sequence relationships between vertebrate 5.8 S ribosomal RNAs. Nucleic Acids Res. 1977 Jul;4(7):2495–2505. doi: 10.1093/nar/4.7.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel D. A., Maizel J. V., Jr, Leder P. The evolution and sequence comparison of two recently diverged mouse chromosomal beta--globin genes. Cell. 1979 Nov;18(3):865–873. doi: 10.1016/0092-8674(79)90138-7. [DOI] [PubMed] [Google Scholar]
- Maden B. E., Forbes J. M., Stewart M. A., Eason R. 18S coding sequences in amplified ribosomal DNA from Xenopus laevis oocytes are highly homogeneous, unmethylated, and lack major open reading frames. EMBO J. 1982;1(5):597–601. doi: 10.1002/j.1460-2075.1982.tb01214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Moss M., Salim M. Nucleotide sequence of an external transcribed spacer in Xenopus laevis rDNA: sequences flanking the 5' and 3' ends of 18S rRNA are non-complementary. Nucleic Acids Res. 1982 Apr 10;10(7):2387–2398. doi: 10.1093/nar/10.7.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maden B. E., Vaughan M. H., Warner J. R., Darnell J. E. Effects of valine deprivation on ribosome formation in HeLa cells. J Mol Biol. 1969 Oct 28;45(2):265–275. doi: 10.1016/0022-2836(69)90104-1. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Michot B., Bachellerie J. P., Raynal F., Renalier M. H. Homology of the 5'-terminal sequence of 28 S rRNA of mouse with yeast and Xenopus. Implication for the secondary structure of the 5.8 S--28 S RNA complex. FEBS Lett. 1982 Apr 19;140(2):193–197. doi: 10.1016/0014-5793(82)80892-2. [DOI] [PubMed] [Google Scholar]
- Moss T., Boseley P. G., Birnstiel M. L. More ribosomal spacer sequences from Xenopus laevis. Nucleic Acids Res. 1980 Feb 11;8(3):467–485. doi: 10.1093/nar/8.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazar R. N. A 5.8 S rRNA-like sequence in prokaryotic 23 S rRNA. FEBS Lett. 1980 Oct 6;119(2):212–214. doi: 10.1016/0014-5793(80)80254-7. [DOI] [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Rubtsov P. M., Musakhanov M. M., Zakharyev V. M., Krayev A. S., Skryabin K. G., Bayev A. A. The structure of the yeast ribosomal RNA genes. I. The complete nucleotide sequence of the 18S ribosomal RNA gene from Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Dec 11;8(23):5779–5794. doi: 10.1093/nar/8.23.5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence of Xenopus laevis 18S ribosomal RNA inferred from gene sequence. Nature. 1981 May 21;291(5812):205–208. doi: 10.1038/291205a0. [DOI] [PubMed] [Google Scholar]
- Skriabin K. G., Kraev A. S., Rubtsov P. M., Baev A. A. Polnaia posledovatel'nost' nukleotidov speisernoi oblasti, raspolozhennoi mezhdu genami 18S i 5.8S RNK drozhzhei. Dokl Akad Nauk SSSR. 1979;247(3):761–765. [PubMed] [Google Scholar]
