Abstract
Completion of the human genome a decade ago laid the foundation for: using genetic information in assessing risk to identify individuals and populations that are likely to develop cancer, and designing treatments based on a person's genetic profiling (precision medicine). Genome-wide association studies (GWAS) completed during the past few years have identified risk-associated single nucleotide polymorphisms that can be used as screening tools in epidemiologic studies of a variety of tumor types. This led to the conduct of epigenome-wide association studies (EWAS). This article discusses the current status, challenges and research opportunities in GWAS and EWAS. Information gained from GWAS and EWAS has potential applications in cancer control and treatment.
KEYWORDS : biomarkers, cancer, epidemiology, epigenetics, genomics, omics technologies, outcome, personalized medicine, pharmacogenomics
In the last decade, an enormous amount of information has been available regarding genomic and epigenomic alterations and their use in cancer risk assessment, detection, diagnosis, prognosis and follow-up survival. The description below includes information about the current status of genome-wide association studies (GWAS) and epigenome-wide association studies (EWAS) and their potential implication in cancer control. The selection of literature related with GWAS and EWAS in different tumor types is based on the potential of variations identified in different studies in clinical implementation. Another point of consideration of inclusion is those studies which moved the field of GWAS and EWAS and provided potential markers for cancer diagnosis, prognosis and survival. Those studies that were not validated, either at the analytic level or clinical level have been omitted. I have summarized key studies in GWAS and EWAS in Tables 1–5.
Table 1. . The Cancer Genomic Atlas supported genome-wide association studies and epigenome-wide association studies conducted in different tumor types.
| Cancer type | SNPS and mutations | DNA methylation | Copy number variations | Other features and/or characteristics |
|---|---|---|---|---|
| Breast cancer [1] |
Spicing mutation analysis was observed in 988 samples out of 5206 samples analyzed which led to identify new pathways in breast cancer development [2] |
Histone alterations involvement in breast cancer progression [1] |
Expressed SNVs were reported in ER+ breast cancer tumor samples [3] |
The activity of histone deacetylase 8 was affected during progression of breast cancer as tested in more than 500 tissue samples of highly characterized biospecimens collected in the TCGA program [1] |
| Colorectal cancer [4] |
Mutation in APC, KRAs and BRFF [4] |
CIMP phenotype, and correlation of microsatellite instability affecting colorectal cancer associated genes in samples acquired in the TCGA program [4] |
Copy number variations associated with 18p11.32 [5] The role of these variations in tumor development is being investigated [5] |
After CIMP phenotype was shown in colorectal cancer, it was reported later on in other tumor types also |
| Glioblastoma [6] |
Mutations in ATRX and TERT |
DNA methylation profiling Hypermethylation as aprognostic biomarker after radiotherapy of IDH1 wild-type non-CIMP glioblastoma [7] |
Multiple sites |
Telomerase length and telomere Integrated molecular analysis of progression from low-grade to high-grade glioblastoma 1p/19q deletion [6] |
| Head and neck cancer [8] |
Mutations in PIK3CA in HPV-positive samples, loss of TRAF3 and amplification of the cell cycle gene E2F1 in smokers |
|
CDKN2A inactivation with frequent copy number alterations including amplification of 3q26/28 and 11q13/22 |
Genomic alterations contribute to the activation of NF-κB pathway [8] |
| Melanoma [9] |
Mutations in BRAF, RAS [N,H,K], and NF1 |
CpG Island Mutator Phenotype (CIMP phenotype) was associated with IDHA1 and ARID2EZH2-based silencing of tumor suppressor genes [10] |
Significant 4q12 focal amplification containing the oncogene KIT Somatic copy number amplification [10] |
Molecular characterization identified BRAF, RAS, and NF1 subtypes Co-amplification of oncogenes PDGFRA and KDR, located adjacent to KIT [9] |
| Oral cancer [11] |
PIK3CA mutations at the rate of 4% [11] |
Methylation in the enhancer region interrupted binding of chromatin looping factors and role of chronic inflammation in tumor predisposition was demonstrated in oral cell squamous carcinoma [12] |
Amplification of MIR30B which correlated with has-miR30b [13] |
SNPs rs114587137 and rs1784907 in intron 9 of PIK3CA in advanced stages of oral squamous cell carcinoma [11] |
| Prostate cancer [14] |
Mutations in SPOP, FOXA1, and IDH1 helped in identifying seven subtypes of prostate cancer |
Hypermethylation of IDH1 Promoter and distal gene hypermethylation marks for prediction of prostate cancer recurrence [15] |
Multiple (indolent and low Gleason tumors have fewer alterations compared with more aggressive tumors |
Actionable lesions in the PI3K, MAPK, and DNA repair pathway provided crucial information for targeted therapeutics [14] AR/TP53 pathway was identified as the major pathway of survival in one TCGA study on samples from prostate cancer patients [15] |
| Renal cell carcinoma [16] |
SNP array analysis indicated loss of one copy of the entire chromosome (on chromosome 1, 2, 6, 10, 13, and 17) |
Methylation array analysis indicated alterations at 64,000 sites |
Multiple copy number variations Recurrent structural breakpoints within TERT promoter mtDNA copy number changes were also observed in 50% samples |
Along with main genome, mitochondrial genome was also characterized in 66 kidney cancer cases [16] Based on the data from TCGA it was observed that exonic MAT variant rs11762213 is an independent predictor of adverse time of recurrence in renal cancer and should be included in prognostic disease stratification [17] |
| Thyroid cancer [18] | Mutations in cancer driver genes FIF1AX, PPM1D, and CHEK2 in 496 samples | Several oncomiRs in less differentiated subgroups | Somatic copy number alterations were identified in 27% of tumors (mostly enriched in cases with no driver mutations) | Molecular characterization by GWAS and EWAS helped in reclassification of papillary thyroid cancer [18] |
EWAS: Epigenome-wide association studies; GWAS: Genome-wide association studies; TCGA: The Cancer Genomic Atlas.
Table 2. . Genome-wide association studies and epigenome-wide association studies conducted in cancers regulated by hormones.
| Cancer type | EWAS | Mutations and susceptibility loci | SNPs and copy number variations |
|---|---|---|---|
| Breast cancer |
Histone analysis [19] Estrogen-mediated epigenetic repression of metallothionine-1 gene cluster [20] Hypermethylation of LHX2, WT1, and OTP in genome-wide analysis [21] |
Several loci [22–24] 11q24.2 [25] |
Polymorphism rs11249433 (1p11.2), rs13387042 (2q35), rs4973768 (3q24.1), rs4415084 (5p12), rs889312 (5q11.2), rs2180341 (6q22.33), rs2046210 (6q25.1), rs13281615 (8q24), rs2981582 and rs1219648 (10q26)rs6504950 (17q23) [110–112] Polymorphisms rs1219648, rs3757318, rs1926657, rs6556756, rs2046210, and rs4973768 [26] Polymorphism rs2059614 was associated with breast cancer survival in estrogen receptor-negative patients [25] Several polymorphism reported in a putative mammographic density locus [27] LRES [19] Polymorphism rs12325489C>T disrupts the binding site for miRNA-370; can be used for risk assessment [28] SNPS rs3757318, rs2046210, and rs4973768 useful for risk assessment [26] |
| Prostate cancer | miRNA profiling, mRNA profiling, gene-expression profiling [29] Epigenetic dysregulation of cellular processes relevant to TNF-α-dependent apoptosis [30] |
Several loci [22] More than 70 genes correlated with the tumor grade and helped in diagnosis [29] 2p15 [22] |
SNPs rs1465618 (2p21), rs12621278 (2q31), rs2660753 (3q12), rs10934853 (3q21), rs17021918 (4q22), rs7679673 (4q24), rs9364554 (6q25), rs12155172 (7p15), rs10486567 (7p15.2), ra6465657 (7q21.3), rs16901979 (8q24)rs10993994 (10q11.2)rs4430796 (7q12), rs8102476 (19q13.2), rs9623117 (22Q13) [22] Polymorphism rs721048 [22] |
EWAS: Epigenome-wide association studies; SNP: Single nucleotide polymorphism.
Table 3. . Genome-wide association studies and epigenome-wide association studies conducted in gynecological cancers.
| Cancer type | EWAS | Mutations and susceptibility loci | SNPs and copy number variations |
|---|---|---|---|
| Cervical cancer |
Methylation profiling [31] Genome-wide histone profiling [34] Late genes L1 and L2 and the host gene DAPK showed progressive methylation corresponding with disease advancement [31] Increased histone alterations (mainly H3K36me3 and H3K9Ac) in the HPV genome correlated with increased HPV16 gene expression [34] |
Chromosomal loss at 3p11-p14 contribute to cancer development [32] PIK3CA and p53 mutations in adenocarcinoma, squamous cell carcinoma, and high-grade intraepithelial neoplasia of cervix [35] |
Copy number gain (CNG) at chromosome 3q26 (spans from q26 to q29) containing 200 protein coding genes including SOX2, ECT2, PRKCI and PI3KCA [33] Deletion of Xq24 encoding a mitochondrial transporter protein [35,36] Copy number gain at 3q, 1q, 19q and loss at 11q, 4q and 13q [37] |
| Ovarian cancer | Genome-wide methylation analysis identified RUNX3 and CAMK2N1 hypermethylation as prognostic markers for serous ovarian cancer [38] 12 loci associated with serous epithelial ovarian cancer [41] Several loci [22,40] Identified HOXB2, HOXB5, HOXB6, HOXB7 at 17q21.32 and HOXD1, HOXD3 at 2q31 [41] |
Mutations in RAS/RAF/ERBB2 in 82% cases in genome-wide analysis [39] A wide spectrum of new BRCA1/2 mutations [42] |
Polymorphism rs2072590 (HOXD-AS1), rs2665390 (TIPARP), rs10088218 and rs10098821 (8q24), rs3814113 (9p22), rs9303542 (SKAP1) and rs2363956 (ANKLE1) for epithelial ovarian cancer in Polish population [22,40] |
EWAS: Epigenome-wide association studies; SNP: Single nucleotide polymorphism.
