Activation of cancer-associated fibroblasts (CAFs) promotes tumorigenesis. Kim et al. show that ATF3 and CSL converge in negative regulation of CAF activation through long-distance chromatin control. Bromodomain and extra-terminal (BET) inhibitors counteract the effects of ATF3 and CSL loss in CAF activation and cancer–stromal cell expansion.
Abstract
Cancer-associated fibroblasts (CAFs) are important for tumor initiation and promotion. CSL, a transcriptional repressor and Notch mediator, suppresses CAF activation. Like CSL, ATF3, a stress-responsive transcriptional repressor, is down-modulated in skin cancer stromal cells, and Atf3 knockout mice develop aggressive chemically induced skin tumors with enhanced CAF activation. Even at low basal levels, ATF3 converges with CSL in global chromatin control, binding to few genomic sites at a large distance from target genes. Consistent with this mode of regulation, deletion of one such site 2 Mb upstream of IL6 induces expression of the gene. Observed changes are of translational significance, as bromodomain and extra-terminal (BET) inhibitors, unlinking activated chromatin from basic transcription, counteract the effects of ATF3 or CSL loss on global gene expression and suppress CAF tumor-promoting properties in an in vivo model of squamous cancer–stromal cell expansion. Thus, ATF3 converges with CSL in negative control of CAF activation with epigenetic changes amenable to cancer- and stroma-focused intervention.
Introduction
Epithelial–mesenchymal interactions play an important role in the control of normal tissue homeostasis and carcinogenesis (Bissell and Hines, 2011; Junttila and de Sauvage, 2013). Changes in tumor stroma, such as induction of cancer-associated fibroblasts (CAFs) and associated chronic inflammation, are most frequently viewed as secondary to changes in the epithelium. However, recent evidence indicates that they may play a concurrent or even primary role in cancer initiation and progression (Dotto, 2014).
Field cancerization—multifocal and recurrent tumors with broader changes of surrounding tissues—is a frequent clinical occurrence in many organs, such as the skin, head/neck, lung, breast, prostate, and colon (Dotto, 2014). It is very difficult to treat, and greater insights into how it develops are urgently needed. We have developed a mouse model of skin field cancerization based on mesenchymal deletion of the Csl gene, the key effector of canonical Notch signaling (Hu et al., 2012). CSL, a DNA-binding protein with intrinsic transcription repressive function, negatively regulates many genes with CAF effector functions, encoding mitogenic growth factors, proinflammatory cytokines, and matrix remodeling proteins (Hu et al., 2012; Procopio et al., 2015). In addition, CSL physically interacts with p53, suppressing its activity, and escape from p53-induced stromal cell senescence, resulting from loss of CSL, is part of a cancer-associated stromal cell evolution process (Procopio et al., 2015). The transcription/chromatin modification events that converge with compromised CSL function in the early steps of CAF activation are as yet unknown.
ATF3, a stress response transcription factor of the AP1/CREB family, has been extensively studied in cells of immune and inflammatory systems (Thompson et al., 2009). Increased Atf3 expression plays an important role in oncogenic conversion of keratinocytes, as it occurs in the skin of organ graft recipients being treated with calcineurin inhibitor drugs (Wu et al., 2010). The role of ATF3 in dermal fibroblasts has not yet been explored. We report herein that ATF3 converges with CSL in negative control of CAF activation. Compromised expression of either gene results in strikingly similar chromatin modifications and changes in gene expression, which can be counteracted by bromodomain and extra-terminal (BET) inhibitors in vitro and in vivo.
Results
Atf3 deficiency promotes dysplastic skin tumors and CAF activation
The role of ATF3 in dermal fibroblasts has not been investigated. UVA, a main cause of skin cancer, can directly affect cells of the dermal compartment (Bachelor and Bowden, 2004). Atf3 expression was induced in primary human dermal fibroblasts (HDFs) soon after UVA treatment at doses equivalent to 2 h of sunlight exposure (Fig. S1, A and B; Balasaraswathy et al., 2002), a finding validated by immunofluorescence (IF) analysis of UVA-treated human skin explants (Fig. S1 C). Atf3 up-regulation was transient and inversely related to Csl expression, which is down-modulated by UVA exposure of HDFs (Fig. S1 D; Menietti et al., 2016).
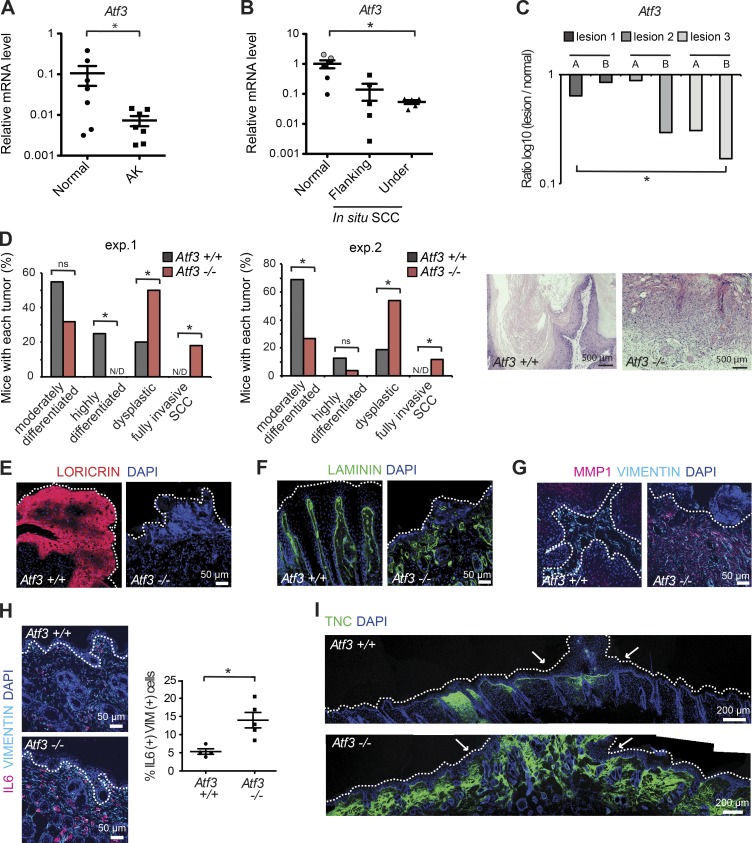
Actinic keratoses (AKs) are precancerous lesions that develop in photo-damaged skin. Laser capture microdissection (LCM) followed by RT-qPCR analysis showed significant down-modulation of Atf3 expression in AK-underlying dermal cells versus cells from surrounding unaffected skin, similarly to decreased Csl (Fig. 1 A; Procopio et al., 2015). In situ skin squamous cell carcinomas (SCCs) have greater cellular abnormalities than AKs, which extend to all epidermal layers. Although there is not yet invasion, alterations of dermal tissue are likely to occur as part of expanding cancer fields (Dotto, 2014). LCM analysis showed decreased Atf3 expression in stromal fibroblasts underlying in situ SCCs relative to those in flanking skin of the same patients and, to a greater extent, of other individuals (Fig. 1 B). Leukocyte and endothelial cell contaminations in tested samples were ruled out by previous analysis with corresponding markers and the use of anti-PDGFRα antibodies for fibroblast identification before capture (Procopio et al., 2015). Results were further validated in mice with mesenchymal deletion of the Csl gene (Hu et al., 2012). LCM and RT-qPCR analysis showed significant down-modulation of stromal Atf3 expression in keratinocyte lesions that developed in these mice at 3 mo of age relative to distant unaffected skin (Fig. 1 C).
Figure 1.
Atf3 deficiency promotes dysplastic keratinocyte tumor development and widespread stromal alterations. (A) Laser-captured microdissection (LCM) and RT-qPCR analysis of Atf3 expression in actinic keratosis (AK)–underlying stromas (n = 7) and matched flanking normal skin stromas (normal; n = 7) from patients using 36β4 normalization. Data are mean ± SEM, two-tailed paired t test, *, P < 0.05. (B) LCM followed by RT-PCR analysis of Atf3 expression in PDGFRα-positive cells from stroma underlying in situ SCC lesions (n = 5) versus flanking skin (n = 5) and dermis (n = 6) from other individuals (face and abdomen derived; black and gray circles) using β−actin for normalization. Data are mean ± SEM, two-tailed unpaired t test, *, P < 0.05. (C) LCM and RT-qPCR analysis of Atf3 expression in stromal cells from three skin lesions (two areas per lesion: A and B) versus matching stroma of unaffected skin in mice with mesenchymal Csl deletion. n = 6 affected regions and three unaffected regions, ratio log10(lesion/unaffected), two-tailed one-sample t test, *, P < 0.05. The same samples were previously analyzed for possible leukocyte contamination (Procopio et al., 2015). (D) Two independent experiments with mice with and without Atf3 gene deletion subjected to a multistep skin chemical carcinogenesis protocol over 25 wk. Time of appearance, incidence, and number of tumors are shown in Fig. S1 E. n = 20 tumors from eight Atf3+/+ mice n = 22 tumors from 7 Atf3−/− mice for the first experiment; n = 32 tumors from 8 Atf3+/+ mice and n = 26 tumors from 9 Atf3−/− mice for the second experiment. Two-tailed χ2 test, *, P < 0.05. N/D, not detected; ns, not significant. The left panels are a quantification of the histological results expressed as percentages of the total number of analyzed tumors per genotype. The right panels are representative images of a highly differentiated versus dysplastic papilloma. (E-G) IF analysis of tumors and underlying tissue from Atf3+/+ versus −/− mice with antibodies (Abs) against the indicated proteins. (H) IF analysis and quantification of double-positive cells in vimentin and IL6 expression in nontumor-bearing skin areas of Atf3−/− versus Atf3+/+ mice. n = 5 fields from two mice for each genotype, counting 40–100 vimentin-positive cells per field. Data are mean ± SEM, two-tailed unpaired t test, *, P < 0.05. (I) IF analysis of entire mouse back skin with Ab against TNC in the skin of Atf3−/− versus Atf3+/+ mice at tumor areas (bracketed by arrows). Additional representative images for D, H, and I are shown in Fig. S1 (F–H).
To assess the functional significance of decreased Atf3 expression, we assessed susceptibility of mice with and without deletion of the Atf3 gene (Hartman et al., 2004) to skin chemical carcinogenesis. The fraction of Atf3−/− mice that developed tumors was slightly less than that of the controls in one experiment, with no significant difference in another (Fig. S1 E). Importantly, Atf3−/− mice developed a much higher fraction of dysplastic lesions than controls and, over the tested time frame, were the only ones to have invasive, at times ulcerative, SCCs (Fig. 1 D and Fig. S1 F). Expression of terminal differentiation markers such as loricrin was strongly reduced in tumors formed in Atf3−/− mice (Fig. 1 E), with basement membrane fragmentation and massive up-regulation of matrix metalloprotease 1 (MMP1) in tumor stroma (Fig. 1, F and G). IL6, a proinflammatory and protumorigenic cytokine, and tenascin C (TNC), a cancer-associated matrix protein, were highly expressed in the tumor-surrounding stroma of Atf3−/− mice and also in the dermis farther away (Fig. 1, H and I; Fig. S1, G and H).
