Alloatti et al. show that Sec22b-dependent antigen cross-presentation is critical to developing effective antitumor CD8+ T cell responses. Conditional deletion of Sec22b in dendritic cells decreases immune response against dead cells and promotes resistance to immunotherapy with anti–PD-1.
Abstract
CD8+ T cells mediate antigen-specific immune responses that can induce rejection of solid tumors. In this process, dendritic cells (DCs) are thought to take up tumor antigens, which are processed into peptides and loaded onto MHC-I molecules, a process called “cross-presentation.” Neither the actual contribution of cross-presentation to antitumor immune responses nor the intracellular pathways involved in vivo are clearly established because of the lack of experimental tools to manipulate this process. To develop such tools, we generated mice bearing a conditional DC-specific mutation in the sec22b gene, a critical regulator of endoplasmic reticulum–phagosome traffic required for cross-presentation. DCs from these mice show impaired cross-presentation ex vivo and defective cross-priming of CD8+ T cell responses in vivo. These mice are also defective for antitumor immune responses and are resistant to treatment with anti–PD-1. We conclude that Sec22b-dependent cross-presentation in DCs is required to initiate CD8+ T cell responses to dead cells and to induce effective antitumor immune responses during anti–PD-1 treatment in mice.
Introduction
DCs are a specialized population of immune cells that excel in antigen presentation and induce adaptive immune responses (Mellman and Steinman, 2001). Like other cells, DCs can present peptides derived from cytosolic antigens loaded on MHC class I to CD8+ T cells and to both endogenous and exogenous antigens bound to MHC class II molecules for recognition by CD4+ T cells. In addition, DCs can take up exogenous antigens and process and load them onto MHC class I molecules to be presented to CD8+ T cells, a process called antigen “cross-presentation” (the resulting induction of a CD8+ T cell response is referred to as “cross-priming”; Joffre et al., 2012).
Several pathways of antigen cross-presentation that involve membrane trafficking through different intracellular compartments were reported in cultured DCs (Savina et al., 2006, 2009; Jancic et al., 2007; Cebrian et al., 2011; Nair-Gupta et al., 2014; Alloatti et al., 2015). One of the described cross-presentation pathways requires transfer of ER resident proteins, including the machinery for MHC class I loading with peptides (TAP1/2 transporters, tapasin, calreticulin, etc.), to the endocytic and phagocytic pathways, a traffic step controlled by the SNARE family member Sec22b (Cebrian et al., 2011).
The actual contribution of different antigen cross-presentation pathways to immune responses in vivo remains unclear. The K. Murphy group (Hildner et al., 2008) has shown that certain subsets of cross-presenting DCs (i.e., Batf3-dependent DCs) have a critical role in antiviral immune responses and in the rejection of established solid tumors by CD8+ T cells. Recently, the R. Germain group (Castellino et al., 2006; Eickhoff et al., 2015) showed that CD8+ DCs act as “cellular platforms” to support CD4+ T cell help for CD8+ responses, a role that goes beyond their cross-presentation capacities. In contrast, increasing examples of CD8− DCs cross-presenting antigen in vivo are being reported (den Haan et al., 2000; Kamphorst et al., 2010). The actual contribution of antigen cross-presentation by DCs to specific immune responses is, therefore, a critical unknown.
This is particularly true in the context of immunotherapies that attempt to harness the immune system to treat cancer, including those using checkpoint inhibitors. Expression of programmed cell death protein-1 (PD-1) on the surface of tumor-specific lymphocytes, and interaction with its corresponding ligands (PD-L1 and PD-L2, respectively) on the tumor- or antigen-presenting target cells is a key immune checkpoint that inhibits T cell function. Seminal studies in mouse models of cancer and diverse clinical studies have established that mAbs blocking the PD-1/PD-L1 pathway, as well as other checkpoints, such as CTLA-4, can unleash the immune system to fight cancer (Leach et al., 1996; Iwai et al., 2002). These therapies can mediate tumor regression in patients with metastatic melanoma, non–small cell lung cancer and renal cell carcinoma, among others (Hodi et al., 2010; Topalian et al., 2012; Lebbé et al., 2014). In mice, anti-immune, checkpoint-based treatments have been analyzed with success in several tumor models. The Melero laboratory (Sánchez-Paulete et al., 2016) has shown recently that Batf3-dependent DCs actively contribute to rejection of tumors during anti–PD-1 and anti-CD137 immunotherapies.
To define the contribution of antigen cross-presentation to CD8+ T cell responses, we generated a mouse line in which the expression of Sec22b was conditionally depleted in DCs. Reduced Sec22b expression in DCs impairs antigen cross-presentation and cross-priming of cell-associated antigens in vivo. Sec22b-defective mice also failed to mount effective antitumor immune responses, to control the growth of immunogenic tumors, and to respond to anti–PD-1–based immunotherapy. These results show that Sec22b-dependent antigen cross-presentation is required during cross-priming of CD8+ T cell responses with dead cell–derived antigens and for anti–checkpoint-tumor immunotherapy in mice.
Results and discussion
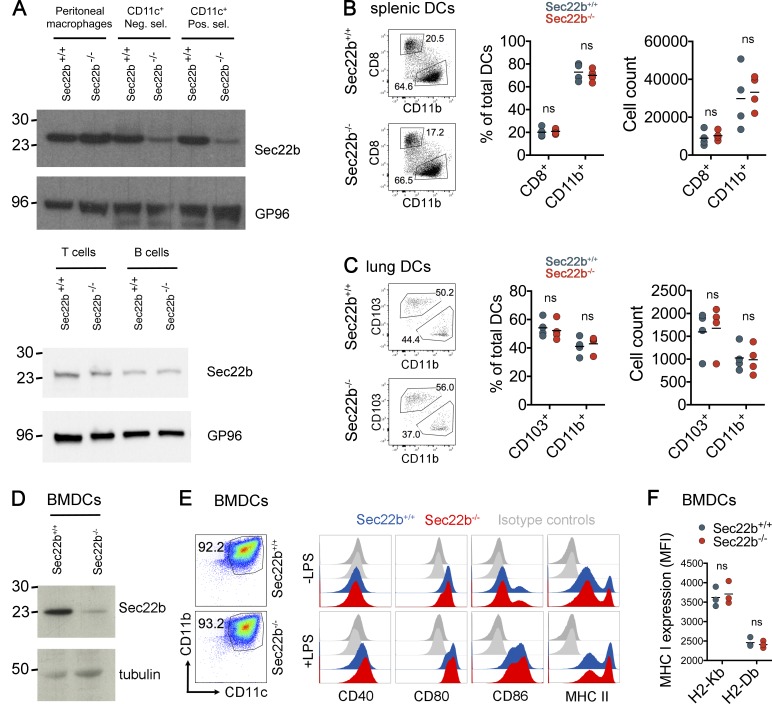
To investigate the role of Sec22b-dependent cross-presentation in vivo, we generated floxed sec22b knock-in mice and crossed them to CD11c-specific Cre-deleter mice (Caton et al., 2007). We thus obtained mice bearing a selective deletion of the sec22b gene in DCs (Sec22b−/−). As controls, throughout the study, we used littermates expressing the Cre recombinase, and WT alleles of the sec22b gene (Sec22b+/+). Western blot analysis of splenic CD11c+ cells isolated by two rounds of selection (negative and then positive) confirmed that Sec22b expression was reduced in primary DCs purified from Sec22b−/−, but not Sec22b+/+ mice (Fig. 1 A, top). Sec22b expression in peritoneal macrophages (Fig. 1, top), as well as in splenic B and T cells (Fig. 1 A, bottom) was not affected, confirming that Sec22b−/− mice bear a conditional defect in Sec22b expression in DCs.
Figure 1.