Table 4. . A comparison of DNA methylation changes, mutations, single nucleotide polymorphism, and copy number variations in genome-wide association studies and epigenome-wide association studies conducted in aerodigestive tumor types.
| Tumor type | GWAS/EWAS | Mutations | SNPs | CNVs |
|---|---|---|---|---|
| Colorectal cancer |
EWAS by profiling genome-wide mRNA and miRNA expression [43] Identified several potential regulatory networks; targeting relevant mRNA–miRNA networks as a potential therapeutic approach for CRC (for example is targeting EZH2) [43] Genome-wide epigenetic profiling [48] Deregulation of the prostaglandin synthase pathway. More specifically, PTGIS, PTGER3, PTGFR, and AKR1B1 were inactivated due to hypermethylation, along with other prostaglandins and their receptors in tumors [48] |
Several mutations at 23 susceptibility loci [44] mutations in KRAS and BRFF along with high microsatellite instability [46] |
Polymorphism rs17716310 Histone modifications are predicted [45] rs11987193 in Han Chinese population; may be useful in colorectal cancer pathology [47] Out of 22 polymorphism identified, rs1321311 was found associated with survival; suggested to be a prognostic marker [49] Polymorphism rs16892766 (8q23.3), rs10505477 (8q24.21), rs10795668 (10p14), rs3802842 (11q23), rs929218 (16q22.11), rs10411210 (19q13.11) [22,50,51] |
Copy number variations at 5q31.1 [45] Variation at 8q12 [47] Meta-analysis from 1000 genome project; at 1p36.2 marked by rs72647484 and at 16q24.1 marked by rs16941835 within the lncRNA [44] |
| Esophageal cancer |
Genome-wide methylation [52]; the major genes affected were PTK2, RND1, and UBL3 which regulate recurrence of cancer [52] |
Sensitive locus for mutations 4q21 [22] |
4q21 [22] SNP rs1229984 in ADH1B [22] SNP rs7922612 related with survival [22] |
Germline copy number loss of UGT28 and gain of PLEC [53] |
| Gastric cancer |
FAT4, a novel tumor suppressor identified by exome sequencing gets inactivated due to hypermethylation (field canerization) [54] Epigenetic silencing of GLDC [57] |
Susceptibility locus 8q24 [55] 8q24.3 [58] |
SNP rs4733616 [55] SNP rs2294008 was associated with decreased risk of duodenal cancer [55] SNP rs2294008 was found to be associated with increased risk of gastric cancer and decreased risk of duodenal cancer [58] SNP rs2976392 [22] |
Loss of CNV at 5q22 region surrounding APC [56] |
| Hepatocellular carcinoma (liver cancer) |
Epigenetic silencing of FOXD3 involved in proliferation, invasion and metastasis [59] Hypermethylation of p15, p16, p21, p27, and RASSF1A [62] |
Susceptibility locus 1p36.22 [60] |
SNP rs17401966 in KIF1B on chromosome 1p36.22 that was highly associated with HBV-related HCC [60] |
Loss of DNA copy number affecting expression of RGS17 and NR2E1 in liver cancer [61] |
| Lung cancer |
miRNA profiling and HOXA9 promoter methylation as biomarkers of lung cancer recurrence as determined by EWAS [63] Genomic loss of KAT6B coding histone H3 lysine acetyltransferase [66] |
COPD with lung cancer Sensitive loci at 15q25, 4q31, 4q22, 6q21, and 1q23 [64] Several loci, 5p15.33 and 3q28 [22,69] 15q25, 20q13.2, 22q12.2, and 5p15.33 [71] Different loci [73] |
In a study with 1400 participants sub-phenotyped for the presence of COPD and matched for smoking exposure [64] Polymorphism rs7963551 (study conducted in Chinese population) [67] Polymorphism rs2736100 and rs7727912 in 5p15.33, rs805297 and rs1802127 in 6p21.33, and rs8034191 and rs12440014 in 15q25.1 [70] Gene promoter hypermethylation in smokers (in sputum samples); polymorphism rs73371737 and rs7179575 drove gene methylation resulting in inactivation of GRBAB3 (epigenetic silencing) [72] SNPs rs2395185, rs4488809, and rs4600802 were found associated with lung cancer in never smoker Chinese females exposed to coal [69] Polymorphism rs401681 (5p15.33), rs3117582 (6p21.33), rs8034191 (15q24), rs75388767 (1p36.13), rs7023329 (9q210, rs1126809 (11q14), rs11170164 (12q12), rs910873 and rs1885120 (20q11.22) [22] 20q13.2 (rs4809957G >A), 22q12.2 (rs36600C >T) and 5p15.33 (rs401681C >T) linked to patient survivor [71] |
High copy number variation of cancer-related miRNAs and DICER1 [65] Somatic genomic rearrangements of TP73 resulting its oncogenic activities [68] |
| Oral cancer | Lymph node metastasis of oral squamous cell carcinoma prediction by hypomethylation of WISP1 [74] | 4q23 [34] | SNP rs991316, located in the ADH gene region of 4q23 indicated susceptibility to oral cancer [34] Polymorphism rs1412115 on chromosome 10 affecting NRP1 expression associated with increased risk of oral cancer [76] |
CNV at 11q14.3 and 6p21.3 associated with cancer pre-disposition [75] |
CNV: Copy number variation; COPD: Chronic obstructive pulmonary disease; CRC: Colorectal cancer; EWAS: Epigenome-wide association studies; HBV: Hepatitis B virus; HCC: Hepatocellular carcinoma; GWAS: Genome-wide association studies; SNP: Single nucleotide polymorphism.
Table 5. . DNA methylation changes, mutations, single nucleotide polymorphism, and copy number variations in genome-wide association studies and epigenome-wide association studies conducted in other tumor types.
| Tumor type | Susceptibility loci or type of profiling conducted | Comments | Ref. |
|---|---|---|---|
| Bladder cancer |
19q12 |
rs8102137 within the cyclin E coding region 19q12 |
[77] |
| Bladder cancer |
GWAS |
TERTC/T and SLC14A1C/T associated with risk of bladder cancer in Indian population |
[78] |
| Bladder cancer |
Several loci |
Polymorphism rs710521 (3q28), rs9642880 (8q24.21), rs2294008 (8q24.2) |
[22] |
| Glioma |
5p15.33 |
Expression of TERT affected due to the SNP rs2736100 |
[22] |
| HCC (liver cancer) |
1p36.22 |
SNP rs17401966 in KIF1B on chromosome 1p36.22 that was highly associated with HBV-related HCC |
[60] |
| Lymphoblastic leukemia |
Several loci |
Polymorphism rs4132601 (7p12.2), rs10994982 (10q21.2), rs10821936 (10q11.22), rs2239633 (14q11.2) |
[22] |
| Lymphocytic leukemia |
2q13 |
Polymorphism rs17483466 |
[22] |
| Myeloproliferative neoplasm |
9p24.1 |
JAK2 contains rs10974944 |
[22] |
| Nasopharyngeal carcinogenesis |
Genome-wide methylation analysis |
Aberrant disruption of the Wnt, MAPK, TGF-β, and Hedgehog signaling pathways |
[79] |
| Neuroblastoma |
6p22.3 |
Polymorphism rs4712653 associated with gene FLJ22536 |
[22] |
| Pancreatic cancer |
9q34 |
SNP rs505922 |
[80,22] |
| Pancreatic cancer |
GWAS |
SNP rs10818684 of the PTGS1 gene showed an interaction with diabetes in pancreatic cancer patients |
[81] |
| Pancreatic cancer |
Several loci More than 90 non-redundant familial pancreatic cancer specific CNVs were found in 50 of 120 cases |
Polymorphism rs3790844 (1q32.1), rs401681 (5p15.33), rs9543325 (13q22.1) |
[82,22] |
| Renal cancer |
Genome-wide methylation |
Differential expression of five genes PITX1, FOXE3, TWF2, EHBPL1, and RIN1 associated with survival |
[83] |
| Testicular cancer |
Several loci |
Polymorphism rs4699052 (4q24), rs4324715 (5q31.3), rs210138 (6q21.3), rs995030 (12q22) |
[22] |
| Thyroid cancer |
Various loci |
Polymorphism rs116909374, rs965513, rs944289, rs966423, and rs2439302 (polymorphism rs966423 linked to mortality) |
[84] |
| Thyroid cancer |
10q26.12 |
Heterogeneity in genetic susceptibility; rs2997312, rs10788123 and rs1254167 |
[85] |
| Thyroid cancer | 9q22.33 | Polymorphism rs965513 | [22] |
CNV: Copy number variation; EWAS: Epigenome-wide association studies; GWAS: Genome-wide association studies; HBV: Hepatitis B virus; HCC: Hepatocellular carcinoma; SNP: Single nucleotide polymorphism.
GWAS in cancer risk assessment
Genetic epidemiology is widely used in identifying natural variations in the genome, namely single nucleotide polymorphisms (SNPs) and copy number variants, to investigate their association with cancer risk, prognosis and pharmacogenomics [86]. GWAS are population-based epidemiologic studies designed to identify genomic segments in which genetic variants may contribute to disease risk. Prior to GWAS, the candidate gene approach was applied to identify cancer-associated genetic variations. These initial studies provided limited information because the selected variants did not tag sufficient variability in the evaluated genes and the studies were underpowered. The introduction of GWAS demonstrated that the functional consequences of candidate causal variants generally affect transcriptional regulation rather than translation. GWAS have resulted in the identification of an impressive and growing number of disease- and trait-associated genetic variants. For example, in the case of thyroid cancer, the status of the germline variant rs966423 can be determined before treatment begins and this variant is a potential candidate for managing mortality risk via treatment modification [84]. Post-GWAS analyses, including pathway-based analyses and functional characterization of associated variants, have provided new insights into the etiology and pathogenesis of different cancers.
GWAS on different cancers
GWAS have been conducted on a number of tumor types, including head and neck cancer [87], breast cancer [88], cervical cancer [33], lung cancer [71], gastric cancer [89], bladder cancer [77], head and neck cancer [8], prostate cancer [90], pancreatic cancer [80,91], thyroid cancer [18,84,86], melanoma [9], renal cell carcinoma [16], glioblastoma [6] and colorectal cancer (CRC) [92]. GWAS conducted in different ethnic populations have identified susceptibility variants for different cancers. The National Cancer Institute [93] initiated a program called The Cancer Genome ATLAS or TCGA [94] where well preserved and highly characterized cancer tissue samples were analyzed for genomic (mutations, SNPs and copy number variations) and epigenomic variations (DNA methylation and in a few cases histone modifications) using the latest technologies. At different points I have discussed about the results coming from the TCGA project. Results from this program are likely to be used for the Precision Medicine Initiative [95]. Examples of GWAS in different tumor types are discussed below.
• Lung cancer
Lung cancer is the second most common cancer in the USA, with about 221,200 new cases and about 158,040 deaths per year. Genetic variations have been shown to affect the length of survival in cancer patients, and investigators have identified susceptibility loci for lung cancer in the genomic region 15q25, which covers a cluster of nicotinic acetylcholine receptors, and three loci (20q13.2, 22q12.2 and 5p15.33) that have been associated with length of survival in lung cancer patients [71]. A significant association was observed in smokers and male patients. In another independent study, both genomic and epigenomic approaches were applied to evaluate epigenetic reprogramming during carcinogenesis. A functional 15q12 variant was identified as a risk factor for gene methylation when two SNPS at 15q12 (rs73371737 and rs7179575) driving gene methylation were used to analyze sputum samples collected from healthy controls and lung cancer patients [72]. Results of this study also indicated DNA double-stranded break repair by homologous recombination as a major pathway affecting susceptibility to gene methylation. Young et al. demonstrated that susceptibility to chronic obstructive pulmonary disease (COPD) is associated with lung cancer [64] in a study with 1400 participants subphenotyped for the presence of COPD and matched for smoking exposure. The loci analyzed in this study included 15q25, 4q31, 4q22, 6q21 and 1q23.
• Breast cancer
Breast cancer is the most common cancer among females in the USA, with about 231,840 new cases and about 40,290 deaths per year. In breast cancer, high-, moderate- and low-penetrance genetic variants can explain more than 40% of familial cancer risk [88]. More than 90 risk loci have been identified for breast cancer using GWAS, and hundreds more are expected to be identified [88]. Chen et al. established multiple interactions among risk variants identified in several breast cancer GWAS [26]. These investigators used random forest (RF), multifactor dimensionality reduction (MDR) and logistic regression approaches to evaluate the association of these variants with breast cancer. The main risk variants that were evaluated in 477 breast cancer patients and 534 healthy controls were rs1219648, rs3757318, rs1926657, rs6556756, rs2046210 and rs4973768. To investigate whether genetic determinants affect patients after they are diagnosed with breast cancer, Guo et al. conducted the largest study to date using genomic and clinical information from 37,954 patients [25]. This study included 6881 estrogen receptor-negative (ER-) patients and 23,059 estrogen receptor-positive (ER+) patients. The study identified one new locus on chromosome 11q24.2 (rs2059614) as being associated with breast cancer survival in ER- patients. This study also showed that germline genotyping can provide prognostic information for breast cancer patients. In another GWA study, a putative mammographic density locus was found at 1q12-q21 [27]. Mammographic density is considered an intermediate phenotype for breast cancer.
