The activation mechanism of ZBP1/DAI to regulate virus-induced programmed cell death is not known. Kesavardhana et al. show that ZBP1 senses viral ribonucleoproteins to induce cell death upon influenza A virus infection. Apical activation of RIG-I–IFNAR signaling to upregulate ZBP1 and influenza-induced ZBP1 ubiquitination are critical events for ZBP1 activation.
Abstract
Innate sensing of influenza virus infection induces activation of programmed cell death pathways. We have recently identified Z-DNA–binding protein 1 (ZBP1) as an innate sensor of influenza A virus (IAV). ZBP1-mediated IAV sensing is critical for triggering programmed cell death in the infected lungs. Surprisingly, little is known about the mechanisms regulating ZBP1 activation to induce programmed cell death. Here, we report that the sensing of IAV RNA by retinoic acid inducible gene I (RIG-I) initiates ZBP1-mediated cell death via the RIG-I–MAVS–IFN-β signaling axis. IAV infection induces ubiquitination of ZBP1, suggesting potential regulation of ZBP1 function through posttranslational modifications. We further demonstrate that ZBP1 senses viral ribonucleoprotein (vRNP) complexes of IAV to trigger cell death. These findings collectively indicate that ZBP1 activation requires RIG-I signaling, ubiquitination, and vRNP sensing to trigger activation of programmed cell death pathways during IAV infection. The mechanism of ZBP1 activation described here may have broader implications in the context of virus-induced cell death.
Introduction
Innate immune sensors activate programmed cell death pathways as an antiviral and antibacterial mechanism to exert host defense (Man and Kanneganti, 2016; Jorgensen et al., 2017). Influenza A virus (IAV) is a negative sense single-stranded RNA virus that belongs to the family Orthomyxoviridae. IAV infection initiates when the virus infects epithelial lining of the respiratory tract. Various innate immune receptors play a central role in the recognition of IAV infection (Iwasaki and Pillai, 2014; Kuriakose and Kanneganti, 2017). IAV sensing by innate receptors trigger concerted activation of intracellular signaling pathways that regulate type I IFN induction, proinflammatory responses, and induction of cell death (Iwasaki and Pillai, 2014; Kuriakose and Kanneganti, 2017). Lung epithelial cell death is a consequence of IAV infection to control virus replication; however, uncontrolled epithelial cell death compromises lung function (Sanders et al., 2013). Apoptosis and necroptosis in lung epithelial cells affect pneumonia, morbidity, and mortality during IAV infection (Herold et al., 2008; Rodrigue-Gervais et al., 2014). Although IAV RNA recognition by innate sensors is known to induce proinflammatory cytokine responses, the innate receptors and viral ligands that contribute to innate sensor-induced cell death are not known. We have recently identified ZBP1, also known as DAI (DNA-dependent activator of IFN regulatory factors), as an innate sensor of IAV infection (Kuriakose et al., 2016). We showed that ZBP1 regulates NLRP3 inflammasome activation, apoptosis, necroptosis, and pyroptosis in response to IAV infection (Thomas et al., 2009; Kuriakose et al., 2016). Two recent studies also demonstrate that ZBP1-induced cell death contributes to perinatal lethality in RIPK1-RHIM (RIP homotypic interaction motif) mutant mice (Lin et al., 2016; Newton et al., 2016).
The ZBP1 constitutes RHIM domain, which enable its physical interaction with other RHIM-containing proteins. Unlike other RHIM-containing proteins, ZBP1 also contains nucleic acid–binding domains at its N terminus called Zα domains. Although the critical role of ZBP1 in inducing programmed cell death is well established, the activation mechanisms of ZBP1 are not established. Moreover, the involvement of other known IAV sensors in regulating cell death responses has not been investigated in detail. In this study, we show that retinoic acid inducible gene I (RIG-I) acts as an apical regulator of ZBP1-mediated cell death during IAV infection. Importantly, viral RNP (vRNP) complexes of IAV generated during virus replication and ubiquitination activate ZBP1 for execution of cell death in infected cells.
Results and discussion
RIG-I–MAVS signaling regulates ZBP1-induced cell death in response to IAV infection
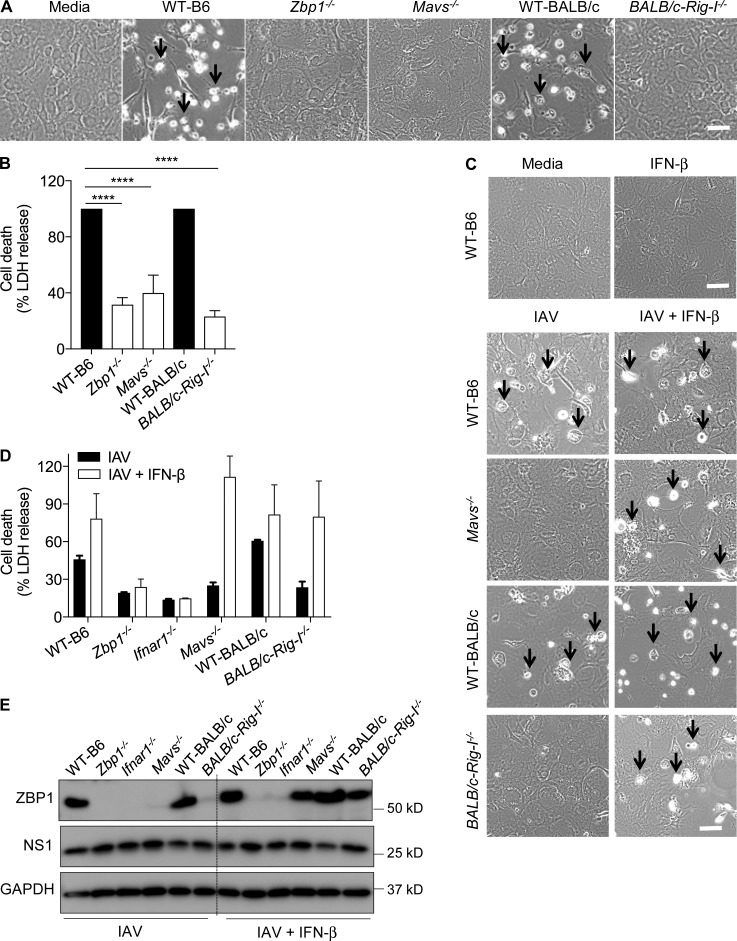
Our previous study demonstrated that type I IFN is essential for ZBP1-mediated cell death during IAV infection (Kuriakose et al., 2016). RIG-I and TLR signaling pathways regulate type I IFN response and programmed cell death pathways such as necroptosis and apoptosis (Blander, 2014; Pasparakis and Vandenabeele, 2015). However, contribution of these receptors to IAV-induced cell death is not thoroughly investigated. Fibroblasts lacking MyD88 (myeloid differentiation primary response gene 88) and TRIF (TIR domain–containing adapter inducing IFN-β) underwent cell death similar to WT fibroblasts after mouse-adapted influenza A/Puerto Rico/8/34 (PR8; H1N1) virus infection (Fig. S1, A and B). Interestingly, cells lacking MAVS (mitochondrial antiviral signaling protein) were resistant to cell death, suggesting a role for MAVS in IAV-induced cell death (Fig. 1, A and B). The cells lacking ZBP1 or IFNAR1 (type I IFN receptor I) were also resistant to IAV-induced cell death (Fig. 1 A and Fig. S1 A). RIG-I is an upstream innate sensor that engages the adaptor MAVS at mitochondrial outer membrane upon IAV RNA recognition (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). Resistance of Mavs−/− fibroblasts to cell death prompted us to investigate the role of RIG-I in IAV-induced cell death. Whereas fibroblasts generated from WT-BALB/c mice were susceptible to IAV-induced cell death, fibroblasts from BALB/c-Rig-I−/− mice were completely resistant (Fig. 1, A and B). These results suggest that RIG-I–MAVS signaling regulates IAV-induced cell death, whereas TLR adaptors (MyD88 and TRIF) are dispensable.
Figure 1.