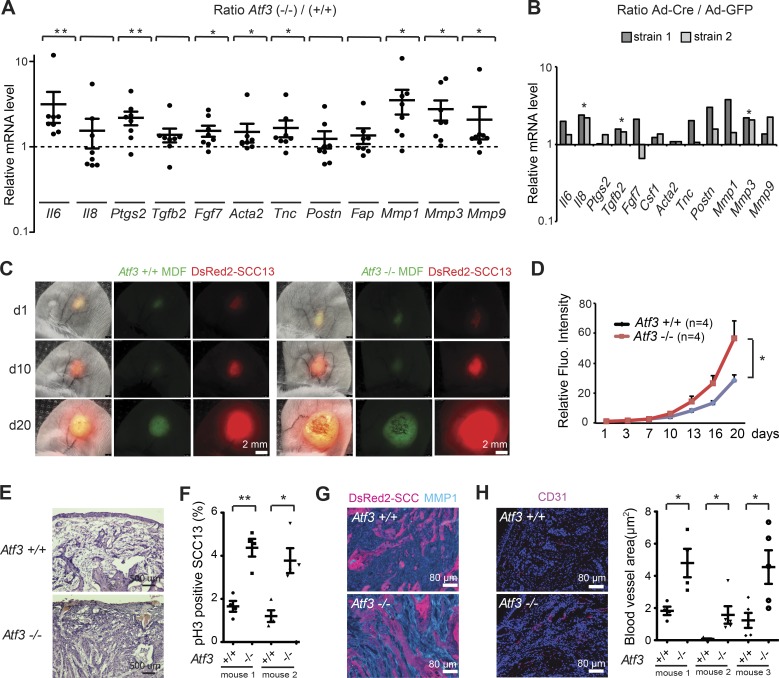
To focus on intrinsic consequences of Atf3 deletion in dermal fibroblasts, we derived these cells from mice with and without Atf3 deletion and from mice with the “floxed” Atf3 gene (Wolford et al., 2013) followed by in vitro deletion via infection with a Cre-expressing adenovirus. In both cases, Atf3 gene deletion resulted in pronounced up-regulation of various CAF effector genes, encoding growth factors and cytokines, matrix proteins, and MMPs (Fig. 2, A and B). For in vivo significance, we used a cancer–stromal cell expansion assay (Procopio et al., 2015) with parallel mouse ear injections of dermal fibroblasts with and without Atf3 deletion admixed with weakly tumorigenic skin SCC cells (SCC13) expressing DsRed2. Analysis of red fluorescence surface area and signal intensity at daily intervals showed greater expansion of SCC13 cells admixed with Atf3-deficient compared with control fibroblasts (Fig. 2, C and D). Two of five mice injected with Atf3−/− dermal fibroblasts developed ulcerative tumors with disruption of the ear epidermis, whereas no such ulcerations arose in contralateral lesions. Histological analysis showed greater tumor cell density in lesions formed together with Atf3−/− fibroblasts with a higher fraction of phospho-H3–positive tumor cells (Fig. 2, E and F; and Fig. S1, I and J). MMP1 expression was much more pronounced in the stromal region of these tumors (Fig. 2 G and Fig. S1 K), and vessel density, as assessed by CD31 immune staining, was significantly increased (Fig. 2 H).
Figure 2.
Atf3 deletion in mouse dermal fibroblasts enhances tumorigenic behavior of keratinocyte-derived SCC. (A) RT-qPCR analysis of the indicated genes in multiple parallel cultures of primary dermal fibroblasts derived from either individual or pooled newborn Atf3−/− (n = 8) and Atf3+/+ (n = 5) mice. Values are expressed as log10 ratios in Atf3−/− versus Atf3+/+ cultures. Data are mean ± SEM, one-tailed one-sample t test, *, P < 0.05; **, P < 0.005. (B) RT-qPCR analysis of the indicated genes in two different preparations of primary dermal fibroblasts from Atf3loxP/loxP mice at 3 d after infection with a Cre-expressing adenovirus (Ad-Cre; n = 2) or GFP control (Ad-GFP; n = 2) for Atf3 gene deletion. In both experiments, Atf3 expression was reduced more than 40-fold in Ad-Cre–infected cells. Values are expressed as log10 ratios in cultures with and without Atf3 deletion. One-tailed, one-sample t test, *, P < 0.05. (C and D) Human SCC13 cells (DsRed) were admixed with mouse dermal fibroblasts (MDFs) with and without Atf3 deletion labeled with the PKH67 dye (green) followed by parallel ear injections into NOD/SCID mice. (C) Representative live images of a mouse pair injected with the combinations of cells 10 and 20 d after injection. (D) Quantification of red fluorescence signal (intensity × surface area) corresponding to SCC13 cell expansion. n = 4 lesions per condition, mean ± SEM, two-tailed paired t test, *, P < 0.05. (E) H&E staining of ear lesions formed by SCC13 cells injected with dermal fibroblasts with and without Atf3 deletion. (F) Quantification of phospho-histone 3 (pH3)–positive DsRed2-SCC13 cells in mouse ear lesions. n = 4 fields per lesion, at least 200 cells per field; mean ± SEM, two-tailed unpaired t test, *, P < 0.05; **, P < 0.005. (G) IF analysis of mouse ear lesions with Ab against MMP1 using DsRed2 fluorescent signal for SCC cell identification. (H) Representative images of IF analysis of ear lesions with Ab against CD31 (left panels), together with quantification of the total blood vessel area per field (μm2) of three pairs of ear lesions using ImageJ software (right panel). n = 4 (for mouse 1) and n = 5 (for mice 2 and 3) fields per lesion, mean ± SEM, two-tailed unpaired t test, *, P < 0.05. Additional representative images for E–G are shown in Fig. S1 (I–K).
As a complement, dermal fibroblasts with and without Atf3 deletion were admixed with either SCC13 cells or a mouse skin SCC line (PAM212) followed by intradermal injections into mouse back skin. Even under these conditions, tumors formed in the presence of Atf3-deficient fibroblasts exhibited greater cellularity, with reduced squamous differentiation, an increased proliferative index, and higher IL6 stromal expression (Fig. S1, L–O).
Overlapping transcription repressive function of ATF3 and CSL on CAF effector genes
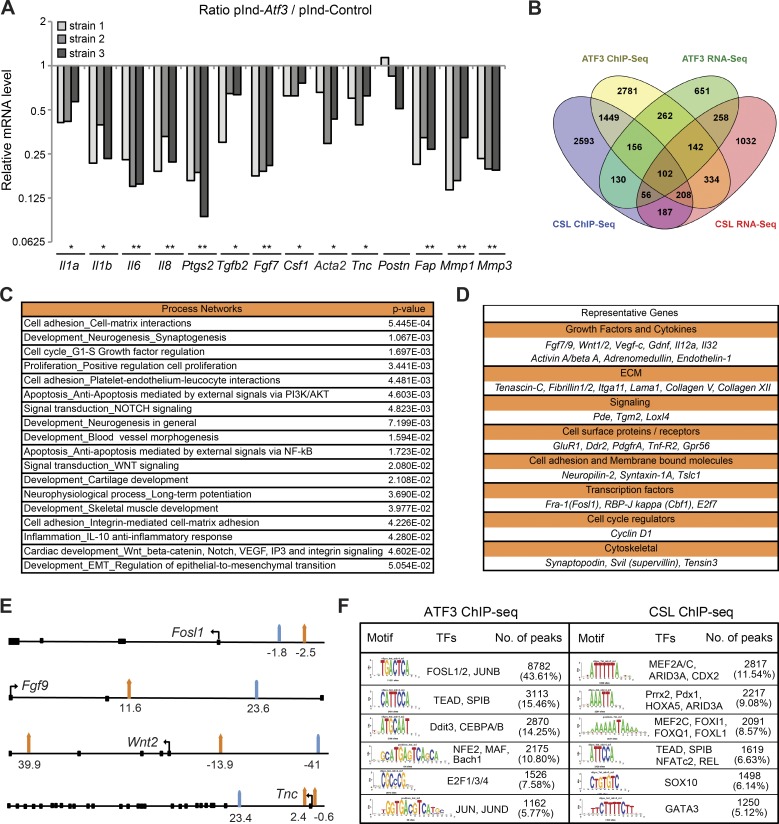
The ATF3 transcription repressive function may overlap with that of CSL. Increased Atf3 expression by lentiviral vector infection, to levels comparable to those induced by UVA (Fig. S1 B), caused suppression of various CAF-related genes (Fig. 3 A) that are under negative CSL control (Procopio et al., 2015). For global assessment, we combined RNA-seq and ChIP-seq analysis of HDFs with and without increased ATF3 expression and compared the results with those for CSL (Procopio et al., 2015). In cells with increased ATF3, 662 modulated genes were direct ATF3 targets, with 102 also being under direct CSL control (Fig. 3 B and Table S1). ATF3 and ATF3/CSL–regulated genes were significantly enriched for families related to cell adhesion–matrix interactions, development, and proliferation (P < 0.05; Fig. 3 C and Table S2). Direct ATF3 and CSL targets included many genes of biological significance, coding for growth factors and cytokines, cancer-associated matrix proteins, tyrosine kinase receptors, and transcription factors, including Csl itself (Fig. 3 D).
Figure 3.
Induced Atf3 suppresses CAF effector genes with large overlap of binding targets with CSL. (A) Several HDF strains infected with a lentiviral vector for Doxycycline (Dox)–inducible Atf3 expression (pInd-Atf3; n = 3) versus empty vector control (pInd-Ctr; n = 2) were analyzed with and without Dox treatment (750 ng/ml for 3 d) for expression of the indicated genes. Values are expressed as ratio log10 (pInd-Atf3/pInd-Ctr), two-tailed, one-sample t test, *, P < 0.05, **, P < 0.005. (B) Overlap between ChIP-seq and RNA-seq profiles of genes bound and modulated by increased ATF3 expression in HDFs and those previously identified to be bound and controlled by CSL (Procopio et al., 2015). A complete list of genes is provided in Table S1. (C) GeneGo MetaCore analysis was performed with the data set of common direct target genes regulated by induced ATF3 and CSL. Shown is a list of process networks with statistically significant enrichment (P < 0.05). The complete list is provided in Table S2. (D) Representative list of common directly regulated genes by ATF3 and CSL divided by biological function. (E) Schematic representation of induced ATF3 (orange arrows) and CSL (turquoise arrows) binding peak positions for four genes commonly targeted and regulated by induced ATF3 and CSL listed in D. Coding exons are shown as black squares, and the directions of the transcription start sites are shown as black arrows. A complete list of induced ATF3 and CSL binding peak positions is provided in Table S3. (F) Motif enrichment analysis of nucleotide sequences surrounding elevated ATF3 and CSL binding peaks was performed by the RSAT peak-motifs tool. The number of ATF3 or CSL binding peaks with at least one predicted site of the indicated transcription factors is indicated along with the corresponding frequency (percentage).
ATF3 and CSL binding peaks on common target genes and at the global genomic level were mostly separate from each other (Fig. 3 E and Table S3), suggesting that the two factors do not directly interact but function in concert with other regulators of transcription. In fact, motif enrichment analysis (MEA) showed frequent overlapping of ATF3 and CSL binding with recognition sequences for different sets of transcription factors with which these proteins are known to associate, such as Jun and Fos family members for ATF3 (Thompson et al., 2009) and MEF2C for CSL (Shen et al., 2006; Fig. 3 F). A fraction of CSL or ATF3-binding regions shared recognition sequences for TEAD transcription factors, downstream mediators of YAP/TAZ activation in CAFs (Calvo et al., 2013) and AP1 family coregulators (Zanconato et al., 2015), and SPIB, an ETS family member (Schotte et al., 2004; Dontje et al., 2006).