• Colorectal cancer
The estimated number of new colorectal cancer (CRC) cases is 132,700 and about 49,700 deaths per year in the USA. About 50 loci have been reported in the literature as being associated with CRC [96]. In a study of Asian consortia participants, six new CRC susceptibility loci were identified in 4725 cases and 9969 controls [97]. The value of association mapping in populations of African, European and Japanese ancestry was studied by Wang et al., who identified susceptibility locus VTI1A [98]. This study may help to explain CRC etiology in trans-ethnic populations. Functional validation of susceptibility locus 11q23 correlated with the expression of two genes (COLCA1 and COLCA2) by the immune system [99]. Post-GWAS analyses, mainly pathway-based analysis and functional characterization of associated variants, have provided new insights into the pathogenesis of different cancers. Generally, GWAS are expensive, but Gaj et al. proposed conducting pooled sample-based GWAS that are cost effective and offer useful information [100]. As a pilot study, these investigators identified a new susceptibility locus in the Polish population when pooled sample-based GWAS were conducted.
• Gastric cancer
The estimated number of new gastric cancer cases is 24,590 and about 10,720 deaths per year in the USA. Analysis of a gastric cancer GWAS dataset indicated the presence of susceptibility loci associated with different pathways in cancer progression [89]. Chromosome locus 8q24 was reported to be associated with different cancers including gastric cancer [55], and SNP rs4733616 was specifically associated with gastric cancer. Gastric cancer GWAS facilitated the identification of two tumor subtypes – gastric cardia and gastric noncardia – in a population study [101]. In another independent study, PSCA rs2294008 was found to be associated with increased risk of gastric cancer and decreased risk of duodenal cancer [58].
• Prostate cancer
The estimated number of new prostate cancer cases is 220,800 and about 27,540 deaths per year in the USA. More than 70 SNP loci have been reported to be associated with prostate cancer risk [102,103]. To achieve population risk stratification for clinical studies, a GWAS was conducted using DNA from 25,074 prostate cancer cases and 24,272 controls from the PRACTICAL consortium [103]. This study identified 23 new prostate cancer susceptibility loci. Agalliu et al. used a different approach and studied the effects of cancer family history and cumulative SNP risk prediction in men of Ashkenazic descent [90]. Results from different GWAS have implications for identifying high-risk individuals at a younger age so that interventions and therapeutic approaches can be planned. Prostate tumors do not all grow at the same rate, and some tumors are very aggressive in nature. Prostate cancer aggressiveness was explored in a study of GWAS of African–Americans and European Americans [104].
• Bladder cancer
Bladder cancer is the sixth most common cancer in the USA, with about 70,000 new cases and about 15,000 deaths per year. Three stages of this cancer are nonmuscle invasive (NMIBC), muscle invasive (MIBC) and metastatic disease. MIBC is considered genomically unstable and aggressive [105]. An association with aggressive bladder cancer was demonstrated at locus 19q12, and the functional analysis indicated overexpression of cyclin E [77]. This study analyzed data from two GWAS with more than 5000 cases and more than 10,000 controls. The most significant SNPs identified were rs810237 and rs7257330, which were located in the 19q12 region. In prostate cancer, cyclin gene CCNE1 is the only functional gene located within the associated linkage disequilibrium block for which functional analysis has shown promising results in risk prediction.
• Pancreatic cancer
The estimated number of new pancreatic cancer cases is 48,960 and about 40,650 deaths per year in the USA (the incidence and mortality rate is almost the same for this cancer). Smoking, alcoholism, diabetes and K-Ras mutations are considered risk factors for pancreatic cancer, the fourth leading cause of cancer deaths in the USA. A limited number of GWAS have been conducted for pancreatic cancer [80,91,106]. Amundadottir et al. identified an association between a locus on 9q34 and pancreatic cancer [80]. The SNP rs505922, identified in this study, falls in the first intron of the ABO blood group gene. Based on the data, the authors have suggested that people in the O group have a lower risk of developing pancreatic cancer than those in blood groups A or B. In another study, analysis of GWAS data in about 2000 cases and 2000 controls (from the Pancreatic Case Control Cancer Consortium, PC4) indicated an altered SLIT/ROBO pathway in pancreatic cancer [91]. Although smoking was considered in this study, the results could not be generalized because only data from pack years were available for this study. The authors indicated that misclassification of pack years due to imputation may have impacted their results. The small sample size used to study gene–environment interactions in this study was identified as another possible limitation. Copy number variations were observed in 60% of familial pancreatic cancer in one study [82].
Components of the epigenome and EWAS
Epigenetics involves alterations in gene expression without structural changes in the DNA [107,108]. Cancer cells are characterized by epigenetic dysregulation, including global genome hypomethylation, regional hypo- and hyper-methylation, altered histone modifications, altered miRNA and noncoding RNA profiling, chromatin accessibility and disturbed genomic imprinting. DNA methylation is a potential tumor marker for several cancers because of its heritable and stable characteristics [109]. DNA methylation at specific loci reflects the characteristic clinicopathological features [52]. Genome-wide analysis of DNA methylation has revealed that epigenetic regulation is not only a site-specific event but also spans long stretches of chromosome regions consisting of clusters of contiguous CpG islands [110,111]. Hypermethylation of the HOXA gene cluster was reported in breast and lung cancers [112]. Histones have four basic subunits, and in the native state they exist as an octamer. DNA winds around this octamer. Acidic histones neutralize DNA charge and maintain chromatin stability. The octamer histone binding is independent of surrounding DNA sequence and the N-terminal region of histones is subject to phosphorylation, acetylation and other modifications. Multiple histone modifications can take place within a short stretch of amino acids of histone tails. These modifications regulate transcription, DNA replication and DNA repair, and thus are part of the transformation process. Modification of histones may occur in large regions of chromatin, including coding and nonpromoter sequences, termed global histone modifications. The histone acetyltransferases (HATs) add the acetyl group to histones whereas the histone deacetylases (HDACs) removes the acetyl group. The opposing activities of HATs and HDACs tightly regulate gene expression through chromatin modifications. HATs activity results in chromatin relaxation, whereas HDACs make chromatin compact. A number of modifications in the tail region of histones have been reported, such as acetylation, deacetylation, phosphorylation, poly-ADP ribosylation, methylation, ubiquitination, sumoylation, carbonylation, citrullination and glycosylation [108,113]. The functional importance of epigenetics lies in its ability to regulate gene expression. By altering chromatin conformation, epigenetic changes can modulate gene expression. In addition to DNA promoter methylation, DNase-I hypersensitive site mapping, whole-genome epigenetic mapping and ChIP-seq for histone characterization have enabled the identification of promoters, enhancers, insulators, shores and shells [114]. miRNAs, which are integral components of the epigenetic regulatory machinery, actively participate in cancer development [115]. miRNAs act like tumor suppressors or oncogenes, depending on the tumor type and the stage of carcinogenesis [116–118]. During the past few years, high-throughput technologies have been developed to determine miRNA profiling in biospecimens from healthy individuals and people with cancer. EWAS aim to systematically identify epigenetic variants associated with different cancers [108]. EWAS can provide information that is complementary to the information obtained from GWAS and can be useful in identifying high-risk populations and in designing novel cancer treatment approaches. Compared with genomic changes, which are static, epigenetic changes are dynamic. Current literature supports the idea that interindividual epigenetic alterations contribute to disease development, including carcinogenesis [119]. Similar to GWAS, EWAS focus on common variations in the population – rather than on rare alterations in the epigenome – and might be more powerful in identifying risk-associated biomarkers. Recently, small-molecule inhibitors of selected epigenetic targets have been identified that cover classes of chromatin-associated epigenomic writers, erasers and readers [120]. Targeting epigenetic alterations may be more effective for therapeutic purposes than using a selective inhibitor of a single signaling pathway. The possibility of reversing epigenetic alterations may facilitate additional cancer treatment options. Epigenetic alterations are affected by environmental and other exogenous factors (Figure 1).
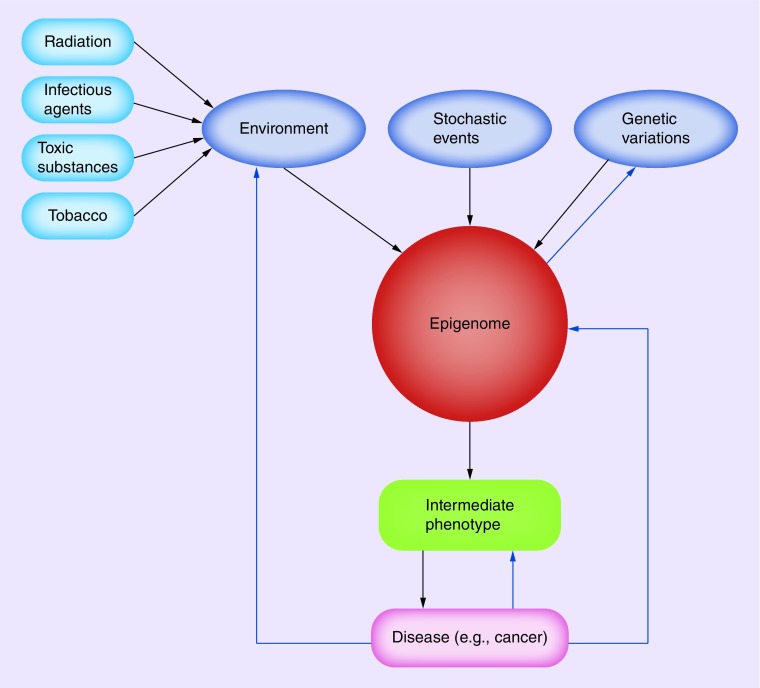
Figure 1. . Factors affecting the epigenome.
Environmental factors (such as radiation, infectious agents, toxic substances), stochastic events and genetic variations can alter the components of the epigenome, resulting in an intermediate phenotype and finally disease development. In turn, a disease also can influence the epigenome and the environment. There is evidence of the influence of intermediate phenotypes on genetic variations.
• EWAS on different cancers
Although the number of EWAS conducted to date is much smaller than GWAS, selected examples are discussed below. In a genome-wide screening study in a Gambian population, scientists identified the epiallele VTRNA 2–1 and found it to be sensitive to environmental changes and involved in innate immunity [121]. In another genome-wide methylation study, two alleles in the imprinted region were found to be regulated differently, one by methylation and the other independent of methylation [122]. Because the epigenome is tissue specific and genomic context-dependent, and because interindividual epigenetic variations are considered to be etiologic factors, Lowe et al. proposed that buccal cells would be more informative than blood for use in EWAS [123]. In two studies that analyzed functional genomic data consisting of histone modifications and DNase-I hotspots to determine whether there is any correlation between such sites and previously identified GWAS hits, a correlation was observed [124,125].