RIG-I–MAVS signaling regulates ZBP1-dependent cell death in response to IAV infection. (A) Microscopic analysis of cell death in unprimed primary fibroblasts infected with IAV (MOI, 10). Microscopic images were collected 20 h after IAV infection (n = 4). Arrows indicate dead cells after IAV infection. Bar, 100 µm. (B) Quantification of cell death by LDH release in unprimed primary fibroblasts infected with IAV (MOI, 10) after 20 h. LDH release was normalized to IAV-infected WT cells, which was considered 100% in individual experiments (n = 4). ****, P = 0.0001 (one-way ANOVA). (C) Microscopic analysis of cell death in unprimed primary fibroblasts infected with IAV (MOI, 10) or IAV in combination with 100 U/ml IFN-β after 20 h. For IFN-β supplementation experiments, IFN-β was added to the cells after 2 h of IAV infection (n = 3). Arrows indicate dead cells after IAV infection. Bars, 100 µm. (D) Quantification of cell death by LDH release in unprimed primary fibroblasts infected with IAV (MOI, 10) or IAV in combination with 100 U/ml IFN-β after 20 h. LDH release was normalized to IFN-β–supplemented, IAV-infected WT cells (n = 3). (E) Immunoblot analysis of ZBP1, NS1 proteins of IAV and GAPDH (loading control) in fibroblasts infected with IAV or IAV in combination with 100 U/ml IFN-β (n = 3). Data are representative of three independent experiments (mean ± SEM).
IAV infection up-regulates ZBP1 expression to induce cell death (Fig. S1 C). MyD88 and TRIF were dispensable for ZBP1 up-regulation after IAV infection (Fig. S1 C). Ifnar1−/− cells lacked ZBP1 expression, demonstrating the critical role of type I IFN signaling in ZBP1 production (Fig. S1 C). Moreover, lack of MAVS or RIG-I expression abolished IAV-induced up-regulation of ZBP1 expression (Fig. S1 C). The absence of ZBP1 induction in RIG-I– and MAVS-deficient cells was not a result of defective IAV replication because similar levels of IAV NS1 protein were observed among the compared genotypes (Fig. S1 C). These results further confirm the specific role of RIG-I–MAVS signaling in regulating ZBP1 expression.
RIG-I, upon sensing IAV RNA, engages its adaptor protein, MAVS, to drive IFN-β expression (Hornung et al., 2006; Pichlmair et al., 2006; Baum et al., 2010; Rehwinkel et al., 2010; Iwasaki and Pillai, 2014). We observed a complete lack of IFN-β production by Mavs−/− and Rig-I−/− fibroblasts during IAV infection, confirming the requirement of RIG-I/MAVS signaling for type I IFN responses during IAV infection (Fig. S1 D). However absence of MyD88 and TRIF did not abolish IFN-β production. Absence of ZBP1 also did not alter IFN-β production in response to IAV infection (Fig. S1 D). Consistent with the requirement of IFN feedback signaling for type I IFN production (Honda et al., 2005), lack of IFNAR1 expression significantly reduced IFN-β release compared with WT cells upon IAV infection (Fig. S1 D).
Based on these observations, we hypothesized that RIG-I–MAVS–dependent regulation of cell death is mediated through IFN-β production. To confirm this, we supplemented IFN-β to the cells lacking MAVS or RIG-I after IAV infection. Addition of IFN-β did not affect cell death in WT, Ifnar1−/−, and Zbp1−/− cells and indeed increased cell death in WT cells (Fig. 1, C and D). Interestingly, Mavs−/− and Rig-I−/− cells, which were resistant to IAV-induced cell death, underwent robust cell death that was comparable to WT cells when exogenous IFN-β was provided during IAV infection (Fig. 1, C and D). Furthermore, addition of IFN-β restored IAV-dependent activation of caspase-8 and caspase-3 in Mavs−/− and Rig-I−/− cells similar to WT cells after IAV infection (Fig. S1 E). We conclude that addition of IFN-β to the Mavs−/− or Rig-I−/− fibroblasts bypasses the requirement of RIG-I/MAVS signaling for the up-regulation of ZBP1 expression. We observed restoration of ZBP1 expression in Mavs−/− or Rig-I−/− cells, but not in Zbp1−/− or Ifnar1−/− cells, when IFN-β was supplemented during IAV infection (Fig. 1 E). These results indicate that the activation of RIG-I by IAV RNA is an apical event, which promotes type I IFN production to license ZBP1-mediated cell death.
To examine whether RIG-I–MAVS–dependent cell death is specific to PR8 virus, we further tested the role of RIG-I–MAVS signaling in cell death induced by other mouse-adapted and human strains of IAV. Fibroblasts lacking MAVS or RIG-I expression did not undergo cell death in response to mouse-adapted influenza A/HK/X31 (H3N2) or non–mouse-adapted seasonal strain influenza A/Brisbane/59/2007 (H1N1; Fig. S1 F). These results indeed suggest that RIG-I–MAVS signaling broadly regulates IAV-induced cell death.
Nogusa et al. (2016) reported that RIG-I/MAVS signaling and IFNAR1 are dispensable for IAV-induced cell death (Nogusa et al., 2016). Our results demonstrate that the absence of RIG-I–MAVS abolishes IAV-induced cell death and that RIG-I–mediated type I IFN response is required for ZBP1 up-regulation. Indeed, lack of IFNAR1 expression abolished IAV-induced cell death corroborating the RIG-I–MAVS data and our previous study (Kuriakose et al., 2016). The striking differences between Nogusa et al. (2016) and our studies with regard to the role of RIG-I–MAVS–IFNAR1 could be due to differences in experimental conditions or type of cells used for infections. Nogusa et al. (2016) performed IAV infections in the presence of zVAD, a pan-caspase inhibitor, which biases IAV-induced cell death toward necroptosis. However, we infected cells with IAV without any additional components in an unbiased manner and showed that IAV infection triggers apoptosis, necroptosis, and pyroptosis (Kuriakose et al., 2016). It is well established that IAV induces apoptosis and necroptosis in vivo (Herold et al., 2008; Rodrigue-Gervais et al., 2014; Kuriakose et al., 2016; Nogusa et al., 2016; Thapa et al., 2016). Multiple studies, including ours, have demonstrated the critical role of IFN signaling in up-regulation of ZBP1 in primary fibroblasts, macrophages, and keratinocytes (Takaoka et al., 2007; Kuriakose et al., 2016; Lin et al., 2016; Newton et al., 2016). Our results also demonstrate that RIG-I–MAVS–induced type I IFN production is essential to induce ZBP1-dependent cell death during IAV infection in primary cells.