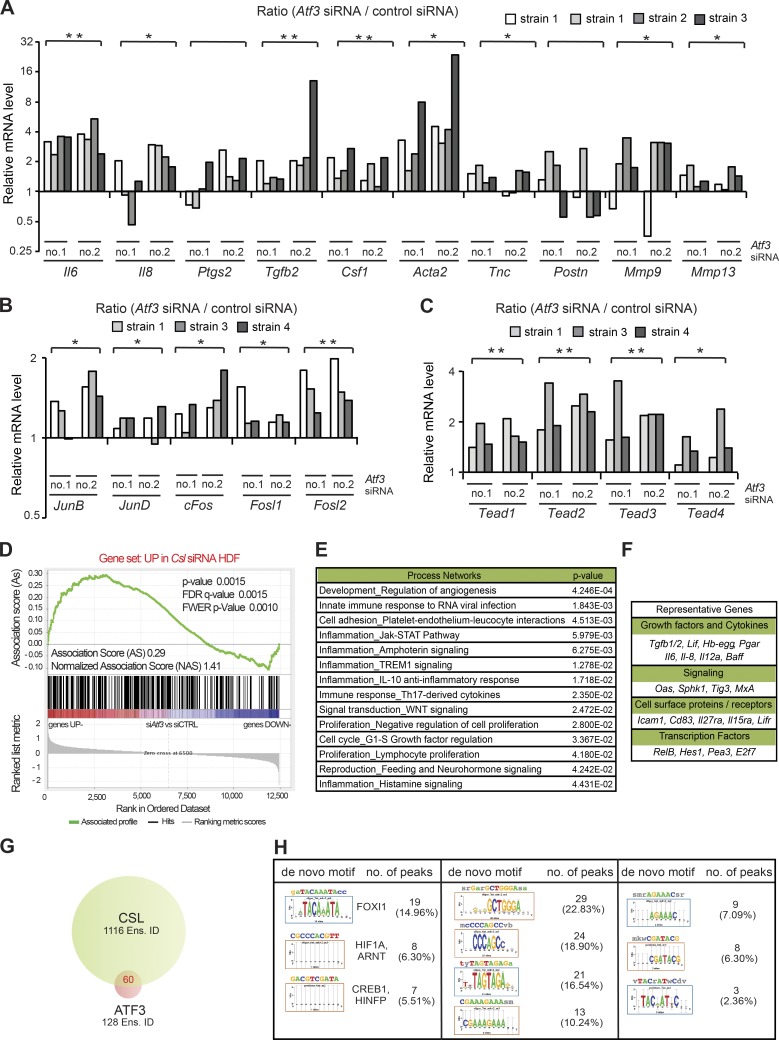
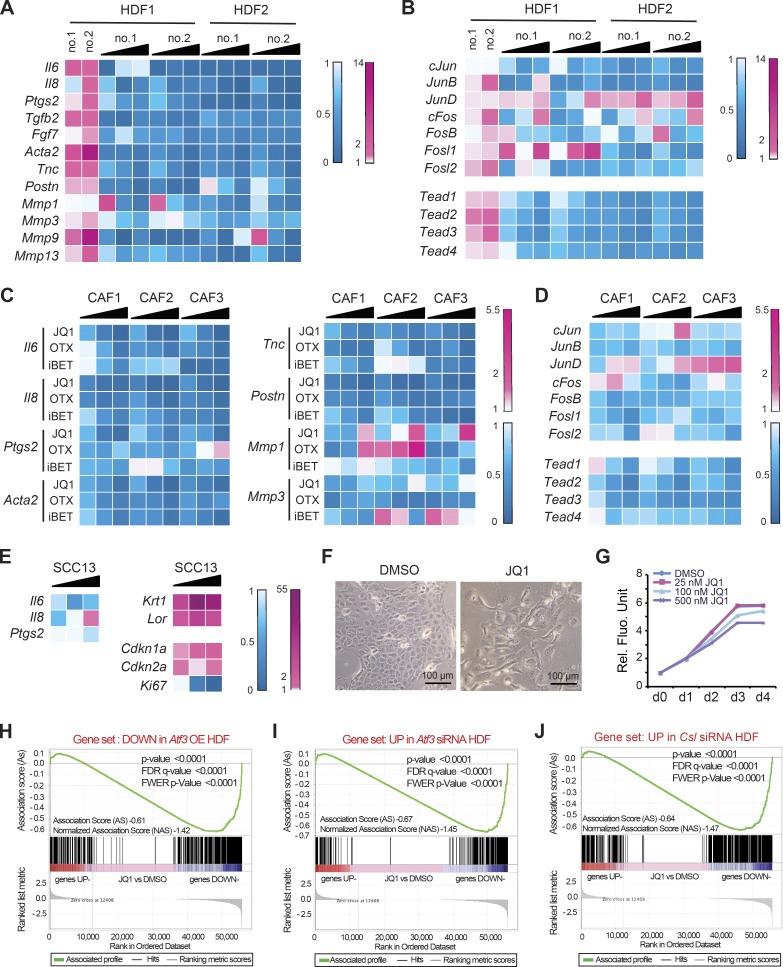
In contrast to stress responses, the role of ATF3 under basal conditions is much less studied. The mouse gene deletion studies indicated that low levels of Atf3, like Csl, are also involved in negative control of CAF effector genes. In fact, up-regulation of these genes was not limited to mouse dermal fibroblasts but was also observed with several HDF strains of independent origin upon Atf3 gene silencing (Fig. 4 A). Genes such as Il6, Il8, Ptgs2 (Cox2), Acta2 (αSma), Tgfb2, Csf1, Tnc, and Postn, suppressed by increased ATF3 expression (Fig. 3 A), were induced by its silencing (Fig. 4 A). Concomitantly, positive regulators of CAF effector gene expression such as specific AP1 family members and TEAD transcription factors were up-regulated (Fig. 4, B and C).
Figure 4.
Overlapping transcription repressive function of low levels of ATF3 and CSL. (A) RT-qPCR analysis of CAF effector genes in several independent strains of HDFs (strain 1 tested twice at different passages) with and without Atf3 silencing by two different siRNAs (no. 1 and no. 2), using 36β4 for normalization. n (HDF strains) = 8 siAtf3 no.1 and 2; 4 control siRNA. Values in A–C are expressed as log2 ratios (Atf3 siRNA/control siRNA), one-tailed, one-sample t test, *, P < 0.05; **, P < 0.005. (B) RT-qPCR analysis of AP1 family genes in independent HDF strains. n = 6 (siAtf3 no. 1 and 2), n = 3 (control siRNA). (C) RT-qPCR analysis of TEAD family members in independent HDF strains (strain 1 tested twice at different passages). n (HDF strains) = 6 siAtf3 no.1 and 2; 3 control siRNA. (D) Plot for gene set enrichment analysis (GSEA) using RNA-seq expression profiles of two HDF strains with and without silencing of Atf3 by two different siRNA against genes up-modulated in HDF by Csl silencing (Procopio et al., 2015). Genes are ranked by signal-to-noise ratio based on their differential expression in control versus ATF3-silenced HDFs; position of genes in the CSL gene set is indicated by black vertical bars, and the enrichment score is shown in green. (E) List of significantly affected process networks (P < 0.05) as identified by GeneGo MetaCore analysis of genes within the core enrichment resulting from GSE analysis in D. The complete list is provided in Table S5. (F) List of representative genes within the core enrichment of GSE analysis in D divided by biological function. (G) Venn diagram illustrating the number of Ensembl gene/transcript identifiers (Ens. ID) bound by endogenous ATF3 and CSL in HDFs, as identified by ChIP-seq analysis with anti-ATF3 Ab of HDFs under basal conditions in comparison with the previously obtained ChIP-seq binding profile of endogenous CSL (Procopio et al., 2015). A complete list of binding peaks and corresponding genes is provided in Table S6. (H) Motif enrichment analysis of nucleotide sequences surrounding endogenous ATF3 binding peaks using the RSAT peak-motifs tool. The number of ATF3 binding peaks with at least one predicted site of the indicated transcription factors is indicated, along with the corresponding percentage.
For global assessment, two HDF strains with and without ATF3 silencing were analyzed by RNA-seq. Several gene families of significance related to development, cell adhesion, cell cycle, inflammation/immune response, and reproduction/sex hormone signaling were affected by both increased and suppressed ATF3 expression (Table S4). Other families of relevance related to Notch and Wnt signaling and EMT were selectively affected by silencing of the gene, consistent with all other evidence discussed later in the paper that ATF3 at basal levels fulfills a constitutive function not affected by increases of its expression (Table S4).
Results were extended by gene set enrichment analysis (GSEA; Subramanian et al., 2005). Using this approach, we found a highly significant correlation between up-regulated genes in HDFs with Atf3 versus Csl gene silencing (Fig. 4 D) and with gene signatures related to development/angiogenesis and inflammation (Fig. 4 E and Table S5). Similarly up-regulated genes included many for growth factors/cytokines, membrane proteins/receptors, signaling, and transcription (Fig. 4 F). Significant correlation was also found between genes down-regulated by Atf3 versus Csl silencing, with families related to developmental pathways including Notch and Wnt signaling (Fig. S2 A and Table S5).
We further tested whether there was any detectable binding of low levels of ATF3 to specific target genes, which may account for observed changes in gene expression. Using ChIP-seq of HDFs under basal conditions, we found only 133 peaks of ATF3 binding with statistical significance equal to that of CSL binding peaks, which mapped within 128 Ensembl gene/transcript identifiers (Ens. ID: http://www.ensembl.org/info/data/ftp/index.html). 60 of these gene identifiers were also CSL targets (Fig. 4 G and Table S6). MEA showed, next to ATF3 binding sites, recognition sequences for possibly relevant transcription factors, including HIF1A and CREB1 (Fig. 4 H).
The Csl gene itself was among the direct ATF3 targets, which were, however, only slightly up-regulated or not significantly modulated by Atf3 silencing in various HDF strains. The other ATF3 target sites were within protein-coding genes of little-known functions with flanking or embedded lncRNAs, pseudogenes, and miRNAs and/or consisted exclusively of noncoding sequences (Table S6). Interestingly, however, ATF3 binding was within 0.2–2 Mb of several genes of relevance, such as Il6 and Tead4, a finding that was confirmed by ChIP assays with corresponding specific primers in two other HDF strains different from those analyzed by ChIP-seq (Fig. S2 B).
Convergent control of chromatin configuration by endogenous ATF3 and CSL
ATF3 functions in association with various chromatin-modifying enzymes, and loss of ATF3 at specific binding sites may result in altered chromatin configuration even of loci at a large distance. To assess this possibility, we performed ChIP-seq analysis of HDFs with and without Atf3 gene silencing for specific histone modifications (H3K27ac, H3K27me3) and RNA polymerase II (Pol II) recruitment. We used the Integrative Genomics Viewer (IGV; Thorvaldsdóttir et al., 2013) to visualize the ChIP-seq peaks in HDFs with and without silencing of Atf3 or Csl, individually and in combination.
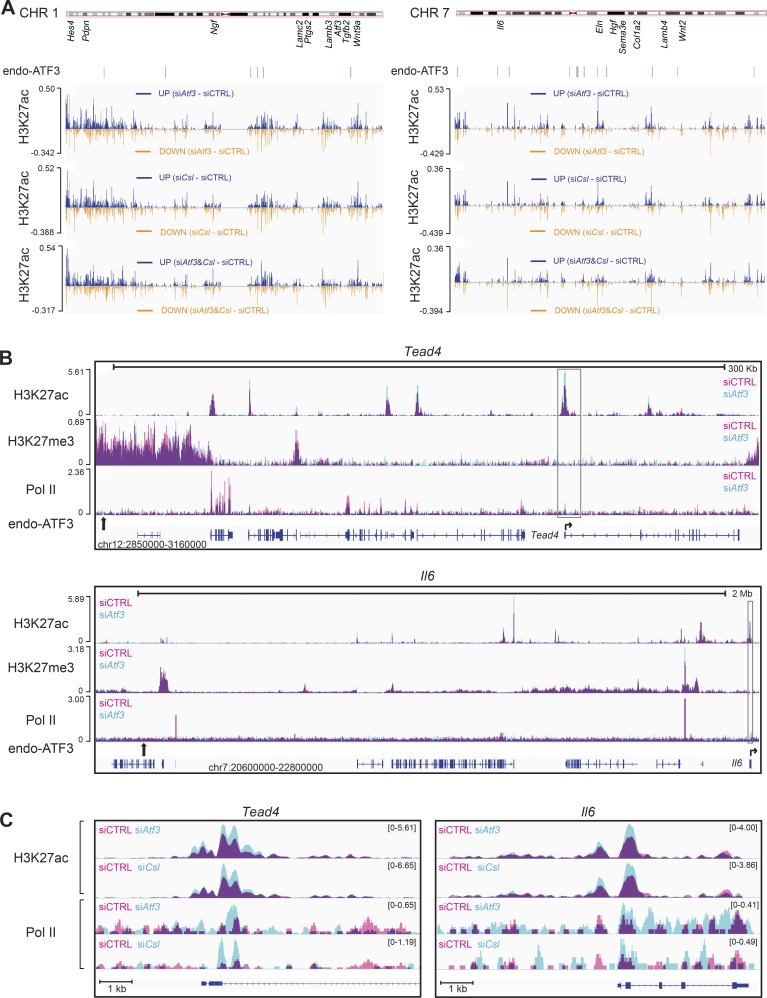
H3K27 acetylation is a general indicator of open chromatin configuration. We found pronounced changes in H3K27ac levels at various regions along chromosomes upon silencing of ATF3, even at a very large distance from its binding sites (Fig. 5 A and Fig. S2, C and D). Interestingly, there was an intermingling of increased and decreased H3K27ac binding peaks, which, at this resolution, could not be resolved. The H3K27ac changes caused by Atf3 silencing were strikingly similar to those resulting from Csl silencing, with concomitant silencing of the two genes having no obvious additive effects (Fig. 5 A).
Figure 5.