• Lung cancer
Smoking has been identified as a risk factor for lung and bladder cancers and other diseases, including respiratory and cardiovascular diseases. In the past, the effects of tobacco on health have been observed and long-term or short-term changes due to tobacco use have been studied. Compared with never smokers, the risk of lung cancer is much higher in former smokers. However, the evolution of epigenetic alterations after stopping smoking has not been studied in detail. In such projects, EWAS should be conducted on hundreds of participants to better characterize smoking-related biological effects and their influence on disease development and to identify epigenetic biomarkers for future use. In a study of more than 700 women, Guida et al. demonstrated that genome-wide methylation profiling could distinguish between those who had stopped smoking and normal healthy women [126]. This series of EWAS was the largest to date conducted in two European populations. The investigators identified two classes of CpG sites. The first class consisted of reversible sites that lose statistical significance following smoking cessation. The second class consisted of persistent sites that remain differentially methylated for a considerable length of time (after 35 years) following smoking cessation. A homogeneous distribution of methylation levels across both sites was observed during a visual inspection of about 750 CpG sites in never and current smokers. The approach used by Guida's group relied on a binary recording of smoking status as a function of time since stopping smoking, which was useful in determining the time taken for potential epigenetic smoking-associated marks to disappear. Guida's group also used genome-wide gene expression data to identify transcripts associated with specific CpG sites. In previous work, Guida et al. identified numerous CpG sites that were differentially methylated in current smokers only. Data on the reversal of methylation in individuals who quit smoking also are being generated. Another study explored F2RL3 gene-specific methylation changes in never smokers, former smokers and smokers [73]. Wan et al. evaluated genome-wide methylation and cigarette smoking and reported three differentially methylated sites between the first and last quartiles of time since individuals quit smoking [127]. This study has potential for determining the risk profiling of smoking-induced chronic diseases, including lung cancer.
Genome-wide methylation analysis of more than 100 individuals with non-small-cell lung cancer and matching controls identified 2414 genomic positions that were differentially methylated between tumor and nonmalignant tissue samples [128]. More than 70 differentially methylated genes were found to be involved in transcription regulation, which was confirmed by epigenetics-based inhibitor analysis. Further research confirmed HOXA2 and HOXA10 as epigenetic markers of squamous cell carcinoma prognosis in people with lung cancer. A few epigenetic variations have been established before gastrulation, in the very early embryonic stage. EWAS can be conducted at this stage to evaluate early stage epigenetic alterations. In another study, variation in cancer associated miRNA genes was observed in lung cancer [65].
• Nasopharyngeal carcinoma
Genome-wide methylation analysis of nasopharyngeal carcinoma tumors showed aberrant disruption of the Wnt, MAPK, TGF-β and Hedgehog signaling pathways [79]. These pathways play a central role in the carcinogenesis process. The examples cited here emphasize the significance of association studies and the utility of epigenetic markers in cancer control. Nawaz et al. identified nasopharyngeal carcinoma patients by screening populations in Morocco using methylation approaches [129]. These investigators also compared methylation profiling in samples from different geographic locations and different ethnicities.
• Colorectal cancer
A stepwise accumulation of genetic and epigenetic alterations results in the transformation of normal colon cells to cancer cells. Karpinski et al. reported three distinct methylation clusters (high-, medium- and low-methylation epitypes) in CRC, which may be useful as a reference in the epigenotyping of people with CRC in the future [130]. More than 500 tumor tissues (adenoma and carcinoma) and 222 adjacent normal tissues were analyzed for epigenetic profiling. The results indicated deregulation of the prostaglandin synthase pathway. More specifically, PTGIS, PTGER3, PTGFR and AKR1B1 were inactivated due to hypermethylation, along with other prostaglandins and their receptors in tumors [48]. Combining methylation epigenotypes with gene mutations and cytogenetic alterations (copy number variants and microsatellite instability) occurring in CRC has enabled the characterization of different CRC types. Vishnubalaji et al. looked for epigenetic therapeutic targets for CRC by profiling genome-wide mRNA and miRNA expression, which resulted in their identifying several potential regulatory networks [43]. These investigators suggested targeting relevant mRNA–miRNA networks as a potential therapeutic approach for CRC. An example is targeting EZH2, which was regulated by hsa-miR26a-5p and members of the let-7 family of miRNAs, with the expectation of the reduction in H3K27me3 resulting in reduced cell proliferation.
• Cervical cancer
During the progression of cervical cancer, the human papillomavirus (HPV) genome and cellular genes are regulated by both genetic and epigenetic mechanisms [131]. When 104 samples of HPV-positive cancer biospecimens and their control counterparts were analyzed by methylation, HPV-late genes L1 and L2 and the host gene DAPK showed progressive methylation corresponding with disease advancement [31]. Also for cervical cancer, genome-wide methylation analysis identified a group of genes that were associated with cancer invasiveness [132]. The implications of epigenetic drugs in drug-resistant cervical cancer cells also were explored recently. The authors suggested that global methylation is associated with the development of drug resistance and could serve as a therapeutic biomarker [133]. The interaction between genomics and epigenomics in cervical cancer was studied by Lando et al., who demonstrated that promoter hypermethylation and chromosomal loss at 3p11-p14 contribute to cancer development [32]. In another study, increased histone alterations (mainly H3K36me3 and H3K9Ac) in the HPV genome correlated with increased HPV16 gene expression [34]. Another component of epigenetic regulation, miRNA expression, also was found to be involved in the development of cervical cancer [134].
• Prostate cancer
Several dietary components interact with epigenetic machinery and are known to have potential to prevent cancer. Genome-wide methylation profiling was studied to evaluate the effects of natural food components with demethylating and acetylation inhibitory properties in prostate cancer cells [135]. This study demonstrated the utility of genome-wide methylation studies in therapeutics. Chemopreventive agents also were evaluated for their effects on decreasing prostate cancer events using whole-genome methylation approaches [136]. When prostate cancer tumor samples collected from different patients at different stages of disease development were analyzed for miRNA profiling, mRNA profiling, gene-expression profiling and mutation and SNP analysis, 74 genes were found to be correlated with tumor grade [29]. miR-34a played a key role in the development of prostate cancer, along with the p53 pathway and MET oncogene variant T992I. Based on epigenetics information, another group of investigators started to include epigenetic inhibitors with androgen-receptor inhibitors as a novel therapeutic approach to prostate cancer treatment [137]. In another study, a gene-methylation prediction model was proposed that could predict the recurrence of prostate cancer with 80% sensitivity and 85% specificity [138].
• Renal cancer
Renal cell carcinoma represents about 3% of all human malignancies. Genome-wide methylation contributed to the development of stratification criteria for renal cell carcinoma [83]. CpG island-based assays were conducted in formalin-fixed samples and a correlation between methylation patterns and survival was observed. Based on CpG island methylation patterns, subjects could be categorized into five lower risk to higher risk groups that showed differential expression of five genes in renal cancer: PITX1, FOXE3, TWF2, EHBPL1 and RIN1.
• Esophageal cancer
A combination of genome-wide methylation analysis and gene expression was applied to identify genes and pathways that contribute to the development of esophageal cancer in a northeast Indian population [139]. Altered methylation and gene expression were observed in pathways for cell adhesion, integrin signaling and cytoskeleton organization. The major genes affected were PTK2, RND1 and UBL3. The ‘integrome’ proposed by these authors represents integration of pathways, methylation status and gene expression in esophageal cancer. Other investigators identified epigenetic regulation of the transmembrane AJAP-1 and its association with recurrence of esophageal cancer [52].
• Breast cancer
Genome-wide methylation analysis was conducted in breast cancer patients and controls, which led to the identification of large DNA methylation contiguous gene clusters of about 70 kb that might serve as prognostic markers for breast cancer [20]. These clusters were reported in both ER+ and ER- breast cancer tumors. Other investigators used a quantitative approach based on statistical methods and machine learning algorithms to quantify methylation differences in cases and controls and identify differentially methylated regions from genome-wide methylation profiles [140,141]. To elucidate the role of estrogen in the coordinated repression of gene clusters located at 16p11.2 in breast cancer, the concept of long-range epigenetic silencing (LRES) was studied. The results showed that persistent estrogen-mediated LRES recruited an H3K27me3-repressive chromatin mark that may be useful in diagnosing breast cancer [19].
In Table 1, results from several studies of GWAS and EWAS have been summarized. Examples of those geneomic and epigenomic variations have been included which are well studied by several investigators.
Challenges & research opportunities
Information on environmental, epidemiologic, genetic, epigenetic and modifiable factors can be combined to develop clinically useful risk-prediction models. Each of these factors alone may be weakly or moderately informative when considered individually; however, a combination of these components may provide better approaches for risk assessment, screening and therapy in cancer.
GWAS have advantages in identifying populations at high risk of developing cancer. For example, GWAS: do not require an initial hypothesis; use digital and additive data that can be mined and augmented without data degradation; provide data on the ancestry of each subject, which assists in matching case subjects with control subjects; provide data on both sequence and copy-number variations; and encourage the formation of collaborative consortia, which tend to continue for subsequent analyses.
GWAS have limitations, however, including the need for samples from a large number of cases and controls, which can be challenging to organize. Also in GWAS, finding loci, not genes, can complicate the identification of pathogenic changes on an associated haplotype, and the detection of alleles that are common (>5%) in a population. Many SNPs are located in the susceptibility loci, making it challenging to identify and prioritize variants that influence the regulatory sequences that actually contribute to a disease phenotype. Although GWAS provide one avenue to identify genomic regions containing potential common risk alleles of cancer, the increased risk attributable to such regions is typically modest. In addition, the actual SNPs identified by GWAS rarely have obvious effects on protein coding regions, nor are they linked to clear causal variants in adjacent genes. The lack of validation cohorts for risk variants identified in different GWAS is an additional challenge. In general, GWAS have not been able to identify genetic loci that are effective classifiers of a disease, limiting their application in the clinic for genetic testing (clinicians still depend on histopathological data in diagnosing cancer). Nonetheless, the integration of GWAS and EWAS data has the potential to explain the functional consequences of genetic variation. In a GWA study on ovarian cancer, SNP rs6674079 located on 1q22 was found to be associated with histone modifications [142]. Findings from GWAS such as this may be useful in understanding clinical outcomes in ovarian cancer.
Another major problem is that several SNPs reported in different GWAS could not be reproduced by independent investigators. Efforts are undergoing to increase the number of participants in these studies and improving the research design so that validated SNPs can be identified.
As noted above, sample size is a critical element in both GWAS and EWAS. The small size reduces the statistical power to detect loci with a moderate effect (false-negative results) and increases the risk of spurious findings (false-positive results). Furthermore, because negative findings are difficult to publish, there is a possibility of bias in the direction of false-positive findings whenever new association results are published. Many GWAS and EWAS results vary across ethnicities, implying that findings in one population should be replicated in another population. Utilizing cohorts, consortia and international collaboration may resolve some of these issues.
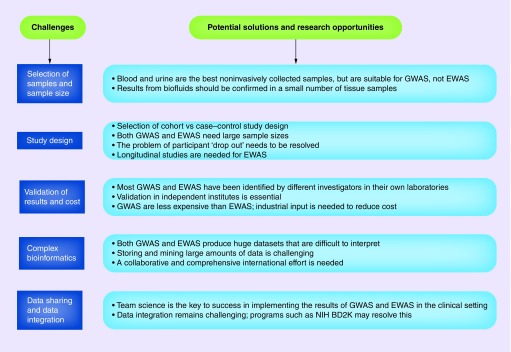
EWAS are associated with a range of issues that generally are not encountered in GWAS. One such issue involves the specific tissue on which EWAS are performed. Surrogate tissues sometimes are preferred for EWAS. Platform selection presents another challenge because a balance must be maintained between cost–effectiveness and genome coverage. A few general challenges and research opportunities in GWAS and EWAS are presented in Figure 2.