RIG-I–MAVS and TLR signaling regulates ZBP1-induced inflammasome activation and pyroptosis in macrophages during IAV infection
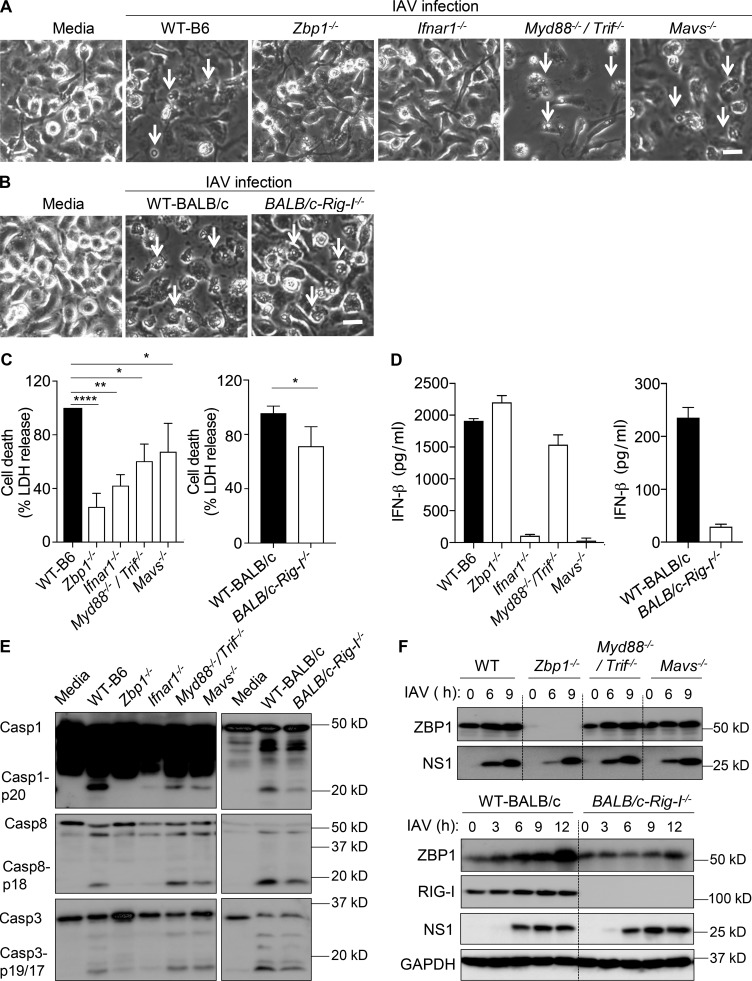
Our previous study demonstrated that ZBP1 also induces NLRP3 inflammasome activation and pyroptosis in primary BMDMs after IAV infection (Kuriakose et al., 2016). It was also suggested that type I IFN signaling triggers NLRP3 inflammasome activation during IAV infection (Pothlichet et al., 2013). These studies prompted us to study specific roles of RIG-I and TLR signaling in ZBP1-induced inflammasome activation and cell death in BMDMs. Absence of ZBP1 or IFNAR1 conferred complete protection from IAV induced cell death in BMDMs (Fig. 2, A and C). We observed a modest but significant reduction in cell death of BMDMs lacking MyD88–TRIF or RIG-I–MAVS signaling compared with WT BMDMs after IAV infection (Fig. 2, A–C). However, BMDMs lacking MAVS, RIG-I, or MyD88–TRIF still exhibited robust cell death compared with the BMDMs lacking ZBP1 (Fig. 2, A–C). Importantly, lack of MAVS or RIG-I, but not MyD88–TRIF, abolished IFN-β production in BMDMs in response to IAV infection (Fig. 2 D). Immunoblot analysis of caspase-1 cleavage suggested that absence of RIG-I–MAVS or MyD88–TRIF signaling pathways significantly reduced caspase-1 activation in BMDMs after IAV infection (Fig. 2 E). In addition, activation of caspase-8 was partly reduced in the absence of RIG-I (Fig. 2 E). Similar expression levels of NS1 proteins of IAV indicate that the lack of RIG-I–MAVS signaling or MyD88–TRIF signaling did not affect IAV replication (Fig. 2 F). These results suggest that RIG-I and TLR signaling in BMDMs are indispensable for ZBP1-mediated inflammasome activation and consequent pyroptosis. Although type I IFN expression was abolished in the absence of RIG-I–MAVS, a significant up-regulation of ZBP1 was observed in BMDMs lacking these molecules (Fig. 2 F). This suggests that apart from RIG-I–MAVS signaling pathway, other signaling cascades might exist in BMDMs for up-regulating ZBP1 expression during IAV infection. Overall, both RIG-I and TLR signaling contribute to the activation of NLRP3 inflammasome and pyroptosis during IAV infection, whereas RIG-I–MAVS signaling regulates IAV-induced type I IFN responses.
Figure 2.
TLR and RIG-I–MAVS signaling regulate inflammasome activation and cell death in response to IAV infection. (A and B) Microscopic analysis of cell death in BMDMs infected with IAV (MOI, 25). Microscopic images were collected 20 h after IAV infection (n = 3). Arrows indicate dead cells after IAV infection. Bars, 45 µm. (C) Quantification of cell death by LDH release in BMDM cultures infected with IAV after 20 h. LDH release was normalized to IAV-infected WT cells, which was considered 100% in individual experiments (n = 3). *, P = 0.016; **, P = 0.0012; and ****, P = 0.0001 (one-way ANOVA or two tailed t test). (D) Levels of IFN-β in cell culture supernatants 16 h after infection with IAV (n = 3). (E) Immunoblot analysis of the pro- and cleaved forms of caspase-1, caspase-8, and caspase-3 in BMDMs 20 h after infection with IAV (n = 3). (F) Immunoblot analysis of ZBP1, RIG-I, NS1, and GAPDH (loading control) in BMDMs infected with IAV (n = 3). Data are representative of three independent experiments (mean ± SEM).
IAV infection triggers ubiquitination of ZBP1
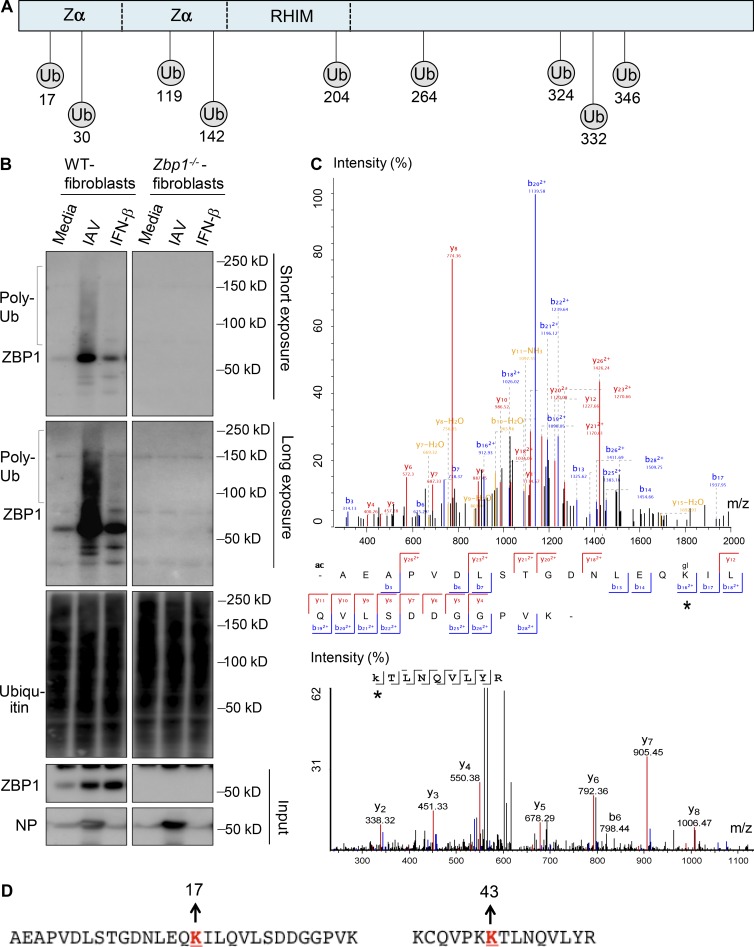
Although RIG-I activation by IAV regulates ZBP1 up-regulation, the posttranslational modifications required for ZBP1 activation are not known. Protein modifications, such as ubiquitination, create docking sites for association of other signaling molecules to modulate their function. RHIM-containing proteins, such as RIPK1 and RIPK3, are known to undergo extensive ubiquitination and phosphorylation to regulate proinflammatory and programmed cell death functions. To understand the role of protein modifications in ZBP1 activation, we characterized the ubiquitination pattern of ZBP1 in response to IAV infection. Using bioinformatics, we predicted that ZBP1 has multiple potential ubiquitination sites (Fig. 3 A). To confirm that ZBP1 is ubiquitinated during IAV infection, we isolated ubiquitinated protein fraction from whole-cell lysates after IAV infection. Immunoblotting with ubiquitin specific antibody confirmed the enrichment of ubiquitinated protein fraction after isolation (Fig. 3 B). We indeed observed ubiquitinated ZBP1 in the polyubiquitinated protein fraction from IAV infected cells, but not in uninfected cells (Fig. 3 B). IFNAR signaling is essential for IAV-induced ZBP1 up-regulation. Although IFN-β stimulation in WT cells induced robust ZBP1 expression, a marginal induction of ZBP1 ubiquitination was observed compared with IAV infection (Fig. 3 B). This suggests that both type I IFN induced up-regulation of ZBP1 and recognition of IAV ligands induces ubiquitination of ZBP1. Mass spectrometry analysis demonstrated that K17 and K43 positions of the ZBP1 were ubiquitinated after IAV infection (Fig. 3, C and D). Mass spectrometry analysis corroborated the prediction tool by identifying ubiquitination at K17 within ZBP1 (Fig. 3, A and C). These results demonstrate that IAV infection induces polyubiquitination of ZBP1, which might be required for the assembly of cell death signaling complex. K17 and K43 are located in Zα region of ZBP1. We speculate that ubiquitination of ZBP1 at Zα region might trigger conformational changes in ZBP1 to expose its RHIM domain for induction of cell death. Collectively, RIG-I–MAVS signaling–induced type I IFN signaling and ubiquitination of ZBP1 are required for IAV-induced cell death.
Figure 3.