Phenotypic analysis of Sec22b−/− mice. (A) Western Blotting of Sec22b expression in peritoneal macrophages and CD11c+ splenic DCs (top: purified by negative (neg. sel.) and positive (pos. sel.) selection for CD11c+ cells), as well as B and T cells (bottom), isolated from Sec22b+/+ and Sec22b−/− mice. Shown is one representative experiment of three independent experiments. (B) Phenotypic characterization of DC subsets in spleens and lungs from Sec22b−/− and Sec22b+/+ mice. Gating strategy are shown for splenic DCs (left), percentages of CD8+ and CD11b+ DCs subpopulations in spleen (middle), and cell numbers (right). Data shown are means of four independent biological replicates. DC populations were analyzed by gating on CD11chigh, MHC class IIhigh/mid and CD8/CD11b. (C) Gating strategy are shown for lung DCs (left), percentages of CD103+ and CD11b+ DCs subpopulations in spleen (middle), and cell counting (right). Data shown are means of four independent biological replicates and each value. DC populations were analyzed by gating on CD11chigh, MHC class IIhigh, then in Siglec Flow (to discriminate from alveolar macrophages), and finally in CD103/CD11b. (D) Phenotypic analysis of BMDCs. Western blotting analysis of Sec22b expression in BMDCs generated from Sec22b+/+ and Sec22b−/− mice. (E) Shown are percentages of CD11chighCD11bhigh cells (left) and expression of the costimulatory molecules CD40, CD80, CD86, and MHC class II upon TLR4 engagement with LPS (right). (F) Expression of MHC class I H2-Kb and Db. For all of the BMDCs results, shown are the pooled data from at least three independent experiments (with the exception of LPS histograms showing one representative experiment of four independent experiments). All results in this figure were analyzed by two-way ANOVA with Bonferroni’s multiple comparisons test.
Phenotypic analysis of DC subsets in spleen (Fig. 1 B) and lungs (Fig. 1 C) from Sec22b−/− and Sec22b+/+ mice did not show any significant differences, neither in the composition of cell subpopulations nor in total cells numbers. Neither the percentages nor the numbers of other immune cell subpopulations from spleen, thymus, blood, lungs, and different lymph nodes were affected in Sec22b-defective mice (Fig. S1, A–F). Sec22b depletion did not modify the capacity of splenic CD8+ DCs to respond to different stimuli, such as LPS, IFN-γ, and TNF, as measured by expression of MHC II and CD86 (Fig. S1 G). We also confirmed by Western blot that Sec22b expression was reduced in BMDCs generated from Sec22b−/− mice, as compared with littermates (Fig. 1 D). Sec22b depletion did not affect the percentages of CD11chigh CD11bhigh cells in culture (Fig. 1 E, left) or LPS-mediated activation, as detected by expression of costimulatory molecules, (CD40, CD80, and CD86 and MHC class II; Fig. 1 E, right). IFN-γ and TNF induced an equivalent augment in the expression of CD86 and MHC II in both Sec22b+/+ and Sec22b−/− mice (Fig. S1 H). Finally, levels of MHC class I expression (both H2-Kb and H2-Db) were not affected by Sec22b depletion (Fig. 1 F). Therefore, defective Sec22b expression in DCs does not affect their development, survival, or activation capacity in vitro or in vivo.
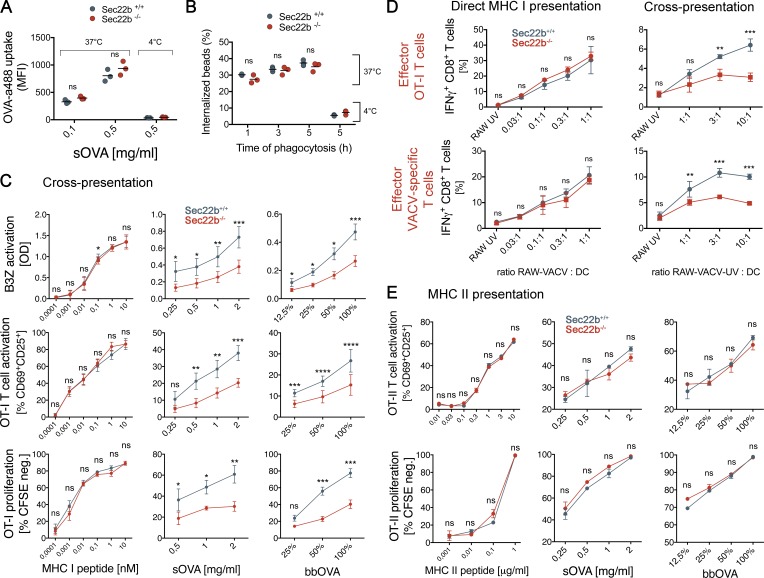
As a first analysis pertaining to DC function, we assessed the phagocytic and endocytic capacities of BMDCs. Both endocytosis and phagocytosis were similar in bone marrow-derived DCs (BMDCs) generated from Sec22b+/+ or Sec22b−/− mice (Fig. 2, A and B, respectively). To investigate whether Sec22b-defective DCs bear a defect in antigen cross-presentation, we first generated BMDCs from Sec22b−/− and control littermates. As shown in Fig. 2 C, BMDCs from Sec22b-deficient mice bear a partial defect in cross-presentation of soluble and bead-bound OVA, as detected by B3Z activation (top; Kurts et al., 1996), CD69 and CD25 expression (middle), and proliferation of OT-I CD8+ T cells, measured by dilution of carboxyfluorescein succinimidyl ester (CFSE; bottom). The synthetic MHC class I–restricted OVA peptide (SIINFEKL) was presented with equal efficacy by BMDCs generated from Sec22b−/− and Sec22b+/+ mice (Fig. 2 C, left).
Figure 2.
In vitro functional analysis of BMDCs generated from Sec22b−/− mice. Analysis of OVA–Alexa Fluor488 endocytosis (A) and bead-bound OVA phagocytosis by BMDCs (B); MFI, mean fluorescence intensity. Data shown are means of three independent biological replicates and each value. The results were analyzed by paired t test (P > 0.05). (C) Antigen cross-presentation capacity of BMDCs generated from Sec22b+/+ and Sec22−/− mice. (top) B3Z hybridoma T cell activation for (from left to right) MHC I–restricted peptides, soluble OVA, and bead-bound OVA. (Middle) OT-I T cell activation measured by CD69 and CD25 expression. (Bottom) OT-I T cell proliferation followed by dilution of CFSE dye. Shown are the pooled data of at least three independent experiments, with each experiment measured in triplicate. Results were analyzed by two-way ANOVA with Bonferroni’s multiple comparisons test for statistical significance. Shown are the means ± SEM. (D) Direct MHC class I presentation (left) and cross-presentation (right) capacity of BMDCs generated from Sec22b+/+ and Sec22b−/− mice, measured by IFN-γ production of CD44+ CD8+ effector T cells obtained from OT-I mice (top) and VACV WT-immunized mice (bottom) after co-culture for 4 h. Shown are the pooled data of three independent experiments. Results were analyzed by two-way ANOVA with Bonferroni’s multiple comparisons for statistical significance. (E) MHC class II antigen presentation efficacy for BMDCs obtained from Sec22b+/+ and Sec22b−/− mice, analyzed by OT-II T cell activation (top) and proliferation (bottom). Shown are the pooled data of three independent experiments, with each experiment analyzed in triplicate. Results were analyzed by two-way ANOVA as stated in C for statistical significance. For all analyses, *, 0.01 < P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
In addition, as an alternative source of antigen, we used vaccinia virus (VACV)–OVA–infected RAW macrophages (H2-Kd; RAW-VACV), which transmit virus and infect DCs that can direct present antigens in H2-Kb MHC class I molecules. Alternatively, infected RAW cells were treated with UV light (RAW-VACV-UV) to inactivate the virus, blocking direct infection of DCs and leaving available only the cross-presentation route, as previously described (Iborra et al., 2012). We also used uninfected RAW cells treated with UV (RAW-UV) to test antigen specificity. BMDCs generated from Sec22b+/+ and Sec22b−/− mice were exposed to VACV-infected or control cells for 4 h and used to stimulate preactivated OT-I T cells or CD8+ T cells purified from WT mice previously infected with VACV WT. Production of IFN-γ by OT-I (Fig. 2 D, top) or vaccinia-specific effector CD8+ T cells (Fig. 2 D, bottom) was not affected by Sec22b-depletion in DCs directly infected with the virus (Fig. 2 D, left) but was impaired in DCs co-cultured with RAW-VACV-UV (Fig. 2 D, right). Moreover, MHC class II–restricted antigen presentation, as measured by activation and proliferation of OT-II CD4+ OVA-specific TCR transgenic T cells, was not affected in Sec22b-deficient BMDCs (Fig. 2 E). Therefore, in agreement with our previous results in Sec22b knockdown BMDCs (Cebrian et al., 2011), BMDCs generated from Sec22b-defective mice display impaired cross-presentation, but conventional MHC class I and II antigen presentation is not affected.