Figure 2. . Potential challenges and research opportunities.
Both GWAS and EWAS have challenges and need improvement. Because the field is still developing, addressing these challenges and finding new research opportunities will be beneficial in the long term.
EWAS: Epigenome-wide association studies; GWAS: Genome-wide association studies.
Large-scale projects that include the genome and epigenome (e.g., Encyclopedia of DNA Elements [ENCODE], Roadmap Epigenomics, International Human Epigenome Consortium [IHEC] and Functional Annotation of the Mammalian Genome [FANTOM]) have generated genome-wide maps of functional elements in diverse human tissue types (normal and cancerous) [111,143–145]. Chen et al. supported the identification of functional SNPs by incorporating high-throughput sequencing of epigenetic and transcription factor datasets [146]. Use of high-quality specimens from the Cancer Genome Atlas (TCGA) [94] for genomic and epigenomic analysis has provided excellent results in identifying tumor-associated diagnostic markers [147]. TCGA recently reported that DNA methylation inhibitors trigger antiviral immune response in cancer [148]. Such studies could be possible only after completing genome-wide methylation analysis of highly purified tumor samples [149].
To address intra-individual variation in EWAS, Shvetsov et al. measured short-term temporal trends in a genome-wide methylation study in blood samples and observed significant variation in trends in 10.9% of CpG loci before cell type adjustment and in 3.4% of loci after adjustment [150]. The results of this study also showed that genomic location at or near CpG islands or functional regions has little effect on the inter- and intra-individual variability of a CpG locus. Variation within an individual tends not to exceed a certain threshold.
GWAS and EWAS information can be utilized in designing precision medicine approaches that require integration of omics technologies, patient exposure history, next-generation sequencing, computational biology, genome medicine, medical informatics, disease-specific dynamic biomarkers and networks, efficacy-dependent therapies and predictive prognosis [151–153]. Understanding and interpreting data from epigenomics, metabolomics, proteomics and microbiomics also are required for precision medicine. One of the goals of precision medicine is to provide patients with customized diagnoses, prognoses and therapy [153]. Basically, establishing and integrating clinical informatics, molecular mechanisms of cellular biology and function, and individual data analysis and mining are needed to develop the precision medicine approach. Efforts are under way to develop new methods and systems to translate clinical descriptive information into the formats needed for digitally named clinical informatics.
Conclusion
Genetic and epigenetic regulations are involved in almost all steps of tumor initiation, development, progression and metastasis. Both genomic and epigenomic alterations occur almost simultaneously during carcinogenesis. As a result, understanding both is essential for cancer diagnosis and prognosis. GWAS are useful in screening populations at high risk of developing cancer, and EWAS can provide therapeutic targets. However, the modest increased risk of cancer associated with the known genomic and epigenomic variations, for the most part, not medically actionable.
Future perspective
It is expected that better powered GWAS, EWAS and genome-wide sequencing projects will continue to identify new cancer causal variants. These studies will facilitate increased understanding of the underlying mechanisms of cancer development and improved treatment strategies. It is further expected that cost-effective GWAS and EWAS approaches will be available and that the information from GWAS and EWAS will be useful in screening for and managing cancer. Also expected is the growth of epidemiologic data that link dietary, environmental and lifestyle exposures to the different genomic and epigenomic signatures of different cancers. This also will facilitate the design of preventive strategies to counteract the onset of genetic or epigenetic changes in healthy individuals and foster development of novel therapeutic strategies that target altered epigenetic components. To achieve this, the approach has to be personalized/personalized with inclusion of the latest information (profiling from genomics, epigenomics, metabolomics and transcriptomics) in diagnosis and prognosis. The day is not far when personalized medicine in cancer can facilitate tailoring preventive, diagnostic and therapeutic approaches to molecular profiling and other technologies applicable to an individual or population. There seems an interest in academicians, government and industry to enhance efforts in this direction as all these sectors are planning to enhance molecular profiling based personalized medicine in improving health. It is also expected that personalized medicine will bring novel therapeutic strategies with emphasis on gene-oriented treatment. This is the right time to validate what we know in the field and start educating clinicians and other health professionals in the field of GWAS and EWAS so that they can interpret results easily and imply them for improving health.
EXECUTIVE SUMMARY.
It is anticipated that some of the genetic and epigenetic markers identified by genome-wide association studies (GWAS) and epigenome-wide association studies (EWAS) will contribute to an improved understanding of the relationship between cancer susceptibility and molecular mechanisms of cancer as well as to developing new approaches in risk prediction and cancer control.
Also anticipated is the growth of epidemiologic data that link dietary, environmental and lifestyle exposures to the different genomic and epigenomic signatures in different cancers.
Differences in tumor behavior arise due to genomic and epigenomic changes. These changes can be measured quantitatively and can be used in cancer diagnosis and prognosis, as well as in following survival.
Acknowledgements
The author thanks J Brodsky of The Scientific Consulting Group, Inc., for reading the manuscript and providing suggestions.
Footnotes
Financial & competing interests disclosure
The author has no relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Hsieh CL, Ma HP, Su CM, et al. Alterations in histone deacetylase 8 lead to cell migration and poor prognosis in breast cancer. Life Sci. 2016 doi: 10.1016/j.lfs.2016.02.092. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 2.Dorman SN, Viner C, Rogan PK. Splicing mutation analysis reveals previously unrecognized pathways in lymph node-invasive breast cancer. Sci. Rep. 2014;4:7063. doi: 10.1038/srep07063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang X, Baheti S, Shameer K, et al. The eSNV-detect: a computational system to identify expressed single nucleotide variants from transcriptome sequencing data. Nucleic Acids Res. 2014;42(22):e172. doi: 10.1093/nar/gku1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim JH, Bae JM, Cho NY, Kang GH. Distinct features between MLH1-methylated and unmethylated colorectal carcinomas with the CpG island methylator phenotype: implications in the serrated neoplasia pathway. Oncotarget. 2016 doi: 10.18632/oncotarget.7374. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masson AL, Talseth-Palmer BA, Evans TJ, et al. Copy number variants associated with 18p11.32, DCC and the promoter 1B region of APC in colorectal polyposis patients. Meta Gene. 2016;7:95–104. doi: 10.1016/j.mgene.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceccarelli M, Barthel FP, Malta TM, et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164(3):550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mock A, Geisenberger C, Orlik C, et al. LOC283731 promoter hypermethylation prognosticates survival after radiochemotherapy in IDH1 wild-type glioblastoma patients. Int. J. Cancer. 2016 doi: 10.1002/ijc.30069. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Network. Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiffen J, Wilson S, Gallagher SJ, Hersey P, Filipp FV. Somatic copy number amplification and hyperactivating somatic mutations of EZH2 correlate with DNA methylation and drive epigenetic silencing of genes involved in tumor suppression and immune responses in melanoma. Neoplasia. 2016;18(2):121–132. doi: 10.1016/j.neo.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shah S, Shah S, Padh H, Kalia K. Genetic alterations of the PIK3CA oncogene in human oral squamous cell carcinoma in an Indian population. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2015;120(5):628–635. doi: 10.1016/j.oooo.2015.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Planello AC, Singhania R, Kron KJ, et al. Pre-neoplastic epigenetic disruption of transcriptional enhancers in chronic inflammation. Oncotarget. 2016 doi: 10.18632/oncotarget.7513. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shao C, Yu Y, Yu L, et al. Amplification and up-regulation of microRNA-30b in oral squamous cell cancers. Arch. Oral Biol. 2012;57(8):1012–1017. doi: 10.1016/j.archoralbio.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research N. The molecular taxonomy of primary prostate cancer. Cell. 2015;163(4):1011–1025. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Jadhav RR, Liu J, et al. Roles of distal and genic methylation in the development of prostate tumorigenesis revealed by genome-wide DNA methylation analysis. Sci. Rep. 2016;6:22051. doi: 10.1038/srep22051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis CF, Ricketts CJ, Wang M, et al. The somatic genomic landscape of chromophobe renal cell carcinoma. Cancer Cell. 2014;26(3):319–330. doi: 10.1016/j.ccr.2014.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakimi AA, Ostrovnaya I, Jacobsen A, et al. Validation and genomic interrogation of the MET variant rs11762213 as a predictor of adverse outcomes in clear cell renal cell carcinoma. Cancer. 2016;122(3):402–410. doi: 10.1002/cncr.29765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research N. Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159(3):676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu PY, Hsu HK, Singer GA, et al. Estrogen-mediated epigenetic repression of large chromosomal regions through DNA looping. Genome Res. 2010;20(6):733–744. doi: 10.1101/gr.101923.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jadhav RR, Ye Z, Huang RL, et al. Genome-wide DNA methylation analysis reveals estrogen-mediated epigenetic repression of metallothionein-1 gene cluster in breast cancer. Clin. Epigenetics. 2015;7(1):13. doi: 10.1186/s13148-015-0045-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim MS, Lee J, Oh T, et al. Genome-wide identification of OTP gene as a novel methylation marker of breast cancer. Oncol. Rep. 2012;27(5):1681–1688. doi: 10.3892/or.2012.1691. [DOI] [PubMed] [Google Scholar]
- 22.Stadler ZK, Vijai J, Thom P, et al. Genome-wide association studies of cancer predisposition. Hematol. Oncol. Clin. North Am. 2010;24(5):973–996. doi: 10.1016/j.hoc.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 23.Thomas G, Jacobs KB, Kraft P, et al. A multistage genome-wide association study in breast cancer identifies two new risk alleles at 1p11.2 and 14q24.1 (RAD51L1) Nat. Genet. 2009;41(5):579–584. doi: 10.1038/ng.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stacey SN, Manolescu A, Sulem P, et al. Common variants on chromosomes 2q35 and 16q12 confer susceptibility to estrogen receptor-positive breast cancer. Nat. Genet. 2007;39(7):865–869. doi: 10.1038/ng2064. [DOI] [PubMed] [Google Scholar]
- 25.Guo Q, Schmidt MK, Kraft P, et al. Identification of novel genetic markers of breast cancer survival. J. Natl Cancer Inst. 2015;107(5):djv081. doi: 10.1093/jnci/djv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W, Song H, Zhong R, et al. Risk of GWAS-identified genetic variants for breast cancer in a Chinese population: a multiple interaction analysis. Breast Cancer Res. Treat. 2013;142(3):637–644. doi: 10.1007/s10549-013-2775-8. [DOI] [PubMed] [Google Scholar]