IAV infection induces ZBP1 ubiquitination. (A) Schematic representation of predicted ubiquitination (Ub) sites in ZBP1. UbPred and UbiSite servers were used to predict potential ubiquitination sites in ZBP1. (B) ZBP1 was ubiquitinated after IAV infection. WT and Zbp1−/− cells were infected with IAV or treated with IFN-β (100U/ml). Whole cell lysates were harvested after 8 h of infection, to purify polyubiquitinated protein fraction by using TUBEs (tandem ubiquitin-binding entities). Purified fractions were subjected to Western blot analysis for ubiquitin and ZBP1 to detect its ubiquitination (n = 4). Poly-Ub, polyubiquitinated protein. (C) Peptide spectrum match of the ubiquitinated peptide at K17 and K43 positions. ZBP1 was immunoprecipitated from IAV infected cells and subjected to protein fragmentation. Peptides were separated by a reverse phase liquid chromatography using a Proxeon Nano-UPLC system and analyzed on a Q-Exactive HF mass spectrometer. Asterisk represents ubiquitination detected by mass spectrometry. (D) Amino acid sequence context for detected ubiquitinated ZBP1 fragments indicating K17 and K43 positions as ubiquitination sites.
Purified IAV RNA or NP ligands are not sufficient to trigger ZBP1-dependent cell death
Although RIG-I sensing of IAV genomic RNA induces type I IFN response, the identity of a specific ligand for ZBP1 activation has remained ambiguous. We demonstrated that ZBP1 interacts with the IAV proteins nuclear protein (NP) and polymerase (PB1; Kuriakose et al., 2016). However, Thapa et al. (2016) reported that ZBP1 recognizes IAV RNA genome to promote RIPK3 necrosome formation. To resolve this ambiguity, we immunoprecipitated ZBP1 from IAV-infected cells and probed for coprecipitated molecules. Consistent with our previous study, pull-down of ZBP1 precipitated the IAV proteins NP and PB1 after IAV infection (Fig. S2 A).
Structural studies suggest that N-terminal Zα domains of ZBP1 interact with Z-DNA and double-stranded Z-RNA (Schwartz et al., 2001; Placido et al., 2007; Ha et al., 2008; Kim et al., 2011). We predicted the RNA interaction potential of ZBP1 protein based on its amino acid sequence because of the lack of structural information for full-length ZBP1 (Wang and Brown, 2006 Agostini et al., 2013; Li et al., 2014). Predictions indicate that the different regions of ZBP1 protein show high RNA-binding propensities (Fig. S2 B). All the prediction servers indicated Zα domains as RNA-binding domains (Fig. S2 B). We immunoprecipitated endogenous ZBP1 in IAV-infected cells, and RNA species were purified from ZBP1 immunoprecipitates. Immunoprecipitation of ZBP1 from WT fibroblasts resulted in robust association with IAV RNA in comparison to Zbp1−/− cells, indicating specificity of IAV-RNA and ZBP1 interaction (Fig. S2 C). The amount of IAV RNA isolated from ZBP1 was significantly higher than the RNA associated with the control IgG pull-down (Fig. S2, D and E). Overall, these results suggest that IAV NP and PB1 proteins and the RNA genome are functional interacting partners of ZBP1 during IAV infection.
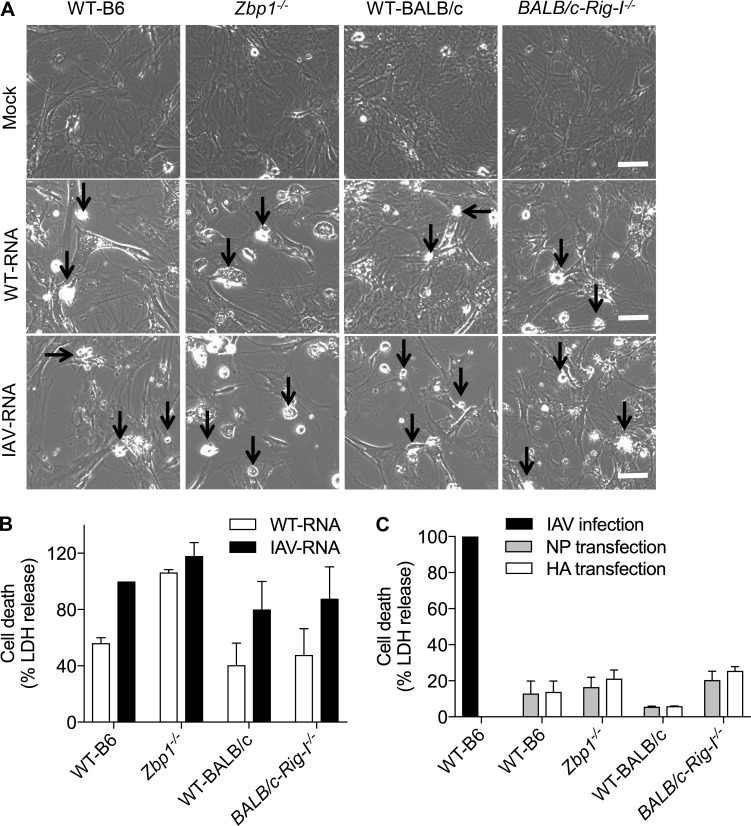
To investigate whether RNA or viral proteins act as the ligands for ZBP1, we performed in vitro ligand transfection assays and assessed whether the cell death was ZBP1 dependent. Cells were treated with IFN-β to up-regulate ZBP1 levels before RNA transfection. WT cells transfected with either WT-RNA (RNA isolated from untreated cells) or IAV-RNA (RNA isolated from IAV-infected cells) showed increased cell death compared with mock-transfected cells (Fig. 4, A and B). Cell death was increased after IAV-RNA transfection in all genotypes compared with WT-RNA transfections (Fig. 4, A and B). However, there was no repression of cell death in the cells lacking ZBP1 after transfection with either RNA (Fig. 4, A and B). In addition, absence of RIG-I did not alter the cell death response induced by both WT-RNA and IAV-RNA in comparison to WT cells (Fig. 4, A and B). These observations suggest that RNA isolated from IAV-infected cells did not engage ZBP1-mediated cell death. Although IAV-RNA serves as a vital pathogen-associated molecular pattern for the type I IFN response, it is not sufficient to drive ZBP1-dependent cell death during IAV infection.
Figure 4.
IAV RNA or IAV protein alone is not sufficient to trigger ZBP1-dependent cell death. (A) Microscopic analysis of cell death in IFN-β primed primary fibroblasts transfected with total RNA isolated from uninfected cells (WT-RNA) or IAV-infected cells (IAV-RNA). Arrows indicate dead cells after IAV infection. Bars, 100 µm. (B) Quantification of cell death by LDH release in IFN-β primed primary fibroblasts transfected with WT-RNA or IAV-RNA. LDH release was normalized for IAV RNA-transfected WT cells (n = 4). (C) Quantitation of cell death by LDH release in IFN-β primed primary fibroblasts transfected with IAV NP or IAV HA protein. LDH release was normalized to IAV-infected WT cells (n = 3). Data are representative of three independent experiments (mean ± SEM).
We also transfected NP of IAV into the cytosol to investigate whether the protein can activate ZBP1-dependent cell death (Fig. 4 C). As a negative control, we transfected IAV HA (hemagglutinin) protein separately, which does not interact with ZBP1 (Kuriakose et al., 2016). Transfection of NP or HA did not induce cell death in WT cells and the cells lacking ZBP1 or RIG-I (Fig. 4 C). These results demonstrated that neither IAV RNA nor viral proteins alone could engage ZBP1-mediated cell death during IAV infection.
ZBP1 spatially colocalizes with vRNP complexes in IAV-infected cells
vRNP complexes are composed of IAV RNA genome packed with multiple copies of NP and a trimeric polymerase complex (PB1, PB2, and PA; Eisfeld et al., 2015). These vRNP complexes display IAV RNA, NP, and PB1 in a single scaffold. vRNP complexes are generated in the nucleus after replication of IAV RNA genome and exported from nucleus to the cytosol for assembly of progeny virions (Eisfeld et al., 2015). To investigate the requirement of IAV replication and vRNP generation for ZBP1-dependent cell death, we made replication incompetent IAV by UV irradiation. We observed a significant decrease in IAV-induced cell death when cells were infected with UV-radiated IAV in comparison to untreated IAV (Fig. S2 F). Nuclear export of vRNP complexes is a critical step to assemble viral particles. Previous studies showed that leptomycin-B (LMB), a nuclear export inhibitor, inhibits nuclear export of vRNP complexes during IAV infection (Elton et al., 2001; Paterson and Fodor, 2012). To study the contribution of vRNP export from the nucleus to activate ZBP1-mediated cell death, cells were subjected to LMB treatment after IAV infection. The cells treated with LMB showed significant decrease in IAV-induced cell death in comparison to untreated cells (Fig. S2 G). These results suggest that replication of IAV to generate vRNP complexes and nuclear export of the vRNP complexes are crucial for ZBP1-mediated cell death.
