Abstract
Purpose
Breast cancer predisposing mutations PALB2 c.1027C>T (p.Gln343*) and PALB2 c.2167_2168delAT have each been observed multiple times in breast cancer families of Italian ancestry. More recently, the c2167_2168delAT mutation was identified in unrelated breast cancer cases of various ancestries. For each mutation, we investigated whether the origin was multiple mutational events (a “hot-spot”) or a single event (a founder allele).
Methods
We genotyped and reconstructed haplotypes for 36 participants of Italian, Italian-American, Hispanic, and Nigerian ancestries, using seven short tandem repeat (STR) markers that covered 3 Megabases within and flanking PALB2 on chromosome 16.
Results
For PALB2 c.1027C>T, a shared haplotype of minimum size 150 kb was shared by all 19 carriers investigated, all of Italian ancestry. This result suggests that this allele arose as a single event in a shared ancestor. For PALB2 c.2167_2168delAT, all 12 carriers from American/Italian and Italian families shared a 1 MB haplotype, the 3 Hispanic carriers shared a different haplotype of size 2MB, and the Nigerian carrier had different alleles at all 7 STR markers. These results suggest that PALB2 c.2167_2168delAT arose multiple times, but that within each population, PALB2 c.2167_2168delAT likely represents a single mutational event.
Conclusion
We identified two PALB2 mutations that are founder alleles in Italian families, one of which is, independently, also a founder mutation in American-Hispanic breast cancers.
Keywords: breast cancer, PALB2, founder mutations, haplotype
Introduction
In 2006, PALB2 (partner and localizer of BRCA2), also known as FANCN, was identified as a key player in homologous recombination [1]. It was first described as a breast cancer-predisposing gene with identification of pathogenic mutations in familial cases [2]. Subsequent studies found carriers of truncating mutations in familial cases of almost all populations investigated with confirmation that pathogenic mutations were associated with breast cancer risk [3]. So far, more than 300 different mutations of PALB2 have been reported (http://databases.lovd.nl/shared/genes/PALB2). and it is ncluded on clinical multigene breast cancer panels. We had previously identified a recurrent mutation – the c.1027C>T (p.Gln343*) in Northern Italy; it was detected in 6 of 113 familial cases and 2 of 477 controls all from the Bergamo province [4]. A second PALB2 mutation, c.2167_2168delAT (p.Met723Valfs*21), has been identified multiple times in individuals from geographically diverse areas by independent research groups. This mutation was found in an Italian and an Italian-American families, three Hispanic probands from Southern California, and a Nigerian proband ([4–6]; S. Neuhausen/J. Weitzel, personal communications; F. Olopade, personal communication). Because PALB2 c.1027C>T and c.2167_2168delAT mutations have been encountered in a number of breast cancer cases from either the same or from different ancestries, we investigated haplotypes to assess whether these recurrent mutations originated from independent or single mutational events.
Participants and methods
Subjects
Women with breast cancer and female controls were enrolled into research studies to identify breast cancer susceptibility genes. For the specific individuals in this study, they either carried or were a family member of a carrier with either a PALB2 c.1027C>T mutation (16 individuals from 4 independently ascertained families, 6 singleton carriers) or a PALB2 c.2167_2168delAT mutation (16 individuals from 2 independently ascertained families and 4 singleton probands). The 44 individuals were ascertained at the following centers: The Unit of Medical Oncology of the Azienda Ospedaliera Papa Giovanni XXIII in Bergamo, Italy; the Medical Genetic Units of the Fondazione IRCCS Istututo Nazionale dei Tumori in Milan, Italy; the Associazione Volontari Italiani Sangue (AVIS), Bergamo, Italy; the Departments of Medical Genetics and Genome Sciences, University of Washington, Seattle, WA, US, the Clinical Cancer Genomics Community Research network at City of Hope, and the University of Chicago (Table 1). All individuals participating in this study signed an informed consent to the use of their biological samples for research projects. The study was approved by the Ethics Committee of each participating centers.
Table 1.