- 27.Fernandez-Navarro P, Gonzalez-Neira A, Pita G, et al. Genome wide association study identifies a novel putative mammographic density locus at 1q12-q21. Int. J. Cancer. 2015;136(10):2427–2436. doi: 10.1002/ijc.29299. [DOI] [PubMed] [Google Scholar]
- 28.Li N, Zhou P, Zheng J, et al. A polymorphism rs12325489C>T in the lincRNA-ENST00000515084 exon was found to modulate breast cancer risk via GWAS-based association analyses. PLoS ONE. 2014;9(5):e98251. doi: 10.1371/journal.pone.0098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sethi S, Kong D, Land S, Dyson G, Sakr WA, Sarkar FH. Comprehensive molecular oncogenomic profiling and miRNA analysis of prostate cancer. Am. J. Transl. Res. 2013;5(2):200–211. [PMC free article] [PubMed] [Google Scholar]
- 30.Kim SJ, Kelly WK, Fu A, et al. Genome-wide methylation analysis identifies involvement of TNF-alpha mediated cancer pathways in prostate cancer. Cancer Lett. 2011;302(1):47–53. doi: 10.1016/j.canlet.2010.12.010. [DOI] [PubMed] [Google Scholar]
- 31.Kalantari M, Osann K, Calleja-Macias IE, et al. Methylation of human papillomavirus 16, 18, 31, and 45 L2 and L1 genes and the cellular DAPK gene: considerations for use as biomarkers of the progression of cervical neoplasia. Virology. 2014;448:314–321. doi: 10.1016/j.virol.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lando M, Fjeldbo CS, Wilting SM, et al. Interplay between promoter methylation and chromosomal loss in gene silencing at 3p11-p14 in cervical cancer. Epigenetics. 2015;10(10):970–980. doi: 10.1080/15592294.2015.1085140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fields AP, Justilien V, Murray NR. The chromosome 3q26 OncCassette: a multigenic driver of human cancer. Adv. Biol. Regul. 2016;60:47–63. doi: 10.1016/j.jbior.2015.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson C, Jamal Fattah T, Yu H, Nygren J, Mossberg AK, Schwartz S. Acetylation of intragenic histones on HPV16 correlates with enhanced HPV16 gene expression. Virology. 2015;482:244–259. doi: 10.1016/j.virol.2015.02.053. [DOI] [PubMed] [Google Scholar]
- 35.Tornesello ML, Annunziata C, Buonaguro L, Losito S, Greggi S, Buonaguro FM. TP53 and PIK3CA gene mutations in adenocarcinoma, squamous cell carcinoma and high-grade intraepithelial neoplasia of the cervix. J. Transl. Med. 2014;12:255. doi: 10.1186/s12967-014-0255-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tina E, Lindqvist BM, Gabrielson M, Lubovac Z, Wegman P, Wingren S. The mitochondrial transporter SLC25A43 is frequently deleted and may influence cell proliferation in HER2-positive breast tumors. BMC Cancer. 2012;12:350. doi: 10.1186/1471-2407-12-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kuglik P, Smetana J, Vallova V, et al. Genome-wide screening of DNA copy number alterations in cervical carcinoma patients with CGH+SNP microarrays and HPV-FISH. Int. J. Clin. Exp. Pathol. 2014;7(8):5071–5082. [PMC free article] [PubMed] [Google Scholar]
- 38.Hafner N, Steinbach D, Jansen L, Diebolder H, Durst M, Runnebaum IB. RUNX3 and CAMK2N1 hypermethylation as prognostic marker for epithelial ovarian cancer. Int. J. Cancer. 2016;138(1):217–228. doi: 10.1002/ijc.29690. [DOI] [PubMed] [Google Scholar]
- 39.Hunter SM, Anglesio MS, Ryland GL, et al. Molecular profiling of low grade serous ovarian tumours identifies novel candidate driver genes. Oncotarget. 2015;6(35):37663–37677. doi: 10.18632/oncotarget.5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mostowska A, Sajdak S, Pawlik P, et al. Replication study for the association of seven genome- GWAS-identified Loci with susceptibility to ovarian cancer in the Polish population. Pathol. Oncol. Res. 2015;21(2):307–313. doi: 10.1007/s12253-014-9822-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kar SP, Tyrer JP, Li Q, et al. Network-based integration of GWAS and gene expression identifies a HOX-centric network associated with serous ovarian cancer risk. Cancer Epidemiol. Biomarkers Prev. 2015;24(10):1574–1584. doi: 10.1158/1055-9965.EPI-14-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kluska A, Balabas A, Paziewska A, et al. New recurrent BRCA1/2 mutations in Polish patients with familial breast/ovarian cancer detected by next generation sequencing. BMC Med. Genomics. 2015;8:19. doi: 10.1186/s12920-015-0092-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vishnubalaji R, Hamam R, Abdulla MH, et al. Genome-wide mRNA and miRNA expression profiling reveal multiple regulatory networks in colorectal cancer. Cell Death Dis. 2015;6:e1614. doi: 10.1038/cddis.2014.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Tassan NA, Whiffin N, Hosking FJ, et al. A new GWAS and meta-analysis with 1000 Genomes imputation identifies novel risk variants for colorectal cancer. Sci. Rep. 2015;5:10442. doi: 10.1038/srep10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ke J, Lou J, Chen X, et al. Identification of a potential regulatory variant for colorectal cancer risk mapping to chromosome 5q31.1: a post-gwas study. PLoS ONE. 2015;10(9):e0138478. doi: 10.1371/journal.pone.0138478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birgisson H, Edlund K, Wallin U, et al. Microsatellite instability and mutations in BRAF and KRAS are significant predictors of disseminated disease in colon cancer. BMC Cancer. 2015;15:125. doi: 10.1186/s12885-015-1144-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qiao HP, Zhang CY, Yu ZL, Li QM, Jiao Y, Cao JP. Genetic variants identified by GWAS was associated with colorectal cancer in the Han Chinese population. J. Cancer Res. Ther. 2015;11(2):468–470. doi: 10.4103/0973-1482.150346. [DOI] [PubMed] [Google Scholar]
- 48.Cebola I, Custodio J, Munoz M, et al. Epigenetics override pro-inflammatory PTGS transcriptomic signature towards selective hyperactivation of PGE2 in colorectal cancer. Clin. Epigenetics. 2015;7(1):74. doi: 10.1186/s13148-015-0110-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kang BW, Jeon HS, Chae YS, et al. Association between GWAS-identified genetic variations and disease prognosis for patients with colorectal cancer. PLoS ONE. 2015;10(3):e0119649. doi: 10.1371/journal.pone.0119649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tomlinson IP, Webb E, Carvajal-Carmona L, et al. A genome-wide association study identifies colorectal cancer susceptibility loci on chromosomes 10p14 and 8q23.3. Nat. Genet. 2008;40(5):623–630. doi: 10.1038/ng.111. [DOI] [PubMed] [Google Scholar]
- 51.Zanke BW, Greenwood CM, Rangrej J, et al. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24. Nat. Genet. 2007;39(8):989–994. doi: 10.1038/ng2089. [DOI] [PubMed] [Google Scholar]
- 52.Tanaka H, Kanda M, Koike M, et al. Adherens junctions associated protein 1 serves as a predictor of recurrence of squamous cell carcinoma of the esophagus. Int. J. Oncol. 2015;47(5):1811–1818. doi: 10.3892/ijo.2015.3167. [DOI] [PubMed] [Google Scholar]
- 53.Hu L, Wu Y, Guan X, et al. Germline copy number loss of UGT2B28 and gain of PLEC contribute to increased human esophageal squamous cell carcinoma risk in Southwest China. Am. J. Cancer Res. 2015;5(10):3056–3071. [PMC free article] [PubMed] [Google Scholar]
- 54.Yoshida S, Yamashita S, Niwa T, et al. Epigenetic inactivation of FAT4 contributes to gastric field cancerization. Gastric Cancer. 2016 doi: 10.1007/s10120-016-0593-5. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 55.Labrador L, Torres K, Camargo M, Santiago L, Valderrama E, Chiurillo MA. Association of common variants on chromosome 8q24 with gastric cancer in Venezuelan patients. Gene. 2015;566(1):120–124. doi: 10.1016/j.gene.2015.04.081. [DOI] [PubMed] [Google Scholar]
- 56.Tsai PC, Huang SW, Tsai HL, et al. The association between DNA copy number aberrations at chromosome 5q22 and gastric cancer. PLoS ONE. 2014;9(9):e106624. doi: 10.1371/journal.pone.0106624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Min HL, Kim J, Kim WH, Jang BG, Kim MA. Epigenetic silencing of the putative tumor suppressor gene GLDC (glycine dehydrogenase) in gastric carcinoma. Anticancer Res. 2016;36(1):179–187. [PubMed] [Google Scholar]
- 58.Garcia-Gonzalez MA, Bujanda L, Quintero E, et al. Association of PSCA rs2294008 gene variants with poor prognosis and increased susceptibility to gastric cancer and decreased risk of duodenal ulcer disease. Int. J. Cancer. 2015;137(6):1362–1373. doi: 10.1002/ijc.29500. [DOI] [PubMed] [Google Scholar]
- 59.He G, Hu S, Zhang D, et al. Hypermethylation of FOXD3 suppresses cell proliferation, invasion and metastasis in hepatocellular carcinoma. Exp. Mol. Pathol. 2015;99(2):374–382. doi: 10.1016/j.yexmp.2015.06.017. [DOI] [PubMed] [Google Scholar]
- 60.Budhu A, Wang XW. Power play: scoring our goals for liver cancer with better GWAS study design. J. Hepatol. 2011;54(4):823–824. doi: 10.1016/j.jhep.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 61.Shen J, Lefave C, Sirosh I, Siegel AB, Tycko B, Santella RM. Integrative epigenomic and genomic filtering for methylation markers in hepatocellular carcinomas. BMC Med. Genomics. 2015;8:28. doi: 10.1186/s12920-015-0105-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin JC, Wu YC, Wu CC, Shih PY, Wang WY, Chien YC. DNA methylation markers and serum alpha-fetoprotein level are prognostic factors in hepatocellular carcinoma. Ann. Hepatol. 2015;14(4):494–504. [PubMed] [Google Scholar]
- 63.Robles AI, Arai E, Mathe EA, et al. An integrated prognostic classifier for stage I lung adenocarcinoma based on mRNA, microRNA, and DNA methylation biomarkers. J. Thorac. Oncol. 2015;10(7):1037–1048. doi: 10.1097/JTO.0000000000000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young RP, Hopkins RJ, Whittington CF, Hay BA, Epton MJ, Gamble GD. Individual and cumulative effects of GWAS susceptibility loci in lung cancer: associations after sub-phenotyping for COPD. PLoS ONE. 2011;6(2):e16476. doi: 10.1371/journal.pone.0016476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Czubak K, Lewandowska MA, Klonowska K, et al. High copy number variation of cancer-related microRNA genes and frequent amplification of DICER1 and DROSHA in lung cancer. Oncotarget. 2015;6(27):23399–23416. doi: 10.18632/oncotarget.4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simo-Riudalbas L, Perez-Salvia M, Setien F, et al. KAT6B is a tumor suppressor histone H3 lysine 23 acetyltransferase undergoing genomic loss in small cell lung cancer. Cancer Res. 2015;75(18):3936–3945. doi: 10.1158/0008-5472.CAN-14-3702. [DOI] [PubMed] [Google Scholar]
- 67.Han S, Gao F, Yang W, et al. Identification of an SCLC susceptibility rs7963551 genetic polymorphism in a previously GWAS-identified 12p13.33 RAD52 lung cancer risk locus in the Chinese population. Int. J. Clin. Exp. Med. 2015;8(9):16528–16535. [PMC free article] [PubMed] [Google Scholar]
- 68.George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature. 2015;524(7563):47–53. doi: 10.1038/nature14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hosgood HD, Sapkota AR, Rothman N, et al. The potential role of lung microbiota in lung cancer attributed to household coal burning exposures. Environ. Mol. Mutagen. 2014;55(8):643–651. doi: 10.1002/em.21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Weissfeld JL, Lin Y, Lin HM, et al. Lung cancer risk prediction using common SNPs located in GWAS-identified susceptibility regions. J. Thorac. Oncol. 2015;10(11):1538–1545. doi: 10.1097/JTO.0000000000000666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li D, Wei L, Xu B, et al. Association of GWAS-identified lung cancer susceptibility loci with survival length in patients with small-cell lung cancer treated with platinum-based chemotherapy. PLoS ONE. 2014;9(11):e113574. doi: 10.1371/journal.pone.0113574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leng S, Liu Y, Weissfeld JL, et al. 15q12 variants, sputum gene promoter hypermethylation, and lung cancer risk: a GWAS in smokers. J. Natl Cancer Inst. 2015;107(5):djv035. doi: 10.1093/jnci/djv035. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Integration of genomic and epigenomic information in detecting lung cancer.