- Skriabin K. G., Zakhar'ev V. M., Rubtsov P. M., Baev A. A. Posledovatel'nost' nukleotidov predpolagaemoi oblasti initsiatsii transkriptsii ribosomnogo operona drozhzhei. Dokl Akad Nauk SSSR. 1979;247(5):1275–1277. [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. A., Hall L. M., Maden B. E. Multiple heterogeneities in the transcribed spacers of ribosomal DNA from Xenopus laevis. Nucleic Acids Res. 1983 Feb 11;11(3):629–646. doi: 10.1093/nar/11.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiegler P., Carbon P., Ebel J. P., Ehresmann C. A general secondary-structure model for procaryotic and eucaryotic RNAs from the small ribosomal subunits. Eur J Biochem. 1981 Dec;120(3):487–495. doi: 10.1111/j.1432-1033.1981.tb05727.x. [DOI] [PubMed] [Google Scholar]
- Subrahmanyam C. S., Cassidy B., Busch H., Rothblum L. I. Nucleotide sequence of the region between the 18S rRNA sequence and the 28S rRNA sequence of rat ribosomal DNA. Nucleic Acids Res. 1982 Jun 25;10(12):3667–3680. doi: 10.1093/nar/10.12.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Brand R. C., Klootwijk J., Planta R. Some characteristics of processing sites in ribosomal precursor RNA of yeast. Nucleic Acids Res. 1980 Jul 11;8(13):2907–2920. doi: 10.1093/nar/8.13.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., van Heerikhuizen H., Planta R. J. The nucleotide sequence of the intergenic region between the 5.8S and 26S rRNA genes of the yeast ribosomal RNA operon. Possible implications for the interaction between 5.8S and 26S rRNA and the processing of the primary transcript. Nucleic Acids Res. 1981 Oct 10;9(19):4847–4862. doi: 10.1093/nar/9.19.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T. A., Johnson K. D., Olsen G. J., Peters M. A., Pace N. R. Enzymatic and chemical structure mapping of mouse 28S ribosomal ribonucleic acid contacts in 5.8S ribosomal ribonucleic acid. Biochemistry. 1982 May 11;21(10):2320–2329. doi: 10.1021/bi00539a008. [DOI] [PubMed] [Google Scholar]
- Warner J. R., Soeiro R. Nascent ribosomes from HeLa cells. Proc Natl Acad Sci U S A. 1967 Nov;58(5):1984–1990. doi: 10.1073/pnas.58.5.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
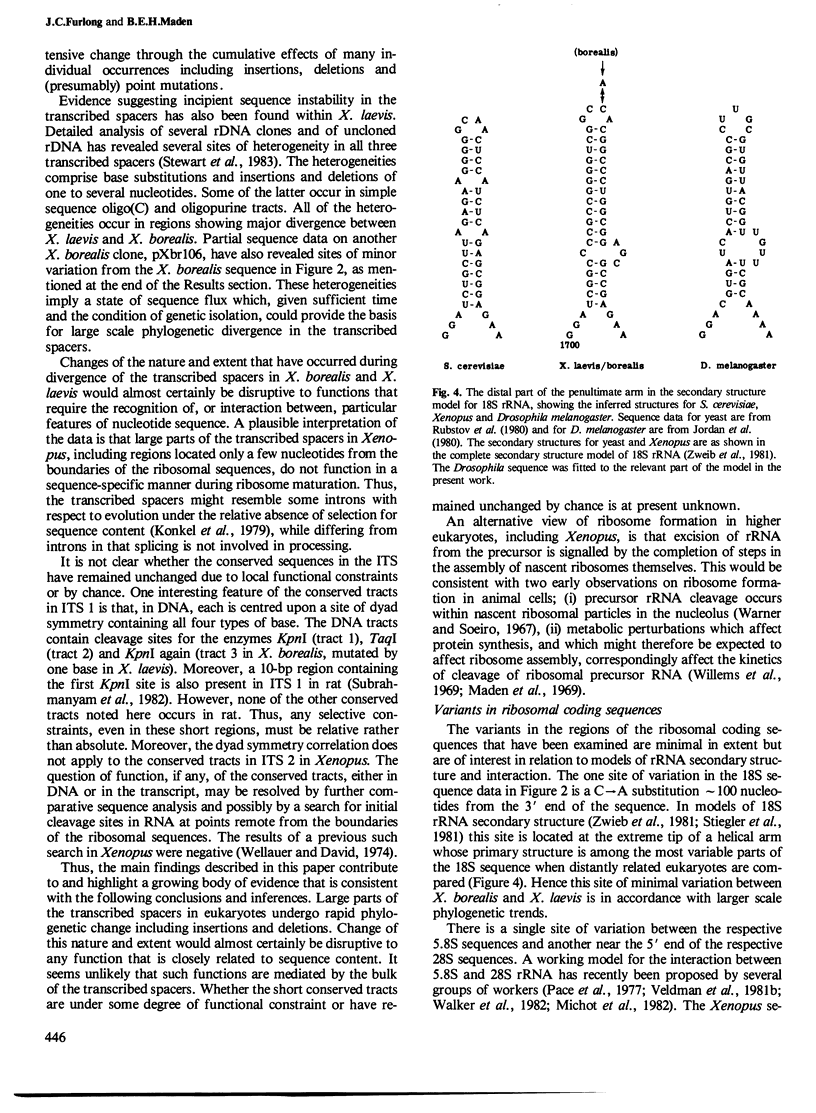
- Wellauer P. K., Dawid I. B. Secondary structure maps of ribosomal RNA and DNA. I. Processing of Xenopus laevis ribosomal RNA and structure of single-stranded ribosomal DNA. J Mol Biol. 1974 Oct 25;89(2):379–395. doi: 10.1016/0022-2836(74)90526-9. [DOI] [PubMed] [Google Scholar]
- Willems M., Penman M., Penman S. The regulation of RNA synthesis and processing in the nucleolus during inhibition of protein synthesis. J Cell Biol. 1969 Apr;41(1):177–187. doi: 10.1083/jcb.41.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. A., Steitz J. A. Complementary sequences 1700 nucleotides apart form a ribonuclease III cleavage site in Escherichia coli ribosomal precursor RNA. Proc Natl Acad Sci U S A. 1978 Aug;75(8):3593–3597. doi: 10.1073/pnas.75.8.3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C., Glotz C., Brimacombe R. Secondary structure comparisons between small subunit ribosomal RNA molecules from six different species. Nucleic Acids Res. 1981 Aug 11;9(15):3621–3640. doi: 10.1093/nar/9.15.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]



