Overview
Natural products are important secondary metabolites produced by bacterial and fungal species that play important roles in cellular growth and signaling, nutrient acquisition, intra- and interspecies communication, and virulence. A subset of natural products is produced by nonribosomal peptide synthetases (NRPSs), a family of large, modular enzymes that function in an assembly line fashion. Because of the pharmaceutical activity of many NRPS products, much effort has gone into the exploration of their biosynthetic pathways and the diverse products they make. Many interesting NRPS pathways have been identified and characterized from both terrestrial and marine bacterial sources. Recently, several NRPS pathways in human commensal bacterial species have been identified that produce molecules with antibiotic activity, suggesting another source of interesting NRPS pathways may be the commensal and pathogenic bacteria that live on the human body. The ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp.) have been identified as a significant cause of human bacterial infections that are frequently multidrug resistant. The emerging resistance profile of these organisms has prompted calls from multiple international agencies to identify novel antibacterial targets and develop new approaches to treat infections from ESKAPE pathogens. Each of these species contains several NRPS biosynthetic gene clusters. While some have been well characterized and produce known natural products with important biological roles in microbial physiology, others have yet to be investigated. This review catalogs the NRPS pathways of ESKAPE pathogens. The exploration of novel NRPS products may lead to a better understanding of the chemical communication used by human pathogens and potentially to the discovery of novel therapeutic approaches.
Graphical abstract
This review describes the peptide natural products produced by NRPS biosynthetic gene clusters from the ESKAPE pathogens
1. The biochemistry and structural biology of the nonribosomal peptide synthetases
1.1 NRPS modular architecture
The history of peptide natural products from marine and terrestrial microbial sources offers a view of the parallel advances in the fields of chemistry, biochemistry, microbiology and genetics. Following the identification of diverse natural products with antibiotic and anticancer activity in the 1960s and 1970s, the following decades saw the unraveling of the enzymes that were responsible for the ribosome- and RNA-independent production of certain peptide products. Early work from Lipmann and colleagues with the high molecular weight proteins that catalyzed synthesis of gramicidin demonstrated that specific amino acids are activated through an adenylation reaction1 and subsequently covalently bound to a thiol on the enzyme.2 The observation that phosphopantethenic acid was present on the gramicidin synthetase3 and that peptide intermediates bound this cofactor as a thioester4 led to a protein thiotemplate model for nonribosomal peptide synthesis in which the amino acid and growing peptide are delivered by a carrier domain to neighboring catalytic domains.
The subsequent genomic revolution identified many genes that are responsible for peptide production by nonribosomal peptide synthetases (NRPSs).5, 6 The encoded proteins were organized into domains7 whose relationship to known proteins enabled the prediction of domain function.8 The confirmation of biochemical activity catalyzed by the core domains9, 10 established the remarkable catalytic strategy used by the multidomain NRPSs.11-16
NRPSs are organized into modules; generally, each module contains the catalytic domains that are responsible for the incorporation of a single amino acid into the peptide product. Some NRPS pathways, often within fungal organisms,17, 18 encode all of the modules on a single gene that encodes a single, large multimodular protein. More commonly, NRPS pathways contain multiple large NRPS proteins that interact through a series of both intra- and intermolecular transfers in the synthesis of the peptide product. A standard NRPS module contains a single peptidyl carrier protein (PCP), a small 8 kDa domain that is homologous to the acyl carrier proteins used in fatty acid transport and synthesis.19, 20 The PCP domains contain a conserved serine residue that is post-translationally modified through the activity of a phosphopantetheinyl transferase (PPTase) with a phosphopantetheine cofactor that is derived from Coenzyme A.21 This phosphopantetheine serves as a flexible linker that harbors a thiol onto which the amino acid and peptide intermediates are installed as a thioester. A module generally will also contain an adenylation domain, most commonly positioned upstream of the PCP, that catalyzes a two-step reaction to activate and then load the amino acid onto the pantetheine cofactor. In the initial adenylate-forming partial reaction, the amino acid substrate and ATP react to form an amino acyl-adenylate and pyrophosphate. In a second step, the pantetheine thiol attacks the carbonyl carbon of the amino acid substrate to displace the AMP, which serves as a facile leaving group for this reaction.22, 23 The final catalytic domain within standard NRPS modules is a condensation domain. Two PCPs from upstream and downstream modules meet in the active site of the condensation domain to allow the upstream amino acid or peptide to be transferred to the amino acid that is loaded onto the downstream PCP. The condensation domain is responsible for the extension of the growing peptide and is therefore the true peptide synthetase domain that is.
Because the peptide is covalently bound to the PCP domains during synthesis, NRPSs require a domain that catalyzes release of the completed peptide into solution. Most commonly, this activity resides in a C-terminal thioesterase domain that contains a catalytic serine, or less commonly a cysteine, that forms a stable acyl-enzyme intermediate.24 This intermediate can then be resolved through hydrolysis to release the free linear peptide, or via cyclization with the N-terminal amine.25 Additional variations on this theme include the cyclization through a nucleophilic side chain present on the peptide such as an internal lysine in the siderophore pyoverdine26 and antibiotics tyrocidin and bacitracin27, or the β-hydroxyl moiety of the N-terminal acyl chain in surfactin.28 The presence of the acyl-enzyme intermediate also allows for the oligomerization of the peptide product in certain instances where a peptide that has been completed through one cycle of synthesis by the NRPS modules, can serve as the nucleophile to attack a second peptide intermediate. The joining of two pentapeptides in gramicidin29 or of three units in enterobactin30, for example, allow for the production of the final peptide oligomer.
Enhancing these core activities of the NRPS domains, and further diversifying the nature of the peptide products, are integrated auxiliary domains that catalyze additional chemical modifications.31 Among the most common of these supplementary activities are S-adenosylmethionine (SAM)-dependent methyltransferases,32, 33, epimerases,34, 35 and domains that catalyze the heterocyclization of serine, threonine, and cysteine residues to form oxazoline and thiazoline rings. The latter are variant condensation domains that share 25-30% sequence identity with peptide bond-forming condensation domains.27, 36 Additionally, some NRPS proteins terminate not in a thioesterase domain but rather in a reductase domain that catalyzes the NADPH dependent formation of a terminal aldehyde that itself may be subject to further spontaneous or enzyme catalyzed modification.37, 38
The organization of NRPS domains and modules might suggest that a straightforward bioinformatic approach would allow the identification of the final peptide product from gene sequence. However, the facile prediction of peptide product from domain and module organization is further challenged by the observation that catalytic domains often perform unexpected chemical transformations. For example, the condensation domain of the didomain protein PacN catalyzes the peptide chain reversing incorporation of a ureido dipeptide linkage in the production of the nucleoside antibiotic pacidomycin.39 The condensation domain of the final module of the nocardicin NRPS cluster, NocB, catalyzes the formation of the β-lactam moiety within this antibiotic.40 Thioesterase domains also have been shown to catalyze unusual activities. Several fungal NRPSs contain thioesterase domains that catalyze the condensation of two β-ketoacids that subsequently cyclize to form furanone cores of several pigment molecules.41, 42 The thioesterase domains have also been implicated in the Dieckmann cyclization in bacterial tetramate natural products43, 44 as well as β-lactam formation in the production of sulfazecin.45 Although not quite as chemically diverse as the chemical reactions catalyzed above, adenylation domains also have been observed to catalyze unexpected reactions. Amide formation is catalyzed by an unusual adenylation domain that by-passes the need for a carrier domain or a condensation domain in the elongation of the L-β-lysine oligopeptide of the antibiotic streptothricin.46 Finally, the presence of domains that play a more structural role further complicates the prediction of product from an NRPS sequence.47 Additional characterization of novel NRPS pathways will continue to inform the prediction and confirmation of novel activities.
1.2 Structural dynamics of NRPSs
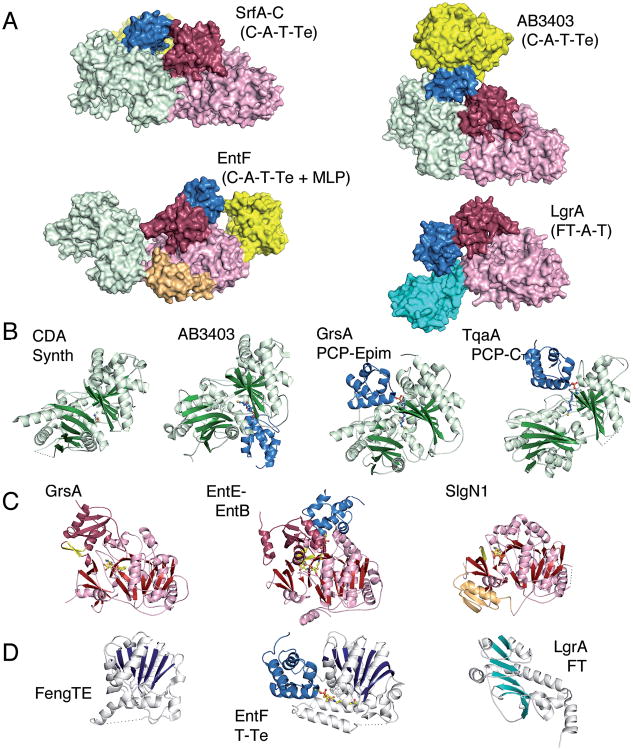
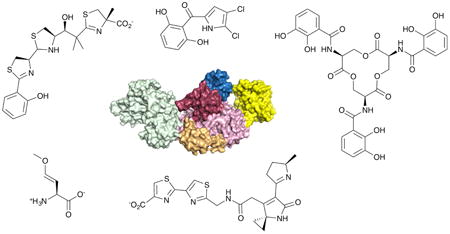
The biochemistry of the NRPS enzymes raises questions about the structural mechanisms used by these large modular proteins that are responsible for their assembly line mechanism. Numerous reviews exist,15, 16, 22 including several that are quite recent,48-55 that detail what is known about the structures of both catalytic domains and larger multidomain and modular proteins. Indeed, this is an exciting time for the structural studies of NRPSs as many structures of multidomain proteins were determined in 2016.56-63 This review will briefly describe the structures of individual domains and the implications of the recent structures on the necessary dynamics to transfer the substrates and intermediates between different catalytic domains (Fig. 1).
Fig. 1.
Structures of NRPS enzymes. A. Surface representation of multidomain NRPS enzymes indicating domain architecture. Complete modules of SrfA-C (PDB 2VSQ), AB3403 (4ZXI), EntF (5JA1), and LgrA (5ES9). Surfaces are colored to highlight condensation (green), adenylation N-terminal (pink) and C-terminal (red) subdomains, PCP (blue), thioesterase (yellow) and formyltransferase (cyan) domains. The EntF structure also illustrates a bound MLP, YbdZ (orange) B. Ribbon diagrams of condensation domains from CDA Synthetase (5DU9) and AB3403, as well as the homologous epimerization (5ISX) and cyclization (5EJD) domains. C. Ribbon diagrams of adenylation domains showing GrsA in the adenylate-forming conformation (1AMU), the functional interaction of the EntE adenylation domain with the EntB carrier protein domain in the thioester-forming conformation (4IZ6), and the interaction between the MLP and N-terminal subdomain of the adenylation domain of SlgN1 (4GR5). D. Ribbon diagrams of the thioesterase domain from fengycin NRPS FenB (2CB9) and the functional interaction between the PCP and thioesterase of EntF (3TEJ). Additionally shown is the ribbon diagram from the formyltransferase domain of LgrA.
NRPS carrier protein domains are members of the family of acyl-carrier protein domains that play many roles in acyl group transport.19, 20 The PCP domains are 70-80 residues in length and are composed of four α-helices and intervening loops. Helices α1, α2, and α4 are longer in length and mostly parallel, whereas helix α3 is shorter and oriented roughly perpendicular to the other three helices. The serine, onto which the phosphopanetheine cofactor is placed is part of a conserved Gx(D/H)S motif, where the second position is most commonly a hydrophobic residue ranging in size from glycine to larger aromatic residues.19
The NRPS condensation domains are 400-450 residues in length and contain two lobes that surround a large central cleft containing the active site.55 The lobes have been observed in multiple conformations (Fig. 1B).64-66 The open cleft contains the binding sites for the upstream (donor) PCP domain, as well as the downstream (acceptor) PCP. The substrates bound to the pantetheine arms then meet for transfer of the upstream amino acid or peptide to the amino acid that has previously been loaded onto the downstream PCP. Recent structures have been obtained with pantetheine chains bound to the acceptor site.56, 58 While no structures exist of a condensation domain bound to a donor PCP, structures of TqaA, the homologous terminal cyclization domain of fungal NRPSs,62 and an epimerization domain of the gramicidin synthetase GrsA57 provide models for the binding of the PCP to the condensation domain donor site.
The NRPS adenylation domain is a member of the structurally homologous ANL superfamily of adenylate-forming enzymes. This superfamily contains three subfamilies, the NRPS adenylation domains, the acyl- and aryl-CoA synthetases, and the beetle luciferase enzymes.22 Members of this enzyme family are 500-700 residues in length and contain two subdomains. A larger N-terminal subdomain contains 400-600 residues, while the smaller C-terminal subdomain is less variable in size at 120-140 residues in length. The larger and smaller subdomains have also been referred to as the Acore and Asub domains.50 Members of this enzyme family catalyze two partial reactions, the initial adenylation step followed by a second thioester-forming reaction. In the case of luciferase, the second reaction is an oxidative decarboxylation leading to the light-producing intermediate. The active site is positioned between the two subdomains and early structures of members of this enzyme family pointed to the possibility that the C-terminal subdomain would adopt two distinct conformations to catalyze the two partial reactions.67-70 This was subsequently confirmed through biochemical and structural studies of multiple family members.22 This large conformational change was also proposed to play a role in transporting the PCP between different catalytic domains of the modular NRPSs.71
In addition to the carrier protein and the core condensation and adenylation domains, structures exist for the additional NRPS domains, including the chain terminating thioester72, 73 and reductase37, 38, 74, 75 domains, as well as the initiating formyltransferase domain (Fig. 1D).60
While structures of individual NRPS domains allow for understanding the reaction mechanisms of a single step in the NRPS biosynthesis, structures of multidomain NRPS proteins provide more detailed views of the modular architecture. Didomain structures of the PCP-thioesterase76, 77 and the PCP-adenylation domains78-80 illustrate the domain interfaces that occur between catalytic domains and the PCP. These structures show that the PCP helix α2, which starts with the pantetheinylated serine, as well as the loop that joins helices α1 and α2 form the PCP interface with catalytic domains. Further, in the structures containing the holo-PCP, the cofactor is directed into the active site of the enzyme. Crystallization of the adenylation-PCP complexes78-80 was achieved with a mechanism based inhibitor that captured the attack of the pantetheine thiol on a vinylsulfonamide analog of the adenylate intermediate within the active site of the adenylation domain.81 In these structures, the C-terminal subdomain is rotated into the thioester-forming conformation, forming the pantetheine tunnel that enables the pantetheine to reach the active site.
The structures of multidomain NRPS proteins provides further understanding of the domain interactions and the dynamic nature of the NRPS structural cycle. The determination in 2008 of the structure of SrfA-C, the terminal module from the surfactin biosynthetic cluster provided the first view of how a complete NRPS module was organized (Fig. 1A).82 This landmark structure showed a significant interface was formed between the condensation domain and the larger N-terminal adenylation subdomain. Much more limited interactions were made between these domains and the C-terminal thioesterase domain. The structure was consistent with the hypothesis that movement of the adenylation C-terminal subdomain could accompany transport of the PCP to the condensation domain and adenylation domain active sites.82
This proposal has been supported by recent structures of two holo-NRPS modules of the same domain architecture as SrfA-C.58, 59 The structure of the A. baumannii NRPS protein AB3403 illustrated the interaction of the PCP with the upstream condensation domain while a structure of the E. coli EntF protein showed the interaction of the PCP with the adenylation domain (Fig. 1A). Accompanying the movement of the PCP from the condensation domain to the adenylation domain, the adenylation domain C-terminal subdomain rotates through the two catalytic conformations.22 In AB3403, the adenylation domain adopts the adenylate-forming conformation to allow the PCP to engage the condensation domain. Upon completion of the adenylation reaction, the C-terminal subdomain rotates to the thioester-forming conformation as seen in EntF to deliver the PCP to the adenylation domain active site and form the conformation that is used to catalyze the thioesterification reaction.
While the above structural studies on modular NRPS proteins used termination modules, equally exciting observations were observed in the recent structures of an initiation module from the gramicidin synthetase.60 The first module of LgrA contains a formyltransferase domain upstream of adenylation and PCP domains, forming a tridomain protein that was structurally characterized in multiple catalytic states. The LgrA structures also demonstrate the domain alternation of the adenylation domain C-terminal subdomain upon movement of the PCP between the adenylate- and thioester-forming conformations. Further, the C-terminal subdomain adopts an extended conformation, projecting away from the N-terminal subdomain, to allow the substrate bound to the PCP to reach to the formyltransferase domain.60
These recent structural observations58 provide snap shots that help to piece together the complete structural cycle. Because the NRPS module can adopt the conformation to catalyze simultaneously peptide bond formation in the condensation domain while also catalyzing substrate adenylation within the adenylation domain, the enzyme is able to prime the next cycle of synthesis while the PCP is engaged with the condensation domain.51 In comparison to the the magnificent rate enhancements of many enzymes, the modular NRPS enzymes are rather slow, with turnover constants ranging from 1 to 100 turnovers per minute.30, 83, 84 This is true for many secondary metabolites and perhaps reflects limited, or none at all, evolutionary pressure to accelerate natural product biosynthesis. Nonetheless, these new structural results suggest that the efficiency of the catalytic cycle could be enhanced by the ability of two NRPS domains within a module to be active at the same time.
Also apparent from the structures of the three terminal modules was the lack of conservation of the positions of the downstream thioesterase domain.58, 59, 82 The lack of interactions between the thioesterase domain and the remaining catalytic domains suggests that there may be limited multimodular organization that govern interactions between adjacent modules. Indeed the Schmeing group has recently successfully crystallized the inter-module structure of a fragment of DhbF, a bimodular protein that is used to produce the siderophore bacillibacatin. The NRPS fragment contains an adenylation-PCP-condensation domain, crossing the border between modules and extending to the downstream condensation domain.63 The structure shows that no interactions exist between the adenylation domain and the condensation domain of the subsequent module. Instead the condensation domain cradles the “back face” of the carrier protein domain. If one considers the downstream thioesterase domain to be part of the “next module”, this structure and the termination modules suggest limited interactions may exist between the core condensation-adenylation domains of one module and the domains of the downstream modules.