- 73.Zhang Y, Yang R, Burwinkel B, Breitling LP, Brenner H. F2RL3 methylation as a biomarker of current and lifetime smoking exposures. Environ. Health Perspect. 2014;122(2):131–137. doi: 10.1289/ehp.1306937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Clausen MJ, Melchers LJ, Mastik MF, et al. Identification and validation of WISP1 as an epigenetic regulator of metastasis in oral squamous cell carcinoma. Genes Chromosomes Cancer. 2016;55(1):45–59. doi: 10.1002/gcc.22310. [DOI] [PubMed] [Google Scholar]
- 75.Low JS, Chin YM, Mushiroda T, et al. A genome wide study of copy number variation associated with nasopharyngeal carcinoma in malaysian chinese identifies CNVs at 11q14.3 and 6p21.3 as Candidate Loci. PLoS ONE. 2016;11(1):e0145774. doi: 10.1371/journal.pone.0145774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ma L, Chen J, Song X, et al. Evidence that the genetic polymorphism rs1412115 on chromosome 10 is associated with risk for oral squamous cell carcinoma. Gene. 2015;560(2):137–139. doi: 10.1016/j.gene.2015.01.049. [DOI] [PubMed] [Google Scholar]
- 77.Fu YP, Kohaar I, Moore LE, et al. The 19q12 bladder cancer GWAS signal: association with cyclin E function and aggressive disease. Cancer Res. 2014;74(20):5808–5818. doi: 10.1158/0008-5472.CAN-14-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singh V, Jaiswal PK, Mittal RD. Replicative study of GWAS TP63C/T, TERTC/T, and SLC14A1C/T with susceptibility to bladder cancer in north Indians. Urol. Oncol. 2014;32(8):1209–1214. doi: 10.1016/j.urolonc.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 79.Li L, Zhang Y, Fan Y, et al. Characterization of the nasopharyngeal carcinoma methylome identifies aberrant disruption of key signaling pathways and methylated tumor suppressor genes. Epigenomics. 2015;7(2):155–173. doi: 10.2217/epi.14.79. [DOI] [PubMed] [Google Scholar]
- 80.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat. Genet. 2009;41(9):986–990. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tang H, Wei P, Duell EJ, et al. Genes-environment interactions in obesity- and diabetes-associated pancreatic cancer: a GWAS data analysis. Cancer Epidemiol. Biomarkers Prev. 2014;23(1):98–106. doi: 10.1158/1055-9965.EPI-13-0437-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Al-Sukhni W, Joe S, Lionel AC, et al. Identification of germline genomic copy number variation in familial pancreatic cancer. Hum. Genet. 2012;131(9):1481–1484. doi: 10.1007/s00439-012-1183-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wei JH, Haddad A, Wu KJ, et al. A CpG-methylation-based assay to predict survival in clear cell renal cell carcinoma. Nat. Commun. 2015;6:8699. doi: 10.1038/ncomms9699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Swierniak M, Wojcicka A, Czetwertynska M, et al. Association between GWAS-derived rs966423 genetic variant and overall mortality in patients with differentiated thyroid cancer. Clin. Cancer. Res. 2015;22(5):1111–1119. doi: 10.1158/1078-0432.CCR-15-1746. [DOI] [PubMed] [Google Scholar]
- 85.Mancikova V, Cruz R, Inglada-Perez L, et al. Thyroid cancer GWAS identifies 10q26.12 and 6q14.1 as novel susceptibility loci and reveals genetic heterogeneity among populations. Int. J. Cancer. 2015;137(8):1870–1878. doi: 10.1002/ijc.29557. [DOI] [PubMed] [Google Scholar]
- 86.Gudmundsson J, Sulem P, Gudbjartsson DF, et al. Discovery of common variants associated with low TSH levels and thyroid cancer risk. Nat. Genet. 2012;44(3):319–322. doi: 10.1038/ng.1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.D'Souza G, Carey TE, William WN, Jr, et al. Epidemiology of head and neck squamous cell cancer among HIV-infected patients. J. Acquir. Immune Defic. Syndr. 2013;65(5):603–610. doi: 10.1097/QAI.0000000000000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fachal L, Dunning AM. From candidate gene studies to GWAS and post-GWAS analyses in breast cancer. Curr. Opin. Genet. Dev. 2015;30C:32–41. doi: 10.1016/j.gde.2015.01.004. [DOI] [PubMed] [Google Scholar]; •• Study demonstrating the significance of genome-wide association studies (GWAS) and post-GWAS analysis in risk assessment, diagnosis and potential in prognosis.
- 89.Lee JH, Kim Y, Choi JW, Kim YS. Genetic variants and risk of gastric cancer: a pathway analysis of a genome-wide association study. Springerplus. 2015;4:215. doi: 10.1186/s40064-015-1005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Correlation of genetic analysis with pathways involved in cancer development.
- 90.Agalliu I, Wang Z, Wang T, et al. Characterization of SNPs associated with prostate cancer in men of Ashkenazic descent from the set of GWAS identified SNPs: impact of cancer family history and cumulative SNP risk prediction. PLoS ONE. 2013;8(4):e60083. doi: 10.1371/journal.pone.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tang H, Wei P, Duell EJ, et al. Axonal guidance signaling pathway interacting with smoking in modifying the risk of pancreatic cancer: a gene- and pathway-based interaction analysis of GWAS data. Carcinogenesis. 2014;35(5):1039–1045. doi: 10.1093/carcin/bgu010. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Correlation of modifiable environmental factors in contributing disease development.
- 92.Dai J, Gu J, Huang M, et al. GWAS-identified colorectal cancer susceptibility loci associated with clinical outcomes. Carcinogenesis. 2012;33(7):1327–1331. doi: 10.1093/carcin/bgs147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.National Cancer Institute. www.cancer.gov
- 94.The Cancer Genome Atlas. http://cancergenome.nih.gov
- 95.The Precision Medicine Initiative. www.whitehouse.gov/precision-medicine
- 96.Lemire M, Qu C, Loo LW, et al. A genome-wide association study for colorectal cancer identifies a risk locus in 14q23.1. Hum. Genet. 2015;134(11–12):1249–1262. doi: 10.1007/s00439-015-1598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schumacher FR, Schmit SL, Jiao S, et al. Genome-wide association study of colorectal cancer identifies six new susceptibility loci. Nat. Commun. 2015;6:7138. doi: 10.1038/ncomms8138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang H, Burnett T, Kono S, et al. Trans-ethnic genome-wide association study of colorectal cancer identifies a new susceptibility locus in VTI1A. Nat. Commun. 2014;5:4613. doi: 10.1038/ncomms5613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Peltekova VD, Lemire M, Qazi AM, et al. Identification of genes expressed by immune cells of the colon that are regulated by colorectal cancer-associated variants. Int. J. Cancer. 2014;134(10):2330–2341. doi: 10.1002/ijc.28557. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Contribution of host-susceptibility factors, such as immunological background, in cancer development.
- 100.Gaj P, Maryan N, Hennig EE, et al. Pooled sample-based GWAS: a cost-effective alternative for identifying colorectal and prostate cancer risk variants in the Polish population. PLoS ONE. 2012;7(4):e35307. doi: 10.1371/journal.pone.0035307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hu N, Wang Z, Song X, et al. Genome-wide association study of gastric adenocarcinoma in Asia: a comparison of associations between cardia and non-cardia tumours. Gut. 2015 doi: 10.1136/gutjnl-2015-309340. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chen X, Mcclelland M, Jia Z, et al. The identification of trans-associations between prostate cancer GWAS SNPs and RNA expression differences in tumor-adjacent stroma. Oncotarget. 2015;6(3):1865–1873. doi: 10.18632/oncotarget.2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Olumi AF. Commentary on “identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array.”. Eeles RA, Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, Dadaev T, Neal DE, Hamdy FC, Donovan JL, Muir K, Giles GG, Severi G, Wiklund F, Gronberg H, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Lindstrom S, Kraft P, Hunter DJ, Gapstur S, Chanock SJ, Berndt SI, Albanes D, Andriole G, Schleutker J, Weischer M, Canzian F, Riboli E, Key TJ, Travis RC, Campa D, Ingles SA, John EM, Hayes RB, Pharoah PD, Pashayan N, Khaw KT, Stanford JL, Ostrander EA, Signorello LB, Thibodeau SN, Schaid D, Maier C, Vogel W, Kibel AS, Cybulski C, Lubinski J, Cannon-Albright L, Brenner H, Park JY, Kaneva R, Batra J, Spurdle AB, Clements JA, Teixeira MR, Dicks E, Lee A, Dunning AM, Baynes C, Conroy D, Maranian MJ, Ahmed S, Govindasami K, Guy M, Wilkinson RA, Sawyer EJ, Morgan A, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As NJ, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatananon A, Cox A, Southey MC, Hopper JL, English DR, Aly M, Adolfsson J, Xu J, Zheng SL, Yeager M, Kaaks R, Diver WR, Gaudet MM, Stern MC, Corral R, Joshi AD, Shahabi A, Wahlfors T, Tammela TL, Auvinen A, Virtamo J, Klarskov P, Nordestgaard BG, Roder MA, Nielsen SF, Bojesen SE, Siddiq A, Fitzgerald LM, Kolb S, Kwon EM, Karyadi DM, Blot WJ, Zheng W, Cai Q, McDonnell SK, Rinckleb AE, Drake B, Colditz G, Wokolorczyk D, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Sellers TA, Lin HY, Slavov C, Mitev V, Lose F, Srinivasan S, Maia S, Paulo P, Lange E, Cooney KA, Antoniou AC, Vincent D, Bacot F, Tessier DC. COGS-Cancer Research UK GWAS-ELLIPSE (part of GAME-ON) Initiative; Australian Prostate Cancer Bioresource; UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons’ Section of Oncology; UK ProtecT (Prostate testing for cancer and Treatment) Study. Urol. Oncol. 2014;32(2):211. doi: 10.1016/j.urolonc.2013.08.019. [DOI] [PubMed] [Google Scholar]
- 104.Bensen JT, Xu Z, Smith GJ, Mohler JL, Fontham ET, Taylor JA. Genetic polymorphism and prostate cancer aggressiveness: a case-only study of 1,536 GWAS and candidate SNPs in African–Americans and European–Americans. Prostate. 2013;73(1):11–22. doi: 10.1002/pros.22532. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Study demonstrating aggressiveness of the disease.