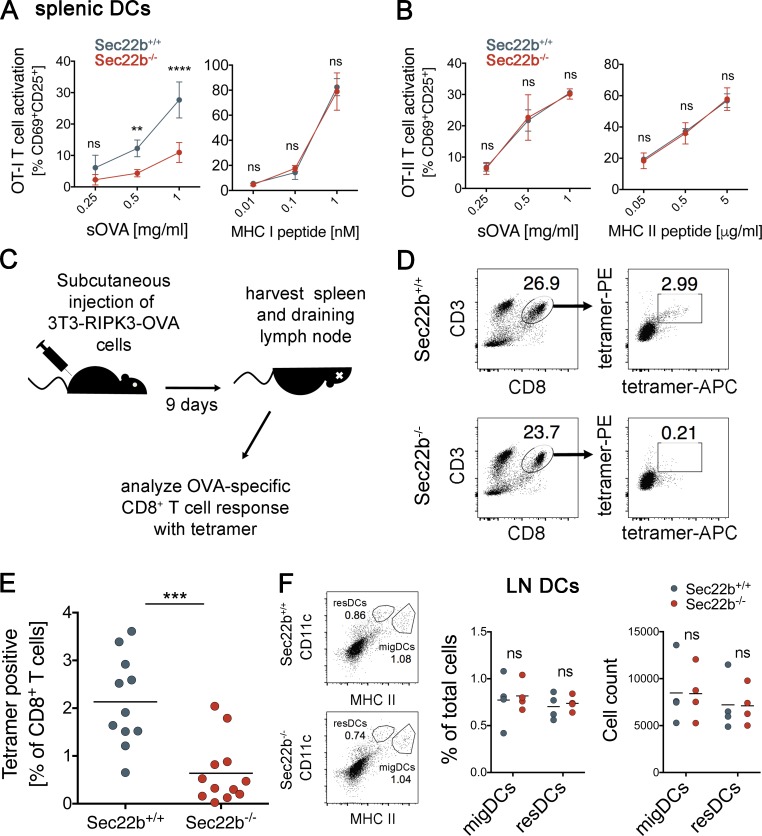
Similar to BMDCs, CD11c+ splenic DCs isolated from Sec22b−/− mice also cross-present OVA to CD8+ T cells less efficiently than do those purified from littermates (Fig. 3 A), whereas MHC class II–restricted presentation to CD4+ T cells is unaffected (Fig. 3 B). To investigate the role of Sec22b depletion in DCs in cross-priming, we first tested whether CD8+ T cells responses were normal in Sec22b−/− mice. We immunized Sec22b+/+ and Sec22b−/− mice with IFA-CpG-SIINFEKL in the footpad and analyzed anti-OVA and anti-SIINFEKL responses by IFN-γ ELISPOT in draining popliteal and inguinal lymph nodes. CD8+ T cells from Sec22b+/+ and Sec22b−/− were equally restimulated with SIINFEKL (Fig. S2 A), whereas the restimulation with OVA was impaired in Sec22b−/− mice. These results indicate that Sec22b−/− mice have a functional CD8+ T cells compartment and that the SIINFEKL-specific repertoire is normal.
Figure 3.
Relevance of antigen cross-presentation to elicit CD8+ T cell responses against dead cell-derived antigen in vivo. Antigen cross-presentation (A) and MHC II presentation by splenic DCs (B) isolated from Sec22b+/+ and Sec22b−/− mice, as well as their peptide controls (right). Shown are the means ± SEM of three independent experiments. Statistical analysis was performed using two-way ANOVA with Bonferroni’s multiple comparisons test. **, P < 0.01; ****, P < 0.0001. (C) Schematic representation of the protocol for the in vivo experiments with necroptotic 3T3-RIPK3-OVA cells. (D) Gating strategy to measure endogenous CD8+ T cell responses with OVA-specific tetramers. (E) Analysis of endogenous OVA-specific CD8+ T cell responses generated against necroptotic 3T3-RIPK3-OVA cells in Sec22b+/+ and Sec22b−/− mice; shown are pooled data from three independent experiments (n = 11–12). ***, P = 0.0001 for the Welch’s t test used herein. (F) Gating strategy for lymph nodes (LN) DCs (left), percentages of migratory DCs (migDCs) and resident DCs (resDCs) subpopulations in LN (middle), and cell counting (right). Data shown are means of four independent biological replicates. DC populations were analyzed by gating on CD11chigh, MHC class IIhigh/mid, and finally in CD103/CD11b. sOVA, soluble OVA.
To determine whether the defect in antigen cross-presentation in Sec22b-depleted DCs results in defective cross-priming in vivo, we injected necroptotic, OVA-expressing 3T3-RIPK3-OVA cells (Yatim et al., 2015) subcutaneously. The endogenous CD8+ T cell response against OVA was analyzed using MHC-I multimers (Yatim et al., 2015) after 9 d (Fig. 3 C). In Sec22b−/− mice, the endogenous cytotoxic CD8+ T cell response was significantly decreased from 2.134 ± 0.2835 to 0.6381 ± 0.1887%, as compared with control littermates (Fig. 3 D for gating strategy; Fig. 3 E). The proportion of migratory and resident DCs in lymph nodes was not affected by Sec22 depletion (Fig. 3 F, left, for gating; middle, for percentages; right, for cell numbers), indicating that the differences in cross-priming capacity are not the consequence of impaired migration capacity of Sec22b−/− DCs from the periphery. In addition, the number and percentages of the putative cross-presenting subsets CD8αα+ resident DCs and CD103+ migratory DCs were not modified by depletion of Sec22b (Fig. S2, B and C). We conclude that cross-priming of dead cell–associated antigens in vivo requires Sec22b expression in DCs.