2. NRPSs in ESKAPE pathogens
2.1. ESKAPE pathogens
A 2008 editorial commentary85 introduced the phrase “ESKAPE bugs” to represent a panel of human pathogens that were increasingly resistant to many of the most common antibiotics. The frequency of nosocomial infections caused by Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp., particularly in older and immunocompromised patients in health care settings, illustrated a need to develop novel antimicrobial agents. The use of the term ESKAPE highlighted the ability of these organisms to “escape” from the antimicrobial activities of the commonly used antibiotics. A growing consensus of the importance of these species has led to a call to action from the Infectious Diseases Society of America as well as the development of public/private partnerships across the globe.86, 87 Indeed, concern has been raised for the relative lack of novel treatments in the pipeline that are needed to reverse the growing threat posed by multidrug resistant ESKAPE infections.88, 89
Beyond the significant clinical impact of these pathogens, they also serve as “paradigms of pathogenesis, transmission, and resistance.”85 It is therefore instructive to understand the fundamental basis of microbial physiology and interaction with the host. There is value in understanding the unique properties that make these organisms particularly suited to survive in the host and, further, in exploring the features encoded within their genomes that promote their virulence.
As many NRPS products have been identified as virulence factors or as novel signaling molecules, a survey of the NRPS biosynthetic gene clusters that are present in ESKAPE pathogens could offer insight into the fundamental microbiology of these organisms and could potentially identify novel targets for antimicrobial discovery.
2.2. NRPS biosynthetic gene clusters
Several recent reviews have focused on the small molecules produced by the human microbiota.90-94 The combined tools of mass spectrometry and the revolution in genomic sequencing revealed much of the biosynthetic potential of the bacteria that co-exist upon and within the human body. This information has in turn led to the identification of novel biosynthetic pathways for products with important biological activity.95
This review will focus specifically on the NRPS products that are produced by the ESKAPE pathogens. For each species, bioinformatic approaches were used to identify known and uncharacterized biosynthetic gene clusters (BGCs).96 Recent efforts at generating curated and publicly available databases such as the Minimal Information about a Biosynthetic Gene Cluster (MIBiG),97 and the Antibiotics and Secondary Metabolite Analysis Shell (antiSMASH),98 as well as additional online tools,99-101 facilitate the identification of and prediction of the products of novel BGCs.
The boundaries of a BGC can be difficult to predict. Multiple genes of a biosynthetic pathway are often expressed on a single operon derived from a single mRNA transcript containing multiple genes that are separated by fewer than 50 nucleotides. However, sometimes the genes of a BGC are separated into multiple operons. The pyoverdine BGC of P. aeruginosa, for example, contains 15 genes on multiple operons that are separated by genes that play no role in pyoverdine biosynthesis.102 We therefore also examined the surrounding genes of any uncharacterized BGCs described below and, if discovered, comment on the proximity of additional genes that may be involved in the biosynthetic pathway.
Herein, the NRPS clusters within the ESKAPE pathogen genomes are described. Where possible, benchmark strains that contain the most common NRPSs for a species are used. BGCs that are present in less common strains are also described. Where known, the product is identified and any associated biology is presented, including the role of the product in microbial physiology or pathogenesis.
3. Enterococcus faecium
The gram-positive streptococci include the genus Enterococcus, which includes both E. faecalis and E. faecium. Both strains are very common in the human gastrointestinal tract as commensal bacteria and have been observed to cause human infections.103 The two species differ in their metabolic profile and, more significantly, in their antibiotic resistance profiles. E. faecium is much more commonly resistant to both ampicillin and vancomycin.104 Vancomycin-resistant enterococci (VREs) are the second most common cause of antibiotic resistant nosocomial infection behind only methicillin resistant S. aureus (MRSA).105
NRPS clusters are rare in the sequenced strains of E. faecium and there are no characterized NRPS clusters or products. Indeed, the first strain of E. faecium that was sequenced, designated DO or TX0016, does not contain any NRPS sequences, nor does the first complete sequence of a vancomycin resistant strain, Aus0004.106 Further, searching a panel107 of three faecal and four clinical samples fails to identify genes encoding NRPS proteins. Interestingly, E. faecium strains lack acyl-CoA synthetase genes as a whole and the only members of the adenlylate forming ANL superfamily22 that they contain appears to be a D-alanyl-carrier protein ligase needed for cell wall biosynthesis. No NRPS BGCs have been identified in the AntiSMASH database.
The Basic Local Alignment Search Tool (BLAST),108 available at the NCBI, was used to identify multiple NRPS protein sequences from different E. faecium strains. Four strains contained the larger cluster described below, while one strain contained an additional NRPS cluster. Two of the identified strains were from unpublished clinical isolates while two, including the strain KAC16100 described in Table 1, were isolated from fermented soybean samples.109
Table 1. NRPS clusters within Enterococcus faecium.
| Locus ID (Accession) | Length (AA) | Predicted Function* |
|---|---|---|
| Strain KAC16100 (Genbank Accession NZ_LDNJ01000053) | ||
|
| ||
| WP_047937824 (NpsA) | 5296 | Five module NRPS |
| WP_002343040 | 131 | Aspartate decarboxylase |
| WP_053002117 | 303 | HAD Hydrolase |
| WP_047929904 (NpsB) | 808 | A-T-Te |
| WP_053002116 | 211 | PPTase |
| Strain KAC16100 (Genbank Accession NZ_LDNJ01000090) | ||
|
| ||
| ACH93_RS12695 (WP_002318470) | 135 | Hydrolase |
| ACH93_RS12690 | 213 | Hypothetical |
| ACH93_RS12685 (WP_047937945) | 222 | Hypothetical |
| ACH93_RS12680 (WP_047937944) | 239 | Dehydrogenase |
| ACH93_RS12675 (WP_047937943) | 987 | A-T-C |
| ACH93_RS12670 (WP_047937942) | 400 | Hypothetical |
| ACH93_RS12665 (WP_047937941) | 346 | Hypothetical |
3.1. Cluster 1
The largest NRPS protein identified in E. faecium is a five module protein that is 5296 residues in length. This gene was present in four strains: KAC16100, KAC16106, EnGen0026, and Hp_22-12. In the latter two strains, the gene is reported to be divided into three separate genes. Whether this represents sequencing errors or the introduction of true stop codons is unknown. However, the proteins encoded by the smaller genes are identical to the full length gene of the two KAC strains and the protein boundaries are both positioned within a condensation domain leading to the suspicion that the truncated genes are more likely a result of sequencing errors.
This BGC contains two NRPS proteins, designated NpsA and NpsB in strain KAC16100. The smaller NpsB contains a single module that contains an adenylation, PCP, and thioesterase domain. The larger NpsA appears to be the initiating protein, as the first module of NpsA contains only an adenylation and carrier protein. The remaining four complete modules harbor condensation, adenylation, and carrier domains. The fourth module contains an additional epimerization domain. After the fifth module of NpsA is a condensation domain that may hand the peptide off to the PCP of the shorter protein. (We note with caution here that the generic “Nps” nomenclature has been used for proteins in multiple species to designate uncharacterized NRPS proteins.) Also present in the operon are two additional biosynthetic enzymes showing homology to aspartate decarboxylase and HAD family hydrolases. Finally, the cluster also contains a PPTase to convert the apo to holo PCP domains.
3.2. Cluster 2
The second cluster from E. faecium appears only in a single strain of E. faecium. The operon contains seven genes, including a gene encoding a single NRPS module. The cluster also encodes four hypothetical proteins of unknown function as well as a hydrolase and an NAD(P)H-dependent dehydrogenase. A search of the NCBI protein sequence database with BLAST finds no homologs above 40% sequence identity for any of the final five proteins of this cluster demonstrating that this is a unique, orphan BGC. The 135 residue protein matches other sequences, as does the initial 110 residues of protein ACH93_RS12690, which shows 100% identity with other E. faceium strains. In strain EnGen0038, accession AHXW01000024, the neighboring genes are transposon and integrase genes suggesting the presence of mobile genetic elements and raising the possibility that the first two genes in this cluster are not involved in the BGC.
4. Staphylococcus aureus
S. aureus is a gram-positive bacterium and a common source of nosocomial infections, occuring in a wide variety of care facilities. Roughly 60% of humans carry S. aureus intermittently and another 20% carry S. aureus strains at all times.89 S. aureus is the primary cause of epidermal and soft-tissue infections and can also cause a number of invasive infections, including abscesses, pneumoniae, and sepsis.110 The prevalence of antibiotic resistant S. aureus, particularly methicillin resistant (MRSA) strains that can represent as much as 50% of S. aureus isolates in some regions,89 has risen over the past decade. This startling trend only raises the importance of efforts to develop new treatment options.111
4.1. Aureusimine
4.1.1. Structure of aureusimine
The S. aureus genome encodes a single NRPS BGC that has been assigned to the natural product aureusimine, a cyclic dipeptide formed through the condensation of either tyrosine or phenylalanine with valine to form pyrazinones (1) and (2). The cluster is conserved in S. aureus and also S. epidermidis, S. lugdunensis, and S. capitus.112 The molecule is thought to be produced through the reductive release of a dipeptide aldehyde, which spontaneously cyclizes. The aureusimine biosynthetic gene cluster contains two genes, named ausA and ausB.113 A search of other S. aureus strains identifies NRPS genes of shorter lengths that are ∼99% identical to AusA, suggesting either sequencing errors or bonafide mutations that have separated the NRPS gene into two multidomain proteins. Aureusimine was identified nearly simultaneously by the Fischbach112 and Magarvey113 groups. It was also noted that the phenylalanine isoform had been identified previously and called phevalin.114
4.1.2. Aureusimine biosynthesis
Aureusimine has been shown to be produced through the activity of a single, dimodular NRPS protein called AusA (Fig. 2).38, 115 The aus operon also contains ausB, encoding a PPTase to convert the AusA protein to the holo form. The first module of AusA encodes an adenylation and PCP domain, while the second module contains adenylation, PCP, and reductase domains. Although the adenylation domains have not been biochemically analyzed in isolation, the sequence of the adenylation domain pockets suggest that the first module activates valine while the second module activates the aromatic substrates. The full length protein has been probed for adenylation activity both in the presence and absence of phenylalanine- and valine-specific inhibitors, supporting the promiscuous activity of the adenylation domain of module 1.115
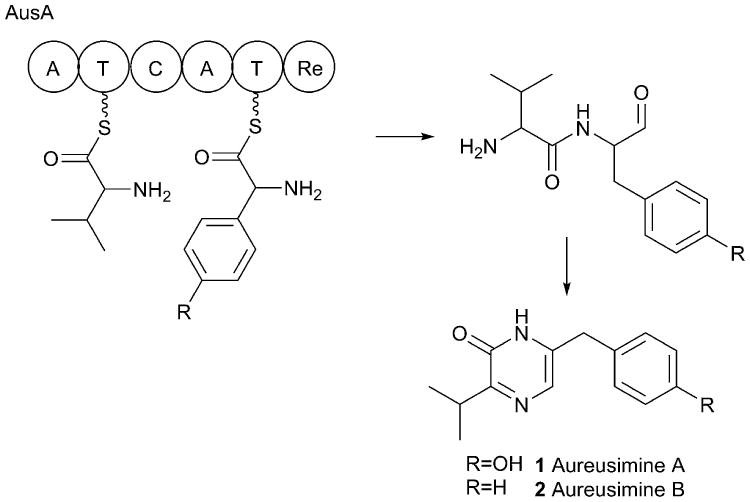
Fig. 2.
Aureusimine biosynthesis. The production of aureusimine has been demonstrated in biochemical experiments with holo-AusA. Cyclization of the dipeptide aldehyde is believed to occur spontaneously. Multidomain NRPS proteins are represented here and in subsequent figures as circles and labeled as described in Table I.
The NRPS protein AusA is relatively straight-forward with two modules that contribute to the production of the dipeptides (Fig. 2), along with a minor production of pyrazinone derived from a Val-Leu dipeptide. The C-terminal reductase domain catalyzes the release of the dipeptide as an aldehyde, which is believed to cyclize spontaneously in the energetically favorable reaction that benefits from the aromatic stabilization of the pyrazinone product.38, 115 The reductase domain has been structurally characterized38 demonstrating that the domain adopts a standard Rossmann fold shared by other short chain dehydrogenase enzymes.
4.1.3. Biological role of aureusimine
As some cyclized dipeptides display antibacterial properties, aureusimine was assayed for growth inhibition against E. coli, B. subtilis, and a panel of actinomycetes from the human skin microbiome. No effect on the tested bacteria was identified.112 Although originally identified as being related to the virulence of S. aureus,113 subsequent analysis demonstrated that the mutant strain used in the initial studies contained a second mutation in the saeS gene, a regulator of known virulence factors.116, 117 Performing the genetic array analysis with an ausA mutant strain showed the regulation of a variety of genes encoding proteins involved with electron transfer properties and redox signaling.
Further investigation of the role of the aureusimines in the biology of S. aureus and human keratinocytes demonstrated that the aureusimines were more highly expressed under biofilm conditions compared to planktonic growth, and that the phenylalanine isoform (2) was more prevalent than the tyrosine isoform.118 This same study concluded that the addition of exogenous aureusimine had no effects on S. aureus growth and no dramatic effects on the extracellular metabolome. Further, the presence of aureusimine had modest effects on the gene expression profile of 24 genes in human keratinocytes.
S. aureus is an intracellular pathogen that is able to withstand the environment of the phagolysosome by disruption of lysosomal membranes through the use of the so-called phenol soluble modulins, which are necessary but not sufficient for cell lysis and escape from the phagosome.119 A search for additional genes that play a role in intracellular survival and phagosomal escape identified ausA and ausB.120 While the authors concluded that the aureusimines are not “major virulence factors”, they raised the possibility that they may contribute to S. aureus virulence in strain- and tissue-specific manners.120
Finally, a recent report suggests the dipeptide aldehyde, and not the cyclic pyrazinone of aureusimine, could be the bioactive compound.121 Examination of NRPS BGCs from microbiome DNA identified clusters that are present in >90% of stool samples from the Human Microbiome Project and almost exclusively within gut bacteria. These NRPSs produce multiple dipeptides, including Val-Phe- and Phe-Phe-aldehyde. These linear aldehydes of aureusimine A and B were shown to be potent inhibitors of cathepsins, a family of lysosomal cysteine proteases that play a role in antigen presenting within macrophages and dendritic cells. As the cyclization is an aerobic step in the biosynthesis of the pyrazinones, the aldehydes may have a sufficiently long half-life in the anaerobic environment of the gut in which they are produced to play a biological role.121
5. Klebsiella pneumoniae
Klebsiella pneumoniae is a common human pathogen that has been identified as the cause of many health-care associated infections of the lung, urinary tract, abdominal cavity, and eye.122, 123 Several reports note the presence and rapid dissemination of carbapenem-resistant K. pneumoniae in long-term care facilities.124, 125 Perhaps even more concerning, hypervirulent strains of K. pneumoniae (designated hvKP)126, 127 that were originally identified in Asia appear to be spreading across the globe. The hypervirulent strains exhibit the ability to infect a younger, healthier population as well as the ability to spread metastatically from the initial sites of infection. To date, most of the strains appear to be sensitive to antibiotics; however, the potential for the acquisition of a drug-resistance phenotype by these hvKP strains is high,128, 129 raising the possibility of a pathogen that is both highly virulent and highly antimicrobial resistant. K. pneumoniae contains several NRPS BGCs that are common to many gram-negative enteric bacteria.
5.1. Enterobactin
5.1.1. Structure of enterobactin
Enterobactin (8) was discovered almost 50 years ago, where it was identified from both S. typhimurium130 and E. coli131. Enterobactin is a trilactone composed of three molecules serine that are cyclized through ester linkages between their carboxylates and the side chain hydroxyls. Each of the three amines of this trilactone is joined via an amide linkage to a molecule of 2,3-dihydroxybenzoic acid (7, DHB).132-134 The six catechol oxygens form the ligands for the Fe3+ ion, providing an extremely high affinity binding motif with a proton-independent formation constant reported as 1052 for the iron-siderophore complex.135
5.1.2. Biosynthesis of enterobactin
Enterobactin is produced through the activities of five proteins encoded in the ent operon.136-139 EntA, EntB, and EntC are responsible for the production of the DHB molecule, while the NRPS proteins are housed on EntE, a freestanding adenylation domain, and EntF, which contains the module for the incorporation of serine. As one of the oldest characterized products of an NRPS, as well as a relatively simple NRPS product from E. coli, enterobactin biosynthesis served as a model system (within the lab of Professor Christopher Walsh) and was utilized for much of the foundational biochemical studies to understand the function of NRPS enzymes. Further, the five biosynthetic proteins from E. coli have all been crystallized and their structures determined, including the three enzymes required for DHB synthesis140-142 and the NRPS proteins in varying functional complexes58, 59, 79, 80, providing further insight into the structural mechanisms. Although limited structural and biochemical data exist on the enterobactin enzymes from K. pneumoniae, the enzymes are 70 to 90% identical to the E. coli proteins. The biosynthesis will therefore focus on data obtained with E. coli enzymes.
5.1.2.1. The chorismate utilizing enzymes
Many siderophores utilize DHB (7) or salicylate (5) capping groups that play critical roles in iron interaction.143 A series of chorismate-utilizing enzymes converts chorismate to the aryl moieties incorporated into the products.133, 144 This section will focus on these pathways (Fig. 3) and set the stage for their use in not only enterobactin, but also yersiniabactin, acinetobactin, fimsbactin, and pyochelin biosynthesis in the sections to follow.
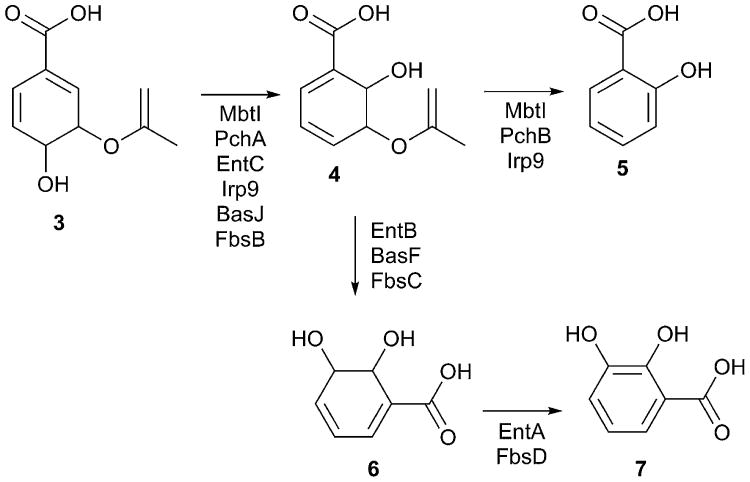
Fig. 3.
Chorismate utilizing enzymes in NRPS siderophore pathways. Members of the MST family of chorismate-utilizing enzymes convert chorismate to isochorismate. MbtI and Irp9 are able to perform a second reaction to convert isochorismate 4 to salicylate 5, while PchB is a monofunctional salicylate synthase. EntB, BasF, and FbsC are isochorismatase enzymes. EntA and FbsD are dehydrogenases that produce 2,3-dihydroxybenzoate 7.