Atf3 and Csl gene silencing cause similar overlapping changes in global chromatin configuration. ChIP-seq analysis of histone marks (H3K27ac and H3K27me3) and RNA Pol II in HDFs upon siRNA-mediated silencing of Atf3 and Csl compared with the scrambled siRNA control. (A) Difference in H3K27 acetylation along the length of two representative chromosomes (CHR 1 and 7) in HDFs with silencing of Atf3 (siAtf3) and Csl (siCsl), individually and in combination (siAtf3 and Csl) relative to control cells (siCtrl). The approximated location of individual CAF-related genes and endogenous ATF3 binding position as assessed by ChIP-seq are shown below the chromosomal maps. Individual patterns of H3K27ac along the length of the two chromosomes in A are shown in Fig. S2 C. Differences in H3K27ac for the other two chromosomes (CHR 2 and 4) are shown in Fig. S2 D. (B) Changes in H3K27 acetylation and trimethylation together with Pol-II recruitment caused by Atf3 silencing at genomic loci encompassing endogenous ATF3 binding sites and flanking Tead4 and Il6 genes. Peaks of histone modification or Pol-II present in control HDFs (siCTRL) are shown in pink, peaks in HDFs with Atf3 gene silencing (siAtf3) are shown in turquoise, and overlapping areas present in both cells are shown in purple. For each panel, the chromosomal locations of the shown loci and scale bars are depicted at the bottom and top, respectively. Positions of ATF3 binding sites (calculated by MACS; Table S6) are indicated by vertical black arrows, and their zoom-in binding peaks are shown in Fig. S3 B. Lower lines are transcribed and spliced regions (blue boxes = coding exons) as predicted by Ensembl, together with the position and direction of transcription start sites (horizontal arrows). (C) “Zoomed in” chromatin modifications of the Tead4 and Il6 loci corresponding to the boxed region in B. Individual profiles for each condition were visualized in IGV and merged using Adobe Photoshop.
We then “zoomed in” on genomic regions encompassing deregulated relevant genes that map within 2 Mb from ATF3 binding sites to include distal enhancer elements (Merkenschlager and Odom, 2013). As previously mentioned, the coding region for the Tead4 transcription factor lies within 0.2 Mb of an ATF3 binding site on chromosome 12. ATF3 silencing resulted in a substantial increase of H3K27ac at several proximal regions of the Tead4 locus and at its transcription start site, to which Pol II recruitment was also enhanced (Fig. 5, B and C). Increases in H3K27ac and Pol II recruitment were similar in HDFs with Csl gene silencing (Fig. 5 C).
Among CAF effector genes, Il6 also maps within 2 Mb of an ATF3 binding site. Even in this case, silencing of the Atf3 and Csl genes caused similar pronounced changes of H3K27ac and Pol II recruitment at the promoter region (Fig. 5, B and C). This similarity of the effects was also found for other transcription regulatory or CAF effector genes with altered expression in HDFs with and without Atf3 silencing located far away from ATF3 binding sites (Fig. S3).
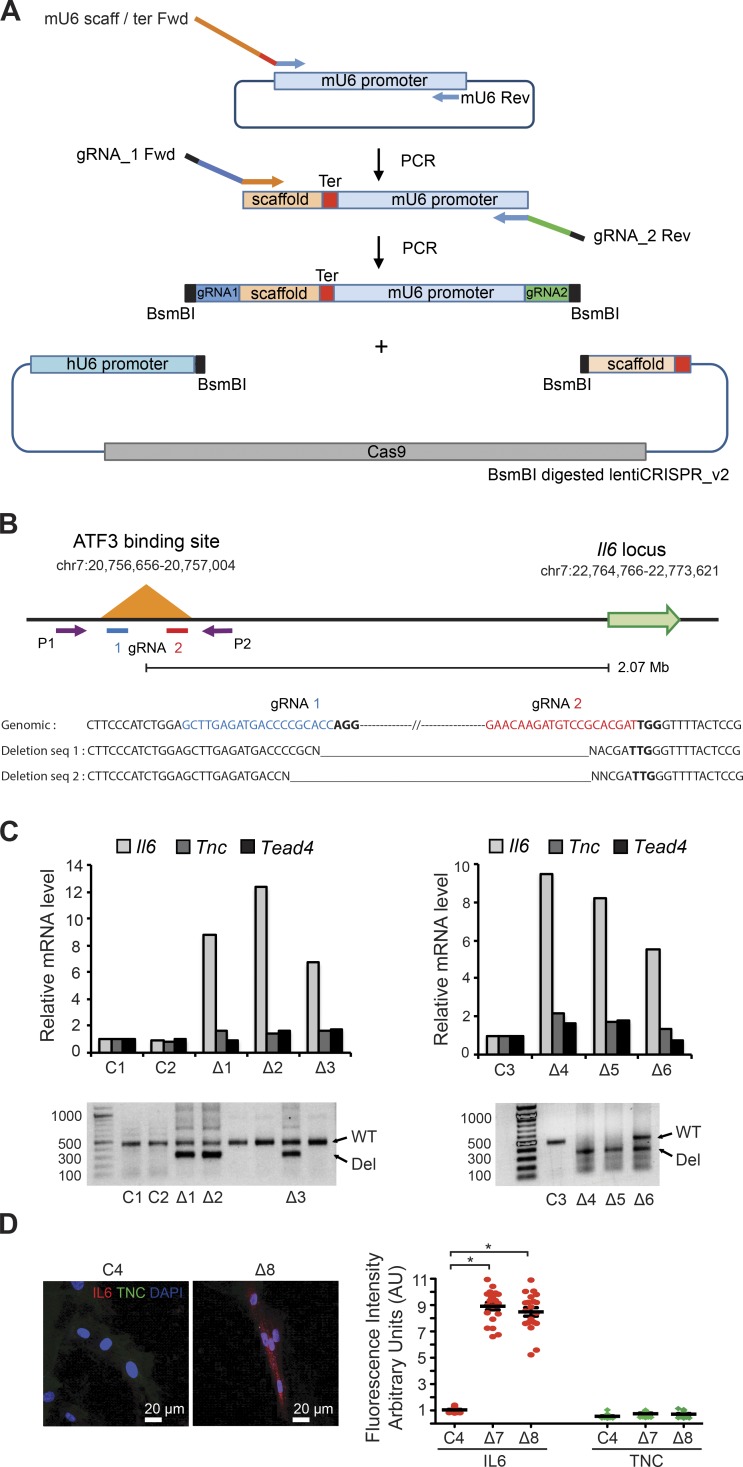
To directly assess whether ATF3 binding plays a specific cis-regulatory function for genes at a large distance, we focused on the Il6 locus and resorted to CRISPR-mediated deletion of the ATF3 binding region located 2 Mb upstream (Fig. 6, A and B). Several independent HDF clones were confirmed to have undergone the expected deletion by genomic PCR and nucleotide sequencing of the flanking region (Fig. 6, B and C). RT-qPCR analysis of these clones showed that Il6 expression was significantly induced, whereas that of two other genes under ATF3 control, Tnc and Tead4, was marginally affected (Fig. 6 C). Consistent with these results, immunofluorescence analysis showed up-regulation of IL6 protein levels in HDFs with deletion of the ATF3 binding site (Fig. 6 D).
Figure 6.
Deletion of an ATF3 binding region 2 Mb upstream of the IL6 gene results in specific induction of IL6 expression. (A) Overview of the construction of a dual-guide RNA (gRNA)–Cas9 expression vector. A two-step PCR amplification followed by insertion into the lentiCRISPR_v2 vector backbone was used to create a dual-expression cassette with mouse (mU6) and human (hU6) U6 promoters driving expression of two gRNAs targeting the ATF3 binding region upstream of the Il6 locus. For details, see Materials and methods. (B, top) Schematic representation of ATF3 binding region 2.07 Mb upstream of the Il6 locus, with the position of the dual gRNAs (gRNA 1 and 2) chosen for CRISPR/Cas9–mediated deletion and of the two primers (P1 and P2) used for genomic PCR analysis. (B, bottom) Nucleotide sequence of the genomic locus targeted by the two gRNAs (blue and red, respectively) together with PAM motifs (bold lines); below are the nucleotide-sequencing results of 300-bp genomic PCR products derived from two HDF clones expected to harbor the deletion (as shown in the panel below). (C) Small clusters/colonies of HDFs infected with the dual gRNAs lentiCRISPR vector (▵) and empty vector control (C) were analyzed by genomic PCR analysis with the P1 and P2 primers indicated above. Clones harboring the deletion in either a heterozygous or homozygous state, on the basis of PCR products of 300 bp versus 500 bp in size, were further analyzed by RT-qPCR for levels of Il6, Tnc, and Tead4 expression. (D) Two additional deletion-harboring clones were analyzed together with controls by IF with Ab against IL6 and TNC. Shown are representative images together with a quantification of the fluorescence intensity signal in individual cells of deletion-harboring clones (▵) versus controls (C). n = 20 cells per condition, mean ± SEM, two-tailed unpaired t test, *, P < 0.0001.
BET inhibitors suppress CAF effector genes, with overlapping impact with ATF3- and CSL-dependent gene transcription
To assess the translational significance of the findings, we tested the impact of pharmacological inhibitors of BET proteins, which link acetylation of histones and other proteins and the basal apparatus (Wang and Filippakopoulos, 2015). Treatment of two different HDF strains with silenced Atf3 with the BET inhibitor JQ1 suppressed expression of all CAF marker genes (Fig. 7 A) and positive regulators of CAF gene expression such as TEAD transcription factors and cJun, JunB, and Fosl2 (Fig. 7 B). Expression of other AP1 family members was less uniformly suppressed, and JunD was induced (Fig. 7 B). Treatment with JQ1 and iBET-762 and OTX-015, two compounds with similar activity in clinical trials (Filippakopoulos and Knapp, 2014), down-modulated all tested CAF effector genes in several SCC-derived CAF strains (Fig. 7 C) in parallel with regulators of CAF gene expression, including various TEAD and AP1 family members; however, JunD, as in HDFs, was induced (Fig. 7 D).
Figure 7.
BET inhibitors function as global suppressors of CAF gene transcription, with overlapping impact with ATF3- and CSL-dependent gene transcription. (A and B) Two independent HDF strains with Atf3 gene silencing by two siRNAs (no. 1 and 2) were treated 24 h after transfection with increasing doses of JQ1 (25, 100, and 500 nM) or DMSO vehicle for an additional 48 h. As a reference, a parallel set of cells was transfected with anti-Atf3 siRNAs versus scrambled siRNA control. Expression of the indicated CAF effector (A) and positive regulator (B) genes was assessed by RT-qPCR. Results are shown as a heat map of ratios of gene expression (folds of down- or up-regulation in blue and magenta, respectively) in drug-treated versus DMSO control HDF and in cells with and without Atf3 silencing. (C) Three independent CAF strains from surgically excised skin SCCs were treated with increasing amounts (25, 100, and 500 nM) of three different inhibitors—JQ1, OTX-015 (OTX), and iBET 762 (iBET)—in parallel with DMSO alone for 48 h followed by RT-qPCR analysis of the indicated genes shown as a heat map plot. (D) CAF strains were treated with JQ1 (25, 100, and 500 nM) or DMSO control for 48 h, followed by RT-qPCR analysis of the indicated CAF-positive regulator genes shown as a heat map plot. (E) SCC13 cells were treated with JQ1 at increasing amounts (25, 100, and 500 nM) versus DMSO vehicle alone for 48 h, followed by RT-qPCR analysis of the indicated genes and heat map plotting of the results. (F) Morphological changes induced in SCC13 cells by JQ1 treatment (500 nM) versus DMSO vehicle for 48 h. (G) Alamar blue cell density assays of SCC13 cells in triplicate wells treated with JQ1 at the indicated doses or DMSO alone, with daily measurements for 4 d. (H–J) Plot of gene set enrichment analysis (GSEA) using RNA-seq expression profile of three CAF strains with and without JQ1 treatment (500 nM for 2 d) versus vehicle (DMSO) against (H) set of genes down-modulated by induced ATF3, (I) set of genes up-regulated by siRNA-mediated Atf3 silencing, and (J) set of genes up-regulated by siRNA-mediated Csl silencing. Genes are ranked by the signal-to-noise ratio based on their differential expression in CAFs with JQ1 versus vehicle treatment; positions of the genes from the gene set are indicated by vertical bars, and the enrichment score is shown in green. The complete list of significantly enriched process networks (P < 0.05) identified by GeneGo MetaCore analysis for genes within the core enrichment resulting from GSEA analysis is provided in Table S8.
BET inhibitors are not ubiquitous inhibitors of gene expression but exhibit gene- and cell-type specificity that can be of therapeutic interest (Filippakopoulos and Knapp, 2014). In fact, JQ1 treatment of SCC13 cells exerted differential effects on expression of three key proinflammatory genes: Il6 was suppressed, Il8 was inhibited at low concentrations and induced at high concentrations, and Ptgs2 (Cox2) was unaffected (Fig. 7 E). Importantly, JQ1 treatment of SCC cells caused strong induction of squamous differentiation markers, such as Keratin-1 (K1) and Loricrin (Lor), and CDK inhibitors, such as Cdkn1a and Cdkn2b, in parallel with flattening and enlargement of cells and slowed proliferation (Fig. 7, E–G).