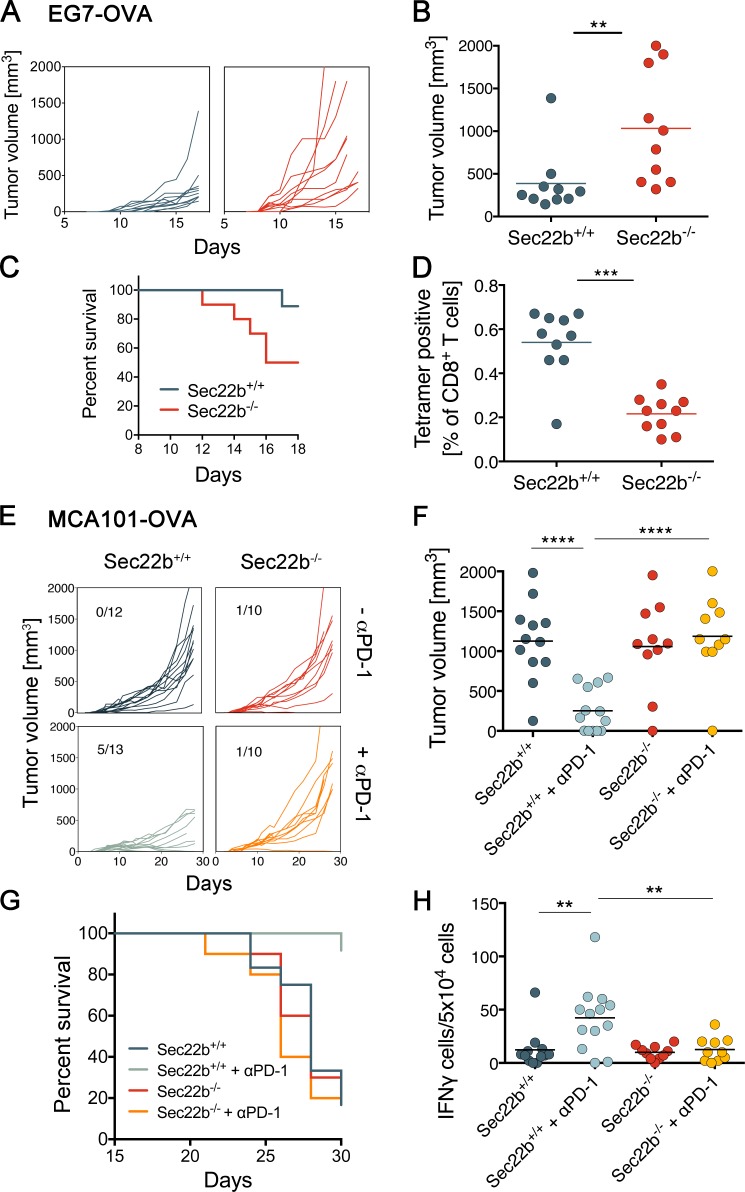
The results presented thus far indicate that Sec22b−/− mice represent a suitable model to investigate the contribution of antigen cross-presentation to immune responses. To evaluate the role of cross-presentation in antitumor immunity, we first analyzed the growth of a highly immunogenic OVA-expressing tumor cell line, EG7 (Moore et al., 1988; Helmich and Dutton, 2001). EG7 tumors grew faster in Sec22b−/− mice (Fig. 4, A and B), causing decreased survival (Fig. 4 C), as compared with littermates. That insufficiency to control tumors correlated with impaired OVA-specific CD8+ T cell responses (Fig. 4 D and Fig. S3 A). These results suggest that impaired cross-presentation of tumor antigens by Sec22-deficient DCs causes reduced antitumor immune responses and exacerbated tumor growth.
Figure 4.
Relevance of antigen cross-presentation to elicit CD8+ T cell responses against tumor-derived antigens in vivo. (A) Tumor growth curves for EG7-OVA injected subcutaneously in Sec22b+/+ (n = 10) and Sec22b−/− (n = 10) mice. (B) Tumor volumes in mm3 (on the day of sacrifice; see Materials and methods for sacrifice criteria). (C) Survival curves for age- and sex-matched Sec22b+/+ and Sec22b−/− mice injected with the EG7-OVA tumor cell line. (D) Endogenous CD8+ T cell response measured with OVA-specific tetramers in total blood cells on d 10 after tumor injection. (E) Tumor growth curves for the tumor cell line MCA101-OVA in Sec22b+/+ and Sec22b−/− mice treated or not treated with anti–PD-1 (αPD-1; n = 10–13). Pooled results from three independent experiments are shown. Statistical significance was analyzed by Welch’s t tests. Numbers refer to rejected tumors out of total mice analyzed. (F) Tumor sizes (on the day of sacrifice). (G) Survival of age- and sex-matched Sec22b+/+ and Sec22b−/− mice injected with the MCA101-OVA–expressing tumor cell line, treated or not treated with αPD-1. (H) Blood cells from Sec22b+/+ and Sec22b−/− mice were restimulated ex vivo with OVA MHC class I–restricted peptide or with nonrelated protein human serum albumin as control. The percentage of OVA-specific T cells producing IFN-γ per 5 × 104 blood cells was determined by ELISPOT analysis. **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 for Welch’s t tests (B, D, F, and H) or log-rank test (C and G). Data in B, D, F, and H are from three independent experiments with at least three independent biological replicates (means ± SEM). Data in A, C, E, and G are pooled from three independent experiment.
To address the possible role of Sec22b-dependent antigen cross-presentation in tumor immunotherapy by checkpoint blocking antibodies, we used a less-immunogenic tumor, OVA-secreting MCA-101 (Zeelenberg et al., 2008; Sedlik et al., 2014). This tumor is well controlled by antibodies against CTLA-4 or PD-1 (Gubin et al., 2014). Sec22b−/− and Sec22b+/+ mice were injected with tumor cells and received or did not receive anti–PD-1 treatment, as described previously (Gubin et al., 2014). As expected, treatment with anti–PD-1 induced tumor rejection in Sec22b+/+ littermates (Fig. 4 E). In contrast, Sec22b−/− mice failed to respond to anti–PD-1 treatment, both in terms of tumor growth and survival (Fig. 4, F and G). Consistently, treatment with anti–PD-1 induced an anti-OVA immune response in littermates, but not in Sec22b−/− mice, when analyzed by T cell restimulation and subsequent secretion of IFN-γ (Fig. 4 H and Fig. S3 B).
To further investigate the mechanism of resistance to anti–PD-1, we measured IFN-γ production and proliferation capacity of CD8+ T cells isolated from spleen (Fig. S3 C) and tumor (Fig. S3 D) of mice bearing MCA101-OVA tumors and treated with anti–PD-1. CD8+ T cells from Sec22b−/− produce IFN-γ and proliferate to the same extent as CD8+ T cells from control mice. Expression of PD-1 in T cells from Sec22b+/+ and Sec22b−/− was similar (Fig. S3 E). The expression of PD-L1 and PD-L2 in CD8+ and CD8−/CD11b+ DCs from spleen was not reduced in Sec22b-deficient DCs (Fig. S3 F). These results show a critical, nonredundant role for Sec22b-dependent antigen cross-presentation in the onset of efficient antitumor immune responses induced by checkpoint blockade.
Although cross-priming of allogeneic antigens in vivo was reported >30 yr ago (Bevan, 1976), the actual contribution of antigen cross-presentation (vs. other mechanisms such as cross-dressing (Wakim and Bevan, 2011; Yewdell and Dolan, 2011) or Gap junction–mediated peptide transfer (Neijssen et al., 2005) to cross-priming is still unclear. In the case of cross-presentation, multiple intracellular pathways have been reported in vitro. Each of those pathways is under the control of master regulators that control key intracellular traffic steps specific for each pathway. Here, we developed tools to perform loss of function experiments for Sec22b-dependent antigen cross-presentation in vivo. Our results show that Sec22b-dependent antigen cross-presentation by DCs has a critical role in the cross-priming of cell-associated antigens and antitumor immune responses in vivo (even though we did not exhaustively investigate all of the cell populations that might express CD11c, notably activated B cells that can help priming CD8+ T cell responses). It is possible that alternative cross-presentation pathways independent of Sec22b exist, but their contribution to the priming of CD8+ T cell responses in vivo needs to be addressed. Our results show that the induction of tumor-specific CD8+ T cells by anti–PD-1 requires Sec22b expression in DCs, suggesting that, in addition to acting on effector cytotoxic T cells by releasing tumor-imposed immunosuppression, anti–PD-1 may also facilitate the priming of CD8+ T cells. Whether Sec22b turns out to be a suitable target to modulate antigen cross-presentation therapeutically will need to be addressed in future studies.