The DHB moiety of enterobactin is produced from chorismate (3) by the sequential activites of EntC, EntB, and EntA. EntC is a Mg2+-dependent isochorismate synthase that converts chorismate to isochorismate (4).141, 145 The isochorismate is then hydrolyzed by the isochorismatase domain of the bifunctional EntB to produce 2,3-dihydro-2,3-dihydroxybenzoate (6) and pyruvate,146 which is then converted to DHB by the activity of the short-chain dehydrogenase/reductase protein EntA.136, 147
EntC is a member of the MST family of enzymes that initiate these chorismate conversion steps involved in the production of menaquinone, siderophores, or tryptophan.148, 149 Additional members of this family include PchA of pyochelin biosynthesis, BasJ of acinetobactin biosynthesis, and Irp9 of yersiniabactin biosynthesis. Interestingly while the initial step in the reaction—the isomerization to isochorismate—is shared, different members of the family form different products. Thus, the common isochorismate intermediate can be converted to salicylate (5) through a second activity within the MST protein, as in the case of Irp9 of yersiniabactin biosynthesis, or a separate isochorismate lyase protein as with PchB. Alternatively, some BGCs use a separate enzyme to retain the hydroxyl at the meta position to produce (6). Such enzymes include EntB and BasF, which both contain the isochorismatase domain as part of a fusion with an NRPS acyl-carrier protein domain.140, 146 In enterobactin biosynthesis, the final step in the production of DHB (7) is catalyzed by EntA, which catalyzes the aromatization of the product of the EntB reaction.136, 142, 150
5.1.2.2. The enterobactin NRPSs
The enterobactin NRPS system encompasses three proteins, EntE, EntB, and EntF, that combine three molecules of DHB and three molecules of serine into the final enterobactin trilactone (Fig. 4).30, 146, 151 Additionally included in the full biosynthetic cluster is EntD, the PPTase that converts the apo to holo carrier protein domains.146 The freestanding adenylation domain EntE first loads a molecule of DHB onto the aryl carrier protein (ArCP) domain of the bifunctional protein EntB. As described above, EntB also contains the isochorismatase domain that is responsible for DHB synthesis. EntF is a four domain NRPS protein with a condensation-adenylation-PCP-thioesterase architecture. The EntF adenylation domain loads a molecule of serine onto the downstream PCP. The loaded PCP then migrates to the condensation domain where it awaits delivery of the DHB from a loaded EntB ArCP domain. The condensation domain catalyzes amide bond formation via nucleophilic attack of the loaded serine amino group on the carbonyl of the DHB-ArCP thioester of EntB. The EntF PCP then delivers the resulting N-(2,3-dihydroxybenzyl)serine to the EntF thioesterase domain.
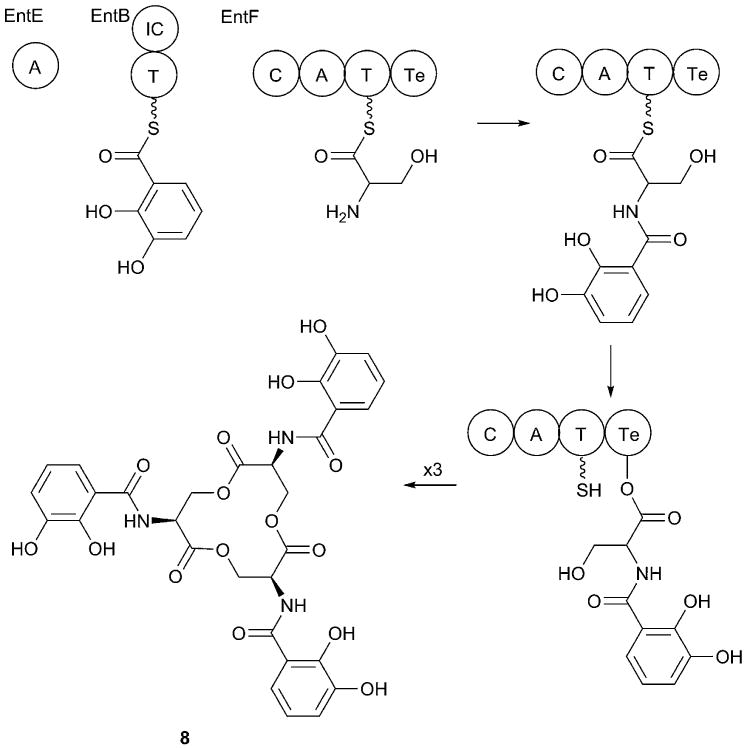
Fig. 4.
Enterobactin biosynthesis. Enterobactin biosynthesis has been demonstrated biochemically for the homologous enzymes from E. coli. EntE and EntB form a single module for incorporation of DHB, while EntF forms a single module for serine. EntB contains the thiolation domain as well as the isochorismatase domain (IC) used in DHB biosynthesis. The N-(2,3-dihydroxybenoyl)serine is bound to a catalytic serine within the thioesterase domain of EntF for two subsequent cycles of synthesis that iteratively complete the trilactone.
The catalytic serine (Ser1138) of the EntF thioesterase domain attacks the thioester linkage to form a covalent enzyme-peptide intermediate at the thioesterase domain active site. A second cycle then provides a second N-(2,3-dihydroxybenzyl)serine to the thioesterase domain. In the second cycle, the serine side chain hydroxyl from the first synthetic cycle, not the Ser1138 side chain, is used as the nucleophile in the transacylation reaction. A final iteration results in the covalently bound linear trimer of N-(2,3-dihydroxybenzyl)serine moieties, joined through ester linkages between the serine side chain and carboxylate, that is loaded on the catalytic serine of the thioesterase domain. Closure of the ring within the thioesterase domain releases enterobactin (8) and frees the catalytic serine to begin a new cycle of enterobactin biosynthesis.30
5.1.3. Biological role of enterobactin
The association of enterobactin production with K. pneumoniae growth in low iron conditions has been known for almost 40 years.152, 153 The redundancy of siderophore production by pathogens reflects not only the significance of iron acquisition but also may be related to the ability of specific siderophores to be involved in bacterial growth in particular microenvironments. Because many studies of K. pneumoniae siderophores consider this functional interplay between the different siderophore systems, the biological role of enterobactin is described along with yersiniabactin in more detail below.
5.2. Yersiniabactin
5.2.1. Structure of yersiniabactin
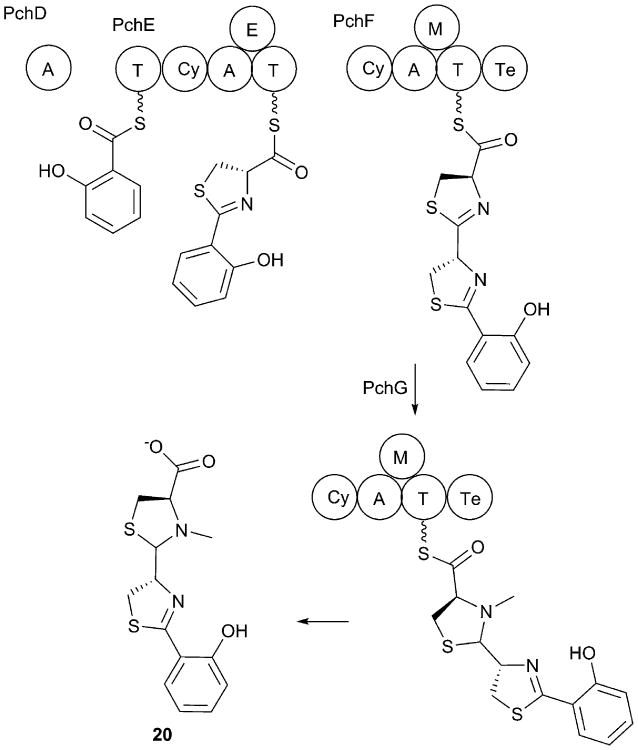
Yersiniabactin (9) was purified in 1993154 and structurally defined several years later155, 156 as a siderophore in Yersinia species and later in additional bacterial species. Like enterobactin, it contains an aryl cap composed of a salicylate moiety connected to a peptide chain (Fig. 5). The final molecule is assembled from building blocks of salicylate, three cysteine residues, and a malonic acid group that is contributed by an integrated PKS domain.83 The three cysteine residues are all cyclized at varying oxidation states and present as either thiazolidine or thiazoline rings. Additionally, methyl groups derived from three molecules of S-adenosylmethionine (SAM) are utilized by integrated methyltransferase domains within the PKS and terminal NRPS module to produce the final product.
Fig. 5.
Yersiniabactin biosynthesis. Yersiniabactin biosynthesis has been reconstituted for the homologous enzymes from Y. pestis. HMWP2 contains condensation/cyclization domains (Cy) that catalyze peptide bond formation and thiazoline formation. HMWP1 contains a PKS module containing ketosynthase (KS), acyltransferase (AT), methyltransferase (M), ketoreductase (KR) and acyl carrier protein (ACP) domains in addition to other domains described earlier.
5.2.2. Biosynthesis of yersiniabactin
As with enterobactin, to our knowledge, there are no biochemical studies on the synthesis of yersiniabactin using the proteins from K. pneumoniae. Most of the studies, again elegantly performed by the Walsh lab, were done with the homologous proteins from Yersinia pestis. As the sequence identities between K. pneumoniae and Y. pestis are greater than 95%, we again describe the biosynthetic pathway using the Y. pestis model system.
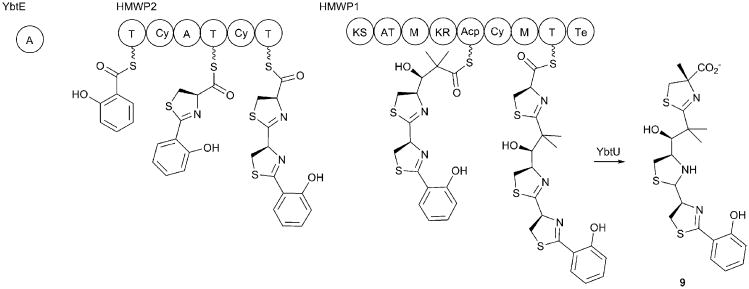
The yersiniabactin BGC encodes five proteins (Fig. 5). YbtS is a salicylate synthase of the MST superfamily discussed above that carries out both the conversion of chorismate to isochorismate and the salicylate synthase reaction.157 The NRPS proteins include YbtE, a freestanding adenylation domain that activates the initial salicylate molecule, and two large multidomain NRPSs named for high molecular weight protein 1 and 2 (HMWP1 and HMWP2), as well as an auxiliary reductase protein called YbtU. The Ybt protein and gene names are used in the Y. pestis literature; in Y. enterocolitica, the two large NRPS proteins are named Irp1 and Irp2, while YbtE, YbtS, and YbtU are called Irp5, Irp9, and Irp3 respectively.
HMWP2 contains two NRPS modules and six domains organized as an ArCP-Cyclization/Condensation-Adenylation-PCP-Cyclization/Condensation-PCP architecture. HMWP1 contains a PKS module containing five domains necessary to incorporate and dimethylate a malonyl moiety, followed by a terminal NRPS module containing cyclization/condensation-methyltransferase-PCP-thioesterase domains.83 The NRPS pathway for yersiniabactin synthesis begins with the activation of salicyate by YbtE and the loading of the aryl substrate onto the N-terminal ArCP domain of HMWP2. The single adenylation domain of HMWP2 loads a cysteine residue onto the two downstream PCP domains within this protein in cis, as well as the PCP domain of the HMWP1 protein in trans.157 The bifunctional cyclization/condensation domains sequentially catalyze the amide bond formation and cyclization of the cysteine residues to form the thiazolinyl rings. The freestanding reductase YbtU then catalyzes the conversion of the second thiazoline ring, but not the others, to a thiazolidine moiety, while the immature siderophore is still bound to the final PCP domain.83, 158 Hydrolysis of the the mature peptide results in the release of yersiniabactin (9).
5.2.3 Biological role of yersiniabactin and other K. pneumoniae siderophores
The yersiniabactin and enterobactin pathways are present on the chromosome in most strains of K. pneumoniae. In contrast, hypervirulent strains of K. pneumoniae (hvKP) contain a large 220 kilobase plasmid that contains genes related to the virulent phenotype.159 Included on the plasmid are the iuc genes responsible for the biosynthesis and uptake of the NRPS-independent siderophore aerobactin, as well as the iroB gene that produces glucosylated variants of enterobactin called salmochelin.160 The reason for this redundancy of siderophore systems is unclear; however, this may reflect the differential utility and regulation of preferred siderophores under certain conditions. Specifically, the pH dependence of iron affinity and the desire to evade eukaryotic lipocalin 2,161, 162 a siderophore binding protein that is present in serum, may dictate which siderophores become most effective in certain pathogenic environments.163
Examination of the roles of different K. pneumoniae siderophores can be done with genetic mutations that are unable to produce one or more of the compounds. In a murine intranasal infection model with a classical K. pneumoniae strain, a yersiniabactin mutant was less virulent than the wild-type or a enterobactin mutant. However, a double mutant defective for both siderophores, the only two present in this strain, was completely avirulent, demonstrating the importance of iron acquisition.164
A more recent study examined the relationship of the four siderophores, enterobactin, yersiniabactin, aerobactin, and salmochelin, that are present in an hvKP strain. The aerobactin BGC is encoded on the hvKP virulence plasmid and is highly over-expressed165 in the hypervirulent strains. In murine pneumonia and subcutaneous infection models, mutants that are deficient for the production of aerobactin were significantly less virulent than wild-type cells or a mutant strain that was defective for the production of the three other siderophores.166
5.3. Colibactin
5.3.1. Colibactin structure
Some pathogenic E. coli strains have been observed to induce megalocytosis in eukaryotic host cells upon infection. This process is characterized by enlargement of the nucleus and the cell without associated cell division. Transposon mutagenesis allowed the mapping of this cluster to a large, 54-kilobase region encoding a hybrid PKS/NRPS BGC that is common to strains that share this cytotoxic phenotype.167 The colibactin pathway has additionally been identified in both classical and hypervirulent K. pneumoniae.168-170
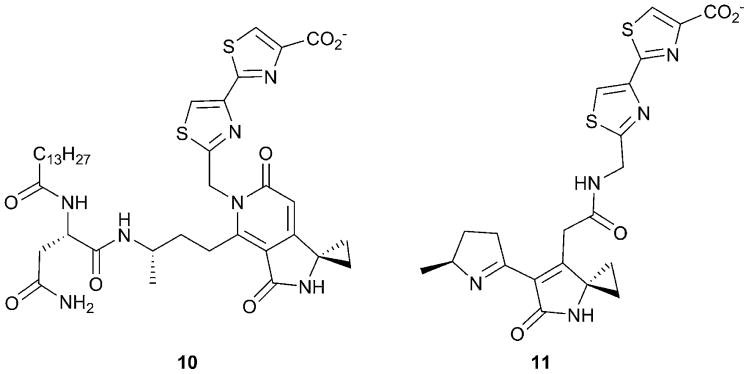
The precise identity of the bioactive product of this complex biosynthetic pathway has not yet been elucidated. However, shared features have been identified that may be responsible for the mechanism of action.171 Because of the highly reactive intermediates and the potential for multiple cyclization steps, different intermediates have been identified in studies that used mutant strains to isolate biosynthetic intermediates. Consensus exists that the PKS/NRPS product is produced through a “prodrug” mechanism that requires proteolysis to form the final active product.172, 173 The peptidase ClbP is responsible for cleavage of an acylasparagine moiety from a precursor during product maturation. The final colibactin molecule is expected to contain an electrophilic warhead that can cross-link DNA as well as heterocyclic thiazoline rings that could facilitate interaction with DNA (Fig. 6).174-176 On-going biochemical analysis of the biosynthetic enzymes is providing insight into the exact structure of the colibactin molecule. A recent proposal suggests that the timing of the hydrolysis of the acylasparagine leader may in fact lead to different precolibactin molecules, including pyridone forms such as (10) that do not react with DNA, as well as lactam (11) that is able to cross-link DNA.174, 177
Fig 6.
Precolibactin molecules. The bioactive product of the colibactin pathway has not yet been identified. Several precolibactin metabolites, including 10 containing the acyl-Asn peptide leader and 11 derived after cleavage of the leader, have been identified.
5.3.2. Colibactin biosynthesis
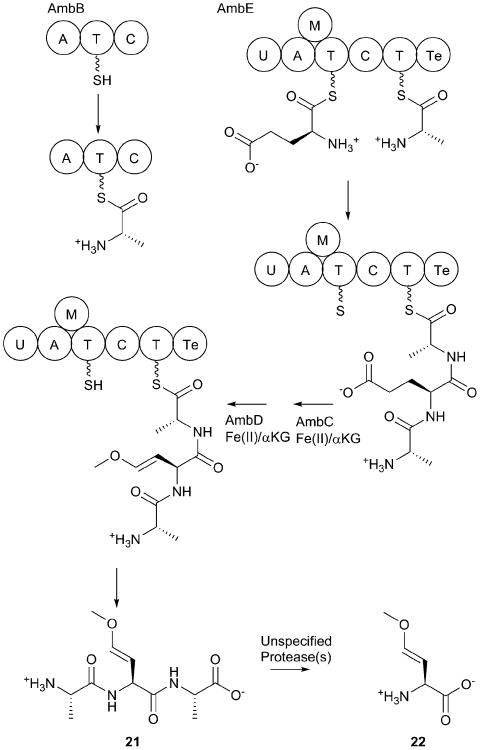
The clb BGC encodes 3 PKSs, 3 NRPSs, and two hybrid PKS/NRPS proteins. Additionally, seven other enzymes are required for the genotoxicity.167 Also encoded within the cluster is ClbM, a transporter that is responsible for export of precolibactin across the cytoplasmic membrane.178
While the complete biosynthetic steps are unknown, some features have been biochemically explored. The acyl-Asn “protecting group” that is removed as part of the pro-drug strategy is installed by the activity of ClbN, a four domain NRPS that activates and epimerizes the asparagine residue.172 The N-terminal condensation domain adds the acyl chain, which is most commonly a myristate fatty acid, however monounsaturated myristate or palmitate moieties have also been observed.179 Elegant bioinformatic analysis allowed the prediction of the next module in the pathway to involve the NRPS portion of the hybrid ClbB protein, which incorporates an alanine residue, affording the production of the acyl-Asn-Ala peptide.172 In the periplasmic space, the capping acyl-Asn moiety is cleaved by the ClbP protease.180 The PKS module of ClbB as well as the PKS proteins ClbC and ClbI incorporate a malonyl moiety into the growing natural product.