PALB2 mutation carrier phenotypes, ancestry, and demographics.
| Phenotypes in carriersa | |||||||
|---|---|---|---|---|---|---|---|
| Mutation | Family | # carriers | # female breast (age) |
# other cancers (age) |
# unaffected | Ancestry | Ascertainment center |
| c.1027C>T | BG149 | 1 | 1 | Italian, Bergamo | AVIS, Bergamo | ||
| c.1027C>T | BG363 | 1 | 1 | Italian, Bergamo | AVIS, Bergamo | ||
| c.1027C>T | BG02 | 1 | 1 (35) | Italian, Bergamo | Ospedale Papa Giovanni XXIII, Bergamo | ||
| c.1027C>T | BG03 | 1 | 1 (45) | Italian, Bergamo | Ospedale Papa Giovanni XXIII, Bergamo | ||
| c.1027C>T | BG04 | 4a | 1 (57) | 3 | Italian, Bergamo | Ospedale Papa Giovanni XXIII, Bergamo | |
| c.1027C>T | BG05 | 1 | 1 (37) | Italian, Bergamo | Ospedale Papa Giovanni XXIII, Bergamo | ||
| c.1027C>T | BG06 | 1 | 1 (35) | Italian, Bergamo | Ospedale Papa Giovanni XXIII, Bergamo | ||
| c.1027C>T | MI04 | 2 | 1 (37) | 1 | Italian, Bergamo | Istututo Nazionale dei Tumori, Milan | |
| c.1027C>T | MI06 | 2 | 1 (33) | 1 | Italian, Bergamo | Istututo Nazionale dei Tumori, Milan | |
| c.1027C>T | MI12/BG01 | 5a | 4 (29, 41, 42,67) | Lung (57) | 1 | Italian, Bergamo | Istututo Nazionale dei Tumori, Milan |
| c.2167delAT | MI03 | 2 | 2 (28, 45) | 1 | Italian, Marche | Istututo Nazionale dei Tumori, Milan | |
| c.2167delAT | CF1908 | 10a | 6 (36, 49, 49, 59, 62, 69) | Pancreatic (65) Prostate (65) |
2 | Italian-American | University of Washington, Seattle |
| c.2167delAT | CH7088 | 1 | 1 (42) | Hispanic | City of Hope, Los Angeles, CA | ||
| c.2167delAT | CH7777 | 1 | 1 (24) | Hispanic | City of Hope, Los Angeles, CA | ||
| c.2167delAT | CH8877 | 1 | 1 (43) | Hispanic | City of Hope, Los Angeles, CA | ||
| c.2167delAT | N286 | 1 | 1 (34) | Nigerian | Nigeria/University of Chicago, Chicago | ||
Includes obligate mutation carriers.
PALB2 mutations were previously identified through Sanger sequencing or targeted next generation sequencing ([4, 7]; Weitzel/Neuhausen personal communication). When available, relatives of PALB2 mutation carriers were genotyped for their family-specific mutations by Sanger sequencing using primers previously described [4, 8].
Short tandem repeat (STR) marker genotyping
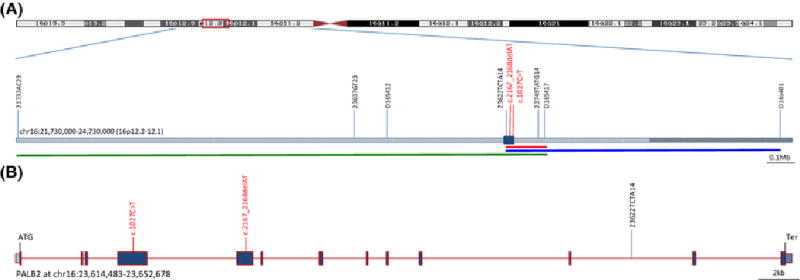
In total, 22 individuals were studied for haplotypes associated with PALB2 c.1027C>T mutation, and 20 individuals for the PALB2 c.2167_2168delAT mutation. Seven STR markers were selected which span a region of approximately 3 Mb on chromosome 16p flanking and intersecting PALB2 (at chr16:23,614,483–23,652,678; hg19) (Figure 1 and Table S1). Four of the seven STR markers were custom developed for genotyping of PALB2 mutation carriers [8] and three were STR markers previously deposited in the Marshfield comprehensive human genetic map [9, 10]. These markers were assayed by fluorescent PCR and capillary electrophoresis on an ABI PRISM 3130 or 3730xL Sequencer using standard methods. Primers and PCR conditions used to genotype the STR markers are reported in Supplementary Table 1. Genotyping was carried out at IFOM, The FIRC Institute of Molecular Oncology (Milan, Italy), Beckman Research Institute of City of Hope (Duarte, CA, USA) and University of Washington (Seattle, WA, USA). Allele sizes were called using the software Gene Mapper (Applied Biosystems/Life Technologies, Foster City, CA, USA). For consistent calling of allele sizes across centers, common controls were used by all centers.