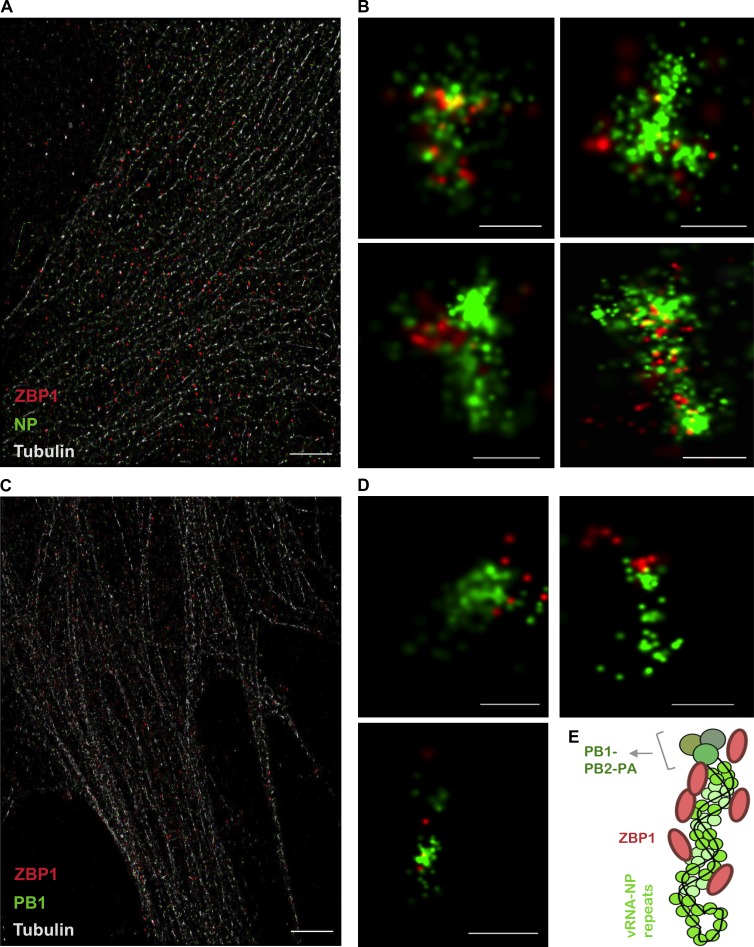
Structural studies of IAV RNP complexes indicate that the IAV vRNPs are stabilized by unique twisted, antiparallel helices of vRNA–NP complexes that could potentially represent an IAV-associated molecular pattern for recognition by innate sensors (Arranz et al., 2012; Moeller et al., 2012; Eisfeld et al., 2015). To further probe and visualize the association of ZBP1 with vRNP complexes, we used three-dimensional stochastic optical reconstruction microscopy (STORM) to visualize vRNP complexes. STORM imaging enables three-dimensional view of IAV vRNP complexes with enhanced spatial resolution upon NP staining (Liedmann et al., 2014). Consistent with previous studies, we observed vRNP complexes of IAV as elongated helical or spiral structures in the cytosol (Fig. 5, A and B; Arranz et al., 2012; Moeller et al., 2012; Liedmann et al., 2014). Staining of ZBP1 further indicated that it is spatially localized to the vRNP complexes (Fig. 5 B and Fig. S3 A). It appeared that ZBP1 was distributed along the elongated structure of the vRNP complex (Fig. 5, B and E; and Fig. S3 A). PB1 was also closely associated with ZBP1 after IAV infection (Fig. 5, C and D), suggesting vRNPs as ligands for ZBP1. The estimated length of the IAV vRNP complex is 110 nm (Arranz et al., 2012; Moeller et al., 2012). Mouse ZBP1 protein constitutes 411 amino acid residues. The mean diameter of the predicted full-length ZBP1 protein structure is 6 to 7 nm, which is much smaller than the IAV vRNP complex (Fig. S3 B). Furthermore, the close proximity of multiple ZBP1 fluorescence puncta on vRNP complexes in STORM images support the notion that multiple ZBP1 monomers or an oligomer of ZBP1 might be required for the sensing of IAV vRNP complex (Fig. 5, B–E; and Fig. S3 A). This collectively suggests that ZBP1 activation requires its spatial association with vRNP complexes during IAV infection. Further studies are needed to understand the precise cellular compartment and other protein cargo required for ZBP1–vRNP complex formation and subsequent modulations in this complex to induce cell death.
Figure 5.
ZBP1 recognizes IAV ribonucleoprotein (vRNP) complexes during IAV infection. (A–D) Primary fibroblasts were infected with IAV and subjected to three-dimensional stochastic optical reconstruction microscopy (STORM) 8 h after infection. STORM revealed the association of ZBP1 (red) with vRNPs (green; n = 3). (A) A wide shot of cellular context of fibroblasts viewed by microtubule tracker (light gray), NP (green), and ZBP1 (red). Bar, 5 µm. (B) Zoom-in version showing NP stained vRNPs (green) in close proximity to ZBP1 (red). Bars, 100 nm. (C) A wide shot of cellular context viewed by microtubule tracker (light gray), PB1 (green), and ZBP1 (red). Bar, 5 µm. (D) Zoom-in images showing PB1 (green) in close proximity to ZBP1 (red). Bars, 100 nm. (E) Model representing vRNP-associated ZBP1.
Previous studies showed that ZBP1 regulates murine cytomegalovirus (MCMV)–induced necroptosis (Upton et al., 2012). MCMV infection in WT primary fibroblasts induced cell death within 2–3 h after infection. In addition, we also observed that fibroblasts lacking ZBP1 expression were resistant to MCMV induced cell death (Fig. S3 C). This suggest that basal expression of ZBP1 is sufficient to recognize MCMV infection where as replication and type I IFN signaling are necessary for ZBP1-dependent cell death during IAV infection.
Zα domain-containing proteins are known to be localized to cytosolic stress granules where translationally repressed RNP complexes are accumulated (Weissbach and Scadden, 2012; Ng et al., 2013). In support of these studies, our results also show that ZBP1 recognizes IAV viral RNP complexes. However, unlike IAV, MCMV is a double-stranded DNA virus. The mechanism of ZBP1 activation during MCMV infection is not known. Based on our observations, we presume that ZBP1 might sense unique patterns in RNP complexes formed during the transcription of MCMV genes to activate cell death. Structural studies showed that Zα domain physically interacts with Z-DNA or Z-RNA (Schwartz et al., 2001; Placido et al., 2007; Ha et al., 2008; Kim et al., 2011). Z-DNA and Z-RNA exist in left-handed form with similar structural features, unlike right-handed DNA and RNA (Rich and Zhang, 2003). In addition, previous studies also suggest that cytosolic RNAs in fixed cells can exist as Z-RNAs (Zarling et al., 1987; Rich and Zhang, 2003). These observations suggest that the viral RNA that appears as vRNP during the replication cycle might attain Z-RNA conformation, and ZBP1 could be recognizing this unique Z-RNA conformation in RNPs. However, knowledge of the existence of Z-RNA in physiological conditions is primitive.
Multiple innate immune receptors are activated in response to IAV infection. Although RIG-I has been studied well for its role in IAV infection, ZBP1 was only recently identified as an innate sensor of IAV. The insights gained from the current study established apical RIG-I signaling and ubiquitination as regulators of ZBP1-mediated cell death. We further showed that vRNPs of IAV activate ZBP1-mediated cell death, which resolves the ambiguity regarding the specific viral ligands activating ZBP1 (Fig. S3 D). ZBP1 contains RHIM domain similar to RIPK1 and RIPK3. Although the mechanisms of RIPK1 and RIPK3 activation in microbial infections are well studied, the activation of ZBP1 is largely unexplored. Identification of the vRNP complexes as ligands for ZBP1 activation and ubiquitination of ZBP1 advances the understanding of molecular mechanisms of innate immune recognition of IAV and creates new therapeutic opportunities to prevent IAV infection.