ClbH is a four domain NRPS protein with an architecture of Adenylation-Condensation-Adenylation-PCP domains. The first adenylation domain activates a serine residue that is transferred in trans to the freestanding carrier protein ClbE.181 The loaded serine residue is then sequentially acted on by two dehydrogenases, ClbD and ClbF, to produce aminomalonyl loaded carrier protein. The genotoxic phenotype of a colibactin-expressing E. coli strain was blocked in mutants for either clbE alone or the entire clbDEF operon, demonstrating that this aminomalonyl group is essential for production of the active colibactin molecule.181 The second adenylation domain of ClbH activates a methionine residue that is believed to be the source of the aminocyclopropylcarboxylate moiety that forms the smaller ring of the warhead. Notably, genetic disruption of clbD, clbE, clbF, clbG, clbJ, clbK, or clbO had no effect on the formation of the warhead, placing the function of these gene products further downstream in colibactin biosynthesis.176
The two module NRPS protein ClbJ then incorporates a glycine and a cysteine, the latter of which is cyclized into a thiazoline moiety. Finally, the NRPS module of the two module PKS/NRPS protein ClbK incorporates a second cysteine residue that is also cyclized.
5.3.3. Colibactin biology
Colibactin was originally identified through its ability to induce megalocytosis on several mammalian cells upon infection.167 Colibactin's ability to alkylate DNA in vitro174 and cause DNA double strand breaks in vivo167 likely plays a role in this effect. The ability of a 0.2 μ membrane between the bacteria and eukaryotic cells to block cytotoxicity suggests a requirement for cell-cell contact between the pathogen and the mammalian cell.167 Despite the tremendous recent progress, the exact nature of the biosynthesis and mode of action of colibactin remains to be fully determined.
6. Acinetobacter baumannii
A. baumannii is a gram-negative bacillus that belongs to the family Moraxellaceae. Infections caused by A. baumannii include both community- and hospital-acquired pneumonia, bacteremia, urinary tract infections, and skin, soft tissue, and burn infections182, 183 and has been responsible for outbreaks in several military hospitals during the wars in Afghanistan and Iraq.184, 185 Increasing the seriousness of nosocomial infections and illustrating the challenge of stopping an outbreak, A. baumannii can live on abiotic surfaces for months and has the ability to form biofilms on both glass and plastic.186 Interestingly, despite a genus name that translates as “non-motile”, A. baumannii exhibits a soft agar motility phenotype that is dependent on Type IV pili and other uncharacterized factors.187-189
Also challenging the treatment of A. baumannii infections, many strains are highly drug resistant to common antibiotics, because of the presence of genomic resistance islands that are acquired through horizontal gene transfer from unrelated species.182, 190 The prevalence of drug resistance has led to the informatic191 and experimental192 search for previously uncharacterized essential genes within A. baumannii that may be targeted through novel antibiotic development campaigns.
While nearly all A. baumannii strains contain an NRPS cluster that encodes the production of an aryl-capped peptide siderophore, acinetobactin, a second peptide siderophore, fimsbactin, has been identified in only a few strains. An additional NRPS cluster that is responsible for an uncharacterized peptide product appears to play a role in motility and biofilm formation.
6.1. Acinetobactin
6.1.1 Acinetobactin structure
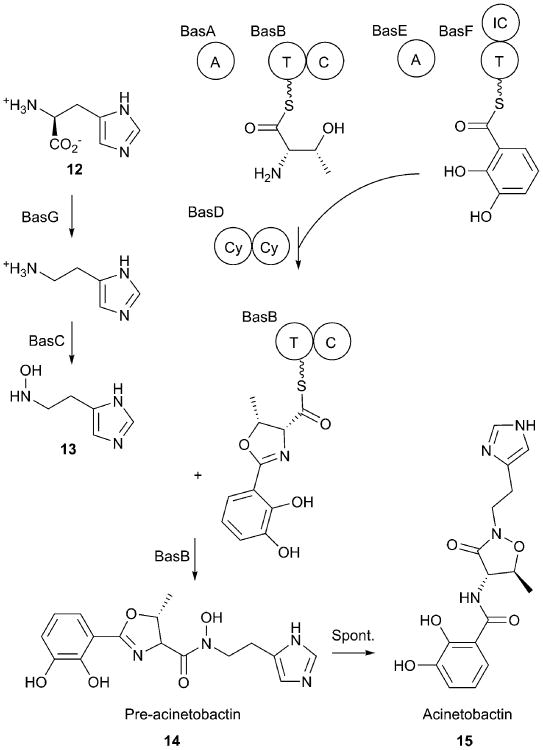
A. baumannii produces a peptide siderophore called acinetobactin (15) containing DHB (7) and N-hydroxyhistamine (13) moieties joined by a heterocyclic moiety (Fig. 7). Originally identified as a oxazoline, acinetobactin is now recognized as being produced in a pre-acinetobactin form (14) that is released from the NRPS, which subsequently isomerizes to mature acinetobactin (15), harboring an isoxazolidinone ring.193 The interconversion of pre-acinetobactin and acinetobactin, both of which can form complexes with ferric iron, is pH-dependent, with the pre-acinetobactin species being more stable at pH < 6.194 Both forms use the same cellular receptor to allow entry of the ferric complexes into the cell, illustrating that a single siderophore may be optimized for different pathogenic environments, allowing A. baumannii to thrive in different sites of infection.
Fig. 7.
Acinetobactin biosynthesis. N-hydroxyhistamine is produced through the activites of BasG and BasC. The intermolecular loading of DHB on the carrier domain of BasF has been demonstrated biochemically while the subsequent reactions are assumed based on the homologous reactions of pseudomonine biosynthesis. The tandem cyclization domains of BasD perform the peptide bond formation and cyclization to form the oxazoline ring. The condensation domain of BasB then transfers the DHB-oxazoline to 13 to result in the formation of pre-acinetobactin 14.
6.1.2 Acinetobactin biosynthesis
The BGC for acinetobactin biosynthesis was identified in 2004 containing a multiple transcriptional operons that express proteins involved in the regulation, uptake, and synthesis of acinetobactin.195, 196 As with enterobactin described above, three proteins are responsible for the conversion of chorismate to DHB: BasJ is an isochorismate synthase, BasF is an isochorismatase that releases pyruvate to form 2,3-didehydro-2,3-dihydrobenzoate, and EntA is the reductase that catalyzes the reductive aromatization to form DHB (Fig. 3). Interestingly, the entA gene is not part of the acinetobactin BGC but is located on a different region of the chromosome.150
The other unusual substrate for acinetobactin is N-hydroxyhistamine (13), which is derived from histidine (12) through the activities of a decarboxylase (BasG) and a hydroxylase (BasC).196 Along with threonine, these building blocks serve as substrates for the acinetobactin NRPS enzymes–BasA, BasB, BasD, BasE, and BasF (Fig. 7). The biochemical reconstitution of acinetobactin biosynthesis has not been reported; however, the biosynthesis of pseudomonine, a Pseudomonas entomophila siderophore that differs from acinetobactin by only the use of a salicylate rather than DHB aryl cap, has been described by the Walsh group.197 BasE is a freestanding adenylation domain that activates DHB and loads it onto the PCP of the isochorismatase-PCP didomain protein BasF. In this system, BasE and BasF are homologs of EntE and EntB of the enterobactin system.198 BasA is a freestanding adenylation domain that loads a molecule of threonine onto the PCP of BasB, a two domain protein containing a PCP-condensation domain architecture.193
The didomain NRPS protein BasD, in analogy to the PmsD protein of pseudomonine biosynthesis, which also contains tandem cyclization domains,197 transfers the DHB moiety from BasF to the loaded threonine of BasB and cyclizes the threonine to form a oxazolinyl moiety on BasB. This loaded 2,3-dihydroxyphenyloxazolinyl group on BasB is then released through the nucleophilic attack with N-hydroxyhistamine, catalyzed by the condensation domain of BasB.197 The combined activity of the five NRPS proteins results in the formation of pre-acinetobactin, which can spontaneously form acinetobactin at neutral or basic pH.
6.1.3 Acinetobactin biology
As discussed above, siderophore biosynthesis represents an increasingly appreciated virulence target. The low iron environment of the host creates a competitive environment and the ability to produce the siderophores that facilitate iron acquisition is an appealing target.199, 200 Acinetobactin production has been examined in a series of ex vivo and in vivo assays, suggesting that this phenotype is required in multiple infection models.201 Specifically, while a basD mutant strain was able to physically interact with human epithelial cells in culture, the mutant showed a reduced ability to persist or induce apoptosis in the eukaryotic cells. Finally, the mutant strain showed reduced virulence in the infection of the larvae of the greater wax moth Galleria mellonella and in a mouse sepsis model.201 A more recent study suggests a critical role for acinetobactin production in the establishment of a traumatic wound infections.202
6.2. Fimsbactin
6.2.1 Fimsbactin structure
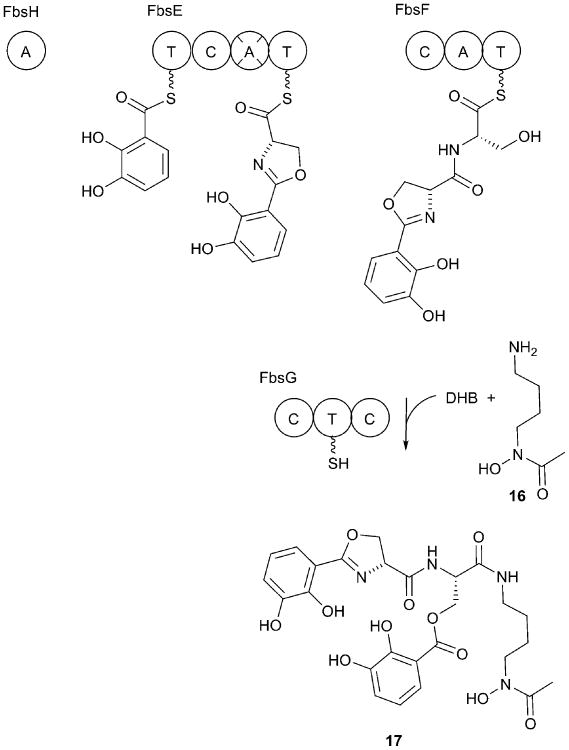
While acinetobactin appears to be the primary siderophore of nearly all A. baumannii strains, a second BGC that encodes a second catecholate siderophore called fimsbactin (17) appears in strain ATCC 17978 (Fig. 8).203 Additional A. baumannii strains, as well as the nonpathogenic species A. baylyi, also contain the identical cluster. Six fimsbactin isoforms were identified that share a DHB moiety as well as an oxazoline ring derived from a serine residue. Attached to this core are two additional residues, a serine residue that has been modified at the side chain through the addition of a second DHB moiety and a hydroxamate containing N-acetyl-N-hydroxyputrescine (16) moiety. Variants contained threonine substitutions for either serine moiety or a 1,3-diaminopropane in place of the putrescine (1,4-diaminobutane).
Fig. 8.
Fimsbactin biosynthesis. The formation of the initial DHB-oxazoline-serine tripeptide on FbsF is catalyzed using standard NRPS biochemistry. The final steps catalyzed by FbsG are not yet characterized. The truncated adenylation domain of FbsE is depicted with the cross-hairs through the domain. The biosynthesis of fimsbactin has not been demonstrated biochemically but the early steps can be confidently assigned on the basis of homologous enzymes.
6.2.2 Fimsbactin biosynthesis
The fimsbactin BGC was characterized for the A. baylyi cluster by the Bode group, 203 who provided annotations and a prediction of the biosynthetic pathway. As the proteins from A. baylyi and A. baumannii all share 60-90% sequence identity and are organized in the same genetic fashion, there is high confidence in the parallel function of the clusters from both members of the acinetobacter genus.203 The DHB (7) building block is built once again through the activities of FbsB, FbsC, and FbsD, which serve as the homologs of EntC, EntB, and EntA, respectively (Fig. 3). The DHB moiety is activated by a freestanding adenylation domain, FbsH, which loads the DHB onto a N-terminal ArCP of FbsE (Fig. 8).
The hydroxamate building block is formed through the activities of FbsJ, an ornithine decarboxylase, FbsI, a hydroxylase, and FbsK, an acetyltransferase. Together, these three proteins likely convert ornithine into putrescine, which is then is hydroxylated and acetylated to the final NRPS substrate. On the basis of homologous proteins from pyochelin or aerobactin biosynthesis, the hydroxylation step likely occurs before the acetylation.204
With the building blocks in place, the function of three multidomain NRPSs are required along with the freestanding adenylation domain FbsH to build the fimsbactin molecule. The three NRPS modules are contained upon FbsE, FbsF, and FbsG. The domain organization suggests that the fimsbactin NRPS operate with a series of both intra- and intermolecular interactions and reactions.
FbsE contains an N-terminal ArCP domain, followed by a condensation domain, a truncated adenylation domain, and a second PCP. FbsF contains a condensation-adenylation-PCP domain architecture. And finally, the three domain FbsG harbors condensation, PCP, and condensation domains.
Several steps can be confidently predicted on the basis of the shared architecture with other NRPS systems, including others discussed herein. The ArCP of FbsE is proposed to be loaded with DHB by the activity of FbsH. The adenylation domain of FbsF could load both the FbsH PCP intramolecularly as well as the PCP of FbsE intermolecularly with serine. The FbsE condensation/cyclization domain then transfers the DHB to the first serine residue on the FbsE PCP and also cyclizes the serine to form the oxazoline ring. The module in FbsF would the be expected to extend the peptide by the addition of the serine residue, which remains uncyclized.
The subsequent steps have not been biochemically shown and are less clear, given the non-linear nature of the reactions. FbsG contains two condensation domains surrounding a PCP. Two additional reactions are required for completion of the fimsbactin molecule, both of which could be carried out by a condensation domain. In one reaction, a DHB moiety is installed on the serine side chain, while the second reaction involves the nucleophilic release of the peptide with the N-acetyl-N-hydroxyputrescine. The source of the DHB in the initial reaction is unclear and could involve a DHB loaded onto the carrier protein of either FbsE or FbsG. The latter possibility would require that the adenylation domain FbsH is able to recognize and load two carrier domains intermolecularly. The unraveling of the unusual NRPS biochemistry of the fimsbactin system will be an interesting addition to the literature on alternate NRPS systems. Similarly, the role of fimsbactin and the redundancy of these siderophore systems in A. baumannii remains to be explored.
6.3. An uncharacterized NRPS cluster involved in biofilm formation and motility
6.3.1. Biological identification of a novel NRPS cluster
Many of the uncharacterized NRPS clusters discussed here and in other studies were identified solely on the basis of bioinformatic cluster mining. However, one NRPS cluster from A. baumannii has been identified repeatedly in a number of genetic screens for important virulence related phenotypes such as motility, biofilm formation, and quorum signaling. These studies suggest an interesting new signaling molecule will be identified in further biochemical and genetic studies.
As noted above, bacteria of the acinetobacter genus were believed to be amotile. However, many strains do exhibit a phenotype known as twitching motility as demonstrated by an ability to form halos around colonies innoculated on soft agar. Twitching motility is mediated by the cyclical extension and retraction of Type IV pili, filaments of proteinacous projections from the poles of the cell. Adherence of the tip of the pilus to the substratum followed by retraction results in the force required for motility.205, 206 Type IV pili genes are required for A. baumannii motility.187, 189
A search for additional factors that are responsible for twitching motility in A. baumannii via transposon mutagenesis identified an NRPS BGC that is present in most strains.187 This cluster, containing eight genes, is located immediately downstream of a gene encoding an acyl homoserine lactone synthase and a luxR type regulator, which are involved in quorum signaling regulation. This A. baumannii NRPS cluster was subsequently identified as being among the most highly upregulated operons in cells grown in biofilms relative to cells from planktonic (liquid culture) growth.207 Finally, mutants within this cluster are defective in forming pellicles, a biofilm structure that forms at the liquid/air interface of cultures.208
6.3.2 Proteins within the uncharacterized NRPS BGC
Given the unknown identity of the product of this NRPS cluster, the proteins have simply been identified by their annotation names in several strains. Much of the biological characterization described above was performed with A. baumannii strain ATCC17978. The protein annotation names of this cluster in ATCC17978, running from A1S_0112 through A1S_0119 are described in Table 2. Our lab has performed initial structural characterization of the enzymes using strain A. baumannii AB307-0294, where the identical proteins are referred to as ABBFA_003406 through ABBFA_003399.
Table 2. Uncharacterized NRPS clusters of A. baumannii.
| Locus ID (Accession) | Length (AA) | Predicted Function |
|---|---|---|
| Strain ATCC17978 (Genbank Accession CP00521) | ||
|
| ||
| A1S_0112 (ABO10599) | 629 | A |
| A1S_0113 (ABO10600) | 597 | Acyl-CoA Dehydrogenase |
| A1S_0114 (ABO10601) | 86 | T |
| A1S_0115 (ABO10602) | 1319 | C-A-T-Te |
| A1S_0116 (ABO10603) | 1216 | RND Exporter |
| A1S_0117 (ABO10604) | 424 | Hypothetical |
| A1S_0118 (ABO10605) | 621 | NAD-dependent Dehydratase |
| A1S_0119 (ABO10606) | 254 | PPTase |
| Strain ATCC17978 (Genbank Accession CP00521) | ||
|
| ||
| A1S_2651 (ABO13068) | 1332 | A-T-Unknown |
The eight proteins encoded by this cluster include three bonafide NRPS proteins, a freestanding adenylation domain (A1S_0112), a freestanding carrier protein (A1S_0114), and a four domain termination module containing a condensation, adenylation, PCP, and thioesterase domain architecture (A1S_0115). These latter two proteins have been structurally characterized from strain AB307-0294, where they are named ABBFA_003404 and ABBFA_003403, respectively.58, 209 Additionally, the cluster contains a PPTase and a transmembrane protein likely involved in product export. The three remaining proteins show homology to an acyl-CoA dehydrogenase (A1S_0113), an NADH dependent dehydratase (A1S_0118), and a protein of unknown function (A1S_0117). The protein encoded upstream at the A1S_0104 (ABBFA_003414) locus is an acyl-CoA synthetase, a family of proteins that produces acyl-CoA thioesters. NRPS clusters often contain members of this family to make starter units that may be incorporated into the product in the absence of a PCP thioester.
Very recently, a proposed structure of the product of this NRPS BGC has been reported, illustrating the presence of a 3-hydroxy long chain fatty acid. Interestingly, the authors note the presence of a Cys-Gly moiety within the structure that may derive from a glutathione adduct of a true product that is further metabolized.210
6.4. Additional uncharacterized NRPSs
A. baumannii strain ATCC 17978 contains an additional multidomain NRPS protein that is present in some, but not all, of the A. baumannii strains. This protein is encoded at locus A1S_2651 and is 1332 residues in length; in the AB307-0294 strain,211 the gene is located at ABBFA_00856, with only five amino acid substitutions over the 1332 residues.