Significant differences are likely to exist among CAFs of various tumor types (Erez et al., 2010). JQ1 treatment was recently reported to suppress CAF effector genes in stromal fibroblasts of pancreatic ductal adenocarcinomas, with inhibition of SHH and TGF-β signaling as the underlying mechanism (Yamamoto et al., 2016). An important question was to what extent the BET inhibitor effects that we found with skin SCC-derived CAFs are linked to ATF3 and/or CSL control of gene expression versus those other pathways. We addressed this question using RNA-seq analysis of three different strains of CAFs with and without JQ1 treatment (Table S7), followed by comparative GSEA against various gene expression signatures. We found a highly significant positive correlation between genes down-modulated by JQ1 treatment and by enhanced Atf3 expression (Fig. 7 H and Table S8), with a similarly significant inverse correlation with genes up-regulated by either Atf3 or Csl gene silencing (Fig. 7, I and J; and Table S8). Closely related process networks and functionally overlapping sets of genes were found in all these cases (Table S8). There was also a very significant correlation between genes down-modulated by JQ1 treatment and a set of 165 CAF signature genes (Table S9), bearing on various aspects of CAF activation (Fig. S4 A). Consistent with previous studies (Brown et al., 2014; Zou et al., 2014), we also found a statistically significant correlation between JQ1-modulated genes and two NF-κB–related gene sets, one from a study with HDFs (Hinata et al., 2003) and another as a common signature from multiple studies (for the noncanonical pathway; Fig. S4 B). In contrast, there was no correlation with either SHH or TGF-β gene sets such as would have been predicted on the basis of the previously mentioned study with pancreatic CAFs (Fig. S4 C; Yamamoto et al., 2016).
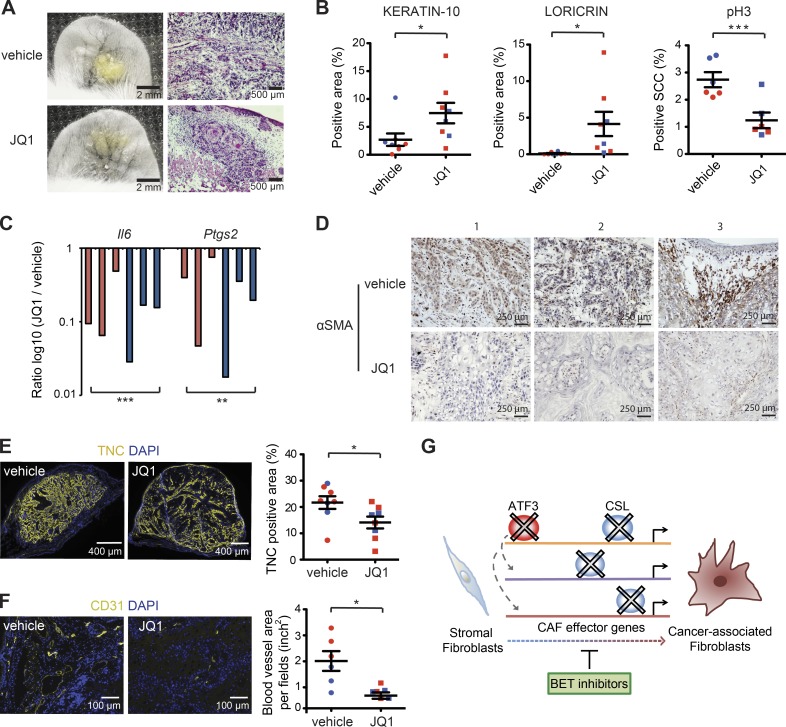
BET inhibitors induce SCC differentiation and suppress CAF activation by in vivo topical treatment
The aforementioned results suggested that BET inhibitors can exert dual beneficial effects in vivo by suppressing CAF activation as well as inducing squamous cancer cell differentiation. As these compounds exert systemic effects of possible concern for treatment (Filippakopoulos and Knapp, 2014), we tested whether they could be used topically using the ear injection assays described previously to assess behavior of SCC13 cells admixed with CAFs. In a first experiment, cells were injected 24 h before topical treatments with JQ1 in parallel with vehicle alone for 1 wk. In a second experiment, previously established ear lesions were treated with the BET inhibitor or vehicle for 1 wk. In both cases, the short time of treatment was not sufficient to affect tumor size. However, histological and immunofluorescence analysis showed enhanced squamous cell differentiation in JQ1-treated tumor lesions in parallel with decreased cancer cell proliferation (Fig. 8, A and B; and Fig. S5, A–D). LCM of stromal fibroblasts (identified by IF with anti-PDGFR-α antibodies) and RT-PCR analysis showed down-modulation of Il6 and Ptgs2 (Cox2) expression using JQ1 treatment (Fig. 8 C), which was paralleled by decreased α-SMA and tenascin C (TNC) marker expression and reduced angiogenesis (as assessed by anti-CD31 staining; Fig. 8, D–F; and Fig. S5, E and F).
Figure 8.
Topical BET inhibitor treatment suppresses expression of CAF-determinant genes and promotes tumor squamous cell differentiation. DsRed2-expressing SCC13 cells admixed with one of the previously analyzed strains of CAFs (strain 2) labeled with the PKH67 dye (green) were injected into the ears of NOD/SCID mice, followed by topical treatment with JQ1 in ethanol (10.93 mM) or ethanol alone, daily for 1 wk, from 24 h after injection. In a second experiment, topical treatment with JQ1 in DMSO (10.93 mM) or DMSO vehicle alone initiated on the 17th day after injection, daily for 1 wk. (A) Macroscopic fluorescence images of an ear pair treated with JQ1 versus vehicle together with corresponding histological analysis at the end of the experiment. (B) Mouse ear lesions from two experiments were analyzed by IF with Ab against the keratin-10 (K10), loricrin (Lor), and phospho-histone 3 (pH3), followed by quantification of the positive area (expressed as a percentage relative to the entire lesion size) by ImageJ software. n = 8 lesions per condition for K10 and Lor quantification, n = 6 lesions per condition for pH3 quantification; mean ± SEM, one-tailed paired t test. (C) LCM-captured PDGFRα-positive fibroblasts of ear lesions treated with JQ1 versus vehicle alone were analyzed by RT-qPCR for Il6 and Cox2 (PTGS2) expression. n = 6 lesions per condition, ratio log10 (JQ1/vehicle), two-tailed, one-sample t test. (D) Immunohistochemical staining with Ab against α-SMA of three different pairs of mouse ear tissues (first two pairs from the first experiment and the last pair from the second experiment). (E) IF analysis with Ab against TNC on JQ1-treated versus control lesions, followed by quantification of positive areas by ImageJ software. n = 8 lesions per condition (first experiment in red, second experiment in blue), mean ± SEM, one-tailed paired t test. (F) IF analysis with Ab against the CD31, followed by quantification of positive total blood vessel area per field (in square inches) in three different fields per lesion using ImageJ software. n = 6 lesions per condition (first experiment in red, second experiment in blue), mean ± SEM, one-tailed paired t test, *, P < 0.05; **, P < 0.005; ***, P < 0.0005. The first experiment is indicated in red, and the second experiment is indicated in blue. Additional representative images for A, B, E, and F from another mouse ear pair are shown in Fig. S5 (A–F). (G) Summarizing scheme: convergent role of ATF3 and CSL in negative control of CAF effector genes, with BET inhibitors exerting opposite effects of ATF3 and CSL loss in CAF activation.
The impact of this compound on cancer stromal cells was further validated by analysis of explants of surgically excised skin SCCs. As shown in Fig. S5 (G and H), immunofluorescence analysis showed that IL6 and MMP1 were significantly reduced in stromal cells of tumor explants treated with JQ1.
Discussion
Conversion of stromal fibroblasts into CAFs can play a primary role in tumor initiation and progression. Several transcription regulatory pathways have been implicated in CAF activation, but the level at which they converge and control each other is mostly unknown (Junttila and de Sauvage, 2013; Dotto, 2014). More importantly, the involvement of chromatin reorganization and epigenetics modification in this context remains to be investigated. This has significant translational potential given the increasing number of compounds developed to target the epigenome, which could be used for preventing and/or reverting CAF activation. We have found that as it is already at low basal levels, ATF3 converges with CSL in negative control of a large battery of CAF-determining genes through maintenance of chromatin configuration. BET-targeting compounds that unlink activated chromatin from the basic transcription apparatus counteract up-regulation of all these genes, with an impact on clinically derived CAFs amenable to in vivo topical treatment.
In contrast to its acute induction by acute UVA exposure, we have found that basal expression of Atf3 is consistently down-modulated in the stromal compartment of photo-damaged skin of premalignant keratinocyte lesions, possibly as a result of epigenetic modifications that also lead to compromised Notch2/Csl expression (Hu et al., 2012; Procopio et al., 2015). Functionally, loss or down-modulation of Atf3, as of Csl, led to induction of CAF activation, whereas increased Atf3 levels exerted opposite effects. This is consistent with a model whereby induction of Atf3 expression plays a role in dampening the impact of acute proinflammatory and cell-damaging signals on dermal fibroblasts, whereas persistent down-modulation of the gene promotes acquisition of proinflammatory and cancer-promoting properties.
Senescence of stromal fibroblasts with associated induction of CAF effector genes has been implicated in increased cancer risk with age (Campisi et al., 2011; van Deursen, 2014). However, CAFs around established tumors often show increased proliferation rather than senescence (Junttila and de Sauvage, 2013; Polanska and Orimo, 2013). We previously showed that CSL functions as a link between control of senescence- and CAF-determinant genes, with loss of Csl function and escape from p53-dependent senescence as a two-step process required for full CAF activation (Procopio et al., 2015). Interestingly, ATF3 is also involved in the control of fibroblast proliferation, which we found to be decreased by Atf3 deletion or silencing and enhanced by increased Atf3 expression. Although Atf3 expression is decreased in stromal fibroblasts adjacent to premalignant keratinocyte lesions, it is possible that as suggested by Buganim et al. (2011), its expression is increased during the later steps of CAF activation. In this case, however, additional genetic and/or epigenetic mechanisms are most likely required to prevent ATF3 from suppressing CAF effector genes involved in the enhancement of tumor formation.
In HDFs, ATF3 at higher levels binds to a significant number of genes in common with CSL, suppressing their transcription. The most surprising finding was that, even at low basal levels, ATF3 plays an essential role in the negative control of these genes, binding to a very restricted number of distant sites that encompass mostly nonprotein coding genes of unknown function. Noncoding RNAs have been implicated in multiple pathologies through a variety of mechanisms, including chromatin deregulation (Betancur, 2016). As a concurrent mechanism, down-modulation or loss of the ATF3 transcriptional repressive complex is poised to cause site-specific chromatin modifications that can trigger a broader downstream cascade. Indicative of a cis-mode of regulation, CRISPR-mediated deletion of a specific ATF3 binding region 2 Mb upstream of the Il6 gene caused induction of IL6 expression but did not affect other ATF3-regulated genes. Deregulation of chromatin regions around ATF3 binding sites extended to genes with roles in transcription/chromatin control, thus accounting for further chromatin modifications of chromosomal domains to which ATF3 does not bind. In fact, we found a striking overlap of H3K27 acetylation and RNA Pol II recruitment elicited by silencing of Atf3 and Csl at both chromosomal and individual gene levels. A similar chromatin profile was also observed in HDFs with concomitant down-modulation of Atf3 and Csl, pointing to a functional convergence of the two proteins with little or no additive effects resulting from their combined loss.
Compounds that interfere with CAF activation can be important for prevention as well as treatment of cancer (Goruppi and Dotto, 2013). Conversion of stromal fibroblasts into CAFs results from transcriptional up-regulation of a large battery of genes with multiple cellular functions, and several transcription regulatory pathways have been identified that control a subset of these genes, sometimes in an opposite manner (Kalluri and Zeisberg, 2006; Augsten, 2014; Dotto, 2014). We have found that BET inhibitors are strikingly effective in suppressing expression of these genes, overcoming the effects of Atf3 or Csl gene silencing and exerting a specific impact on Atf3 and Csl gene expression signatures. Treatment of HDFs with these compounds caused marginal modulation of Csl and Atf3, placing the mechanism of action of BET inhibitors possibly downstream of these proteins.