Materials and methods
Cell culture
Fibroblasts were generated from pinnae of adult mice. Pinnae were minced and digested with 100 mg/ml collagenase type IV (Worthington Biochemical Corporation) for 3 h, followed by filtration through 70-µm strainers to obtain fibroblasts. Cells were cultured in 50% FBS (EMD Millipore) in DMEM (Gibco) supplemented with Hepes, 1% penicillin and streptomycin, l-glutamine, sodium pyruvate, nonessential amino acids, and β-mercaptoethanol for the first 3 or 4 d. Cells were then subcultured in 10% FBS in DMEM supplemented with 1% penicillin and streptomycin. Primary lung fibroblasts were cultured as described previously (Yamamoto et al., 2003; Tzeng et al., 2016). All primary fibroblasts were used before to reaching sixth passage. Fibroblasts were seeded onto six-well plates at a density of 2 × 105 or 12-well plates at a density of 105 cells per well and incubated overnight.
Primary BMDMs were grown for 7 d in DMEM (Gibco) supplemented with 10% FBS (Atlanta Biologicals), 30% L929 conditioned media, and 1% penicillin and streptomycin (Sigma-Aldrich). BMDMs were seeded in antibiotic-free media at a concentration of 106 cells onto 12-well plates and incubated overnight before infection.
IAV infection and cell death studies
Cells were cultured overnight in antibiotic-free media before infection. The PR8 virus generated by an eight-plasmid reverse genetics system was propagated in allantoic cavity of 9- to 11-d-old embryonated SPF (specific pathogen free) chicken eggs and viral titers were enumerated by plaque assays. For cell death studies, primary fibroblasts (MOI [multiplicity of infection], 10) were infected with PR8 virus for 2 h. DMEM media containing 20% FBS was added after 2 h of infection and samples were collected at indicated time points. Cell supernatants were collected at indicated time points of lactate dehydrogenase (LDH) release assays. For IFN-β supplementation studies, 100 U/ml IFN-β was added to the cells after 2 h of IAV infection. For pharmacological studies, 10–20 ng/ml LMB (Sigma-Aldrich) was added to the cells 1 h after IAV infection.
Mice
Zbp1−/−, Ifnar1−/−, Mavs−/−, Myd88−/−, and Trif−/− mice have been described previously (Kuriakose et al., 2016). Rig-I−/− mice (129SvXC57BL/6XICR) were a gift from S. Akira (Osaka University, Osaka, Japan; Kato et al., 2006). These mice were backcrossed with BALB/c mice for 10 generations to generate BALB/c-Rig-I−/− mice. Animal study protocols were approved by the St. Jude Children’s Research Hospital committee on the use and care of animals.
Immunoblotting analysis
For immunoblotting of proteins, cells were lysed in RIPA buffer and sample loading buffer containing SDS and 100 mM DTT after washing with cold PBS. Proteins were separated on 8–12% polyacrylamide gels and transferred onto polyvinylidene difluoride (PVDF) membranes. Membranes were blocked in 5% skim milk followed by incubation with the following primary antibodies and secondary HRP antibodies: caspase-8 (#1492, 1:1,000 dilution; Cell Signaling Technology), cleaved caspase-8 (#8592, 1:1,000 dilution; Cell Signaling Technology), caspase-3 (#9662, 1:1,000 dilution; Cell Signaling Technology), cleaved caspase-3 (#9661, 1:1,000 dilution; Cell Signaling Technology), ZBP-1 (AG-20B-0010-C100, 1:3,000 dilution; Adipogen), IAV NS1 (NS1-23-1: sc-130568, 1:2,000 dilution; Santa Cruz Biotechnology, Inc.), PB1 (sc-17601, 1:500; Santa Cruz Biotechnology, Inc.), NP (PA5-32242, 1:1,000; Thermo Fisher Scientific), GAPDH (#5174, 1:10,000 dilution; Cell Signaling Technology), and HRP antibodies (1:1,000 dilution; Jackson ImmunoResearch Laboratories).
Coimmunoprecipitation
For immunoprecipitation, cell lysates were incubated with 4 µg of indicated primary antibodies on a rocking platform for 4 to 5h at 4°C. Protein A/G PLUS-Agarose (Santa Cruz Biotechnology, Inc.) was added to the samples and incubated for another 2 h on the rocking platform. Agarose was centrifuged and washed three times with lysis buffer. Immunoprecipitates were eluted in sample buffer after three washes in lysis buffer and then subjected to immunoblotting analysis.
Real-time quantitative RT-PCR analysis
RNA was extracted using TRIzol according to the manufacturer’s instructions (Thermo Fisher Scientific). Isolated RNA was reverse transcribed into cDNA using the First-Strand cDNA Synthesis kit (Thermo Fisher Scientific). Real-time quantitative PCR was performed on an ABI 7500 real-time PCR instrument with 2× SYBR Green (Applied Biosystems). Sequences for quantitative RT-PCR primers are as follows: IAV-M1 forward primer, 5′-TGAGTCTTCTAACCGAGGTC-3′; IAV-M1 reverse primer, 5′-GGTCTTGTCTTTAGCCATTCC-3′; IAV-NP forward primer, 5′-CTCGTCGCTTATGACAAAGAAG-3′; IAV-NP reverse primer, 5′-AGATCATCATGTGAGTCAGAC-3′.
RNA or protein transfections
4 µg RNA was incubated with Lipofectamine-2000 transfection reagent and incubated at room temperature for 15 min. Cells were washed in PBS, and reduced serum media was added. The transfection mix was added to the cells, and cell death was monitored. Cell supernatants were collected after 20 h of transfection for LDH release assays. Microscopic pictures were collected after 20 h of transfection. For protein transfection studies, 2–3 µg purified NP or HA proteins of IAV (Sino Biological) were mixed with 20 µl DOTAP (Sigma-Aldrich) transfection reagent following 15 min incubation. The cells were washed three times with HBSS/modified/calcium-magnesium media (Sigma-Aldrich), and the transfection mix was added. Cell supernatants were collected after 15 h of transfection for LDH release assays.
STORM
Primary fibroblasts were seeded 24 h before IAV infection. Cells were infected with IAV (MOI, 5) and incubated at 37°C for 8 h. IFN-β was supplemented after 4 h of infection to increase ZBP1 expression levels. After 8 h of infection, cells were processed for STORM imaging as described previously (Liedmann et al., 2014). The following antibodies were used for labeling: NP (1:2,000; PA5-32242; Thermo Fisher Scientific), PB1 (1:1,000; sc-17601; Santa Cruz Biotechnology, Inc.), ZBP1 (1:1,000; AG-20B-0010; Adipogen Life Sciences), and tubulin (1:1,000; clone YOL1/34; Thermo Fisher Scientific).
Tandem ubiquitin binding entity (TUBE) assays for ubiquitination
Agarose-TUBEs are available through LifeSensors. Primary lung fibroblasts were infected with IAV, and whole-cell lysates were harvested in cell lysis buffer (50 mM Tris-HCl, pH 7.5, 0.15 M NaCl, 1 mM EDTA, and 1% NP-40) supplemented with 10 mM N-ethylmaleimide (Sigma-Aldrich) and a Complete Protease Inhibitor Cocktail tablet (Roche). Lysates were cleared by centrifugation, and protein concentration was determined using BCA assay (Thermo Fisher Scientific). TUBE 1 Agarose (LifeSensors) beads were prewashed in TBS-T buffer (20 mM Tris-HCl, pH 8.0, 0.15 M NaCl, and 0.1% Tween-20) according to the manufacturer’s guidelines and incubated with 1 mg total protein lysate overnight on a rotating platform at 4°C. The next day, beads were washed three times before being resuspended in 1× Laemmli buffer and boiled for 10 min. Eluted samples were analyzed by SDS-PAGE.
Mass spectrometry
Immunoprecipitation samples were run on a 4–20% gradient gel. The bands corresponding to ZBP1 and the putative ubiquitinated form of ZBP1 were cut out, cysteines reduced, blocked with iodoacetamide, and in-gel digested with a cocktail of LysC and trypsin. The digested peptides were separated with a reverse-phase liquid chromatography using a Proxeon Nano-UPLC system and analyzed on a Q-Exactive HF mass spectrometer. Raw data were searched against mouse UniProt protein database using Andromeda running under MaxQuant using default settings, except for peptide modifications (Cox et al., 2011).