The protein contains an N-terminal adenylation domain that spans the first 520 residues followed by a carrier protein domain from 530 through 596. The C-terminal half of the protein is a domain of unknown function. Sequence comparison fails to identify any characterized homologs that allow for prediction of function. The gene encoding this NRPS does not belong to a larger operon. The neighboring genes, while conserved in multiple NRPS strains, do not appear to encode biosynthetic enzymes that are common to other NRPS clusters.
7. Pseudomonas aeruginosa
The gram-negative bacteria P. aeruginosa is an opportunistic human pathogen. P. aeruginosa is a common cause of hospital-acquired pneumonia, particularly with immunocompromised, cancer, and cystic fibrosis patients.212 The metabolic diversity and the ability to produce a variety of virulence factors contribute to the ability of P. aeruginosa to colonize a variety of environments.213 Additionally, P. aeruginosa has been a model organism for the understanding of biofilm formation and quorum signaling, and exhibits a high propensity for antimicrobial resistance.214, 215
The genome of P. aeruginosa strain PA01, a standard laboratory strain, contains three well-characterized NRPS clusters and three additional BGCs encoding NRPS proteins that have not yet been characterized. These three BGCs appear to be induced in quorum signaling conditions and likely produce novel signaling molecules. Additionally, several NRPS clusters will be mentioned that appear in alternate strains of P. aeruginosa, along with the more common biosynthetic clusters shared by most strains.
7.1. Pyoverdine
7.1.1. Structure of pyoverdine
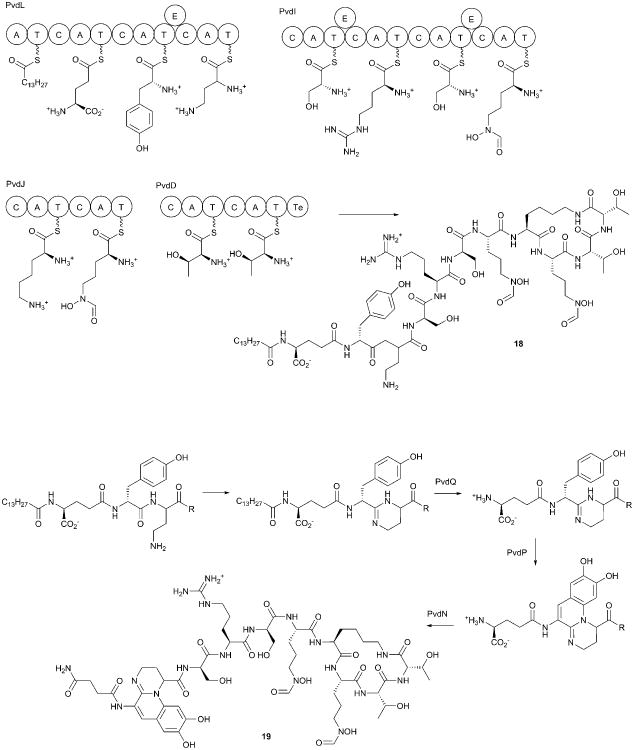
Under iron limiting conditions, P. aeruginosa produces two peptide siderophores that are both produced by NRPSs. Pyoverdine (19) is a larger molecule that is built from an initial acyl-peptide composed of 11 amino acids (Fig. 9), while pyochelin (20, discussed in the next section) is a smaller molecule derived from three building blocks. While both pyochelin and pyoverdine play important roles in different environments, pyoverdine is produced at higher levels and appears to be the primary siderophore under many pathogenic conditions. Multiple reviews describe the siderophores of Pseudomonas;199, 216-218 however, several recent papers have elaborated the steps involved in the final maturation of the pyoverdine molecule.
Fig. 9.
Pyoverdine biosynthesis. Four NRPS proteins are involved in the production of the initial pyoverdine precursor (18). Final steps in the biosynthesis convert precursor (18) to mature pyoverdine (19). The complete biosynthesis of pyoverdine has not been reconstituted in vitro with the NRPS proteins. Several of the steps in chromophore maturation have been demonstrated with precursor or analog substrates.
The pyoverdine molecule can be divided into three components, a variable N-terminal sidearm that can include succinate or succinamide groups. Additionally, a heterocyclic chromophore contains catechol oxygens that contribute to iron binding. Finally, a peptide backbone that varies between different strains of P. aeruginosa.218 In many cases, the peptide tail is cyclized, often through an amide linkage with a lysine side chain and the terminal carboxylate. Pyoverdine often contains the non-proteinogenic amino acids 2,4-diaminobutyrate (DAB) and N5-formyl-N5-hydroxyornithine (fOHOrn) within the peptide.
The sidearm is derived from γ-glutamate, while the chromophore is produced via the oxidation and cyclization of a dipeptide that begins as Tyr3-DAB4. In strain PA01, the pyoverdine peptide chain contains the sequence Ser5-Arg6-Ser7-fOHOrn8-Lys9-fOHOrn10-Thr11-Thr12, with the cyclization occuring between Lys9 and Thr12. The iron chelating groups in pyoverdine are the catechol oxygens of the chromophore and the two oxygens from each of the fOHOrn groups. This octahedral arrangement allows pyoverdine to coordinate Fe(III) with affinity constants as high as 1030 M-1.218
7.1.2. Pyoverdine biosynthesis
The pyoverdine biosynthetic genes in strain PA01 are clustered in one region of the chromosome however the genes are not expressed as a single operon. Instead, the genes are present on both strands of DNA separated by regulatory genes as well as other genes that encode proteins of unrelated function. The biosynthesis of the pyoverdine peptide utililizes four NRPS enzymes and the products of at least nine additional genes (Fig. 9).102 The nonproteinogenic amino acids are provided by PvdH, an aminotransferase that produces DAB from aspartate β- semialdehyde219, and PvdA220, 221 and PvdF222 which convert ornithine to fOHOrn in sequential hydroxylation and acyltransferase steps. The NRPS enzymes that produce the immature pyoverdine peptide are PvdL and PvdI, which each contain four modules, and PvdJ and PvdD, which each contain two modules.
The pyoverdine gene cluster also encodes the protein PA2412223, an MbtH-like protein (MLP).224 These small auxiliary proteins interact with NRPS adenylation domains to enhance adenylation activity.225, 226 Crystal structures of MLPs bound to an adenylation domain227 or to multidomain NRPS proteins59, 63 illustrate how the MLP interacts with the N-terminal subdomain over 15 Å away from the active site. Structures of NRPS proteins in the presence and absence of the partner MLP have not yet identified the basis for activation.
Finally, production of pyoverdine requires the activities of five other proteins that are involved in chromophore maturation. The proteins encoded by pvdM, pvdN, pvdO, pvdP, and pvdQ were originally identified as being necessary for production of wild-type levels of pyoverdine.102 Studies using biochemical analysis and metabolite profiling of mutant strains have enabled the discovery of the final steps in pyoverdine synthesis.
Although the pyoverdine peptide contains 11 amino acids, the four NRPS proteins harbor a total of 12 modules. Bioinformatic prediction of the amino acid specificity of the NRPS adenylation domains,228, 229 allowed module assignments of all four proteins, except for the first module of PvdL. This module was predicted to encode preferences for fatty acids. The lack of a fatty acid in the mature pyoverdine molecule was reconciled with the presence of PvdQ, a protein that showed homology with fatty acid hydrolase enzymes involved in cephalosporin biosynthesis.230 The initial adenylation domain of PvdL was expressed recombinantly and showed specificity for fatty acids of 12 or 14 carbons in length. A mutant pvdQ strain produced an alternate pyoverdine precursor (18) that contains the extra fatty acid component that can be hydrolyzed by PvdQ.231, 232 Thus, the initial module of PvdL incorporates the myristate (or myristoleic233) fatty acid, which PvdQ then removes during pyoverdine maturation. As PvdQ contains a periplasmic signal and is present in the periplasm,231 this suggests that the acylation step might be involved in trafficking the pyoverdine molecule to the periplasm for the final maturation steps. PvdQ is a member of the family of NTN hydrolase enzymes that are formed through the autoproteolysis of a larger protein into an α/β heterodimer, which reveals the catalytic nucleophile at the N-terminal residue of the β chain. Recently, the structure of PvdQ bound to the acyl pyoverdine precursor was determined through the use of a circularly permuted enzyme that enabled the production of a processed but catalytically inactive form of the enzyme.234
Also containing periplasmic signaling sequences are PvdN, PvdO, and PvdP,232 suggesting additional steps in maturation occur in the periplasm after release of the acyl chain by PvdQ. PvdP is homologous to phenoloxidases and tyrosinases suggesting a role in the installation of the second hydroxyl group on the Tyr3 position. PvdP has recently been shown to be a copper-dependent tyrosinase that is able to hydroxylate ferribactin and initiate the proposed oxidative cascade235, 236 to enable the formation of the mature chromophore.237 PvdN has now been shown to catalyze changes to the N-terminal sidearm converting the γ-glutamate into the succinamide moiety that is commonly observed in pyoverdine isoforms.238 The mechanistic details provided for this enzyme, supported by the recent crystal structure of PvdN bound to PLP239, suggest an oxidative decarboxylation that results in the production of the alternate sidearm. The roles of the remaining proteins, PvdM and PvdO, in pyoverdine biosynthesis have not yet been determined. A crystal structure of PvdM (PDB code 3B40) shows structural and sequence homology with dipeptidase enzymes. A PvdO crystal structure shows structural similarity to a formyl-glycine generating enzyme however PvdO lacks key catalytic residues suggesting an alternate function.240
7.1.3. Biological role of pyoverdine
The exact role of pyoverdine production in virulence has been mixed, with studies that both support an important role in establishment and maintenance of an infection as well as others that suggest that pyoverdine production is dispensable.241, 242 A recent meta analysis examined the effect of pyoverdine on both plant, insect, and vertebrate infection models, supporting a limited but consistent contribution to virulence.243 Furthermore, chemical precedent exists for the intervention in the signaling pathways that induce pyoverdine production. The antifungal 5-fluorocytosine has been shown to inhibit the transcriptional regulation of the pyoverdine operon as mediated by the regulator PvdS.244 While treatment reduced virulence of the P. aeruginosa in a murine lung infection model, it did not reduce the pulmonary bacterial load, illustrating a specific impact on virulence caused by pyoverdine production.
7.2. Pyochelin
7.2.1. Structure of pyochelin
A second siderophore produced by P. aeruginosa is pyochelin (20), a smaller molecule derived from three building blocks and consisting of catechol and thiazole moieties (Fig. 10).132, 245 While the redundancy of iron uptake systems may simply reflect the importance of iron acquisition and the need for multiple “backup” systems, alternate functions for secondary siderophores such as pyochelin have been proposed, including induction of further signaling cascades and modulation of host defense mechanisms.246 Pyochelin consists of a salicylate moiety condensed with two thiazolidine rings derived from cysteine residues.247 The iron is coordinated by the phenolic oxygen of the salicylate moiety, the two nitrogens of the thiazolidine rings, and an oxygen from the carboxylate.248, 249
Fig. 10.
Pyochelin biosynthesis. Pyochelin biosynthesis has been demonstrated biochemically using the three NRPS enzymes PchD, PchE, PchF, as well as the redutase PchG, along with building blocks DHB and cysteine, along with necessary substrates ATP, NADPH, and SAM. PchE contains an additional epimerization domain (E), while PchF contains an N-methyltransferase.
7.2.2. Pyochelin biosynthesis
The biosynthetic gene cluster for the synthesis of pyochelin consists of seven genes. Two genes, pchA and pchB encode chorismate utilizing enzymes that are involved in the production of salicylate.250 Three NRPS genes, pchD, pchE, and pchF,251 along with a thiazolinyl reductase encoded by pchG158, 252 catalyze the formation of the pyochelin molecule (Fig. 10). Finally, pchC encodes a proofreading thioesterase that increases the efficiency via the release of mischarged molecules from the PCP domains of PchE and PchF.253
Salicylic acid is produced through the activities of PchA and PchB. PchA is an isochorismate synthase254 while PchB catalyzes the conversion of isochorismate to salicylate.255 The three NRPS enzymes then function to combine the starter units.84, 256 PchD is a freestanding adenylation domain that loads the salicylate onto the N-terminal carrier protein domain of PchE. Following transfer of the salicylate to the first PCP of PchE, the bifunctional cyclization/condensation domain forms the initial peptide linkage between the cysteine and salicylate, followed by cyclization to generate the hydroxyphenylthiazolinyl-enzyme intermediate. The same process is repeated at the third pyochelin module located on the PchF, to result in the formation of the hydroxyphenylthiazyl-thiazolinyl-enzyme intermediate on PchF. Two steps remain for pyochelin production. First, PchG catalyzes the NADPH-dependent reduction in trans to convert the thiazolinyl moiety to a thiazolidinyl group.252 In this reduced form, the nitrogen is then methylated in a SAM-dependent reaction catalyzed by the embedded methyltransferase of PchF. Release of pyochelin by the terminal thioesterase domain of PchF completes the process.
7.2.3. A pyochelin-related cluster for the synthesis of dihydroaeruginoic acid
Although not present in the standard laboratory strain PA01, a four gene cluster appears on a mobile genomic element in the virulent PA14 strain257 of P. aeruginosa. The BGC contains four NRPS genes that synthesize of dihydroaeruginoic acid (DHA), a truncated form of pyochelin containing a salicylate cap joined with a single thiazoline ring of opposite stereochemistry to that observed in pyochelin.258
Present in this BGC is a gene encoding PA14_54940, a bi-domain protein that is an unusual fusion between an adenylation domain, which shows predicted specificity for salicylate over DHB,70 and a salicylate synthase domain that may directly convert chorismate to salicylate, similar to MbtI of M. tuberculosis.259 The cluster then contains two proteins, PA14_54930 and PA14_54920, that each encode one complete module. The two proteins share 36 % sequence identity over the first ∼1000 residues and differ by the presence of a reductase domain at the C-terminus of PA14_54920. Finally, the BGC also contains a proofreading thioesterase domain. The cluster is followed by several other genes, including one encoding a serine O-acetyltransferase that may contribute to sufficient cysteine levels for DHA production.258
The two adenylation domains of PA14_54930 and PA14_54920 show specificity predictions for cysteine using the prediction tools of the antiSMASH database, while the two condensation domains classify as cyclization domains. The cluster therefore appears very similar to the pyochelin NRPS with several notable differences.
First, there are no internal epimerization or methylation domains in either NRPS module as is seen in PchE and PchF. While the epimerization and methylation domains are not necessary for the production of DHA, the cluster also appears to be missing a carrier protein for the salicylate module. While the adenylation domain of PA14_54940 will adenylate the substrate, there does not appear to be an aryl carrier protein that can serve to shuttle the salicylate to the downstream condensation domain. Furthermore, it appears that one of the NRPS modules encoded by PA14_54930 or PA14_54920 is not needed in the production of the truncated pyochelin variant. The PA14_54930 protein was able to complement a pchE mutant in P. putida in the production of enantiopyochelin as it lacks the epimerization domain.258 The ability of PA14_54920 to complement either a pchE or pchF mutant was not tested.
Expression of the four gene operon in both the presence or absence of the downstream operon containing the serine O-acetyltransferase gene resulted in the production of the same product, which was shown by mass spectrometry and thin layer chromatography to be the truncated pyochelin variant, DHA.258 The exact role of the PA14_54920 and the identity of the carrier protein for the salicylate adenylating domain remain to be determined.
7.2.4. Biological role of pyochelin
Although present in all isolates of P. aeruginosa,245 pyochelin appears to be somewhat less significant than pyoverdine in a number of clinically relevant isolates. Indeed, it has been classified among a variety of siderophores with lower affinity for iron that have been labeled “secondary siderophores”.246 For example, while the gene encoding the pyochelin synthetase PchF was expressed in the majority of sputum samples from cystic fibrosis patients, the level of transcripts was much lower than several pyoverdine biosynthetic genes. Further, pyochelin was detected in only 6 of 45 samples tested.242
However, there is also evidence that different siderophores may be exploited by P. aeruginosa under certain environments. As a lower complexity molecule, pyochelin may be more useful under low iron stress conditions. P. aeruginosa would then switch to the more metabolically demanding pyoverdine under extreme iron depletion.260 It has been proposed that in the cystic fibrosis lung, pyoverdine may be required to establish an initial infection. At later stages in the infection, the accompanying damage to the pulmonary tissue may result in inflammation and disruption of the host cells, releasing iron that may make pyochelin the more relevant siderophore.245
7.3. L-2-amino-4-methoxy-trans-3-butenoic acid (AMB)
7.3.1. Structure of AMB
Another NRPS cluster from P. aeruginosa encodes the synthesis of the antimetabolite AMB (22) (Fig. 11). AMB is a substituted vinylglycine that along with other similar molecules have been shown to inhibit growth of bacterial and eukaryotic cells,261-263 possibly through the inhibition of PLP-dependent enzymes.264, 265 The literature on AMB biosynthesis in P. aeruginosa includes an alternate pathway that was proposed to be responsible for a novel quorum signaling molecule based on analysis of the whole cell metabolic profile.266 However, recent biochemical evidence convincingly demonstrates the ability of the AmbBCDE proteins to catalyze production of AMB.267
Fig. 11.
AMB biosynthesis. The AMB residue is produced in two steps from glutamate through the activities of AmbC and AmbD. The formation of the Ala-Glu-Ala tripeptide on AmbE has been shown biochemically to precede conversion to the AMB containing tripeptide, which occurs while the tripeptide is loaded on AmbE if AmbC and AmbD are present.
7.3.2. AMB biosynthetic cluster
Production of AMB by P. aeruginosa can be assayed through growth inhibition of standard E. coli strains. The cluster responsible for AMB production was therefore isolated by screening a mutant library of P. aeruginosa for loss of this growth inhibition phenotype.268 Four mutants were identified that all mapped to the cluster encoded by genes PA2302 through PA2305 in strain PA01. The five gene cluster was named ambABCDE. The ambA gene encodes a LysE type transmembrane transporter, which is proposed to be responsible for the export of, and potentially resistance to, the antimetabolite.267 AmbB and AmbE encode NRPS proteins, while AmbC and AmbD are members of the family269 of Fe(II)/α-ketoglutarate dependent oxygenases. The puzzling NRPS architecture, necessary to produce AMB from what might be expected to be a single amino acid, contains two adenylation, two condensation, and three PCP domains.