Materials and methods
Compounds and antibodies
For flow cytometry, the following antibodies were used: anti–CD69-eFluor450 (clone H1.2F3, 48-0691-82; eBioscience), anti–CD25-FITC (clone 7D4, 553072; BD), anti–CD8a-PerCP-Cy5.5 (clone 53–6.7, 45-0081-82; eBioscience), anti–TCR vβ 5.1-PE (clone MR9-4, 553190; BD), anti–CD4-PE-Cy7 (clone RM4-5, 552775; BD), anti–CD19-eFluor450 (clone 1D3, 48-0193; eBioscience), anti–CD3-eFluor450 (clone 17A2, 48-0032-80; eBioscience), anti–CD11c-FITC (clone HL3, 553801; BD), anti–CD11b-PerCPCyanine5.5 (clone M1/70, eBioscience #45-0112-80), anti-F4/80-FITC (clone BM8, eBioscience #11-4801-81), anti-NKp46-PE (clone 29A1.4, 12-3351-80; eBioscience), anti–MHC I (H-2Kb)-FITC (clone AF6-88.5.5.3, 11-5958-80; eBioscience), anti–MHC II (Iab)-eFluor450 (clone AF120.1, 48-5320-80; eBioscience). For Western blot analysis, the anti-Sec22b antibody was purchased from Santa Cruz Biotechnology, Inc. (29-F7, sc-101267), anti-α-actin from Sigma-Aldrich (A2066), and anti-gp96 from Enzo Life Sciences (spa850). For phenotypic analysis of immune cell populations CD4, CD8, CD25, CD44, CD62L, TCRb, and CD3 were used as T cell markers (in different fluorophores, depending on the tissue analyzed and the panel of antibodies used). B220 and CD19 were used as B cell markers. TCR-γδ was used as the γδ T cell marker. NK1.1 and NKp46 were used as natural killer markers. Gr-1 and Ly6G were used as granulocyte/neutrophil markers. CD11c was generally used as a DC marker, with the exception of lungs: alveolar macrophages (CD11chigh) were discriminated from DCs by analyzing Siglec F expression. CD103, I-Ab, H2-Kb, CD11b, CD8, CD40, and CD86 were used to analyze DCs phenotype (in different fluorophores). F4/80 and Ly6C were used as macrophage markers. As for the direct MHC I antigen-presentation analysis, anti-mouse antibodies to CD8α, CD44, and IFN-γ were used as conjugates to PE, FITC, and APC, and were obtained from eBioscience. Anti–PD-L1-PE and anti–PD-L2-APC were purchased from eBioscience.
Cell lines and cell culture
RAW264.7 macrophages (RAW) were grown in DMEM supplemented with 5 mM glutamine, penicillin, streptomycin, β-mercaptoethanol (all from Invitrogen) and 10% heat-inactivated FBS (Sigma-Aldrich). CD8+ T cells were negatively selected using a cocktail of biotin-conjugated antibodies (anti-CD11c, B220, MHC-II, CD4, NK1.1), followed by incubation with streptavidin microbeads (Miltenyi Biotec). Typical yields by FACS staining were >95% pure. Preactivated OT-I cells were obtained by culturing spleen cells from OT-I transgenic mice (C57BL/6-Tg(TcraTcrb)1100Mjb/J) with 10−9 M of the peptide 257SIINFEKL264 from OVA for 5 d. At that time, ∼90% of the cells were CD8+CD44hi. The 3T3-RIPK3-OVA cells, expressing a nonsecretable form of OVA cells, were obtained from the Matthew Albert laboratory (Yatim et al., 2015) and were cultured in DMEM (Thermo Fisher Scientific) supplemented with 10% FBS (Biowest), 0.1 mM nonessential aa, 1 mM sodium pyruvate, 10 mM Hepes, and 50 µM β-mercaptoethanol (all from Thermo Fisher Scientific). Necroptosis was induced by treatment with a specific drug ligand (AP20187, BB homodimerizer; Takara Bio Inc.). EG7 tumor cells were cultured in DMEM supplemented with 10% FBS (Lonza), as well as 100 IU/ml penicillin, 100 µg/ml streptomycin, 2 mM GlutaMAX, and 50 µM β-mercaptoethanol (all from Thermo Fisher Scientific). MCA-101 OVA-secreting cells were grown in RPMI-1640 containing 10% heat-inactivated FBS (Biowest), 100 IU/ml penicillin, 100 µg/ml streptomycin, 2 mM GlutaMAX, and 50 µM β-mercaptoethanol (all from Thermo Fisher Scientific), as well as hygromycin B 1 mg/ml (Gibco) for selection. All cell lines were tested as mycoplasma-negative by PCR.
DC activation
Maturation of BMDCs was induced by a 16-h treatment with 100 ng/ml of ultrapure LPS from Escherichia coli 0111:B4 (InvivoGen) or, alternatively, with 100 ng/ml of TNF (Protein Service Facility, Inflammation Research Center) or IFN-γ (Invitrogen). DC maturation was controlled by cell-surface expression of costimulatory molecules and MHC class II molecules using specific antibodies. Maturation of splenic CD11c+ DCs was induced by a 4-h treatment with 100 ng/ml of LPS, TNF, or IFN-γ.
Virus strains
The rVACV-OVA and VACV WR strains were gifts from Jonathan W. Yewdell and Jack R. Bennink (National Institutes of Health, Bethesda, MD). Stocks were grown in CV-1 monolayers and used as clarified sonicated cell extracts.
Primary cell isolation and culture
BMDCs were produced by culturing the cells for 10 d in GM-CSF–containing medium (as described in Alloatti et al., 2015), with IMDM (Sigma-Aldrich and Thermo Fisher Scientific) containing 10% heat-inactivated FBS (Biowest), 100 IU/ml penicillin, 100 µg/ml streptomycin, 2 mM GlutaMAX, and 50 µM β-mercaptoethanol (all from Thermo Fisher Scientific). Supernatant from J558 plasmacytoma cells was used as the granulocyte macrophage colony-stimulating factor source (Winzler et al., 1997). To obtain splenic CD11c+ DCs, each mouse spleen was injected with 2 ml of a digestion solution with 0.1 mg/ml Liberase (Roche), 0.1 mg/ml DNase I (Roche), 1× PenStrep in RPMI-1640 (both from Thermo Fisher Scientific), following the Roche instructions, and was cut into small pieces and incubated for 25 min at 37°C. Reaction was stopped by addition of complete medium. Spleens were passed through a cell strainer and centrifuged at 200× g for 5 min. After red blood cell lysis (Sigma-Aldrich), splenic DCs were isolated by performing one round of negative selection for CD11c+ cells (using EasySep Mouse Pan-DC enrichment kit from STEMCELL Technologies), followed by a round of positive selection using CD11c microbeads (Miltenyi Biotec), according to the manufacturer’s instructions. In all experiments, purity of cells was >90%. For phenotypic characterization of immune cells, thymus and lungs were processed by digestion with Liberase/DNase I as stated above and was subsequently analyzed by flow cytometry.
Animals
C57BL/6J mice and C57BL/6J recombination activating gene 1-deficient OT-I and OT-II TCR (Vα2, Vβ5.1) transgenic mice were obtained from Charles River, Janvier, and Centre de Distribution, Typage et Archive Animal. Mice were between 5 and 12 wk old. WT C57BL/6J mice for tumor experiments were always obtained from the aforementioned sources.
Sec22bFlox/Flox (see below) and control mice were originally produced at the Centre d’Immunologie de Marseille (the Malissen group) and were subsequently bred in the animal facility of Institut Curie. CD11c-Cre transgenic mice were obtained from The Jackson Laboratory. All animal procedures were in accordance with the guidelines and regulations of the Institut Curie veterinary department, and all mice used were <6 mo old.