Figure 1.
Haplotype analysis
In families, haplotypes were reconstructed manually according to the known mutation status and the genotypes observed at each marker in the family members. In the families where it was not possible to reconstruct haplotypes with certainty, or in single individuals, haplotypes were reconstructed according to the hypothesis of the maximum conservation of a common mutation haplotype.
Results
PALB2 c.1027C>T haplotype analysis
DNA samples extracted from blood from 16 individuals from 4 families, and 6 single individuals, all of Italian/Bergamo ancestry were genotyped (Table 1; Figure 2A). Enlarged pedigrees with family cancer history were previously published for these individuals except for BG149 and BG363 [4]. Four haplotypes across the seven STR markers were reconstructed. A core haplotype is shared by all 19 mutation carriers (Table 2, Figure 2A). This core haplotype, with a size of ~0.15Mb, is defined by recombination events 3′ of PALB2 between markers D16S412 and 23622TCTA14 and 5′ of PALB2 between markers D16S417 and D16S401 (Figure 1A, Table 2). This shared haplotype indicates that the PALB2 c.1027C>T mutation occurred as a unique event in a single ancestor related to all 10 families in this study with this mutation.
Figure 2.
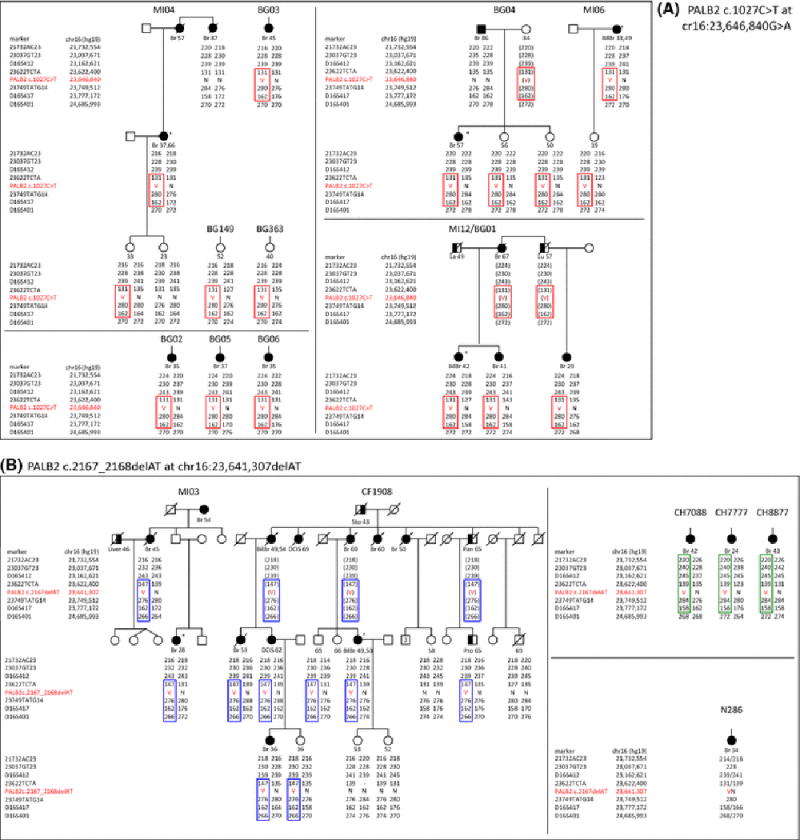
Table 2.