RNA isolation for transfection experiments
L929 cells were infected with IAV in 150-mm plates and incubated for 8 h. Cells were harvested in TRIzol reagent for RNA purification. 200 µl chloroform was added to the tubes, mixed well, and centrifuged at 13,500 rpm for 5 min to separate the organic and aqueous phase. The aqueous phase was gently aspirated out and 1 vol isopropanol was added. The solution was incubated at room temperature for 10 min and centrifuged at 13,500 rpm for 30 min at 4°C to precipitate the RNA. The RNA pellet was washed twice with 75% ethanol, air dried, and solubilized in pure water.
Prediction methods
RNA-binding properties of ZBP1 were predicted using protein-RNA–binding prediction analytical tools BindN, catRAPID, and aaRNA (Wang and Brown, 2006; Agostini et al., 2013; Li et al., 2014). UbPred and UbiSite servers were used to predict potential ubiquitination sites in ZBP1 (Radivojac et al., 2010; Huang et al., 2016). i-TASSER server was used to predict full-length ZBP1 structure (Roy et al., 2010).
Statistical analysis
GraphPad Prism 7.0 software was used for data analysis. Statistical significance was determined by a paired two-tailed t test or one-way ANOVA, where P < 0.05 was considered statistically significant. Data presented mean ± SEM.
Online supplemental material
Fig. S1 shows the role of TLR and RIG-I signaling cascades and type I IFN responses in ZBP1-regulated cell death after IAV infection. It also display the role of the RIG-I–MAVS signaling pathway during cell death induced by human and mouse adapted IAV strains. Fig. S2 shows ZBP1’s association with the RNA genome and the IAV proteins NP and PB1. Fig. S3 shows ZBP1’s spatial localization with vRNPs, the role of ZBP1 during MCMV-induced cell death, and a model representing ZBP1 activation in response to IAV infection to trigger programmed cell death.
Supplementary Material
Acknowledgments
We thank Amanda Burton, Brittany Storey, and Bhesh Raj Sharma for technical support, Dr. Ken J. Ishii (Osaka University) for Zbp1−/− mice, Dr. Vishva Dixit (Genentech) for supplying various mutant mice, Dr. P.G. Thomas (St. Jude) for IAV (X31), and Dr. Richard Webby (St. Jude) for seasonal strains of IAV. We thank Dr. Prajwal Gurung and Dr. Sarang Tartey for critical reading of the manuscript and members of the Kanneganti laboratory for their comments and suggestions.
T.-D. Kanneganti is supported by the National Institutes of Health (grants AI101935, AI124346, AR056296, and CA163507) and the American Lebanese Syrian Associated Charities.
The authors declare no competing financial interests.
Author contributions: S. Kesavardhana and T.-D. Kanneganti conceptualized the study. S. Kesavardhana, T. Kuriakose, P. Samir, R.K. Subbarao Malireddi, C.S. Guy, and T.-D. Kanneganti designed the methodology, performed the experiments and conducted the analysis. S. Kesavardhana and T.-D. Kanneganti wrote the manuscript. T.-D. Kanneganti acquired the funding and provided overall supervision.
Footnotes
Abbreviations used:
- DAI
- DNA-dependent activation of IFN regulatory factor
- HA
- hemagglutinin
- IAV
- influenza A virus
- LDH
- lactate dehydrogenase
- LMB
- leptomycin-B
- MAVS
- mitochondrial antiviral signaling protein
- MCMV
- murine cytomegalovirus
- MOI
- multiplicity of infection
- NP
- nuclear protein
- RHIM
- RIP homotypic interaction motif
- STORM
- stochastic optical reconstruction microscopy
- TUBE
- tandem ubiquitin-binding entity
- vRNP
- viral RNP
References
- Agostini F., Zanzoni A., Klus P., Marchese D., Cirillo D., and Tartaglia G.G.. 2013. catRAPID omics: A web server for large-scale prediction of protein-RNA interactions. Bioinformatics. 29:2928–2930. 10.1093/bioinformatics/btt495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arranz R., Coloma R., Chichón F.J., Conesa J.J., Carrascosa J.L., Valpuesta J.M., Ortín J., and Martín-Benito J.. 2012. The structure of native influenza virion ribonucleoproteins. Science. 338:1634–1637. 10.1126/science.1228172 [DOI] [PubMed] [Google Scholar]
- Baum A., Sachidanandam R., and García-Sastre A.. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. USA. 107:16303–16308. (published erratum appears in Proc. Natl. Acad. Sci. USA 2011. 108:3092) 10.1073/pnas.1005077107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blander J.M. 2014. A long-awaited merger of the pathways mediating host defence and programmed cell death. Nat. Rev. Immunol. 14:601–618. 10.1038/nri3720 [DOI] [PubMed] [Google Scholar]
- Cox J., Neuhauser N., Michalski A., Scheltema R.A., Olsen J.V., and Mann M.. 2011. Andromeda: A peptide search engine integrated into the MaxQuant environment. J. Proteome Res. 10:1794–1805. 10.1021/pr101065j [DOI] [PubMed] [Google Scholar]
- Eisfeld A.J., Neumann G., and Kawaoka Y.. 2015. At the centre: Influenza A virus ribonucleoproteins. Nat. Rev. Microbiol. 13:28–41. 10.1038/nrmicro3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elton D., Simpson-Holley M., Archer K., Medcalf L., Hallam R., McCauley J., and Digard P.. 2001. Interaction of the influenza virus nucleoprotein with the cellular CRM1-mediated nuclear export pathway. J. Virol. 75:408–419. 10.1128/JVI.75.1.408-419.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S.C., Kim D., Hwang H.Y., Rich A., Kim Y.G., and Kim K.K.. 2008. The crystal structure of the second Z-DNA binding domain of human DAI (ZBP1) in complex with Z-DNA reveals an unusual binding mode to Z-DNA. Proc. Natl. Acad. Sci. USA. 105:20671–20676. 10.1073/pnas.0810463106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S., Steinmueller M., von Wulffen W., Cakarova L., Pinto R., Pleschka S., Mack M., Kuziel W.A., Corazza N., Brunner T., et al. . 2008. Lung epithelial apoptosis in influenza virus pneumonia: The role of macrophage-expressed TNF-related apoptosis-inducing ligand. J. Exp. Med. 205:3065–3077. 10.1084/jem.20080201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K., Yanai H., Negishi H., Asagiri M., Sato M., Mizutani T., Shimada N., Ohba Y., Takaoka A., Yoshida N., and Taniguchi T.. 2005. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 434:772–777. 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- Hornung V., Ellegast J., Kim S., Brzózka K., Jung A., Kato H., Poeck H., Akira S., Conzelmann K.K., Schlee M., et al. . 2006. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 314:994–997. 10.1126/science.1132505 [DOI] [PubMed] [Google Scholar]
- Huang C.H., Su M.G., Kao H.J., Jhong J.H., Weng S.L., and Lee T.Y.. 2016. UbiSite: Incorporating two-layered machine learning method with substrate motifs to predict ubiquitin-conjugation site on lysines. BMC Syst. Biol. 10:S6 10.1186/s12918-015-0246-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A., and Pillai P.S.. 2014. Innate immunity to influenza virus infection. Nat. Rev. Immunol. 14:315–328. 10.1038/nri3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen I., Rayamajhi M., and Miao E.A.. 2017. Programmed cell death as a defence against infection. Nat. Rev. Immunol. 17:151–164. 10.1038/nri.2016.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato H., Takeuchi O., Sato S., Yoneyama M., Yamamoto M., Matsui K., Uematsu S., Jung A., Kawai T., Ishii K.J., et al. . 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 441:101–105. 10.1038/nature04734 [DOI] [PubMed] [Google Scholar]
- Kawai T., Takahashi K., Sato S., Coban C., Kumar H., Kato H., Ishii K.J., Takeuchi O., and Akira S.. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988. 10.1038/ni1243 [DOI] [PubMed] [Google Scholar]