Targeting strategy for generation of Sec22b mice
A genomic fragment encompassing exon 2 and 4 of the sec22b gene was isolated from a BAC clone of B6 origin. Using ET recombination, loxP sites were introduced in the introns flanking the 5′ and 3′ ends of exon 2. The loxP site in the intron found at the 3′ end of exon 2 was abutted to a ftr-neoR-frt cassette. The final targeting construct was abutted to a cassette coding for thymidine kinase and linearized with Pme1.
ES clone selection
JM8.F6 C57BL/6N ES cells (Pettitt et al., 2009) were electroporated with the targeting vector. After selection in G418 and ganciclovir, ES cell clones were screened for proper homologous recombination by Southern blot and PCR analysis. A neomycin-specific probe was used to ensure that adventitious, nonhomologous recombination events had not occurred in the selected ES clones. Properly recombined ES cells were injected into FVB blastocysts. Upon germline transmission, mice were then crossed to flipper mice to delete the frt-flanked neoR cassette, and the resulting floxed Sec22b allele (official name B6-Sec22btm1Ciphe, called here Sec22bfl was identified by PCR of tail DNA. The pair of primers: sense 5′-ATGGTAAAAAGCACACCAAATACTTTGC-3′ and antisense 5′-TGAGGTAACCTTGAAGGCTAGAAGA-3′ amplified a 600-bp band in the case of the Sec22bWT allele and a 900-bp band in the case of the Sec22bfl allele. The excised Sec22bfl allele generated a band of 300 bp (animals were screened for breeding and experiments based on the presence of this 300-bp band). Considering that the phase of the introns flanking exon 2 is asymmetrical, deletion of exon 2 results in an out-of-frame Sec22b allele.
Western blotting
Total cell lysates from BMDCs and splenic DCs were subjected to 4–12% gradient gels (Thermo Fisher Scientific) or 10% and 12% gels and were separated by SDS-PAGE. After transfer to nitrocellulose membranes, they were blocked and incubated with primary antibodies and peroxidase-conjugated secondary antibodies. Bound antibodies were revealed using the BM chemiluminescence blotting substrate (POD) from Roche or GE Healthcare, according to the manufacturers’ directions. The intensity of the bands was quantified by densitometry using Quantity One 4.6.6 software (Bio-Rad Laboratories) and was expressed as arbitrary units.
Antigen uptake assay
Phagocytosis and the endocytic capacity of BMDCs were assessed using 3-µm blue latex beads (Polysciences) and OVA Alexa Fluor 488 conjugate (O34781; Thermo Fisher Scientific), respectively.
Viral infections and virus titration.
Mice were infected intradermally in the ears with 5 × 104 PFU of the required VACV WR strain, as previously described (Iborra et al., 2012).
Antigen presentation assays
Proliferation assay
DCs were incubated with Low Endo soluble OVA from Worthington Biochemical Corporation (LS003061) or with 3-µm beads coated with different ratios of OVA and BSA proteins (OVA 10 mg/ml alone; OVA 2.5 mg/ml–BSA 7.5 mg/ml; OVA 5 mg/ml–BSA 5 mg/ml; and BSA 10 mg/ml alone) or different concentrations of the control minimal peptide (OVA SIINFEKL for cross-presentation and OVA323–339 for MHC II presentation). After 5 h, DCs were washed three times with PBS containing 0.1% (vol/vol) BSA and co-cultured with purified CFSE-OT-I CD8+ for 3 d. For monitoring T cell proliferation, diminution of CFSE staining on TCR+ CD8+ populations was measured by flow cytometry.
T cell activation assay
DCs were incubated for 5 h with different concentrations of Low Endo soluble OVA from Worthington Biochemical Corporation (LS003061) or with 3-µm beads coated with different ratios of OVA and BSA proteins (as mentioned above). Minimal peptide OVA257–264 was used as a control for the capacity of DCs to activate T cells. Next, DCs were washed three times with 0.1% (vol/vol) PBS/BSA, fixed with 0.008% (vol/vol) glutaraldehyde during 10 min at 4°C, washed twice with 0.2 M glycine and once with 0.1% (vol/vol) PBS/BSA, and finally, B3Z hybrid T cells were added. After 16 h, T cell activation was measured by detecting β-galactosidase activity by OD at 590 nm using chlorophenol red-β-d-galactopyranoside as the substrate for the reaction. The efficiency of antigen presentation on MHC I and MHC II was also evaluated using OT-I (CD8+) and OT-II (CD4+ T) cells, respectively. Where indicated, DCs were fixed with 0.008% of glutaraldehyde before T cell addition. The percentage of CD25+CD69+ T cells was measured by flow cytometry after 16 h of co-culture (MHC I and II–restricted peptides, OVA257–264 and OVA323–339, respectively, were used as controls).
Direct MHC class I antigen presentation and cross-presentation assay with VACV
DCs were stimulated by co-culture with VACV-OVA–infected RAW cells treated with or without UV irradiation to inactivate the virus. To test DC cross-presenting ability, RAW cells were irradiated with UVC (240 mJ/cm2) either without exposure to VACV-OVA (RAW UV) or after incubation with VACV for 4 h (RAW-VACV-UV). Alternatively, infected RAW cells were left unirradiated (RAW-VACV) to allow direct infection of DCs. 16 h after UV irradiation, RAW cells were co-cultured for 4 h with BMDCs generated from Sec22b+/+ and Sec22b−/− mice. To the co-cultures, we then added CD8+ T cells purified from splenocytes of mice intradermally injected 7 d earlier with WR VACV, or preactivated OT-I cells were added to the cultures for 6 h; then, Brefeldin A (5 µg/ml; Sigma-Aldrich) was added for the last 4 h of culture. Cells were then stained with PE-anti-CD8α and FITC-CD44, fixed in 4% PFA, and incubated with APC-anti-IFN-γ during permeabilization with 0.1% saponin. A mean of 10,000 of each T cell subset was analyzed in each sample. Background activation obtained with CD8+ T cells nonpulsed with any peptide (0–0.3%) was subtracted.
In vivo cross-priming assay with 3T3-RIPK3-OVA necroptotic cells
The cross-priming assay was performed as described in (Yatim et al., 2015). In brief, 3T3-RIPK3-OVA cells were harvested and resuspended in complete media at 5 × 106 cell/ml. Dimerizer was added at 1 µM, and cells were incubated for 10 min at 37°C, gently flicking the tube every 2 min. Ice-cold PBS was added, and cells were washed, counted, and resuspended in cold PBS at 107 cells/ml and were kept on ice until injections. 100 µl of cells (106 cells) were injected intradermally in the flanks of the mice. 9 d later, the spleen and the draining lymph node (inguinal lymph node) were harvested, pooled, and stained for surface markers and Kb-MHC I tetramers specific for OVA presented in the context of MHC class I molecules.
Tumor growth experiments
EG7 tumor assay
Sec22b+/+ and Sec22b−/− mice were injected intradermally with 106 EG7 tumor cells in 100 µl of cold PBS. Tumor cells for injection were recovered from log phase in vitro growth and were injected into the right flank skin of recipient mice. Tumors were clearly visible after 7 d and grew progressively, in an encapsulated fashion. Tumor growth was measured each day and followed until d 20 or when the size reached 1,000 mm3. Afterward, mice were euthanized. 10 mice were used for both Sec22b+/+ and Sec22b−/− groups. To measure CD8+ T cell responses, blood samples were collected from mice 7 d after tumor injections, RBCs were lysed three times with RBC lysis buffer (Sigma-Aldrich), and the nonlysed cells were stained with a couple of Kb-MHC I tetramers specific for OVA presented in the context of MHC class I molecules (provided by the Albert group).