Genotypes and haplotypes identified in carriers of PALB2 c.1027C>T and PALB2 c.2167_2168delAT
| Markers and genotypes and haplotypesa | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mutation | Ancestry | Family | # carriers | 21732AC23 | 23037GT23 | D16S412 | 23622TCTA14 | 23749TATG14 | D16S417 | D16S401 |
| c.1027C>T | Italian | BG03 | 1 | 216/220 | 228 | 239 | 131 | 280 | 162 | 270 |
| c.1027C>T | Italian | BG149 | 1 | 216/218 | 228 | 239/241 | 131 | 280 | 162 | 270/274 |
| c.1027C>T | Italian | BG363 | 1 | 216/224 | 228 | 239 | 131 | 280 | 162 | 270/274 |
| c.1027C>T | Italian | MI04 | 2 | 216 | 228 | 239 | 131 | 280 | 162 | 270 |
| c.1027C>T | Italian | BG04 | 4 | 220 | 228 | 239 | 131 | 280 | 162 | 272 |
| c.1027C>T | Italian | MI06 | 2 | 220 | 228 | 239 | 131 | 280 | 162 | 272 |
| c.1027C>T | Italian | BG02 | 1 | 224/220 | 230/238 | 243/241 | 131 | 280 | 162 | 270/276 |
| c.1027C>T | Italian | BG05 | 1 | 224/220 | 230/236 | 243/239 | 131 | 280 | 162 | 270 |
| c.1027C>T | Italian | BG06 | 1 | 224/220 | 230/228 | 243/241 | 131 | 280 | 162 | 270 |
| c.1027C>T | Italian | MI12/BG01 | 5 | 224 | 230 | 243 | 131 | 280 | 162 | 272 |
| c.2167delAT | Italian | MI03 | 2 | 216 | 232 | 243 | 147 | 276 | 162 | 266 |
| c.2167delAT | Italian | CF1908 | 10 | 218 | 230 | 239 | 147 | 276 | 162 | 266 |
| c.2167delAT | Hispanic | CH7088 | 1 | 220/226 | 240/228 | 245/237 | 139/135 | 284/276 | 158/162 | 268 |
| c.2167delAT | Hispanic | CH7777 | 1 | 220/226 | 240/238 | 245 | 139/123 | 284/280 | 158/176 | 272/264 |
| c.2167delAT | Hispanic | CH8877 | 1 | 220/226 | 240/242 | 245 | 139/131 | 284 | 158 | 272/274 |
| c.2167delAT | Nigerian | N286 | 1 | 214/218 | 228 | 239/241 | 139 | 280 | 158/166 | 268/270 |
Core haplotypes shared among unrelated families are underlined
PALB2 c.2167_2168delAT haplotype analysis
Germline DNA samples from 16 members of 2 families of Italian and American/Italian ancestry, 3 female breast cancer cases of Hispanic ancestry and one female breast cancer case of Nigerian ancestry were genotyped (Table 1, Figure 2B). Haplotype construction suggests that there were three unique events (Table 2). All 12 mutation carriers in the Italian and American/Italian families share a conserved haplotype, of approximate size of 1 Mb, defined by a recombination event 3′ of PALB2 between markers D16S412 and 23622TCTA14 (Figure 1A, Table 2). The three Hispanic individuals share a conserved haplotype of 2 Mb defined by a recombination event 5′ of PALB2 between markers D16S417 and D16S401 (Figure 1A, Table 2). The Nigerian breast cancer case carries alleles which are not part of any of the previously described haplotypes. These unique events, at least in the Italian and Hispanic ancestry cases, resulted in independent founder mutations in these two populations.