The biosynthetic pathway of AMB has recently been proposed267 as follows (Fig. 11). The adenylation domain of AmbB performs two loading steps, attaching an alanine residue intramolecularly to the downstream PCP domain and catalyzing a second intermolecular reaction to load the second PCP domain of AmbE with a second alanine molecule. The adenylation domain of AmbE loads a molecule of glutamate onto the first PCP domain of AmbE. The condensation domain of AmbB transfers the alanine residue to the loaded glutamate, forming the Ala-Glu dipeptide on the first AmbB PCP domain. Subsequently the condensation domain of AmbE transfers this dipeptide to the loaded alanine residue on the second PCP domain of AmbE, resulting in the formation of an Ala-Glu-Ala tripeptide. This intermediate serves as the substrate for the Fe(II)/α-ketoglutarate dependent oxygenases, AmbC and AmbD as well as the methyltransferase domain of AmbE, which together convert the glutamate residue to a molecule of AMB. These steps in the synthesis are believed to occur while the tripeptide is bound to AmbE, and not after peptide release, as the mature Ala-AMB-Ala tripeptide (21) was identified covalently bound to the PCP.267 While the presence of the AMB-containing tripeptide on the AmbE protein suggests the tailoring reactions occur on the PCP-bound peptide, the authors note that the exact timing of the steps in the synthesis have not yet been conclusively demonstrated. Similarly, neither the role of the alanine residues in the Ala-AMB-Ala tripeptide nor the source of peptidase activity needed to release the AMB have been identified. Inhibition of PLP-dependent enzymes would require a free amine on the AMB residue to form an initial external aldimine with the cofactor.264 Potentially, the alanine residues serve as a protective group until the AMB has been exported from the cell.
7.3.3 Biological role of AMB
The lack of an obvious one-to-one correspondence between the modules and the product may have been partly responsible for the misidentification of the product of the amb operon as a new quorum signaling molecule.266 An ambB mutant also showed reduced production of several quorum signaling molecules, including 2-(2-hydroxyphenyl)thiazole-4-carbaldehyde, a molecule that the authors named IQS. The subsequent biochemical reconstitution of AMB biosynthesis confirmed the product shown above and suggests that AMB is part of a signaling cascade that includes the production of multiple quorum signals, iron-acquisition molecules, and the potential metabolite IQS.
Beyond its role as an antimetabolite with the ability to inhibit PLP-dependent enzymes, the precise role of AMB in the microbial physiology of P. aeruginosa is unknown. Several reports correlate the expression of this operon with quorum signaling and biofilm formation. The amb operon is regulated by LasR and RhlR, two transcription factors that sense and activate genes in response to the acylhomoserine lactone quorum signals. Transcriptome analysis showed that the ambBCDE genes are expressed 30- to 130-times more highly in wild-type cells compared to lasRrhlR mutants.270 Furthermore, the amb operon is induced by ∼10-fold by the addition of the these acylHSL signals.271 The amb operon has been more recently included in the core set of genes that are controlled by quorum signals across multiple P. aeruginosa strains.272 Finally, expression of BpiB09, a, NADP-dependent reductase that catalyzes the inactivation of acyl-HSL signals, results in a 30- and 79-fold down-regulation of ambC and ambD expression, respectively.273 While no precise phenotype has been ascribed to AMB production, these signs suggest a link to quorum signaling pathways.
7.4. Pyoluteorin
7.4.1 Pyoluteorin structure
Several P. aeruginosa strains have been annotated to contain a hybrid NRPS/PKS cluster that encodes proteins for the synthesis of the antibiotic pyoluteorin (23), a compound more commonly associated with other Pseudomonads.274 The two sequenced P. aeruginosa strains with pyoluteorin clusters are the Liverpool epidemic strains LESB58 and LES431, two virulent strains from cystic fibrosis patients.275, 276 Pyoluteorin contains a resorcinol moiety fused to a dihalogenated pyrrole derived from proline.
7.4.2. Biosynthesis of Pyoluteorin
The pyoluteorin BGC contains eight biosynthetic genes, two genes involved in regulation, and six genes277 that support metabolite transport.278 The eight biosynthetic genes encode PltA through PltG, and PltL (Fig. 12). PltF is a freestanding adenylation domain that loads the freestanding carrier protein PltL with proline.279 The proline then is oxidized by the FAD-dependent enzyme PltE and chlorinated by the halogenase PltA.280 NMR studies of PltL loaded with proline demonstrate the interaction of the ligand with the core of the PCP domain.281 PltF is specific for L-proline and shows no activity with pyrrolyl-2-carboxylate,279 demonstrating that the subsequent oxidation of the proline derivative occurs while bound to the carrier protein. The dichloropyrrolyl group is then offered to the PKS enzymes PltB, PltC, and PltG for the final steps.
Fig. 12.
Pyoluteorin biosynthesis. The early steps in pyoluteorin biosynthesis have been demonstrated with enzymes from related pseuodomonads. The 4,5-dichloropyrrole has been shown to be produced while tethered to the carrier protein PltL. Subsequent PKS enzymes produce the resorcinol moiety.
7.5. Additional uncharacterized NRPS clusters of P. aeruginosa
P. aeruginosa also contains four NRPS clusters that have not yet been characterized. The first three of these clusters are common to most sequenced P. aeruginosa strains, while some appear only in a few strains (Table 3). They are as yet unnamed and are described below using their protein locus in the P. aeruginosa PA01 strain. Interestingly, the PA1221, PA3327, and PA4078 BGC are present in other gram-negative bacteria. The genes are organized in the same order and share ∼50% sequence identity, sometimes as high as 70%. These do not appear to simply be misclassified genomic samples because of the low overall sequence identities, and likely represent horizontal transfer of the entire BGCs between different species.
Table 3. Uncharacterized NRPS clusters of P. aeruginosa.
| Locus ID (Accession) | Length (AA) | Predicted Function |
|---|---|---|
| Strain PA01 (Genbank Accession AE004091.2) | ||
|
| ||
| PA1221 (NP_249912) | 618 | A-T |
| PA1220 (NP_249911) | 420 | C |
| PA1219 (NP_249910) | 204 | Te |
| PA1218 (NP_249909) | 292 | Fe/αKG-dependent dioxygenase |
| PA1217 (NP_249908) | 455 | Isopropylmalate synthase |
| PA1216 (NP_249907) | 248 | Methyltransferase |
| PA1215 (NP_249906) | 492 | A |
| PA1214 (NP_249905) | 532 | Amidotransferase |
| PA1213 (NP_249904) | 319 | Fe/αKG-dependent dioxygenase |
| PA1212 (NP_249903) | 409 | MFS Transporter |
| PA1211 (NP_249902) | 211 | α/β Hydrolase (Te) |
| Strain PA01 (Genbank Accession AE004091.2) | ||
|
| ||
| PA3327 (NP_252017) | 2352 | C-A-T-C-A-T-Te |
| PA3328 (NP_252018) | 388 | FAD-dep monooxygenase |
| PA3329 (NP_252019) | 442 | C |
| PA3330 (NP_252020) | 304 | NAD-dependent dehydrogenase |
| PA3331 (NP_252021) | 418 | Cytochrome P450 |
| PA3332 (NP_252022) | 141 | Hypothetical Protein |
| PA3333 (NP_252023) | 330 | Ketoacyl-ACP synthase |
| PA3334 (NP_252024) | 79 | ACP |
| PA3335 (NP_252025) | 250 | Hypothetical Protein |
| PA3336 (NP_252026) | 388 | MFS Transporter |
| Strain PA01 (Genbank Accession AE004091.2) | ||
|
| ||
| PA4078 (NP_252767) | 991 | A-T-Re |
| PA4079 (NP_252768) | 229 | NAD-dependent dehydrogenase |
| Strain PA7 (Genbank Accession CP000744) | ||
|
| ||
| PSPA7_2112 (ABR82062) | 258 | Hypothetical protein |
| PSPA7_2111 (ABR85185) | 115 | Cupin domain |
| PSPA7_2110 (ABR85498) | 309 | Amine oxygenase |
| PSPA7_2109 (ABR83084) | 1145 | A-T-Re |
| PSPA7_2108 (ABR84094) | 155 | Hypothetical protein |
Although none of the products of the following BGCs has been identified, these three clusters from strain PA01 frequently appear in transcriptomic studies that aim to understand the genes that are regulated in association with biofilm and virulence related phenotypes, including the production of acyl-homoserine lactones, the Pseudomonas Quinolone Signal (PQS) and phenazines. 270-272, 282, 283
7.5.1. PA1221
The PA1221 cluster encodes 11 proteins. Several have homology to known proteins while several are uncharacterized. There are three NRPS proteins, a didomain adenylation and PCP, and freestanding condensation, thioesterase, and adenylation domains. The didomain protein PA1221 was used for structural studies to identify the functional interactions of the NRPS adenylation and PCP domains. PA1221 showed adenylating activity with valine, among the proteinogenic amino acids tested.78 The cluster contains two Fe/α-ketoglutarate dependent oxygenases; PA1218 shows reasonable homology with phytanoyl-CoA dioxygenases, suggesting it may add a hydroxyl group to an acyl-CoA thioester. The presence of an MFS transporter suggests the production of a novel product that is excreted from the cell.
7.5.2. PA3327
A ten-gene cluster beginning at PA3327 encodes several NRPS proteins, along with other enzymes and proteins likely to be involved in export of the natural product. The NRPS protein PA3327 is a dimodular NRPS, containing two extending modules and a C-terminal thioesterase domain. Additionally, the cluster contains a freestanding condensation domain at PA3329 and a freestanding carrier protein at PA3334. Additional proteins within the cluster include a monooxygenase, an NAD-dependent dehydrogenase, a cytochrome P450, a ketoacyl-ACP synthase, and two hypothetical proteins.
7.5.3. PA4078
In contrast to the other clusters from strain PA01, the NRPS gene PA4078 appears in a much smaller operon, with only one additional gene appearing to be co-transcribed. The PA4078 gene encodes a three-domain adenylation-PCP-reductase gene, while the downstream PA4079 gene encodes an NAD-dependent dehydrogenase. While there are additional enzymes encoded on neighboring genes, located on both strands, none can be confidently assigned to belong to any NRPS associated families such as carrier proteins, acyl-CoA synthetases, or PKS related proteins.
7.5.4. PA7-Cluster
The clinical isolate strain P. aeruginosa PA7257 contains a five-gene operon containing two hypothetical proteins, a cupin domain, an amine oxygenase, and a three-domain NRPS. Upstream is an additional operon containing genes encoding an editing thioesterase and a 528-residue adenylation domain at PSPA7_2121. This upstream operon also encodes a predicted isopropylmalate synthase and an acyl-CoA dehydrogenase, reminiscent of the A1S_0112 operon of A. baumannii.
8. Enterobacter spp
The final group of ESKAPE pathogens belong to genus Enterobacter. Enterobacter species are facultative anaerobic gram-negative bacilli that are a common cause of nosocomial infections, including blood stream and urinary tract infections, and menengitis.89, 284 Enterobacter are increasingly resistant to common antibacterials, a phenotype that is mediated by resistance factors encoded on a transmissible plasmid.
Enterobacter strains are often not classified into individual species because of the difficulty in phenotypic classification and recent genotypic classifications define an Enterobacter cloacae complex composed of multiple heterogeneous subspecies.285 Among the most common causes of human infections are E. hormaechei and E. cloacae.284-286 We therefore focused our search for the NRPS clusters identified within these two species. The genome of E. cloacae strain ATCC 13047, reported as the first complete genome sequence, contains only a single NRPS operon encoding enterobactin.287 The same was true of E. hormaechei strain ATCC 49162.288 Examination of the databases for additional NRPS clusters identifies colibactin and an additional uncharacterized BGCs as noted below.
E. cloacae strain s611 contains a BGC containing six genes. Interestingly, it begins with an MLP, followed by a large NRPS gene. Following this three module NRPS protein is a PPTase, a second MLP, and two additional NRPS proteins. This unusual architecture containing two MLPs that share 79% sequence identity, raises the possibility that the adenylation domains could be differentially regulated by these auxiliary proteins.
9. Conclusions
The past few decades have seen tremendous advances in our understanding of the biosynthetic capabilities of microorganisms and the complicated role that these natural products play both in the environment and in the pathogenic setting of the host. These natural products, by definition, have overcome many of the challenges that face medicinal chemists in the development of novel therapeutics. Specifically, they are bioactive molecules that are able to enter the cell or are able to exert their effect from the environment through the activity of specific receptors. Further, they are soluble in the acqueous biological environment and are stable, at least at the time scale with which they need to operate. It is not surprising then that many natural products have been exploited in the development of pharmaceuticals.
The natural products produced by all bacteria, and by human pathogens in particular, can provide insight into bacterial metabolism and how cells adapt to diverse environments. Identifying the biosynthetic potential of ESKAPE pathogens will be valuable to understand the role these molecules play. The demonstration of the specific conditions under which BGCs are induced is an equally valuable goal that should accompany discovery of the specific products. Understanding the regulatory networks that involve novel signaling molecules will facilitate the establishment of the roles these molecules play.
This is an exciting and important time in the history of antimicrobial discovery. The prevalence of antibiotic resistance and the lack of novel compounds in the pipeline raises concern about the rise of highly pathogenic organisms that are able to overcome the treatments currently in use. This possibility emphasizes the continued need to explore the fundamental biochemistry of natural product biosynthesis.
Table 4. Uncharacterized NRPS cluster of E. cloacae.
| Locus ID (Accession) | Length (AA) | Predicted Function |
|---|---|---|
| Strain s611 (Genbank Accession NZ_AXOM01000000) | ||
|
| ||
| WP_023480884 | 66 | MLP |
| WP_023480734 | 4466 | C-A-T-C-A-T-C-A-T-Te |
| WP_023480967 | 237 | PPTase |
| WP_023480995 | 67 | MLP |
| WP_080263008 | 3632 | C-A-T-C-A-T-C-A-T-Epim |
| WP_051372113 | 2366 | C-A-T-C-A-T-Te |
Acknowledgments
I would like to thank all of the past and present members of my lab, whose hard work has allowed us to contribute to the understanding of the structural biology of NRPS proteins. I would particularly like to thank Daniel C. Bailey and Lisa S. Mydy for helpful comments on this manuscript. Work in my lab is supported by the National Institute of General Medical Sciences (GM-116957) and the National Institute of Allergy and Infectious Diseases (AI-116998) of the National Institutes of Health.