MCA101 OVA-secreting tumor assay and immunotherapy
Sec22b+/+ and Sec22b−/− mice were injected intradermally with 5 × 105 MCA-101 OVA-secreting tumor cells in 100 µl of cold PBS. Tumor cells for injection were recovered from log phase in vitro growth and were injected into the right flank skin of recipient mice. Tumors were clearly visible after 4–5 d and grew progressively, in an encapsulated fashion. Tumor growth was measured every 2 d and was followed until d 30 or until the size reached 1,000 mm3. Afterward, mice were euthanized. 10–15 mice were used for each condition. Anti–PD-L1 treatment consisted of five injections of 200 µg of antibody, delivered on d 0, 3, 6, 9, and 12, i.p. Cold PBS was injected to control groups. Mice were bled on d 13 after tumor injections, and RBCs were lysed as described in the previous section. The unrelated protein human serum albumin, the peptide SIINFEKL, and the PMA/ionomycin were given to nonlysed cells and loaded in MHC I by blood antigen-presenting cells. Subsequent restimulation of OVA-specific T cells was analyzed by IFN-γ ELISpot kit, following the manufacturer’s instructions.
IFN-γ production by CD8+ T cells from spleen and tumors obtained from MCA101-OVA–injected mice
Mice were inoculated with MCA101-OVA tumors and received anti–PD-1 treatment as mentioned above. At d 13, mice were sacrificed, and spleens and tumors were collected and processed with Liberase and DNase I, as described. CD8+ T cells from spleens were isolated with EasySep Mouse Naive CD8+ T Cell isolation kit, following manufacturer’s instruction, and were loaded with Cell Trace Violet (Invitrogen). In addition, total tumor cells were loaded with Cell Trace Violet. 500,000 cells from both spleens and tumors were plated per well in a flat, 96-well plate coated with CD3 (Miltenyi Biotec) at 1 mg/ml with increasing concentrations of CD28 (0, 0.25, 0.5, and 1 mg/ml). After 72 h, plated T cells were treated with Brefeldin A (5 µg/ml; BioLegend) for 3 h to enhance intracellular IFN-γ accumulation. Subsequently, intracellular staining for IFN-γ, as well as CD3, CD4, CD8, and PD-1 staining, was performed.
Analysis of OVA-specific CD8+ T cells in Sec22b+/+ and Sec22b−/− mice
Mice were immunized in the footpad with 50 μl of IFA-CpG (25 µg per mouse)-SIINFEKL (100 µg per mouse). 8 d later, draining popliteal and inguinal lymph nodes were collected and smashed with a cell strainer and subsequently filtered. 500,000 cells were plated per well, and IFN-γ ELISpot analysis was performed as described earlier using OVA and SIINFEKL as antigens for restimulation.
Statistical analysis
All statistical analyses were performed with Prism 7 (GraphPad Software). P-values, as well as statistical tests, are detailed in the figure legends.
Online supplemental material
Fig. S1 shows the phenotypic characterization of immune cell subsets from Sec22b+/+ and Sec22b−/− mice, as well as the expression of costimulatory molecules upon treatment with LPS, TNF, and IFN-γ in DCs from the aforementioned mice. It relates to Fig. 1. Fig. S2 comprises information regarding the immunization of Sec22b+/+ and Sec22b−/− mice to analyze endogenous responses, as well as the phenotypic analysis of putative cross-presenting, migratory and resident DCs. It relates to Fig. 3. Fig. S3 includes a correlative analysis of tumor growth and T cell responses and of the different controls regarding the functionality of CD8+ T cells from Sec22b+/+ and Sec22b−/− mice.
Supplementary Material
Acknowledgments
We would like to thank B. Malissen (Centre d’Immunologie de Marseille, Marseille, France) and Centre d’Immunophénomique for providing us with the Sec22bFloxFlox mice. We would also like to thank I. Cebrian, C. Sedlik, and J. Denizeau for technical assistance.
S. Amigorena received funding from Institut Curie, Institut National de la Santé et de la Recherche Médicale; Centre National de la Recherche Scientifique, la Ligue Contre le Cancer (Equipe labelisée Ligue, grant EL2014.LNCC/SA), Association de Recherche Contre le Cancer, the H2020 European Research Council (grant 2013-AdG 340046 DCBIOX), Institut National Du Cancer (grant PLBIO13-057), and Agence Nationale de la Recherche (grants ANR-11-LABX-0043, ANR-10-IDEX-0001-02 PSL, ANR-16-CE15001801, and ANR-16-CE18002003). A. Alloatti was supported by the European Molecular Biology Organization (grant ALTF 883-2011) and the Agence Nationale de la Recherche (grant ANR-15-CHIN-0002-01).
The authors declare no competing financial interests.
Author contributions: A. Alloatti designed, carried out, and analyzed all experiments, except those detailed below. D.C. Rookhuizen assisted with all T cell assays. L. Joannas performed tumor experiments and assisted with all animal work. J.-M. Carpier assisted with tumor experiments and Western blotting. S. Iborra carried out all vaccinia virus assays. J.G. Magalhaes assisted in assay development and the EG7 tumor experiment. P. Kozik assisted with tumor experiments and cross-presentation assays. N. Yatim assisted with necroptotic cells assays. N. Yatim and M.L. Albert assisted with experimental design on the RipK3 experiments. D. Sancho conceived the vaccinia virus experiments. S. Amigorena conceived and supervised the study, and A. Alloatti and S. Amigorena wrote the manuscript.
Footnotes
Abbreviations used:
- CFSE
- carboxyfluorescein succinimidyl ester
- VACV
- vaccinia virus
References
- Alloatti, A., Kotsias F., Pauwels A.-M., Carpier J.-M., Jouve M., Timmerman E., Pace L., Vargas P., Maurin M., Gehrmann U., et al. 2015. Toll-like receptor 4 engagement on dendritic cells restrains phago-lysosome fusion and promotes cross-presentation of antigens. Immunity. 43:1087–1100. 10.1016/j.immuni.2015.11.006 [DOI] [PubMed] [Google Scholar]
- Bevan, M.J. 1976. Cross-priming for a secondary cytotoxic response to minor H antigens with H-2 congenic cells which do not cross-react in the cytotoxic assay. J. Exp. Med. 143:1283–1288. 10.1084/jem.143.5.1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellino, F., Huang A.Y., Altan-Bonnet G., Stoll S., Scheinecker C., and Germain R.N.. 2006. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature. 440:890–895. 10.1038/nature04651 [DOI] [PubMed] [Google Scholar]
- Caton, M.L., Smith-Raska M.R., and Reizis B.. 2007. Notch-RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J. Exp. Med. 204:1653–1664. 10.1084/jem.20062648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cebrian, I., Visentin G., Blanchard N., Jouve M., Bobard A., Moita C., Enninga J., Moita L.F., Amigorena S., and Savina A.. 2011. Sec22b regulates phagosomal maturation and antigen crosspresentation by dendritic cells. Cell. 147:1355–1368. 10.1016/j.cell.2011.11.021 [DOI] [PubMed] [Google Scholar]
- den Haan, J.M., Lehar S.M., and Bevan M.J.. 2000. CD8+ but not CD8− dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 192:1685–1696. 10.1084/jem.192.12.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff, S., Brewitz A., Gerner M.Y., Klauschen F., Komander K., Hemmi H., Garbi N., Kaisho T., Germain R.N., and Kastenmüller W.. 2015. Robust anti-viral immunity requires multiple distinct T cell-dendritic cell interactions. Cell. 162:1322–1337. 10.1016/j.cell.2015.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubin, M.M., Zhang X., Schuster H., Caron E., Ward J.P., Noguchi T., Ivanova Y., Hundal J., Arthur C.D., Krebber W.-J., et al. 2014. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 515:577–581. 10.1038/nature13988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmich, B.K., and Dutton R.W.. 2001. The role of adoptively transferred CD8 T cells and host cells in the control of the growth of the EG7 thymoma: factors that determine the relative effectiveness and homing properties of Tc1 and Tc2 effectors. J. Immunol. 166:6500–6508. 10.4049/jimmunol.166.11.6500 [DOI] [PubMed] [Google Scholar]