It was recently reported that in the context of fibrotic conversion of liver stellate cells (Ding et al., 2015) or ras oncogene–induced senescence (Tasdemir et al., 2016), Brd4 silencing exerted the same effects as the BET inhibitors. The BET inhibitors that we chose for our studies are acetyl–lysine mimetics that can cause Brd2, Brd3, and/or Brd4 proteins to release their partners (Filippakopoulos and Knapp, 2014). However, more complex mechanisms are possible, such as the reported chromatin association of BET inhibitors with sites of Brd protein occupancy (Anders et al., 2014).
Irrespectively of detailed mechanisms, we have shown that in parallel with their impact on CAFs, BET inhibitors can induce differentiation of squamous carcinoma cells, thus enhancing their potential therapeutic benefit. Despite encouraging phase I responses, safety concerns have been raised for the use of these inhibitors because of the possible unwarranted effects on metabolism or the immune system (Filippakopoulos and Knapp, 2014). These concerns would be bypassed by the topical application we have used. This approach could be of great interest for treatment of early precancerous and cancerous lesions originating in the skin and the surface of internal organs, coupled with methods to overcome epithelia barrier functions.
Materials and methods
Cell/tissue explant preparation and treatments
Mouse dermal fibroblasts were prepared from individual or pooled newborn mice with homozygous disruption of the Atf3 gene (Hartman et al., 2004) versus littermate wild-type controls of the same genetic background (C57BL/68) or from newborn mice with the Atf3 gene flanked by loxP sites (Wolford et al., 2013). Excised mouse back skin was immersed in 0.25% trypsin–EDTA (Gibco) overnight at 4°C, and the dermis compartment was separated from the epidermis. Then, the dermis was minced and digested by collagenase I treatment (0.4 mg/ml in HBSS [Ca2+ and Mg2+ free], Sigma-Aldrich) of the dermis for 30 min at room temperature with gentle agitation, followed by filtering through a 100-µm nylon mesh. After spinning, the cell pellet was resuspended and cultured in DMEM 10% FBS. For in vitro deletion of the Atf3 gene, dermal fibroblasts from Atf3loxP/loxP mice were infected with an adenovirus expressing Cre recombinase and GFP in parallel with a GFP-expressing control virus at multiplicity of infection of 100 for 8 h, which resulted in >99% of GFP-positive infected cells (Rangarajan et al., 2001). HDFs were prepared from discarded human skin samples from abdominoplasty or circumcision at the Department of Surgery or Pediatrics, Lausanne University, with required institutional approvals and informed consent. Normal human skin samples and samples of SCCs were obtained at the Department of Dermatology, Massachusetts General Hospital, as discarded parts not needed for diagnosis. All samples were processed as approved by the institutional review board. After removal of subcutaneous adipose tissue, the skin was minced into small pieces and digested using 3% collagenase I (Sigma-Aldrich) in HBSS (Ca2+ and Mg2+ free) at 37°C for 1 h. After termination of enzymatic digestion by adding an equal volume of DMEM plus 10% FBS, cells were filtered through a 40-μm mesh, collected by centrifugation (1,500 rpm for 5 min), plated, and cultured in DMEM plus 10% FBS until 60–70% confluency before passage. CAF strains were derived from surgically excised skin SCCs obtained from the Department of Dermatology, Massachusetts General Hospital, with institutional approval and informed consent as required. Skin SCC samples were cut in 1- to 2-mm pieces after removal of fat excess, followed by incubation in 2 ml of PBS containing 0.25 mg/ml of Liberase TL (Roche) for 40 min at 37°C with gentle shaking. After terminating the enzymatic digestion, the dissociated tissue was passed through a 10-ml syringe attached to a 70-μm sieve. Cells from the flowthrough were centrifuged, washed three times with DMEM plus 10% FBS, and seeded in a 10-cm tissue culture dish. Adherent cells were expanded for characterization as with HDFs. Normal skin and skin SCC explant cultures were prepared from discarded human skin samples obtained from the Department of Surgery or Pediatrics, Lausanne University, and from surgically excised human skin SCCs obtained from the Department of Dermatology, Massachusetts General Hospital, with institutional approval and informed consent as required. Skin samples sterilized in 70% ethanol and cut after removal of subcutaneous fat into 1 × 1–cm pieces were placed in DMEM supplemented with 20% FBS and 1% MEM vitamin (Gibco) in a 0.25% semisolid agar medium (Sigma-Aldrich). The epidermis was maintained at the air–medium interface. For the UV treatment of HDFs or skin explants, we used a Bio-Link cross-linker UV irradiation system (Vilber Lourmat) equipped with a UVA (375 nm) lamp, with a portable photometer IL1400A (International Light Technologies) measuring for dosage determination. Cells or skin SCC explants were treated with indicated doses of JQ1 (Cayman Chemical), iBET-762 (MedChem Express), and OTX-015 (Active Biochem) dissolved in DMSO for 2 d (cultured cells) or daily for 5 d (SCC explants).
Skin SCC13 (Rheinwald and Beckett, 1981) and PAM212 (Roop et al., 1983) were originally reported and subsequently checked for the presence of oncogenic mutations (https://cansar.icr.ac.uk/cansar/cell-lines/PAM212/mutations/; Wu et al., 2010).They have been routinely tested for mycoplasma and have not been authenticated for STR (short tandem repeat) profiling. None of the used human cell lines is listed in the NCBI BioSample Database of misidentified cell lines.
Genetic manipulations and gene expression analysis
Conditions for viral infection, shRNA- and siRNA-mediated gene silencing, and RT-qPCR were as follows. The pMXs-Atf3 vector was prepared by inserting 543 bp of human ATF3 cDNA into the Xho I site of the retrovirus vector pMXs-neo (Tamura et al., 2005; Wu et al., 2010). For doxycycline-inducible Atf3 expression, the human Atf3 coding sequence was amplified from pMXs-Atf3 by PCR and cloned into a TOPO-pENTR vector, followed by transfer into pINDUCER20 (Meerbrey et al., 2011) using the Gateway LR recombinase system (Thermo Fisher Scientific). pMXs-Atf3 and pINDUCER20 lentivirus were packaged in HEK293T cells and incubated with HDFs for 24 h. Cells were selected, respectively, with 500 μg/ml of G418 for 7 d and 1 μg/ml of puromycin for 5 d for pMXs-Atf3 and pINDUCER20-ATF3 2 d after selection. For the siRNA experiment, HDFs were transfected with 20 nM siRNA (Invitrogen) using INTERFERin reagent (Polyplus) for 12 h according to the manufacturer’s recommendation. HDFs were collected for RT-qPCR analysis 3 d after siRNA transfection, 9 d after constitutive Atf3 expression (pMXs-Atf3), 3 d after doxycycline (750 ng/μl)–inducible Atf3 expression (pIND20-ATF3), and 3 d after Cre-expressing adenovirus infection. HDFs with Atf3 gene silencing were treated with JQ1 for 2 d after 1 d of Atf3 siRNA transfection. 1 µg of total RNA was used to reverse transcribe cDNA using RevertAidH Minus Reverse transcription (Thermo Fisher Scientific) followed by real-time qPCR using the SYBR Fast qPCR master mix (Kapa Biosystems) in the Light Cycler 480 (Roche) according to the manufacturer’s protocols. All RNA samples were analyzed in triplicate with gene-specific primers and 36β4 normalization for human samples and Gapdh for mouse samples. The list of siRNAs and RT-qPCR primers is provided in Table S10.
CRISPR/Cas9-mediated deletion of the ATF3 binding site
The endogenous ATF3 binding region 2.07 Mb upstream of the Il6 locus—as determined by the ChIP-seq analysis provided in Table S6—was deleted using a dual-guide RNA CRISPR/Cas9 gene editing strategy (Fig. 6 A). Specifically, two guide RNAs (gRNA 1 and 2) encompassing this region (listed in Table S10) were identified using the MIT (Massachusetts Institute of Technology) CRISPR design tool (http://crispr.mit.edu). A lentiviral vector with a double CRISPR cassette and Cas9 expression was constructed, starting from lentiCRISPR_v2 (Addgene, 60954) using the following steps: (a) the mouse U6 (mU6) promoter was PCR amplified with a forward primer also containing an upstream scaffold and a terminator sequence (mU6 scaff/ter Fwd) and a reverse primer corresponding to the 3′ end of the mU6 sequence (mU6 Rev); (b) the PCR product was subjected to a second round of PCR with a forward primer containing the gRNA 1 sequence (gRNA_1 Fwd) preceded by a BsmBI recognition sequence and a reverse primer with the gRNA 2 sequence (gRNA_2 Rev) followed by a BsmBI recognition sequence; (c) the PCR product was then digested with BsmBI to generate compatible sticky ends and ligated into the BsmBI sites located between the human U6 (hU6) promoter and scaffold sequence of lentiCRISPR_v2 using NEB Quick Ligase (New England Biolabs, Inc., M2200). A control ‘empty’ lentiCRISPR_v2 vector lacking any gRNAs was generated by BsmBI digestion followed by blunt-end ligation.
The dual gRNAs lentiCRISPR vector and empty vector control were used for viral production and infection of low-passage HDFs. 48 h later, infected cells were diluted, seeded in limited numbers, and subjected to puromycin (2 μg/ml) selection. Puromycin-resistant cells were allowed to grow until they formed small clusters/colonies. Cells from individual clusters were collected for genomic PCR analysis, with primers flanking the targeted ATF3 binding site (listed in Table S10) using Q5 high-fidelity DNA polymerase (New England Biolabs, Inc., M0491). PCR products of the expected smaller deletion size were excised for nucleotide sequencing to confirm deletion. Replated HDF colonies carrying the deletion were analyzed in parallel with colonies without the deletion and/or infected with the lentiCRISPR empty vector control by qRT-PCR analysis or by IF with anti-IL6 and anti-TNC antibodies (Table S11).
Tumorigenesis experiments
Mice with homozygous disruption of the Atf3 gene (Hartman et al., 2004) and wild-type controls in the same genetic background (C57BL/68; 8 wk old, Charles River) were subjected to a skin carcinogenesis protocol (Topley et al., 1999), with a single topical application of 7, 12-Dimethylbenz[a]anthracene (DMBA; 20 μg in 200 μl acetone) followed by biweekly treatments with 12-O-tetradecanoylphorbol 13-acetate (TPA; 10−4 M in acetone) for 25 wk. Tumors larger than 3 mm2 were analyzed and classified by histology: highly differentiated papillomas (differentiation >50% of tumor volume), moderately differentiated papillomas, dysplastic tumors with penetration into superficial dermis, and fully invasive SCC, with tumor invasion into the panniculus carnosus.
Intradermal ear injection assays and live imaging were performed as follows. Mouse dermal fibroblasts with and without Atf3 deletion labeled with PKH67 (Sigma-Aldrich) were admixed in equal numbers (105 cells per injection), with SCC13 cells stably infected with a DsRed2-expressing lentivirus. To minimize individual animal variation, the two combinations of cells were injected in parallel into the left and right ears of mice (8 to 12 wk old NOD/SCID, IL-2 receptor gamma chain negative (Il2rg−/−). Mice were sacrificed 3 wk later for tissue analysis. Quantification of red fluorescence signal (intensity × surface area) corresponding to SCC13 cell expansion was obtained by ImageJ software analysis of digitally acquired images and expressed, at each time point, as relative to the initial signal 24 h after cell injection. Quantification of stromal cell expansion by green fluorescence signal yielded uneven results, due in some cases to disappearance of the lipid dye staining. For in vivo JQ1 assays, similar ear injections were performed with DsRed2-expressing SCC13 cells admixed with clinically derived CAFs (strain 2) labeled with PKH67. 1 or 17 d after injection, mice were treated topically, on the left and right ears, respectively, with JQ1 (100 μg in 20 μl of ethanol or DMSO) or vehicle alone daily for 1 wk. For both assays, cells were loaded into a 33-gauge microsyringe (Hamilton) in 3 μl of sterile HBSS. Live images were acquired with a fluorescence stereomicroscope (Leica M205F) every 2–3 d. ImageJ Lasso Toll was used for quantification of red and green fluorescence signal (intensity × surface area).
Back skin intradermal tumorigenicity assays were performed by admixing mouse dermal fibroblasts with and without Atf3 deletion (106 cells per injection) with weakly transformed mouse Pam212 keratinocyte line or SCC13 cells (2 × 106 cells per injection). To minimize individual animal variation, mice (NOD/SCID) were injected in parallel, in the left and right flanks, respectively, with the two combinations of cells. Mice were sacrificed 4 wk later for tumor tissue retrieval and analysis.