Notes and references
- 1.Gevers W, Kleinkauf H, Lipmann F. Proc Natl Acad Sci U S A. 1968;60:269–276. doi: 10.1073/pnas.60.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gevers W, Kleinkauf H, Lipmann F. Proc Natl Acad Sci U S A. 1969;63:1335–1342. doi: 10.1073/pnas.63.4.1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gilhuus-Moe CC, Kristensen T, Bredesen JE, Zimmer T, Laland SG. FEBS Lett. 1970;7:287–290. doi: 10.1016/0014-5793(70)80183-1. [DOI] [PubMed] [Google Scholar]
- 4.Kleinkauf H, Roskoski R, Jr, Lipmann F. Proc Natl Acad Sci U S A. 1971;68:2069–2072. doi: 10.1073/pnas.68.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mittenhuber G, Weckermann R, Marahiel MA. J Bacteriol. 1989;171:4881–4887. doi: 10.1128/jb.171.9.4881-4887.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weckermann R, Furbass R, Marahiel MA. Nucleic Acids Res. 1988;16:11841. doi: 10.1093/nar/16.24.11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turgay K, Krause M, Marahiel MA. Mol Microbiol. 1992;6:529–546. doi: 10.1111/j.1365-2958.1992.tb01498.x. [DOI] [PubMed] [Google Scholar]
- 8.Marahiel MA. Febs Letters. 1992;307:40–43. doi: 10.1016/0014-5793(92)80898-q. [DOI] [PubMed] [Google Scholar]
- 9.Gocht M, Marahiel MA. J Bacteriol. 1994;176:2654–2662. doi: 10.1128/jb.176.9.2654-2662.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stachelhaus T, Mootz HD, Bergendahl V, Marahiel MA. J Biol Chem. 1998;273:22773–22781. doi: 10.1074/jbc.273.35.22773. [DOI] [PubMed] [Google Scholar]
- 11.Cane DE, Walsh CT, Khosla C. Science. 1998;282:63–68. doi: 10.1126/science.282.5386.63. [DOI] [PubMed] [Google Scholar]
- 12.Marahiel MA. Chem Biol. 1997;4:561–567. doi: 10.1016/s1074-5521(97)90242-8. [DOI] [PubMed] [Google Scholar]
- 13.Walsh CT. Nat Chem Biol. 2015;11:620–624. doi: 10.1038/nchembio.1894. [DOI] [PubMed] [Google Scholar]
- 14.Walsh CT. Nat Prod Rep. 2016;33:127–135. doi: 10.1039/c5np00035a. [DOI] [PubMed] [Google Scholar]
- 15.Marahiel MA, Essen LO. Methods Enzymol. 2009;458:337–351. doi: 10.1016/S0076-6879(09)04813-7. [DOI] [PubMed] [Google Scholar]
- 16.Strieker M, Tanovic A, Marahiel MA. Curr Opin Struct Biol. 2010;20:234–240. doi: 10.1016/j.sbi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 17.Weber G, Schorgendorfer K, Schneider-Scherzer E, Leitner E. Curr Genet. 1994;26:120–125. doi: 10.1007/BF00313798. [DOI] [PubMed] [Google Scholar]
- 18.Wiest A, Grzegorski D, Xu BW, Goulard C, Rebuffat S, Ebbole DJ, Bodo B, Kenerley C. J Biol Chem. 2002;277:20862–20868. doi: 10.1074/jbc.M201654200. [DOI] [PubMed] [Google Scholar]
- 19.Crosby J, Crump MP. Nat Prod Rep. 2012;29:1111–1137. doi: 10.1039/c2np20062g. [DOI] [PubMed] [Google Scholar]
- 20.Mercer AC, Burkart MD. Nat Prod Rep. 2007;24:750–773. doi: 10.1039/b603921a. [DOI] [PubMed] [Google Scholar]
- 21.Lambalot RH, Gehring AM, Flugel RS, Zuber P, LaCelle M, Marahiel MA, Reid R, Khosla C, Walsh CT. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 22.Gulick AM. ACS Chem Biol. 2009;4:811–827. doi: 10.1021/cb900156h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Linne U, Schafer A, Stubbs MT, Marahiel MA. FEBS Lett. 2007 doi: 10.1016/j.febslet.2007.01.066. [DOI] [PubMed] [Google Scholar]
- 24.Keating TA, Ehmann DE, Kohli RM, Marshall CG, Trauger JW, Walsh CT. Chembiochem. 2001;2:99–107. doi: 10.1002/1439-7633(20010202)2:2<99::AID-CBIC99>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 25.Trauger JW, Kohli RM, Mootz HD, Marahiel MA, Walsh CT. Nature. 2000;407:215–218. doi: 10.1038/35025116. [DOI] [PubMed] [Google Scholar]
- 26.Ravel J, Cornelis P. Trends Microbiol. 2003;11:195–200. doi: 10.1016/s0966-842x(03)00076-3. [DOI] [PubMed] [Google Scholar]
- 27.Konz D, Klens A, Schorgendorfer K, Marahiel MA. Chem Biol. 1997;4:927–937. doi: 10.1016/s1074-5521(97)90301-x. [DOI] [PubMed] [Google Scholar]
- 28.Kohli RM, Trauger JW, Schwarzer D, Marahiel MA, Walsh CT. Biochemistry. 2001;40:7099–7108. doi: 10.1021/bi010036j. [DOI] [PubMed] [Google Scholar]
- 29.Krause M, Marahiel MA. J Bacteriol. 1988;170:4669–4674. doi: 10.1128/jb.170.10.4669-4674.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gehring AM, Mori I, Walsh CT. Biochemistry. 1998;37:2648–2659. doi: 10.1021/bi9726584. [DOI] [PubMed] [Google Scholar]
- 31.Walsh CT, Chen H, Keating TA, Hubbard BK, Losey HC, Luo L, Marshall CG, Miller DA, Patel HM. Curr Opin Chem Biol. 2001;5:525–534. doi: 10.1016/s1367-5931(00)00235-0. [DOI] [PubMed] [Google Scholar]
- 32.Burmester J, Haese A, Zocher R. Biochem Mol Biol Int. 1995;37:201–207. [PubMed] [Google Scholar]
- 33.Hacker C, Glinski M, Hornbogen T, Doller A, Zocher R. J Biol Chem. 2000;275:30826–30832. doi: 10.1074/jbc.M002614200. [DOI] [PubMed] [Google Scholar]
- 34.Linne U, Doekel S, Marahiel MA. Biochemistry. 2001;40:15824–15834. doi: 10.1021/bi011595t. [DOI] [PubMed] [Google Scholar]
- 35.Stachelhaus T, Walsh CT. Biochemistry. 2000;39:5775–5787. doi: 10.1021/bi9929002. [DOI] [PubMed] [Google Scholar]
- 36.Marshall CG, Burkart MD, Keating TA, Walsh CT. Biochemistry. 2001;40:10655–10663. doi: 10.1021/bi010937s. [DOI] [PubMed] [Google Scholar]
- 37.Kinatukara P, Patel KD, Haque AS, Singh R, Gokhale RS, Sankaranarayananan R. J Struct Biol. 2016;194:368–374. doi: 10.1016/j.jsb.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 38.Wyatt MA, Mok MC, Junop M, Magarvey NA. Chembiochem. 2012;13:2408–2415. doi: 10.1002/cbic.201200340. [DOI] [PubMed] [Google Scholar]
- 39.Zhang W, Ostash B, Walsh CT. Proc Natl Acad Sci U S A. 2010;107:16828–16833. doi: 10.1073/pnas.1011557107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaudelli NM, Long DH, Townsend CA. Nature. 2015;520:383–387. doi: 10.1038/nature14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Braesel J, Gotze S, Shah F, Heine D, Tauber J, Hertweck C, Tunlid A, Stallforth P, Hoffmeister D. Chem Biol. 2015;22:1325–1334. doi: 10.1016/j.chembiol.2015.08.016. [DOI] [PubMed] [Google Scholar]
- 42.Geib E, Gressler M, Viediernikova I, Hillmann F, Jacobsen ID, Nietzsche S, Hertweck C, Brock M. Cell Chem Biol. 2016;23:587–597. doi: 10.1016/j.chembiol.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Blodgett JAV, Oh DC, Cao S, Currie CR, Kolter R, Clardy J. Proc Natl Acad Sci U S A. 2010;107:11692–11697. doi: 10.1073/pnas.1001513107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F, Du L. J Am Chem Soc. 2011;133:643–645. doi: 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li R, Oliver RA, Townsend CA. Cell Chem Biol. 2016 doi: 10.1016/j.chembiol.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maruyama C, Toyoda J, Kato Y, Izumikawa M, Takagi M, Shinya K, Katano H, Utagawa T, Hamano Y. Nat Chem Biol. 2012;8:791–797. doi: 10.1038/nchembio.1040. [DOI] [PubMed] [Google Scholar]
- 47.Huang T, Li L, Brock NL, Deng Z, Lin S. Chembiochem. 2016;17:1421–1425. doi: 10.1002/cbic.201600156. [DOI] [PubMed] [Google Scholar]
- 48.Kittila T, Mollo A, Charkoudian LK, Cryle MJ. Angew Chem Int Ed Engl. 2016;55:9834–9840. doi: 10.1002/anie.201602614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ladner CC, Williams GJ. Journal of industrial microbiology & biotechnology. 2016;43:371–387. doi: 10.1007/s10295-015-1704-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schmeing TM. Clin Invest Med. 2016;39:E220–E226. doi: 10.25011/cim.v39i6.27490. [DOI] [PubMed] [Google Scholar]
- 51.Gulick AM. Curr Opin Chem Biol. 2016;35:89–96. doi: 10.1016/j.cbpa.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miller BR, Gulick AM. Methods Mol Biol. 2016;1401:3–29. doi: 10.1007/978-1-4939-3375-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weissman KJ. Nat Chem Biol. 2015;11:660–670. doi: 10.1038/nchembio.1883. [DOI] [PubMed] [Google Scholar]
- 54.Sussmuth RD, Mainz A. Angew Chem Int Ed Engl. 2017;56:3770–3821. doi: 10.1002/anie.201609079. [DOI] [PubMed] [Google Scholar]
- 55.Bloudoff K, Schmeing TM. Biochim Biophys Acta. 2017 doi: 10.1016/j.bbapap.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 56.Bloudoff K, Alonzo DA, Schmeing TM. Cell Chem Biol. 2016;23:331–339. doi: 10.1016/j.chembiol.2016.02.012. [DOI] [PubMed] [Google Scholar]
- 57.Chen WH, Li K, Guntaka NS, Bruner SD. ACS Chem Biol. 2016;11:2293–2303. doi: 10.1021/acschembio.6b00332. [DOI] [PubMed] [Google Scholar]
- 58.Drake EJ, Miller BR, Shi C, Tarrasch JT, Sundlov JA, Allen CL, Skiniotis G, Aldrich CC, Gulick AM. Nature. 2016;529:235–238. doi: 10.1038/nature16163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller BR, Drake EJ, Shi C, Aldrich CC, Gulick AM. J Biol Chem. 2016;291:22559–22571. doi: 10.1074/jbc.M116.746297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reimer JM, Aloise MN, Harrison PM, Schmeing TM. Nature. 2016;529:239–242. doi: 10.1038/nature16503. [DOI] [PubMed] [Google Scholar]
- 61.Dowling DP, Kung Y, Croft AK, Taghizadeh K, Kelly WL, Walsh CT, Drennan CL. Proc Natl Acad Sci U S A. 2016;113:12432–12437. doi: 10.1073/pnas.1608615113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, Liu N, Cacho RA, Gong Z, Liu Z, Qin W, Tang C, Tang Y, Zhou J. Nat Chem Biol. 2016;12:1001–1003. doi: 10.1038/nchembio.2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tarry MJ, Haque AS, Bui KH, Schmeing TM. Structure. 2017;25:783–793. doi: 10.1016/j.str.2017.03.014. [DOI] [PubMed] [Google Scholar]
- 64.Bloudoff K, Rodionov D, Schmeing TM. J Mol Biol. 2013;425:3137–3150. doi: 10.1016/j.jmb.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Keating TA, Marshall CG, Walsh CT, Keating AE. Nat Struct Biol. 2002;9:522–526. doi: 10.1038/nsb810. [DOI] [PubMed] [Google Scholar]
- 66.Samel SA, Schoenafinger G, Knappe TA, Marahiel MA, Essen LO. Structure. 2007;15:781–792. doi: 10.1016/j.str.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 67.Conti E, Franks NP, Brick P. Structure. 1996;4:287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- 68.Conti E, Stachelhaus T, Marahiel MA, Brick P. EMBO J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gulick AM, Starai VJ, Horswill AR, Homick KM, Escalante-Semerena JC. Biochemistry. 2003;42:2866–2873. doi: 10.1021/bi0271603. [DOI] [PubMed] [Google Scholar]
- 70.May JJ, Kessler N, Marahiel MA, Stubbs MT. Proc Natl Acad Sci U S A. 2002;99:12120–12125. doi: 10.1073/pnas.182156699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gulick AM, Horswill AR, Thoden JB, Escalante-Semerena JC, Rayment I. Acta Crystallogr D Biol Crystallogr. 2002;58:306–309. doi: 10.1107/s0907444901018832. [DOI] [PubMed] [Google Scholar]
- 72.Bruner SD, Weber T, Kohli RM, Schwarzer D, Marahiel MA, Walsh CT, Stubbs MT. Structure (Camb) 2002;10:301–310. doi: 10.1016/s0969-2126(02)00716-5. [DOI] [PubMed] [Google Scholar]
- 73.Samel SA, Wagner B, Marahiel MA, Essen LO. J Mol Biol. 2006;359:876–889. doi: 10.1016/j.jmb.2006.03.062. [DOI] [PubMed] [Google Scholar]
- 74.Barajas JF, Phelan RM, Schaub AJ, Kliewer JT, Kelly PJ, Jackson DR, Luo R, Keasling JD, Tsai SC. Chem Biol. 2015;22:1018–1029. doi: 10.1016/j.chembiol.2015.06.022. [DOI] [PubMed] [Google Scholar]
- 75.Chhabra A, Haque AS, Pal RK, Goyal A, Rai R, Joshi S, Panjikar S, Pasha S, Sankaranarayanan R, Gokhale RS. Proc Natl Acad Sci U S A. 2012;109:5681–5686. doi: 10.1073/pnas.1118680109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frueh DP, Arthanari H, Koglin A, Vosburg DA, Bennett AE, Walsh CT, Wagner G. Nature. 2008;454:903–906. doi: 10.1038/nature07162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Y, Zheng T, Bruner SD. Chem Biol. 2011;18:1482–1488. doi: 10.1016/j.chembiol.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mitchell CA, Shi C, Aldrich CC, Gulick AM. Biochemistry. 2012;51:3252–3263. doi: 10.1021/bi300112e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sundlov JA, Gulick AM. Acta Crystallogr D Biol Crystallogr. 2013;69:1482–1492. doi: 10.1107/S0907444913009372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sundlov JA, Shi C, Wilson DJ, Aldrich CC, Gulick AM. Chem Biol. 2012;19:188–198. doi: 10.1016/j.chembiol.2011.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Qiao C, Wilson DJ, Bennett EM, Aldrich CC. J Am Chem Soc. 2007;129:6350–6351. doi: 10.1021/ja069201e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tanovic A, Samel SA, Essen LO, Marahiel MA. Science. 2008;321:659–663. doi: 10.1126/science.1159850. [DOI] [PubMed] [Google Scholar]
- 83.Miller DA, Luo L, Hillson N, Keating TA, Walsh CT. Chem Biol. 2002;9:333–344. doi: 10.1016/s1074-5521(02)00115-1. [DOI] [PubMed] [Google Scholar]
- 84.Patel HM, Walsh CT. Biochemistry. 2001;40:9023–9031. doi: 10.1021/bi010519n. [DOI] [PubMed] [Google Scholar]
- 85.Rice LB. J Infect Dis. 2008;197:1079–1081. doi: 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- 86.Boucher HW, Talbot GH, Benjamin DK, Jr, Bradley J, Guidos RJ, Jones RN, Murray BE, Bonomo RA, Gilbert D A. Infectious Diseases Society of. Clin Infect Dis. 2013;56:1685–1694. doi: 10.1093/cid/cit152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Stavenger RA, Winterhalter M. Science translational medicine. 2014;6 doi: 10.1126/scitranslmed.3008605. 228ed227. [DOI] [PubMed] [Google Scholar]
- 88.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, Scheld M, Spellberg B, Bartlett J. Clin Infect Dis. 2009;48:1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 89.Pendleton JN, Gorman SP, Gilmore BF. Expert Rev Anti Infect Ther. 2013;11:297–308. doi: 10.1586/eri.13.12. [DOI] [PubMed] [Google Scholar]
- 90.Donia MS, Fischbach MA. Science. 2015;349:1254766. doi: 10.1126/science.1254766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Garg N, Luzzatto-Knaan T, Melnik AV, Caraballo-Rodriguez AM, Floros DJ, Petras D, Gregor R, Dorrestein PC, Phelan VV. Nat Prod Rep. 2016 doi: 10.1039/c6np00063k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gilbert JA, Quinn RA, Debelius J, Xu ZZ, Morton J, Garg N, Jansson JK, Dorrestein PC, Knight R. Nature. 2016;535:94–103. doi: 10.1038/nature18850. [DOI] [PubMed] [Google Scholar]
- 93.Koppel N, Balskus EP. Cell Chem Biol. 2016;23:18–30. doi: 10.1016/j.chembiol.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 94.Trautman EP, Crawford JM. Curr Top Med Chem. 2016;16:1705–1716. doi: 10.2174/1568026616666151012111046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zipperer A, Konnerth MC, Laux C, Berscheid A, Janek D, Weidenmaier C, Burian M, Schilling NA, Slavetinsky C, Marschal M, Willmann M, Kalbacher H, Schittek B, Brotz-Oesterhelt H, Grond S, Peschel A, Krismer B. Nature. 2016;535:511–516. doi: 10.1038/nature18634. [DOI] [PubMed] [Google Scholar]
- 96.Medema MH, Fischbach MA. Nat Chem Biol. 2015;11:639–648. doi: 10.1038/nchembio.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Medema MH, Kottmann R, Yilmaz P, Cummings M, Biggins JB, Blin K, de Bruijn I, Chooi YH, Claesen J, Coates RC, Cruz-Morales P, Duddela S, Dusterhus S, Edwards DJ, Fewer DP, Garg N, Geiger C, Gomez-Escribano JP, Greule A, Hadjithomas M, Haines AS, Helfrich EJ, Hillwig ML, Ishida K, Jones AC, Jones CS, Jungmann K, Kegler C, Kim HU, Kotter P, Krug D, Masschelein J, Melnik AV, Mantovani SM, Monroe EA, Moore M, Moss N, Nutzmann HW, Pan G, Pati A, Petras D, Reen FJ, Rosconi F, Rui Z, Tian Z, Tobias NJ, Tsunematsu Y, Wiemann P, Wyckoff E, Yan X, Yim G, Yu F, Xie Y, Aigle B, Apel AK, Balibar CJ, Balskus EP, Barona-Gomez F, Bechthold A, Bode HB, Borriss R, Brady SF, Brakhage AA, Caffrey P, Cheng YQ, Clardy J, Cox RJ, De Mot R, Donadio S, Donia MS, van der Donk WA, Dorrestein PC, Doyle S, Driessen AJ, Ehling-Schulz M, Entian KD, Fischbach MA, Gerwick L, Gerwick WH, Gross H, Gust B, Hertweck C, Hofte M, Jensen SE, Ju J, Katz L, Kaysser L, Klassen JL, Keller NP, Kormanec J, Kuipers OP, Kuzuyama T, Kyrpides NC, Kwon HJ, Lautru S, Lavigne R, Lee CY, Linquan B, Liu X, Liu W, Luzhetskyy A, Mahmud T, Mast Y, Mendez C, Metsa-Ketela M, Micklefield J, Mitchell DA, Moore BS, Moreira LM, Muller R, Neilan BA, Nett M, Nielsen J, O'Gara F, Oikawa H, Osbourn A, Osburne MS, Ostash B, Payne SM, Pernodet JL, Petricek M, Piel J, Ploux O, Raaijmakers JM, Salas JA, Schmitt EK, Scott B, Seipke RF, Shen B, Sherman DH, Sivonen K, Smanski MJ, Sosio M, Stegmann E, Sussmuth RD, Tahlan K, Thomas CM, Tang Y, Truman AW, Viaud M, Walton JD, Walsh CT, Weber T, van Wezel GP, Wilkinson B, Willey JM, Wohlleben W, Wright GD, Ziemert N, Zhang C, Zotchev SB, Breitling R, Takano E, Glockner FO. Nat Chem Biol. 2015;11:625–631. doi: 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Blin K, Medema MH, Kottmann R, Lee SY, Weber T. Nucleic Acids Res. 2017;45:D555–D559. doi: 10.1093/nar/gkw960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Dejong CA, Chen GM, Li H, Johnston CW, Edwards MR, Rees PN, Skinnider MA, Webster AL, Magarvey NA. Nat Chem Biol. 2016;12:1007–1014. doi: 10.1038/nchembio.2188. [DOI] [PubMed] [Google Scholar]