Discussion
Overall, PALB2 loss of function mutations have been found in 0.6 to 3.9% of high-risk breast cancer families, with an average cumulative risk of breast cancer of 35% by age 70 years [5]. In this manuscript, we describe the haplotype analysis of two recurrent PALB2 mutations. Five other truncating mutations have been identified to be recurrent in specific populations. The PALB2 c.509_510delGA (p.Arg170Ilefs*14) was found in Poland in 4/648 (0.6%) familial breast cancer cases and in 1/1310 controls (0.08%) [11], with no evidence of shared alleles. This mutation was later detected in approximately 0.25% of unselected breast cancer cases from Central and Eastern Europe [12]. With such a narrow distribution of the mutation, yet an apparent lack of a shared haplotype, it is not clear whether this is a single founder mutation or whether there were multiple mutation events at this site. The PALB2 c.2323C>T (p.Gln775*) was found in approximately 0.5% of tested French-Canadian early onset breast cancer cases and in 0 of 6,000 controls. Genotyping of four STR markers showed that mutation carriers shared common alleles suggesting that the PALB2 c.2323C>T is a founder mutation [13]. In Finland, a recurrent founder mutation PALB2 c.1592delT (p.Leu531Cysfs*30) was identified [14]. They were able to estimate that this mutation was associated with a 40% cumulative risk for developing breast cancer by age 70 years. In Australia, a founder PALB2 c.3113G>A (p.Trp1038* or complete or partial deletion of exon 10) was identified, and they estimated a 91% cumulative risk of developing breast cancer by age 70 years [15]. Analyses of founder mutations allow for determination of mutation-specific risks, which still vary by population and the extent of family history.
The common haplotype shared by all four Italian families and six Italian single individuals with the PALB2 c.1027C>T mutation suggests that this mutation occurred as a single event in an ancestor common to all the families and single individuals studied. This mutation, to our knowledge, has been found only in individuals from Bergamo and represents one of the few examples of population-specific mutation in Italians. Its limited range to this specific area in Northern Italy may reflect limited migration of the local population, possibly due to territory geographical conformation characterized by several secluded valleys that may have caused genetic isolation [4]. Consistent with this hypothesis, the BRCA1 c.190T>C (p.Cys64Arg) founder mutation is only found in breast cancer families from Bergamo, and was estimated to be more than 3,000 years old [16]. There are too few families and family members with the PALB2 c.1027C>T to estimate the age of the mutation. However, the shorter length of the PALB2 c.1027C>T haplotype with respect to the BRCA1 c.190C>T haplotype may indicate that the c.1027C>T is an older mutation.
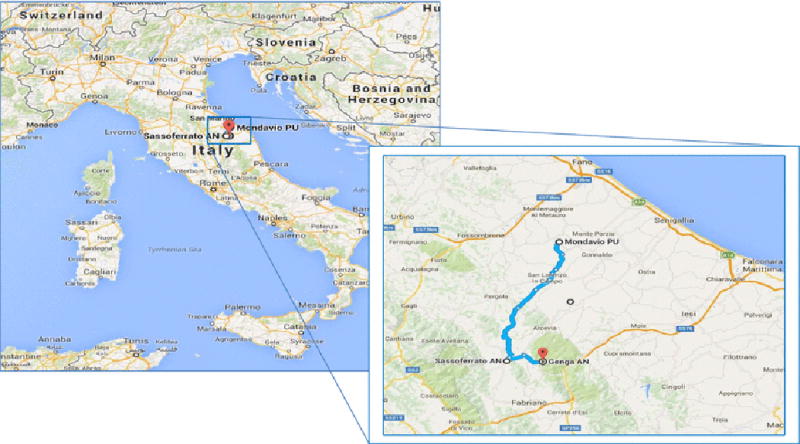
PALB2 c.2167_2168delAT has been observed in an African, Italians, North Americans, and Hispanics. Based on haplotype and allele sharing, these mutations appear to represent different mutational events in each of the three populations. No haplotype sharing was observed among the Nigerian, Hispanic, and Italian carriers. However, a conserved haplotype was found between the North American family who is of Italian ancestry and the Italian family ascertained in Milan, representing evidence of a common ancestor. We investigated whether the Italian-American family (CF1908) and the Milan family (MI03) originated from the same geographic region in Italy. Surnames are often informative with respect to Italian region of origin [17]. The two ancestors in the Italian-American family, who migrated to America at the beginning of the 1900 century, were from Sasso Ferrato (AN) and Genga (AN), two small towns from the province of Ancona in the Italian Appennini Mountains, located at a distance of about 10 kilometers from each other. In the Italian family from Milan, the paternal aunt of the proband was originally from Mondavio, a small town in the province of Pesaro-Urbino, 45 kilometers from the above mentioned locations (Figure 3). A genealogy search also found that, based on actual frequency and distribution, the family names originated from the region Marche in Central Italy, confirming the geographic origin of their families. The three Hispanic breast cancer cases share the same associated-mutation alleles indicating a putative Amerindian mutational event (data not shown). This PALB2 mutation was identified in 3 of 188 unrelated Hispanic breast cancer cases (1.6%), and we have since identified additional Hispanic breast cancer cases with this mutation (personal communication). Interestingly, the PALB2 c.2167_2168delAT appears to have arisen through multiple independent mutation events, yet within populations, it appears likely to be a founder mutation.