- 105.Guey LT, Garcia-Closas M, Murta-Nascimento C, et al. Genetic susceptibility to distinct bladder cancer subphenotypes. Eur. Urol. 2010;57(2):283–292. doi: 10.1016/j.eururo.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Murtaugh LC. Putting GWAS to the functional test: NR5A2 and pancreatic cancer risk. Gut. 2014;63(4):535–536. doi: 10.1136/gutjnl-2013-305030. [DOI] [PubMed] [Google Scholar]
- 107.Verma M. Viral genes and methylation. Ann. NY. Acad. Sci. 2003;983:170–180. doi: 10.1111/j.1749-6632.2003.tb05972.x. [DOI] [PubMed] [Google Scholar]
- 108.Verma M. Epigenome-wide association studies (EWAS) in cancer. Curr. Genomics. 2012;13(4):308–313. doi: 10.2174/138920212800793294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mishra A, Brat DJ, Verma M. P53 tumor suppression network in cancer epigenetics. Methods Mol. Biol. 2015;1238:597–605. doi: 10.1007/978-1-4939-1804-1_31. [DOI] [PubMed] [Google Scholar]
- 110.Clark SJ. Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Hum. Mol. Genet. 2007;16(Spec No 1):R88–R95. doi: 10.1093/hmg/ddm051. [DOI] [PubMed] [Google Scholar]
- 111.Satterlee JS, Beckel-Mitchener A, Mcallister K, et al. Community resources and technologies developed through the NIH Roadmap Epigenomics Program. Methods Mol. Biol. 2015;1238:27–49. doi: 10.1007/978-1-4939-1804-1_2. [DOI] [PubMed] [Google Scholar]
- 112.Novak P, Jensen T, Oshiro MM, et al. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer Res. 2006;66(22):10664–10670. doi: 10.1158/0008-5472.CAN-06-2761. [DOI] [PubMed] [Google Scholar]
- 113.Verma M, Rogers S, Divi RL, et al. Epigenetic research in cancer epidemiology: trends, opportunities, and challenges. Cancer Epidemiol. Biomarkers Prev. 2014;23(2):223–233. doi: 10.1158/1055-9965.EPI-13-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhu X, Liu J, Xu X, Zhang C, Dai D. Genome-wide analysis of histone modifications by ChIP-chip to identify silenced genes in gastric cancer. Oncol. Rep. 2015;33(5):2567–2574. doi: 10.3892/or.2015.3824. [DOI] [PubMed] [Google Scholar]
- 115.Neerincx M, Sie DL, Van De Wiel MA, et al. MiR expression profiles of paired primary colorectal cancer and metastases by next-generation sequencing. Oncogenesis. 2015;4:e170. doi: 10.1038/oncsis.2015.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bucay N, Shahryari V, Majid S, et al. miRNA expression analyses in prostate cancer clinical tissues. J. Vis. Exp. 2015 doi: 10.3791/53123. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hirata H, Ueno K, Shahryari V, et al. Oncogenic miRNA-182-5p targets Smad4 and RECK in human bladder cancer. PLoS ONE. 2012;7(11):e51056. doi: 10.1371/journal.pone.0051056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Majid S, Dar AA, Saini S, et al. miRNA-34b inhibits prostate cancer through demethylation, active chromatin modifications, and AKT pathways. Clin. Cancer Res. 2013;19(1):73–84. doi: 10.1158/1078-0432.CCR-12-2952. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Integration of epigenomics and genomics in controlling prostate cancer.
- 119.Swierczynski S, Klieser E, Illig R, Alinger-Scharinger B, Kiesslich T, Neureiter D. Histone deacetylation meets miRNA: epigenetics and post-transcriptional regulation in cancer and chronic diseases. Expert Opin. Biol. Ther. 2015;15(5):651–664. doi: 10.1517/14712598.2015.1025047. [DOI] [PubMed] [Google Scholar]
- 120.Tanaka M, Roberts JM, Qi J, Bradner JE. Inhibitors of emerging epigenetic targets for cancer therapy: a patent review (2010–2014) Pharm. Pat. Anal. 2015;4(4):261–284. doi: 10.4155/ppa.15.16. [DOI] [PubMed] [Google Scholar]
- 121.Silver MJ, Kessler NJ, Hennig BJ, et al. Independent genomewide screens identify the tumor suppressor VTRNA2–1 as a human epiallele responsive to periconceptional environment. Genome Biol. 2015;16:118. doi: 10.1186/s13059-015-0660-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Court F, Tayama C, Romanelli V, et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24(4):554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lowe R, Gemma C, Beyan H, et al. Buccals are likely to be a more informative surrogate tissue than blood for epigenome-wide association studies. Epigenetics. 2013;8(4):445–454. doi: 10.4161/epi.24362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337(6099):1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Guida F, Sandanger TM, Castagne R, et al. Dynamics of smoking-induced genome-wide methylation changes with time since smoking cessation. Hum. Mol. Genet. 2015;24(8):2349–2359. doi: 10.1093/hmg/ddu751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wan ES, Qiu W, Baccarelli A, et al. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum. Mol. Genet. 2012;21(13):3073–3082. doi: 10.1093/hmg/dds135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Heller G, Babinsky VN, Ziegler B, et al. Genome-wide CpG island methylation analyses in non-small cell lung cancer patients. Carcinogenesis. 2013;34(3):513–521. doi: 10.1093/carcin/bgs363. [DOI] [PubMed] [Google Scholar]
- 129.Nawaz I, Moumad K, Martorelli D, et al. Detection of nasopharyngeal carcinoma in Morocco (North Africa) using a multiplex methylation-specific PCR biomarker assay. Clin. Epigenetics. 2015;7(1):89. doi: 10.1186/s13148-015-0119-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Karpinski P, Walter M, Szmida E, et al. Intermediate- and low-methylation epigenotypes do not correspond to CpG island methylator phenotype (low and -zero) in colorectal cancer. Cancer Epidemiol. Biomarkers Prev. 2013;22(2):201–208. doi: 10.1158/1055-9965.EPI-12-0157. [DOI] [PubMed] [Google Scholar]
- 131.Farkas SA, Milutin-Gasperov N, Grce M, Nilsson TK. Genome-wide DNA methylation assay reveals novel candidate biomarker genes in cervical cancer. Epigenetics. 2013;8(11):1213–1225. doi: 10.4161/epi.26346. [DOI] [PubMed] [Google Scholar]
- 132.Sova P, Feng Q, Geiss G, et al. Discovery of novel methylation biomarkers in cervical carcinoma by global demethylation and microarray analysis. Cancer Epidemiol. Biomarkers Prev. 2006;15(1):114–123. doi: 10.1158/1055-9965.EPI-05-0323. [DOI] [PubMed] [Google Scholar]
- 133.Chen CC, Lee KD, Pai MY, et al. Changes in DNA methylation are associated with the development of drug resistance in cervical cancer cells. Cancer Cell Int. 2015;15:98. doi: 10.1186/s12935-015-0248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ben W, Yang Y, Yuan J, et al. Human papillomavirus 16 E6 modulates the expression of host microRNAs in cervical cancer. Taiwan J. Obstet. Gynecol. 2015;54(4):364–370. doi: 10.1016/j.tjog.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 135.Karsli-Ceppioglu S, Ngollo M, Adjakly M, et al. Genome-wide DNA methylation modified by soy phytoestrogens: role for epigenetic therapeutics in prostate cancer? OMICS. 2015;19(4):209–219. doi: 10.1089/omi.2014.0142. [DOI] [PubMed] [Google Scholar]
- 136.Wong CP, Hsu A, Buchanan A, et al. Effects of sulforaphane and 3,3’-diindolylmethane on genome-wide promoter methylation in normal prostate epithelial cells and prostate cancer cells. PLoS ONE. 2014;9(1):e86787. doi: 10.1371/journal.pone.0086787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lochrin SE, Price DK, Figg WD. BET bromodomain inhibitors-a novel epigenetic approach in castration-resistant prostate cancer. Cancer Biol. Ther. 2014;15(12):1583–1585. doi: 10.4161/15384047.2014.962297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Luo JH, Ding Y, Chen R, et al. Genome-wide methylation analysis of prostate tissues reveals global methylation patterns of prostate cancer. Am. J. Pathol. 2013;182(6):2028–2036. doi: 10.1016/j.ajpath.2013.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Singh V, Singh LC, Vasudevan M, et al. Esophageal cancer epigenomics and integrome analysis of genome-wide methylation and expression in high risk northeast Indian population. OMICS. 2015;19(11):688–699. doi: 10.1089/omi.2015.0121. [DOI] [PubMed] [Google Scholar]; • Study showing population-specific epigenetic marks in specific cancer.
- 140.Gu F, Doderer MS, Huang YW, et al. CMS: a web-based system for visualization and analysis of genome-wide methylation data of human cancers. PLoS ONE. 2013;8(4):e60980. doi: 10.1371/journal.pone.0060980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rakyan VK, Down TA, Thorne NP, et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18(9):1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Johnatty S, Tyrer JP, Kar SP, et al. Genome-wide analysis identifies novel loci associated with ovarian cancer outcomes: findings from the Ovarian Cancer Association Consortium. Clin. Cancer. Res. 2015;21(23):5264–5276. doi: 10.1158/1078-0432.CCR-15-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Siggens L, Ekwall K. Epigenetics, chromatin and genome organization: recent advances from the ENCODE project. J. Intern. Med. 2014;276(3):201–214. doi: 10.1111/joim.12231. [DOI] [PubMed] [Google Scholar]; • Excellent resource of database and analytical tools.
- 144.De Hoon M, Shin JW, Carninci P. Paradigm shifts in genomics through the FANTOM projects. Mamm. Genome. 2015;26(9–10):391–402. doi: 10.1007/s00335-015-9593-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sobhani I, Tap J, Roudot-Thoraval F, et al. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE. 2011;6(1):e16393. doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chen CY, Chang IS, Hsiung CA, Wasserman WW. On the identification of potential regulatory variants within genome wide association candidate SNP sets. BMC Med. Genomics. 2014;7:34. doi: 10.1186/1755-8794-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lasseigne BN, Burwell TC, Patil MA, Absher DM, Brooks JD, Myers RM. DNA methylation profiling reveals novel diagnostic biomarkers in renal cell carcinoma. BMC Med. 2014;12:235. doi: 10.1186/s12916-014-0235-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.DNA methylation inhibitor triggers anti-viral immune response in cancer. http://cancergenome.nih.gov/researchhighlights/researchbriefs/antivirus
- 149.Chiappinelli KB, Strissel PL, Desrichard A, et al. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell. 2015;162(5):974–986. doi: 10.1016/j.cell.2015.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Shvetsov YB, Song MA, Cai Q, et al. Intraindividual variation and short-term temporal trend in DNA methylation of human blood. Cancer Epidemiol. Biomarkers Prev. 2015;24(3):490–497. doi: 10.1158/1055-9965.EPI-14-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ashley EA. The precision medicine initiative: a new national effort. JAMA. 2015;313(21):2119–2120. doi: 10.1001/jama.2015.3595. [DOI] [PubMed] [Google Scholar]; •• Implication of GWAS and epigenome-wide association studies in controlling cancer and other diseases will require longitudinal studies of a number of participants.
- 152.Barlas S. Precision medicine initiative aims for a new generation of diagnostics and treatments: but is the promise of genetic targeting overinflated? PT. 2015;40(5):340–352. [PMC free article] [PubMed] [Google Scholar]
- 153.Collins FS, Varmus H. A new initiative on precision medicine. N. Engl. J. Med. 2015;372(9):793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]