- Kim K., Khayrutdinov B.I., Lee C.K., Cheong H.K., Kang S.W., Park H., Lee S., Kim Y.G., Jee J., Rich A., et al. . 2011. Solution structure of the Zbeta domain of human DNA-dependent activator of IFN-regulatory factors and its binding modes to B- and Z-DNAs. Proc. Natl. Acad. Sci. USA. 108:6921–6926. 10.1073/pnas.1014898107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose T., and Kanneganti T.D.. 2017. Regulation and functions of NLRP3 inflammasome during influenza virus infection. Mol. Immunol. 86:56–64. 10.1016/j.molimm.2017.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose T., Man S.M., Malireddi R.K., Karki R., Kesavardhana S., Place D.E., Neale G., Vogel P., and Kanneganti T.D.. 2016. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 1:aag2045 10.1126/sciimmunol.aag2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Yamashita K., Amada K.M., and Standley D.M.. 2014. Quantifying sequence and structural features of protein-RNA interactions. Nucleic Acids Res. 42:10086–10098. 10.1093/nar/gku681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedmann S., Hrincius E.R., Guy C., Anhlan D., Dierkes R., Carter R., Wu G., Staeheli P., Green D.R., Wolff T., et al. . 2014. Viral suppressors of the RIG-I-mediated interferon response are pre-packaged in influenza virions. Nat. Commun. 5:5645 10.1038/ncomms6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J., Kumari S., Kim C., Van T.M., Wachsmuth L., Polykratis A., and Pasparakis M.. 2016. RIPK1 counteracts ZBP1-mediated necroptosis to inhibit inflammation. Nature. 540:124–128. 10.1038/nature20558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man S.M., and Kanneganti T.D.. 2016. Converging roles of caspases in inflammasome activation, cell death and innate immunity. Nat. Rev. Immunol. 16:7–21. 10.1038/nri.2015.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E., Curran J., Hofmann K., Moradpour D., Binder M., Bartenschlager R., and Tschopp J.. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 437:1167–1172. 10.1038/nature04193 [DOI] [PubMed] [Google Scholar]
- Moeller A., Kirchdoerfer R.N., Potter C.S., Carragher B., and Wilson I.A.. 2012. Organization of the influenza virus replication machinery. Science. 338:1631–1634. 10.1126/science.1227270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K., Wickliffe K.E., Maltzman A., Dugger D.L., Strasser A., Pham V.C., Lill J.R., Roose-Girma M., Warming S., Solon M., et al. . 2016. RIPK1 inhibits ZBP1-driven necroptosis during development. Nature. 540:129–133. 10.1038/nature20559 [DOI] [PubMed] [Google Scholar]
- Ng S.K., Weissbach R., Ronson G.E., and Scadden A.D.. 2013. Proteins that contain a functional Z-DNA-binding domain localize to cytoplasmic stress granules. Nucleic Acids Res. 41:9786–9799. 10.1093/nar/gkt750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogusa S., Thapa R.J., Dillon C.P., Liedmann S., Oguin T.H. III, Ingram J.P., Rodriguez D.A., Kosoff R., Sharma S., Sturm O., et al. . 2016. RIPK3 activates parallel pathways of MLKL-driven necroptosis and FADD-mediated apoptosis to protect against influenza A virus. Cell Host Microbe. 20:13–24. 10.1016/j.chom.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasparakis M., and Vandenabeele P.. 2015. Necroptosis and its role in inflammation. Nature. 517:311–320. 10.1038/nature14191 [DOI] [PubMed] [Google Scholar]
- Paterson D., and Fodor E.. 2012. Emerging roles for the influenza A virus nuclear export protein (NEP). PLoS Pathog. 8:e1003019 10.1371/journal.ppat.1003019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A., Schulz O., Tan C.P., Näslund T.I., Liljeström P., Weber F., and Reis e Sousa C.. 2006. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 314:997–1001. 10.1126/science.1132998 [DOI] [PubMed] [Google Scholar]
- Placido D., Brown B.A. II, Lowenhaupt K., Rich A., and Athanasiadis A.. 2007. A left-handed RNA double helix bound by the Z alpha domain of the RNA-editing enzyme ADAR1. Structure. 15:395–404. 10.1016/j.str.2007.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothlichet J., Meunier I., Davis B.K., Ting J.P., Skamene E., von Messling V., and Vidal S.M.. 2013. Type I IFN triggers RIG-I/TLR3/NLRP3-dependent inflammasome activation in influenza A virus infected cells. PLoS Pathog. 9:e1003256 10.1371/journal.ppat.1003256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radivojac P., Vacic V., Haynes C., Cocklin R.R., Mohan A., Heyen J.W., Goebl M.G., and Iakoucheva L.M.. 2010. Identification, analysis, and prediction of protein ubiquitination sites. Proteins. 78:365–380. 10.1002/prot.22555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehwinkel J., Tan C.P., Goubau D., Schulz O., Pichlmair A., Bier K., Robb N., Vreede F., Barclay W., Fodor E., and Reis e Sousa C.. 2010. RIG-I detects viral genomic RNA during negative-strand RNA virus infection. Cell. 140:397–408. 10.1016/j.cell.2010.01.020 [DOI] [PubMed] [Google Scholar]
- Rich A., and Zhang S.. 2003. Timeline: Z-DNA: The long road to biological function. Nat. Rev. Genet. 4:566–572. 10.1038/nrg1115 [DOI] [PubMed] [Google Scholar]
- Rodrigue-Gervais I.G., Labbé K., Dagenais M., Dupaul-Chicoine J., Champagne C., Morizot A., Skeldon A., Brincks E.L., Vidal S.M., Griffith T.S., and Saleh M.. 2014. Cellular inhibitor of apoptosis protein cIAP2 protects against pulmonary tissue necrosis during influenza virus infection to promote host survival. Cell Host Microbe. 15:23–35. 10.1016/j.chom.2013.12.003 [DOI] [PubMed] [Google Scholar]
- Roy A., Kucukural A., and Zhang Y.. 2010. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5:725–738. 10.1038/nprot.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders C.J., Vogel P., McClaren J.L., Bajracharya R., Doherty P.C., and Thomas P.G.. 2013. Compromised respiratory function in lethal influenza infection is characterized by the depletion of type I alveolar epithelial cells beyond threshold levels. Am. J. Physiol. Lung Cell. Mol. Physiol. 304:L481–L488. 10.1152/ajplung.00343.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz T., Behlke J., Lowenhaupt K., Heinemann U., and Rich A.. 2001. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat. Struct. Biol. 8:761–765. 10.1038/nsb0901-761 [DOI] [PubMed] [Google Scholar]
- Seth R.B., Sun L., Ea C.K., and Chen Z.J.. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 122:669–682. 10.1016/j.cell.2005.08.012 [DOI] [PubMed] [Google Scholar]
- Takaoka A., Wang Z., Choi M.K., Yanai H., Negishi H., Ban T., Lu Y., Miyagishi M., Kodama T., Honda K., et al. . 2007. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 448:501–505. 10.1038/nature06013 [DOI] [PubMed] [Google Scholar]
- Thapa R.J., Ingram J.P., Ragan K.B., Nogusa S., Boyd D.F., Benitez A.A., Sridharan H., Kosoff R., Shubina M., Landsteiner V.J., et al. . 2016. DAI senses influenza a virus genomic RNA and activates RIPK3-dependent cell death. Cell Host Microbe. 20:674–681. 10.1016/j.chom.2016.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P.G., Dash P., Aldridge J.R. Jr., Ellebedy A.H., Reynolds C., Funk A.J., Martin W.J., Lamkanfi M., Webby R.J., Boyd K.L., et al. . 2009. The intracellular sensor NLRP3 mediates key innate and healing responses to influenza A virus via the regulation of caspase-1. Immunity. 30:566–575. 10.1016/j.immuni.2009.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzeng T.C., Schattgen S., Monks B., Wang D., Cerny A., Latz E., Fitzgerald K., and Golenbock D.T.. 2016. A fluorescent reporter mouse for inflammasome assembly demonstrates an important role for cell-bound and free ASC Specks during in vivo infection. Cell Reports. 16:571–582. 10.1016/j.celrep.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton J.W., Kaiser W.J., and Mocarski E.S.. 2012. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe. 11:290–297. 10.1016/j.chom.2012.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., and Brown S.J.. 2006. BindN: a web-based tool for efficient prediction of DNA and RNA binding sites in amino acid sequences. Nucleic Acids Res. 34:W243–W248. 10.1093/nar/gkl298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach R., and Scadden A.D.. 2012. Tudor-SN and ADAR1 are components of cytoplasmic stress granules. RNA. 18:462–471. 10.1261/rna.027656.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L.G., Wang Y.Y., Han K.J., Li L.Y., Zhai Z., and Shu H.B.. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell. 19:727–740. 10.1016/j.molcel.2005.08.014 [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., and Akira S.. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 301:640–643. 10.1126/science.1087262 [DOI] [PubMed] [Google Scholar]
- Zarling D.A., Calhoun C.J., Hardin C.C., and Zarling A.H.. 1987. Cytoplasmic Z-RNA. Proc. Natl. Acad. Sci. USA. 84:6117–6121. 10.1073/pnas.84.17.6117 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.