Figure 3.
In conclusion, we’ve identified two founder mutations of Italian ancestry, of which one is an independent founder in those of Amerindian ancestry.
Supplementary Material
Acknowledgments
Funding: Work in Italy was partially supported by funds from Ministero della Salute, Italy ‘Ricerca Finalizzata (RF-2010-2316374 to PP and CT), AIRC Associazione Italiana per la Ricerca sul Cancro (IG 12821 to PP and “5xmille” n. 12237 to AF) and from Italian citizens who allocated a 5/1000 share of their tax payment in support of the Fondazione IRCCS Istituto Nazionale Tumori (SM and PR). Work at University of Washington was supported by NIH R01CA175716 (M-CK) and R35CA197458 (M-CK). Work at City of Hope (COH) was supported by R01CA184585 (SLN), a COH Excellence Award (SLN) and the Morris and Horowitz Families Professorship (SLN). Work performed in the Integrative Genomics Core was supported by the National Cancer Institute of the National Institutes of Health under award number P30CA033572. Work at University of Chicago was supported by American Cancer Society CRP-10-119-01 –CCE (OIO) and the Susan G. Komen Foundation (OIO). The City of Hope Clinical Cancer Genetics Community Research Network was supported in part by Award Number RC4A153828 (PI: J.Weitzel) from the National Cancer Institute and the Office of the Director, National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Compliance with ethical standards
Conflict of interest. The authors declare that they have no conflicts of interest.
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
References
- 1.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–29. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 2.Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, Reid S, Spanova K, Barfoot R, Chagtai T, Jayatilake H, McGuffog L, Hanks S, Evans DG, Eccles D, Breast Cancer Susceptibility C. Easton DF, Stratton MR. PALB2 which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Southey MC, Teo ZL, Winship I. PALB2 and breast cancer: ready for clinical translation! Appl Clin Genet. 2013;6:43–52. doi: 10.2147/TACG.S34116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catucci I, Peterlongo P, Ciceri S, Colombo M, Pasquini G, Barile M, Bonanni B, Verderio P, Pizzamiglio S, Foglia C, Falanga A, Marchetti M, Galastri L, Bianchi T, Corna C, Ravagnani F, Bernard L, Fortuzzi S, Sardella D, Scuvera G, Peissel B, Manoukian S, Tondini C, Radice P. PALB2 sequencing in Italian familial breast cancer cases reveals a high-risk mutation recurrent in the province of Bergamo. Genet Med. 2014;16:688–94. doi: 10.1038/gim.2014.13. [DOI] [PubMed] [Google Scholar]
- 5.Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkas K, Roberts J, Lee A, Subramanian D, De Leeneer K, Fostira F, Tomiak E, Neuhausen SL, Teo ZL, Khan S, Aittomaki K, Moilanen JS, Turnbull C, Seal S, Mannermaa A, Kallioniemi A, Lindeman GJ, Buys SS, Andrulis IL, Radice P, Tondini C, Manoukian S, Toland AE, Miron P, Weitzel JN, Domchek SM, Poppe B, Claes KB, Yannoukakos D, Concannon P, Bernstein JL, James PA, Easton DF, Goldgar DE, Hopper JL, Rahman N, Peterlongo P, Nevanlinna H, King MC, Couch FJ, Southey MC, Winqvist R, Foulkes WD, Tischkowitz M. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506. doi: 10.1056/NEJMoa1400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanchi KL, Johnson KJ, Lu C, McLellan MD, Leiserson MD, Wendl MC, Zhang Q, Koboldt DC, Xie M, Kandoth C, McMichael JF, Wyczalkowski MA, Larson DE, Schmidt HK, Miller CA, Fulton RS, Spellman PT, Mardis ER, Druley TE, Graubert TA, Goodfellow PJ, Raphael BJ, Wilson RK, Ding L. Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun. 2014;5:3156. doi: 10.1038/ncomms4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walsh T, Lee MK, Casadei S, Thornton AM, Stray SM, Pennil C, Nord AS, Mandell JB, Swisher EM, King MC. Detection of inherited mutations for breast and ovarian cancer using genomic capture and massively parallel sequencing. Proc Natl Acad Sci U S A. 2010;107:12629–33. doi: 10.1073/pnas.1007983107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casadei S, Norquist BM, Walsh T, Stray S, Mandell JB, Lee MK, Stamatoyannopoulos JA, King MC. Contribution of inherited mutations in the BRCA2-interacting protein PALB2 to familial breast cancer. Cancer Res. 2011;71:2222–9. doi: 10.1158/0008-5472.CAN-10-3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63:861–9. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.White JA, McAlpine PJ, Antonarakis S, Cann H, Eppig JT, Frazer K, Frezal J, Lancet D, Nahmias J, Pearson P, Peters J, Scott A, Scott H, Spurr N, Talbot C, Jr, Povey S. Guidelines for human gene nomenclature (1997). HUGO Nomenclature Committee. Genomics. 1997;45:468–71. doi: 10.1006/geno.1997.4979. [DOI] [PubMed] [Google Scholar]
- 11.Dansonka-Mieszkowska A, Kluska A, Moes J, Dabrowska M, Nowakowska D, Niwinska A, Derlatka P, Cendrowski K, Kupryjanczyk J. A novel germline PALB2 deletion in Polish breast and ovarian cancer patients. BMC Med Genet. 2010;11:20. doi: 10.1186/1471-2350-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noskowicz M, Bogdanova N, Bermisheva M, Takhirova Z, Antonenkova N, Khusnutdinova E, Bremer M, Christiansen H, Park-Simon TW, Hillemanns P, Dork T. Prevalence of PALB2 mutation c.509_510delGA in unselected breast cancer patients from Central and Eastern Europe. Fam Cancer. 2014;13:137–42. doi: 10.1007/s10689-013-9684-1. [DOI] [PubMed] [Google Scholar]
- 13.Foulkes WD, Ghadirian P, Akbari MR, Hamel N, Giroux S, Sabbaghian N, Darnel A, Royer R, Poll A, Fafard E, Robidoux A, Martin G, Bismar TA, Tischkowitz M, Rousseau F, Narod SA. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007;9:R83. doi: 10.1186/bcr1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erkko H, Dowty JG, Nikkila J, Syrjakoski K, Mannermaa A, Pylkas K, Southey MC, Holli K, Kallioniemi A, Jukkola-Vuorinen A, Kataja V, Kosma VM, Xia B, Livingston DM, Winqvist R, Hopper JL. Penetrance analysis of the PALB2 c.1592delT founder mutation. Clin Cancer Res. 2008;14:4667–71. doi: 10.1158/1078-0432.CCR-08-0210. [DOI] [PubMed] [Google Scholar]
- 15.Southey MC, Teo ZL, Dowty JG, Odefrey FA, Park DJ, Tischkowitz M, Sabbaghian N, Apicella C, Byrnes GB, Winship I, Baglietto L, Giles GG, Goldgar DE, Foulkes WD, Hopper JL. kConFab for the Beast Cancer Family R. A PALB2 mutation associated with high risk of breast cancer. Breast Cancer Res. 2010;12:R109. doi: 10.1186/bcr2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caleca L, Putignano AL, Colombo M, Congregati C, Sarkar M, Magliery TJ, Ripamonti CB, Foglia C, Peissel B, Zaffaroni D, Manoukian S, Tondini C, Barile M, Pensotti V, Bernard L, Papi L, Radice P. Characterization of an Italian founder mutation in the RING-finger domain of BRCA1. PLoS One. 2014;9:e86924. doi: 10.1371/journal.pone.0086924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavalli-Sforza L, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton, NJ: Princeton University Press; 1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