All mouse work was performed according to the Swiss guidelines and regulations for the care and use of laboratory animals, with approved protocol from the Canton de Vaud veterinary office.
Tissue analysis
LCM with an Arcturus XT microdissection system (Applied Biosystems) was performed as follows. Frozen tissue sections were cut in 8 μm slices and mounted on membrane-coated glass slides (LCM522, Applied Biosystems). After staining in 1% methylene green in DEPC (diethyl pyrocarbonate) water for 10 s and washing three times in DEPC water, sections were immediately used for microdissection. Each capture was collected from the membrane and transferred to a TRI reagent (Sigma-Aldrich), and RNA was extracted using a standard protocol with additional proteinase K and DNase I treatment. For immunofluorescence-guided LCM, fresh frozen block sections were briefly incubated with fluorescein isothiocyanate (FITC)–conjugated antibody against PDGFRα (Santa Cruz Biotechnology, Inc.) and propidium iodide for 30 s. For RNA extraction, an Arcturus PicoPure RNA isolation kit (Applied Biosystems) was used according to the manufacturer’s recommendations. Primers used for RT-qPCR are listed in Table S10. Conditions for IF and immunohistochemistry were as follows. Sections were fixed in 4% paraformaldehyde (PFA) and permeabilized in 0.5% Triton X-100 in PBS followed by blocking in 5% donkey serum. Primary antibodies were stained at 4°C overnight; secondary antibodies were incubated at room temperature for 2 h and mounted in Fluoromount Mounting Medium (Sigma-Aldrich) after nuclear DAPI staining. Antibodies are listed in Table S11. Immunofluorescence images were acquired with a ZEISS LSM510 or ZEISS LSM700 Meta laser scanning.
For fluorescence signal quantification, acquired images for each color channel were imported into either Adobe Photoshop software, followed by Lasso Tool selection, or ImageJ software, using the functions “measurement” or “particle analysis” for selection of areas or cells (tumor or stromal) of interest. All measurements were exported as Microsoft Excel data files.
RNA-seq and GSEA
RNA-seq analysis was performed as follows. Total RNA was extracted 3 d after siRNA transfection for ATF3 knockdown samples and 3 d after doxycycline treatment for ATF3 overexpression samples using the directZol RNA miniprep kit (Zymo Research) with on-column DNase treatment. RNA quality was verified on the bioanalyzer (Agilent Technologies) with an RNA integrity number greater than 8. Four μg of total RNA was used for library preparation using the Truseq kit (Illumina). A single read was done on the Illumina HiSeq 2000 sequencer at the Genomic Technologies Facility at the University of Lausanne. Reads were trimmed using Trimmomatic (version 0.22) and then mapped to the human hg19 reference genome using TopHat (version 2.0.8b). Gene expression levels were evaluated using the HTSeq package (release 0.5.4p1).
GSEA for RNA-seq expression profiles was conducted using the software GSAA-seqSP (gene set association analysis for RNA-seq data with sample permutation; Xiong et al., 2014) from the GSAA platform (version GSAA_2.0, http://gsaa.unc.edu/) with default parameters. Curated gene sets were retrieved from the Molecular Signatures Database (MSigDB version 5.2, http://www.broadinstitute.org/gsea/msigdb/). The pathway gene sets used in this study are listed in Fig. S4 C.
ChIP-seq, ChIP tagmentation, and MEA
ChIP-seq analysis was performed as follows starting from 107 HDFs for ATF3 and ∼5 × 106 HDFs for histone modification marks and RNA Pol II (the antibodies and concentrations used are listed in Table S10). Immunoprecipitated DNA using 5 μg of antibodies per 106 cells was quantified by fluorometry on the Qubit system (Invitrogen). A total of 10 ng of ChIPed DNA was used for library preparation using the NEBNext ChIP-seq Library Prep Reagent Set for Illumina (New England Biolabs, Inc.) as recommended by the manufacturer. A Burrows-Wheeler Aligner (http://bio-bwa.sourceforge.net) was used for FASTQ file alignments, and MACS software (http://liulab.dfci.havard.edu/MACS) with default parameters was used for peak detection.
All statistical analysis of ChIP-seq data was done using statistical computing R (version 3.2.2) and various Bioconductor packages (http://www.Bioconductor.org). Raw FASTQ sequences were aligned to the human reference genome (Ensembl release, version GRCh37) with the software BWA (version 0.7.12; Li and Durbin, 2009). Overall sequencing quality and absence of contaminants were assessed with the tool FASTqc (version 0.11.2, FASTQC, http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Immunoprecipitation efficiency was assessed with the tool CHANCE (version 2.0; Diaz et al., 2012), which showed a cumulative percent enrichment over input >25% in all ChIP-seq samples, each relative to its own input.
ChIP-seq peaks with a false discovery rate and fold change enrichment relative to the background signal were identified with the peak caller software MACS (version 2.1; Zhang et al., 2008) used in treatment control mode with input as background for ATF3 binding; the peak model building mode was disabled for broad histone marker peaks using a global background signal.
To compare peaks in regions of interest across different samples, we generated signal tracks with MACS2. MACS2 builds the signal track, computing the number of fragments found at each genomic position. These values are normalized in millions of reads, to account for the library size, and stored in the bedGraph format. The bedGraph files were subsequently converted to the bigWig format using the UCSC tool begGraphToBigWig (Kent et al., 2010). The advantages of this latter format are the reduced size and the presence of an index for faster access. The resulting tracks were visualized with IGV version 2.3.88 (Thorvaldsdóttir et al., 2013). For direct comparisons, the IGV subtract operator tool was used to combine two data tracks to generate and visualize sample differences. Individual profiles for each condition were visualized in IGV and merged using Adobe Photoshop.
ChIP tagmentation (Schmidl et al., 2015) followed by quantitative PCR analysis was used for endogenous ATF3 ChIP-seq data validation. Chromatin immune precipitation was carried from 4 × 106 HDFs as for the ChIP assay with anti-ATF3 antibody versus nonimmune IgGs. The bead-bound chromatin was tagged with Tn5 transposase (Nextera DNA Sample Prep kit, Illumina). The “tagmented” chromatin was de-cross-linked, purified, and amplified (14 cycles) using tag-specific primers. The amplified products were diluted (1:10), and 1 μl was used as a template for qPCR to determine the enrichment of the indicated sites; the primer sequences and antibodies are listed in Tables S10 and S11, respectively.
MEA was conducted using Regulatory Sequence Analysis Tools (RSAT; Thomas-Chollier et al., 2012). The ATF3 peak coordinates in the bed format were uploaded onto the RSAT web server (http://floresta.eead.csic.es/rsat/peak-motifs_form.cgi), and the fetched sequences were compared with the discovered motifs using JASPAR CORE vertebrates databases.
Statistics
Data are presented as means or ratios ± SD or ± SEM among experimental groups and controls, as indicated in the figure legends. For gene expression and functional assays, statistical significance of differences between experimental groups and controls (as biological replicates) was assessed by one- or two-tailed unpaired or paired t tests, one- or two-sided one-sample t tests, and two-tailed χ2 tests, as indicated in the figure legends. More specifically, a one-tailed t test was used when the null hypothesis was articulated directionally for comparisons of relatively large combinations of samples and genes (such as in Fig. 2, A and B; Fig. 4, A–C; Fig. 8, B, E, and F; Fig. S1, C, E, and N; and Fig. S5, G and H). Parametric tests were used without any assumption for data distribution, and variation within each group of data has not been estimated.
For ear injection and tumorigenicity assays, a sample size of adequate statistical power was ensured by minimizing the individual animal variability issue by parallel ear or back injections of the same animals with control versus experimental combinations of cells.
No statistical method was used to predetermine sample size. No exclusion criteria were adopted for animal studies and samples collection. No randomization was used, and the researchers involved in the study were not blinded during sample obtainment or data analysis.
Accession numbers
The RNA-seq and ChIP-seq data sets generated for this study were deposited in GEO as the GSE81406 super-series dataset.
Reproducibility of experiments
For RT-qPCR in Fig. 2 (A and B), Fig. 3 A, Fig. 4 (A–C), Fig. 6 C, Fig. 7 (A–D), and Fig. S1 D, repeated experiments on several primary cell strains or culture preparations are shown as indicated in the corresponding figure legends.
Additional representative IF and H&E tissue-staining images for Fig. 1 (D and I), Fig. 2 (E and G), and Fig. 8 A are shown in Fig. S1 (F, H, I, and K) and Fig. S5 A, respectively.
In Fig. 1 H, Fig. 2 (C, F, and H), Fig. 6 D, Fig. 8 (B, E, and F), Fig. S1 (C, N, and O), and Fig. S5 (G and H), representative IF images are shown together with signal quantification from multiple samples as specified in the corresponding figure legends.
Findings in Fig. 2 (C–F) were validated by alternative in vivo approaches in two independent experiments with two different tumor cell lines, as shown in Fig. S1 (L–N).
Findings in Fig. 1 D and Fig. 8 (A–F) were obtained in two independent experiments.
For Fig. S1 A, similar results were obtained in Fig. S1 D in two independent experiments and further validated with different approaches, as shown in Fig. S1 (B and C).
Online supplemental material
Fig. S1 shows that Atf3 deficiency promotes dysplastic keratinocyte tumors and CAF activation. Fig. S2 shows that Atf3 and Csl gene silencing cause similar changes in transcriptional profile and global chromatin configuration. Fig. S3 shows that Atf3 and Csl gene silencing cause similar chromatin changes at genomic loci to which endogenous ATF3 does not bind. Fig. S4 shows that JQ1-modulated genes are significantly correlated with NF-κB but not SHH or TGF-β signaling gene sets. Fig. S5 shows that BET inhibitor treatment promotes tumor squamous cell differentiation and suppresses expression of CAF-determinant genes in induced skin lesions and human skin SCC explants. Table S1 provides combined ChIP-seq and RNA-seq analysis of induced ATF3 and CSL in HDF. Table S2 provides GeneGo MetaCore analysis of ATF3/CSL–regulated target genes identified by ChIP-seq and RNA-seq. Table S3 provides the induced ATF3 and endogenous CSL binding peak positions on 102 common regulated target genes. Table S4 provides the RNA-seq profiles of HDF with and without Atf3 gene silencing. Table S5 provides GeneGo MetaCore analysis for GSEA of Atf3-silenced HDFs. Table S6 provides the ChIP-seq profiles of endogenous ATF3 in HDF. Table S7 provides the RNA-seq profiles of CAFs with and without JQ1 treatment. Table S8 provides GeneGo MetaCore analysis for GSEA of JQ1-treated CAFs. Table S9 provides the CAF gene set used for GSEA. Table S10 provides the RT-qPCR primers, guide RNA, and siRNAs used in this study. Table S11 lists the antibodies used in this study. Tables S1–S11 are provided as Excel files.
Supplementary Material
Acknowledgments
We thank Dr. T. Hai for providing ATF3 mutant mice; Drs. W. Raffoul and J.M. Joseph for skin tissues as source of cells; Drs. C. Brisken, C. Missero, and K. Lefort for critical reading of the manuscript; Drs. P. Ostano, P. Provero, and U. Ala for help with the bioinformatics analysis; K. Harshman for RNA-seq and ChIP-seq analysis; and C. Pasche and T. Proust for technical help.
This study was supported by grants from the Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (310030_156191/1), the National Institutes of Health (NIH; R01AR039190 and R01AR064786), and the H2020 European Research Council (26075083). The content does not necessarily represent the official views of the NIH.
The authors declare no competing financial interests.
Author contributions: D.E. Kim, M.-G. Procopio, S. Ghosh, S.-H. Jo, S. Goruppi, F. Magliozzi, and P. Bordignon performed work and contributed to analysis of the results. V. Neel provided clinical samples. P. Angelino conducted bioinformatics. G.P. Dotto designed the study and wrote the manuscript with the help of D.E. Kim and M.-G. Procopio.