- Hildner, K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8α+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100. 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi, F.S., O’Day S.J., McDermott D.F., Weber R.W., Sosman J.A., Haanen J.B., Gonzalez R., Robert C., Schadendorf D., Hassel J.C., et al. 2010. Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med. 363:711–723. 10.1056/NEJMoa1003466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iborra, S., Izquierdo H.M., Martínez-López M., Blanco-Menéndez N., Reis e Sousa C., and Sancho D.. 2012. The DC receptor DNGR-1 mediates cross-priming of CTLs during vaccinia virus infection in mice. J. Clin. Invest. 122:1628–1643. 10.1172/JCI60660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai, Y., Ishida M., Tanaka Y., Okazaki T., Honjo T., and Minato N.. 2002. Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. USA. 99:12293–12297. 10.1073/pnas.192461099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jancic, C., Savina A., Wasmeier C., Tolmachova T., El-Benna J., Dang P.M.-C., Pascolo S., Gougerot-Pocidalo M.-A., Raposo G., Seabra M.C., and Amigorena S.. 2007. Rab27a regulates phagosomal pH and NADPH oxidase recruitment to dendritic cell phagosomes. Nat. Cell Biol. 9:367–378. 10.1038/ncb1552 [DOI] [PubMed] [Google Scholar]
- Joffre, O.P., Segura E., Savina A., and Amigorena S.. 2012. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 12:557–569. 10.1038/nri3254 [DOI] [PubMed] [Google Scholar]
- Kamphorst, A.O., Guermonprez P., Dudziak D., and Nussenzweig M.C.. 2010. Route of antigen uptake differentially impacts presentation by dendritic cells and activated monocytes. J. Immunol. 185:3426–3435. 10.4049/jimmunol.1001205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurts, C., Heath W.R., Carbone F.R., Allison J., Miller J.F., and Kosaka H.. 1996. Constitutive class I-restricted exogenous presentation of self antigens in vivo. J. Exp. Med. 184:923–930. 10.1084/jem.184.3.923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach, D.R., Krummel M.F., and Allison J.P.. 1996. Enhancement of antitumor immunity by CTLA-4 blockade. Science. 271:1734–1736. 10.1126/science.271.5256.1734 [DOI] [PubMed] [Google Scholar]
- Lebbé, C., Weber J.S., Maio M., Neyns B., Harmankaya K., Hamid O., O’Day S.J., Konto C., Cykowski L., McHenry M.B., and Wolchok J.D.. 2014. Survival follow-up and ipilimumab retreatment of patients with advanced melanoma who received ipilimumab in prior phase II studies. Ann. Oncol. 25:2277–2284. 10.1093/annonc/mdu441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman, I., and Steinman R.M.. 2001. Dendritic cells: specialized and regulated antigen processing machines. Cell. 106:255–258. 10.1016/S0092-8674(01)00449-4 [DOI] [PubMed] [Google Scholar]
- Moore, M.W., Carbone F.R., and Bevan M.J.. 1988. Introduction of soluble protein into the class I pathway of antigen processing and presentation. Cell. 54:777–785. 10.1016/S0092-8674(88)91043-4 [DOI] [PubMed] [Google Scholar]
- Nair-Gupta, P., Baccarini A., Tung N., Seyffer F., Florey O., Huang Y., Banerjee M., Overholtzer M., Roche P.A., Tampé R., et al. 2014. TLR signals induce phagosomal MHC-I delivery from the endosomal recycling compartment to allow cross-presentation. Cell. 158:506–521. 10.1016/j.cell.2014.04.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neijssen, J., Herberts C., Drijfhout J.W., Reits E., Janssen L., and Neefjes J.. 2005. Cross-presentation by intercellular peptide transfer through gap junctions. Nature. 434:83–88. 10.1038/nature03290 [DOI] [PubMed] [Google Scholar]
- Pettitt, S.J., Liang Q., Rairdan X.Y., Moran J.L., Prosser H.M., Beier D.R., Lloyd K.C., Bradley A., and Skarnes W.C.. 2009. Agouti C57BL/6N embryonic stem cells for mouse genetic resources. Nat. Methods. 6:493–495. 10.1038/nmeth.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Paulete, A.R., Cueto F.J., Martínez-López M., Labiano S., Morales-Kastresana A., Rodríguez-Ruiz M.E., Jure-Kunkel M., Azpilikueta A., Aznar M.A., Quetglas J.I., et al. 2016. Cancer Immunotherapy with immunomodulatory anti-CD137 and anti-PD-1 monoclonal antibodies requires BATF3-dependent dendritic cells. Cancer Discov. 6:71–79. 10.1158/2159-8290.CD-15-0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savina, A., Jancic C., Hugues S., Guermonprez P., Vargas P., Moura I.C., Lennon-Duménil A.-M., Seabra M.C., Raposo G., and Amigorena S.. 2006. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 126:205–218. 10.1016/j.cell.2006.05.035 [DOI] [PubMed] [Google Scholar]
- Savina, A., Peres A., Cebrian I., Carmo N., Moita C., Hacohen N., Moita L.F., and Amigorena S.. 2009. The small GTPase Rac2 controls phagosomal alkalinization and antigen crosspresentation selectively in CD8+ dendritic cells. Immunity. 30:544–555. 10.1016/j.immuni.2009.01.013 [DOI] [PubMed] [Google Scholar]
- Sedlik, C., Vigneron J., Torrieri-Dramard L., Pitoiset F., Denizeau J., Chesneau C., de la Rochere P., Lantz O., Thery C., and Bellier B.. 2014. Different immunogenicity but similar antitumor efficacy of two DNA vaccines coding for an antigen secreted in different membrane vesicleassociated forms. J. Extracell. Vesicles. 3:24646. 10.3402/jev.v3.24646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topalian, S.L., Hodi F.S., Brahmer J.R., Gettinger S.N., Smith D.C., McDermott D.F., Powderly J.D., Carvajal R.D., Sosman J.A., Atkins M.B., et al. 2012. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 366:2443–2454. 10.1056/NEJMoa1200690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakim, L.M., and Bevan M.J.. 2011. Cross-dressed dendritic cells drive memory CD8+ T-cell activation after viral infection. Nature. 471:629–632. 10.1038/nature09863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzler, C., Rovere P., Rescigno M., Granucci F., Penna G., Adorini L., Zimmermann V.S., Davoust J., and Ricciardi-Castagnoli P.. 1997. Maturation stages of mouse dendritic cells in growth factor-dependent long-term cultures. J. Exp. Med. 185:317–328. 10.1084/jem.185.2.317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatim, N., Jusforgues-Saklani H., Orozco S., Schulz O., Barreira da Silva R., Reis e Sousa C., Green D.R., Oberst A., and Albert M.L.. 2015. RIPK1 and NF-κB signaling in dying cells determines cross-priming of CD8+ T cells. Science. 350:328–334. 10.1126/science.aad0395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yewdell, J.W., and Dolan B.P.. 2011. Immunology: cross-dressers turn on T cells. Nature. 471:581–582. 10.1038/471581a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeelenberg, I.S., Ostrowski M., Krumeich S., Bobrie A., Jancic C., Boissonnas A., Delcayre A., Le Pecq J.-B., Combadière B., Amigorena S., and Théry C.. 2008. Targeting tumor antigens to secreted membrane vesicles in vivo induces efficient antitumor immune responses. Cancer Res. 68:1228–1235. 10.1158/0008-5472.CAN-07-3163 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.