Footnotes
Abbreviations used:
- AK
- actinic keratose
- BET
- bromodomain and extra-terminal
- CAF
- cancer-associated fibroblast
- gRNA
- guide RNA
- HDF
- human dermal fibroblast
- IF
- immunofluorescence
- LCM
- laser capture microdissection
- MEA
- motif enrichment analysis
- SCC
- squamous cell carcinoma
References
- Anders L., Guenther M.G., Qi J., Fan Z.P., Marineau J.J., Rahl P.B., Lovén J., Sigova A.A., Smith W.B., Lee T.I., et al. . 2014. Genome-wide localization of small molecules. Nat. Biotechnol. 32:92–96. 10.1038/nbt.2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augsten M. 2014. Cancer-associated fibroblasts as another polarized cell type of the tumor microenvironment. Front. Oncol. 4:62 10.3389/fonc.2014.00062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachelor M.A., and Bowden G.T.. 2004. UVA-mediated activation of signaling pathways involved in skin tumor promotion and progression. Semin. Cancer Biol. 14:131–138. 10.1016/j.semcancer.2003.09.017 [DOI] [PubMed] [Google Scholar]
- Balasaraswathy P., Kumar U., Srinivas C.R., and Nair S.. 2002. UVA and UVB in sunlight, optimal utilization of UV rays in sunlight for phototherapy. Indian J. Dermatol. Venereol. Leprol. 68:198–201. [PubMed] [Google Scholar]
- Betancur J.G. 2016. Pervasive lncRNA binding by epigenetic modifying complexes—The challenges ahead. Biochim. Biophys. Acta. 1859:93–101. 10.1016/j.bbagrm.2015.10.009 [DOI] [PubMed] [Google Scholar]
- Bissell M.J., and Hines W.C.. 2011. Why don’t we get more cancer? A proposed role of the microenvironment in restraining cancer progression. Nat. Med. 17:320–329. 10.1038/nm.2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J.D., Lin C.Y., Duan Q., Griffin G., Federation A.J., Paranal R.M., Bair S., Newton G., Lichtman A.H., Kung A.L., et al. . 2014. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 56:219–231. 10.1016/j.molcel.2014.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buganim Y., Madar S., Rais Y., Pomeraniec L., Harel E., Solomon H., Kalo E., Goldstein I., Brosh R., Haimov O., et al. . 2011. Transcriptional activity of ATF3 in the stromal compartment of tumors promotes cancer progression. Carcinogenesis. 32:1749–1757. 10.1093/carcin/bgr203 [DOI] [PubMed] [Google Scholar]
- Calvo F., Ege N., Grande-Garcia A., Hooper S., Jenkins R.P., Chaudhry S.I., Harrington K., Williamson P., Moeendarbary E., Charras G., and Sahai E.. 2013. Mechanotransduction and YAP-dependent matrix remodelling is required for the generation and maintenance of cancer-associated fibroblasts. Nat. Cell Biol. 15:637–646. 10.1038/ncb2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J., Andersen J.K., Kapahi P., and Melov S.. 2011. Cellular senescence: A link between cancer and age-related degenerative disease?. Semin. Cancer Biol. 21:354–359. 10.1016/j.semcancer.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A., Nellore A., and Song J.S.. 2012. CHANCE: comprehensive software for quality control and validation of ChIP-seq data. Genome Biol. 13:R98 10.1186/gb-2012-13-10-r98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N., Hah N., Yu R.T., Sherman M.H., Benner C., Leblanc M., He M., Liddle C., Downes M., and Evans R.M.. 2015. BRD4 is a novel therapeutic target for liver fibrosis. Proc. Natl. Acad. Sci. USA. 112:15713–15718. 10.1073/pnas.1522163112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontje W., Schotte R., Cupedo T., Nagasawa M., Scheeren F., Gimeno R., Spits H., and Blom B.. 2006. Delta-like1-induced Notch1 signaling regulates the human plasmacytoid dendritic cell versus T-cell lineage decision through control of GATA-3 and Spi-B. Blood. 107:2446–2452. 10.1182/blood-2005-05-2090 [DOI] [PubMed] [Google Scholar]
- Dotto G.P. 2014. Multifocal epithelial tumors and field cancerization: Stroma as a primary determinant. J. Clin. Invest. 124:1446–1453. 10.1172/JCI72589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erez N., Truitt M., Olson P., Arron S.T., and Hanahan D.. 2010. Cancer-associated fibroblasts are activated in incipient neoplasia to orchestrate tumor-promoting inflammation in an NF-κB-dependent manner. Cancer Cell. 17:135–147. 10.1016/j.ccr.2009.12.041 [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P., and Knapp S.. 2014. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat. Rev. Drug Discov. 13:337–356. 10.1038/nrd4286 [DOI] [PubMed] [Google Scholar]
- Goruppi S., and Dotto G.P.. 2013. Mesenchymal stroma: Primary determinant and therapeutic target for epithelial cancer. Trends Cell Biol. 23:593–602. 10.1016/j.tcb.2013.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman M.G., Lu D., Kim M.L., Kociba G.J., Shukri T., Buteau J., Wang X., Frankel W.L., Guttridge D., Prentki M., et al. . 2004. Role for activating transcription factor 3 in stress-induced β-cell apoptosis. Mol. Cell. Biol. 24:5721–5732. 10.1128/MCB.24.13.5721-5732.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinata K., Gervin A.M., Jennifer Zhang Y., and Khavari P.A.. 2003. Divergent gene regulation and growth effects by NF-κ B in epithelial and mesenchymal cells of human skin. Oncogene. 22:1955–1964. 10.1038/sj.onc.1206198 [DOI] [PubMed] [Google Scholar]
- Hu B., Castillo E., Harewood L., Ostano P., Reymond A., Dummer R., Raffoul W., Hoetzenecker W., Hofbauer G.F., and Dotto G.P.. 2012. Multifocal epithelial tumors and field cancerization from loss of mesenchymal CSL signaling. Cell. 149:1207–1220. 10.1016/j.cell.2012.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila M.R., and de Sauvage F.J.. 2013. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 501:346–354. 10.1038/nature12626 [DOI] [PubMed] [Google Scholar]
- Kalluri R., and Zeisberg M.. 2006. Fibroblasts in cancer. Nat. Rev. Cancer. 6:392–401. 10.1038/nrc1877 [DOI] [PubMed] [Google Scholar]
- Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., and Karolchik D.. 2010. BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics. 26:2204–2207. 10.1093/bioinformatics/btq351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meerbrey K.L., Hu G., Kessler J.D., Roarty K., Li M.Z., Fang J.E., Herschkowitz J.I., Burrows A.E., Ciccia A., Sun T., et al. . 2011. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 108:3665–3670. 10.1073/pnas.1019736108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menietti E., Xu X., Ostano P., Joseph J.M., Lefort K., and Dotto G.P.. 2016. Negative control of CSL gene transcription by stress/DNA damage response and p53. Cell Cycle. 15:1767–1778. 10.1080/15384101.2016.1186317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkenschlager M., and Odom D.T.. 2013. CTCF and cohesin: Linking gene regulatory elements with their targets. Cell. 152:1285–1297. 10.1016/j.cell.2013.02.029 [DOI] [PubMed] [Google Scholar]
- Polanska U.M., and Orimo A.. 2013. Carcinoma-associated fibroblasts: Non-neoplastic tumour-promoting mesenchymal cells. J. Cell. Physiol. 228:1651–1657. 10.1002/jcp.24347 [DOI] [PubMed] [Google Scholar]
- Procopio M.G., Laszlo C., Al Labban D., Kim D.E., Bordignon P., Jo S.H., Goruppi S., Menietti E., Ostano P., Ala U., et al. . 2015. Combined CSL and p53 downregulation promotes cancer-associated fibroblast activation. Nat. Cell Biol. 17:1193–1204. 10.1038/ncb3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangarajan A., Talora C., Okuyama R., Nicolas M., Mammucari C., Oh H., Aster J.C., Krishna S., Metzger D., Chambon P., et al. . 2001. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20:3427–3436. 10.1093/emboj/20.13.3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheinwald J.G., and Beckett M.A.. 1981. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultured from human squamous cell carcinomas. Cancer Res. 41:1657–1663. [PubMed] [Google Scholar]
- Roop D.R., Hawley-Nelson P., Cheng C.K., and Yuspa S.H.. 1983. Expression of keratin genes in mouse epidermis and normal and malignantly transformed epidermal cells in culture. J. Invest. Dermatol. 81:S144–S149. 10.1111/1523-1747.ep12540939 [DOI] [PubMed] [Google Scholar]
- Schmidl C., Rendeiro A.F., Sheffield N.C., and Bock C.. 2015. ChIPmentation: Fast, robust, low-input ChIP-seq for histones and transcription factors. Nat. Methods. 12:963–965. 10.1038/nmeth.3542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schotte R., Nagasawa M., Weijer K., Spits H., and Blom B.. 2004. The ETS transcription factor Spi-B is required for human plasmacytoid dendritic cell development. J. Exp. Med. 200:1503–1509. 10.1084/jem.20041231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen H., McElhinny A.S., Cao Y., Gao P., Liu J., Bronson R., Griffin J.D., and Wu L.. 2006. The Notch coactivator, MAML1, functions as a novel coactivator for MEF2C-mediated transcription and is required for normal myogenesis. Genes Dev. 20:675–688. 10.1101/gad.1383706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., and Mesirov J.P.. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102:15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Hua B., Adachi S., Guney I., Kawauchi J., Morioka M., Tamamori-Adachi M., Tanaka Y., Nakabeppu Y., Sunamori M., et al. . 2005. Stress response gene ATF3 is a target of c-myc in serum-induced cell proliferation. EMBO J. 24:2590–2601. 10.1038/sj.emboj.7600742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdemir N., Banito A., Roe J.S., Alonso-Curbelo D., Camiolo M., Tschaharganeh D.F., Huang C.H., Aksoy O., Bolden J.E., Chen C.C., et al. . 2016. BRD4 connects enhancer remodeling to senescence immune surveillance. Cancer Discov. 6:612–629. 10.1158/2159-8290.CD-16-0217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas-Chollier M., Darbo E., Herrmann C., Defrance M., Thieffry D., and van Helden J.. 2012. A complete workflow for the analysis of full-size ChIP-seq (and similar) data sets using peak-motifs. Nat. Protoc. 7:1551–1568. 10.1038/nprot.2012.088 [DOI] [PubMed] [Google Scholar]
- Thompson M.R., Xu D., and Williams B.R.. 2009. ATF3 transcription factor and its emerging roles in immunity and cancer. J. Mol. Med. (Berl.). 87:1053–1060. 10.1007/s00109-009-0520-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorvaldsdóttir H., Robinson J.T., and Mesirov J.P.. 2013. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 14:178–192. 10.1093/bib/bbs017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topley G.I., Okuyama R., Gonzales J.G., Conti C., and Dotto G.P.. 1999. p21(WAF1/Cip1) functions as a suppressor of malignant skin tumor formation and a determinant of keratinocyte stem-cell potential. Proc. Natl. Acad. Sci. USA. 96:9089–9094. 10.1073/pnas.96.16.9089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen J.M. 2014. The role of senescent cells in ageing. Nature. 509:439–446. 10.1038/nature13193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C.Y., and Filippakopoulos P.. 2015. Beating the odds: BETs in disease. Trends Biochem. Sci. 40:468–479. 10.1016/j.tibs.2015.06.002 [DOI] [PubMed] [Google Scholar]
- Wolford C.C., McConoughey S.J., Jalgaonkar S.P., Leon M., Merchant A.S., Dominick J.L., Yin X., Chang Y., Zmuda E.J., O’Toole S.A., et al. . 2013. Transcription factor ATF3 links host adaptive response to breast cancer metastasis. J. Clin. Invest. 123:2893–2906. 10.1172/JCI64410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Nguyen B.C., Dziunycz P., Chang S., Brooks Y., Lefort K., Hofbauer G.F., and Dotto G.P.. 2010. Opposing roles for calcineurin and ATF3 in squamous skin cancer. Nature. 465:368–372. 10.1038/nature08996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Q., Mukherjee S., and Furey T.S.. 2014. GSAASeqSP: A toolset for gene set association analysis of RNA-Seq data. Sci. Rep. 4:6347 10.1038/srep06347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K., Tateishi K., Kudo Y., Hoshikawa M., Tanaka M., Nakatsuka T., Fujiwara H., Miyabayashi K., Takahashi R., Tanaka Y., et al. . 2016. Stromal remodeling by the BET bromodomain inhibitor JQ1 suppresses the progression of human pancreatic cancer. Oncotarget. 7:61469–61484. 10.18632/oncotarget.11129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Forcato M., Battilana G., Azzolin L., Quaranta E., Bodega B., Rosato A., Bicciato S., Cordenonsi M., and Piccolo S.. 2015. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 17:1218–1227. 10.1038/ncb3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Huang B., Wu X., Zhang H., Qi J., Bradner J., Nair S., and Chen L.F.. 2014. Brd4 maintains constitutively active NF-κB in cancer cells by binding to acetylated RelA. Oncogene. 33:2395–2404. 10.1038/onc.2013.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.