- 100.Skinnider MA, Dejong CA, Rees PN, Johnston CW, Li H, Webster AL, Wyatt MA, Magarvey NA. Nucleic Acids Res. 2015;43:9645–9662. doi: 10.1093/nar/gkv1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang M, Carver JJ, Phelan VV, Sanchez LM, Garg N, Peng Y, Nguyen DD, Watrous J, Kapono CA, Luzzatto-Knaan T, Porto C, Bouslimani A, Melnik AV, Meehan MJ, Liu WT, Crusemann M, Boudreau PD, Esquenazi E, Sandoval-Calderon M, Kersten RD, Pace LA, Quinn RA, Duncan KR, Hsu CC, Floros DJ, Gavilan RG, Kleigrewe K, Northen T, Dutton RJ, Parrot D, Carlson EE, Aigle B, Michelsen CF, Jelsbak L, Sohlenkamp C, Pevzner P, Edlund A, McLean J, Piel J, Murphy BT, Gerwick L, Liaw CC, Yang YL, Humpf HU, Maansson M, Keyzers RA, Sims AC, Johnson AR, Sidebottom AM, Sedio BE, Klitgaard A, Larson CB, Boya PC, Torres-Mendoza D, Gonzalez DJ, Silva DB, Marques LM, Demarque DP, Pociute E, O'Neill EC, Briand E, Helfrich EJ, Granatosky EA, Glukhov E, Ryffel F, Houson H, Mohimani H, Kharbush JJ, Zeng Y, Vorholt JA, Kurita KL, Charusanti P, McPhail KL, Nielsen KF, Vuong L, Elfeki M, Traxler MF, Engene N, Koyama N, Vining OB, Baric R, Silva RR, Mascuch SJ, Tomasi S, Jenkins S, Macherla V, Hoffman T, Agarwal V, Williams PG, Dai J, Neupane R, Gurr J, Rodriguez AM, Lamsa A, Zhang C, Dorrestein K, Duggan BM, Almaliti J, Allard PM, Phapale P, Nothias LF, Alexandrov T, Litaudon M, Wolfender JL, Kyle JE, Metz TO, Peryea T, Nguyen DT, VanLeer D, Shinn P, Jadhav A, Muller R, Waters KM, Shi W, Liu X, Zhang L, Knight R, Jensen PR, Palsson BO, Pogliano K, Linington RG, Gutierrez M, Lopes NP, Gerwick WH, Moore BS, Dorrestein PC, Bandeira N. Nat Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ochsner UA, Wilderman PJ, Vasil AI, Vasil ML. Mol Microbiol. 2002;45:1277–1287. doi: 10.1046/j.1365-2958.2002.03084.x. [DOI] [PubMed] [Google Scholar]
- 103.Lebreton F, Willems RJL, Gilmore MS. In: Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Boston: 2014. [PubMed] [Google Scholar]
- 104.Kristich CJ, Rice LB, Arias CA. In: Enterococci: From Commensals to Leading Causes of Drug Resistant Infection. Gilmore MS, Clewell DB, Ike Y, Shankar N, editors. Boston: 2014. [PubMed] [Google Scholar]
- 105.Chiang HY, Perencevich EN, Nair R, Nelson RE, Samore M, Khader K, Chorazy ML, Herwaldt LA, Blevins A, Ward MA, Schweizer ML. Infect Control Hosp Epidemiol. 2017;38:203–215. doi: 10.1017/ice.2016.254. [DOI] [PubMed] [Google Scholar]
- 106.Lam MM, Seemann T, Bulach DM, Gladman SL, Chen H, Haring V, Moore RJ, Ballard S, Grayson ML, Johnson PD, Howden BP, Stinear TP. J Bacteriol. 2012;194:2334–2341. doi: 10.1128/JB.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Schaik W, Top J, Riley DR, Boekhorst J, Vrijenhoek JE, Schapendonk CM, Hendrickx AP, Nijman IJ, Bonten MJ, Tettelin H, Willems RJ. BMC Genomics. 2010;11:239. doi: 10.1186/1471-2164-11-239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 109.Kim EB, Jin GD, Lee JY, Choi YJ. PLoS One. 2016;11:e0153279. doi: 10.1371/journal.pone.0153279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Miller LS, Cho JS. Nat Rev Immunol. 2011;11:505–518. doi: 10.1038/nri3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Choo EJ, Chambers HF. Infect Chemother. 2016;48:267–273. doi: 10.3947/ic.2016.48.4.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zimmermann M, Fischbach MA. Chem Biol. 2010;17:925–930. doi: 10.1016/j.chembiol.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 113.Wyatt MA, Wang W, Roux CM, Beasley FC, Heinrichs DE, Dunman PM, Magarvey NA. Science. 2010;329:294–296. doi: 10.1126/science.1188888. [DOI] [PubMed] [Google Scholar]
- 114.Alvarez ME, White CB, Gregory J, Kydd GC, Harris A, Sun HH, Gillum AM, Cooper R. J Antibiot (Tokyo) 1995;48:1165–1167. doi: 10.7164/antibiotics.48.1165. [DOI] [PubMed] [Google Scholar]
- 115.Wilson DJ, Shi C, Teitelbaum AM, Gulick AM, Aldrich CC. Biochemistry. 2013;52:926–937. doi: 10.1021/bi301330q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sun F, Cho H, Jeong DW, Li C, He C, Bae T. PLoS One. 2010;5:e15703. doi: 10.1371/journal.pone.0015703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wyatt MA, Wang W, Roux CM, Beasley FC, Heinrichs DE, Dunman PM, Magarvey NA. Science. 2011;333:1381. doi: 10.1126/science.1188888. [DOI] [PubMed] [Google Scholar]
- 118.Secor PR, Jennings LK, James GA, Kirker KR, Pulcini ED, McInnerney K, Gerlach R, Livinghouse T, Hilmer JK, Bothner B, Fleckman P, Olerud JE, Stewart PS. PLoS One. 2012;7:e40973. doi: 10.1371/journal.pone.0040973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Grosz M, Kolter J, Paprotka K, Winkler AC, Schafer D, Chatterjee SS, Geiger T, Wolz C, Ohlsen K, Otto M, Rudel T, Sinha B, Fraunholz M. Cell Microbiol. 2014;16:451–465. doi: 10.1111/cmi.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Blattner S, Das S, Paprotka K, Eilers U, Krischke M, Kretschmer D, Remmele CW, Dittrich M, Muller T, Schuelein-Voelk C, Hertlein T, Mueller MJ, Huettel B, Reinhardt R, Ohlsen K, Rudel T, Fraunholz MJ. PLoS pathogens. 2016;12:e1005857. doi: 10.1371/journal.ppat.1005857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guo CJ, Chang FY, Wyche TP, Backus KM, Acker TM, Funabashi M, Taketani M, Donia MS, Nayfach S, Pollard KS, Craik CS, Cravatt BF, Clardy J, Voigt CA, Fischbach MA. Cell. 2017;168:517–526 e518. doi: 10.1016/j.cell.2016.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Keynan Y, Rubinstein E. International journal of antimicrobial agents. 2007;30:385–389. doi: 10.1016/j.ijantimicag.2007.06.019. [DOI] [PubMed] [Google Scholar]
- 123.Kuehn BM. JAMA. 2013;309:1573–1574. doi: 10.1001/jama.2013.2922. [DOI] [PubMed] [Google Scholar]
- 124.Lee GC, Burgess DS. Annals of clinical microbiology and antimicrobials. 2012;11:32. doi: 10.1186/1476-0711-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Prabaker K, Weinstein RA. Current opinion in critical care. 2011;17:472–479. doi: 10.1097/MCC.0b013e32834a4b03. [DOI] [PubMed] [Google Scholar]
- 126.Shon AS, Bajwa RP, Russo TA. Virulence. 2013;4:107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Shon AS, Russo TA. Future microbiology. 2012;7:669–671. doi: 10.2217/fmb.12.43. [DOI] [PubMed] [Google Scholar]
- 128.Liu Y, Li XY, Wan LG, Jiang WY, Yang JH, Li FQ. Microb Drug Resist. 2014;20:150–155. doi: 10.1089/mdr.2013.0107. [DOI] [PubMed] [Google Scholar]
- 129.Siu LK, Huang DB, Chiang T. BMC infectious diseases. 2014;14:176. doi: 10.1186/1471-2334-14-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pollack JR, Neilands JB. Biochem Biophys Res Commun. 1970;38:989–992. doi: 10.1016/0006-291x(70)90819-3. [DOI] [PubMed] [Google Scholar]
- 131.O'Brien IG, Cox GB, Gibson F. Biochim Biophys Acta. 1970;201:453–460. doi: 10.1016/0304-4165(70)90165-0. [DOI] [PubMed] [Google Scholar]
- 132.Crosa JH, Walsh CT. Microbiol Mol Biol Rev. 2002;66:223–249. doi: 10.1128/MMBR.66.2.223-249.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Walsh CT, Liu J, Rusnak F, Sakaitani M. Chem Rev. 1990;90:1105–1129. [Google Scholar]
- 134.Raymond KN, Dertz EA, Kim SS. Proc Natl Acad Sci U S A. 2003;100:3584–3588. doi: 10.1073/pnas.0630018100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Harris WR, Carrano CJ, Cooper SR, Sofen SR, Avdeef AE, McArdle JV, Raymond KN. J Am Chem Soc. 1979;101:6097–6104. [Google Scholar]
- 136.Liu J, Duncan K, Walsh CT. J Bacteriol. 1989;171:791–798. doi: 10.1128/jb.171.2.791-798.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nahlik MS, Brickman TJ, Ozenberger BA, McIntosh MA. J Bacteriol. 1989;171:784–790. doi: 10.1128/jb.171.2.784-790.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rusnak F, Faraci WS, Walsh CT. Biochemistry. 1989;28:6827–6835. doi: 10.1021/bi00443a008. [DOI] [PubMed] [Google Scholar]
- 139.Rusnak F, Sakaitani M, Drueckhammer D, Reichert J, Walsh CT. Biochemistry. 1991;30:2916–2927. doi: 10.1021/bi00225a027. [DOI] [PubMed] [Google Scholar]
- 140.Drake EJ, Nicolai DA, Gulick AM. Chem Biol. 2006;13:409–419. doi: 10.1016/j.chembiol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 141.Sridharan S, Howard N, Kerbarh O, Blaszczyk M, Abell C, Blundell TL. J Mol Biol. 2010;397:290–300. doi: 10.1016/j.jmb.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 142.Sundlov JA, Garringer JA, Carney JM, Reger AS, Drake EJ, Duax WL, Gulick AM. Acta Crystallogr D Biol Crystallogr. 2006;62:734–740. doi: 10.1107/S0907444906015824. [DOI] [PubMed] [Google Scholar]
- 143.Quadri LE. Mol Microbiol. 2000;37:1–12. doi: 10.1046/j.1365-2958.2000.01941.x. [DOI] [PubMed] [Google Scholar]
- 144.Kerbarh O, Bulloch EM, Payne RJ, Sahr T, Rebeille F, Abell C. Biochem Soc Trans. 2005;33:763–766. doi: 10.1042/BST0330763. [DOI] [PubMed] [Google Scholar]
- 145.Liu J, Quinn N, Berchtold GA, Walsh CT. Biochemistry. 1990;29:1417–1425. doi: 10.1021/bi00458a012. [DOI] [PubMed] [Google Scholar]
- 146.Gehring AM, Bradley KA, Walsh CT. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 147.Sakaitani M, Rusnak F, Quinn NR, Tu C, Frigo TB, Berchtold GA, Walsh CT. Biochemistry. 1990;29:6789–6798. doi: 10.1021/bi00481a006. [DOI] [PubMed] [Google Scholar]
- 148.Meneely KM, Sundlov JA, Gulick AM, Moran GR, Lamb AL. J Am Chem Soc. 2016;138:9277–9293. doi: 10.1021/jacs.6b05134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Meneely KM, Luo Q, Lamb AL. Arch Biochem Biophys. 2013;539:70–80. doi: 10.1016/j.abb.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Penwell WF, Arivett BA, Actis LA. PLoS One. 2012;7:e36493. doi: 10.1371/journal.pone.0036493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ehmann DE, Shaw-Reid CA, Losey HC, Walsh CT. Proc Natl Acad Sci U S A. 2000;97:2509–2514. doi: 10.1073/pnas.040572897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lodge JM, Williams P, Brown MR. J Bacteriol. 1986;165:353–356. doi: 10.1128/jb.165.2.353-356.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Perry RD, San Clemente CL. J Bacteriol. 1979;140:1129–1132. doi: 10.1128/jb.140.3.1129-1132.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Haag H, Hantke K, Drechsel H, Stojiljkovic I, Jung G, Zahner H. J Gen Microbiol. 1993;139:2159–2165. doi: 10.1099/00221287-139-9-2159. [DOI] [PubMed] [Google Scholar]
- 155.Chambers CE, McIntyre DD, Mouck M, Sokol PA. Biometals. 1996;9:157–167. doi: 10.1007/BF00144621. [DOI] [PubMed] [Google Scholar]
- 156.Drechsel H, Stephan H, Lotz R, Haag H, Zahner H, Hantke K, Jung G. Liebigs Ann. 1995:1727–1733. [Google Scholar]
- 157.Gehring AM, DeMoll E, Fetherston JD, Mori I, Mayhew GF, Blattner FR, Walsh CT, Perry RD. Chem Biol. 1998;5:573–586. doi: 10.1016/s1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 158.Meneely KM, Ronnebaum TA, Riley AP, Prisinzano TE, Lamb AL. Biochemistry. 2016;55:5423–5433. doi: 10.1021/acs.biochem.6b00735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Gene. 2004;337:189–198. doi: 10.1016/j.gene.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 160.Müller SI, Valdebenito M, Hantke K. Biometals. 2009;22:691–695. doi: 10.1007/s10534-009-9217-4. [DOI] [PubMed] [Google Scholar]
- 161.Bachman MA, Lenio S, Schmidt L, Oyler JE, Weiser JN. mBio. 2012;3 doi: 10.1128/mBio.00224-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bachman MA, Oyler JE, Burns SH, Caza M, Lepine F, Dozois CM, Weiser JN. Infect Immun. 2011;79:3309–3316. doi: 10.1128/IAI.05114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Valdebenito M, Crumbliss AL, Winkelmann G, Hantke K. International journal of medical microbiology : IJMM. 2006;296:513–520. doi: 10.1016/j.ijmm.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 164.Lawlor MS, O'Connor C, Miller VL. Infect Immun. 2007;75:1463–1472. doi: 10.1128/IAI.00372-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Russo TA, Olson R, Macdonald U, Metzger D, Maltese LM, Drake EJ, Gulick AM. Infect Immun. 2014;82:2356–2367. doi: 10.1128/IAI.01667-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Russo TA, Olson R, MacDonald U, Beanan J, Davidson BA. Infect Immun. 2015 doi: 10.1128/IAI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nougayrede JP, Homburg S, Taieb F, Boury M, Brzuszkiewicz E, Gottschalk G, Buchrieser C, Hacker J, Dobrindt U, Oswald E. Science. 2006;313:848–851. doi: 10.1126/science.1127059. [DOI] [PubMed] [Google Scholar]
- 168.Conlan S, Deming C, Tsai YC, Lau AF, Dekker JP, Korlach J, Segre JA. Genome announcements. 2014;2 doi: 10.1128/genomeA.01332-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Struve C, Roe CC, Stegger M, Stahlhut SG, Hansen DS, Engelthaler DM, Andersen PS, Driebe EM, Keim P, Krogfelt KA. mBio. 2015;6:e00630. doi: 10.1128/mBio.00630-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Putze J, Hennequin C, Nougayrede JP, Zhang W, Homburg S, Karch H, Bringer MA, Fayolle C, Carniel E, Rabsch W, Oelschlaeger TA, Oswald E, Forestier C, Hacker J, Dobrindt U. Infect Immun. 2009;77:4696–4703. doi: 10.1128/IAI.00522-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Balskus EP. Nat Prod Rep. 2015;32:1534–1540. doi: 10.1039/c5np00091b. [DOI] [PubMed] [Google Scholar]
- 172.Brotherton CA, Balskus EP. J Am Chem Soc. 2013;135:3359–3362. doi: 10.1021/ja312154m. [DOI] [PubMed] [Google Scholar]
- 173.Bian X, Fu J, Plaza A, Herrmann J, Pistorius D, Stewart AF, Zhang Y, Muller R. Chembiochem. 2013;14:1194–1197. doi: 10.1002/cbic.201300208. [DOI] [PubMed] [Google Scholar]
- 174.Vizcaino MI, Crawford JM. Nature chemistry. 2015;7:411–417. doi: 10.1038/nchem.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Brotherton CA, Wilson M, Byrd G, Balskus EP. Org Lett. 2015;17:1545–1548. doi: 10.1021/acs.orglett.5b00432. [DOI] [PubMed] [Google Scholar]
- 176.Bian XY, Plaza A, Zhang YM, Muller R. Chemical Science. 2015;6:3154–3160. doi: 10.1039/c5sc00101c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Trautman EP, Healy AR, Shine EE, Herzon SB, Crawford JM. J Am Chem Soc. 2017;139:4195–4201. doi: 10.1021/jacs.7b00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Mousa JJ, Newsome RC, Yang Y, Jobin C, Bruner SD. Biochem Biophys Res Commun. 2017;482:1233–1239. doi: 10.1016/j.bbrc.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 179.Vizcaino MI, Engel P, Trautman E, Crawford JM. J Am Chem Soc. 2014;136:9244–9247. doi: 10.1021/ja503450q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Dubois D, Baron O, Cougnoux A, Delmas J, Pradel N, Boury M, Bouchon B, Bringer MA, Nougayrede JP, Oswald E, Bonnet R. J Biol Chem. 2011;286:35562–35570. doi: 10.1074/jbc.M111.221960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Brachmann AO, Garcie C, Wu V, Martin P, Ueoka R, Oswald E, Piel J. Chem Commun (Camb) 2015;51:13138–13141. doi: 10.1039/c5cc02718g. [DOI] [PubMed] [Google Scholar]
- 182.McConnell MJ, Actis L, Pachon J. FEMS Microbiol Rev. 2013;37:130–155. doi: 10.1111/j.1574-6976.2012.00344.x. [DOI] [PubMed] [Google Scholar]
- 183.Peleg AY, Seifert H, Paterson DL. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Calhoun JH, Murray CK, Manring MM. Clin Orthop Relat Res. 2008;466:1356–1362. doi: 10.1007/s11999-008-0212-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Keen EF, 3rd, Robinson BJ, Hospenthal DR, Aldous WK, Wolf SE, Chung KK, Murray CK. Burns. 2010;36:819–825. doi: 10.1016/j.burns.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 186.Karageorgopoulos DE, Falagas ME. The Lancet infectious diseases. 2008;8:751–762. doi: 10.1016/S1473-3099(08)70279-2. [DOI] [PubMed] [Google Scholar]
- 187.Clemmer KM, Bonomo RA, Rather PN. Microbiology. 2011;157:2534–2544. doi: 10.1099/mic.0.049791-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Eijkelkamp BA, Stroeher UH, Hassan KA, Papadimitrious MS, Paulsen IT, Brown MH. FEMS Microbiol Lett. 2011;323:44–51. doi: 10.1111/j.1574-6968.2011.02362.x. [DOI] [PubMed] [Google Scholar]
- 189.Harding CM, Tracy EN, Carruthers MD, Rather PN, Actis LA, Munson RS., Jr mBio. 2013;4 doi: 10.1128/mBio.00360-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Poirel L, Bonnin RA, Nordmann P. IUBMB life. 2011;63:1061–1067. doi: 10.1002/iub.532. [DOI] [PubMed] [Google Scholar]
- 191.Kim HU, Kim TY, Lee SY. Mol Biosyst. 2010;6:339–348. doi: 10.1039/b916446d. [DOI] [PubMed] [Google Scholar]
- 192.Umland TC, Schultz LW, Macdonald U, Beanan JM, Olson R, Russo TA. mBio. 2012;3 doi: 10.1128/mBio.00113-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sattely ES, Walsh CT. J Am Chem Soc. 2008;130:12282–12284. doi: 10.1021/ja804499r. [DOI] [PubMed] [Google Scholar]
- 194.Shapiro JA, Wencewicz TA. ACS Infect Dis. 2016;2:157–168. doi: 10.1021/acsinfecdis.5b00145. [DOI] [PubMed] [Google Scholar]
- 195.Dorsey CW, Tomaras AP, Connerly PL, Tolmasky ME, Crosa JH, Actis LA. Microbiology. 2004;150:3657–3667. doi: 10.1099/mic.0.27371-0. [DOI] [PubMed] [Google Scholar]
- 196.Mihara K, Tanabe T, Yamakawa Y, Funahashi T, Nakao H, Narimatsu S, Yamamoto S. Microbiology. 2004;150:2587–2597. doi: 10.1099/mic.0.27141-0. [DOI] [PubMed] [Google Scholar]
- 197.Wuest WM, Sattely ES, Walsh CT. J Am Chem Soc. 2009;131:5056–5057. doi: 10.1021/ja900815w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Drake EJ, Duckworth BP, Neres J, Aldrich CC, Gulick AM. Biochemistry. 2010;49:9292–9305. doi: 10.1021/bi101226n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Lamb AL. Biochim Biophys Acta. 2015;1854:1054–1070. doi: 10.1016/j.bbapap.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Miethke M, Marahiel MA. Microbiol Mol Biol Rev. 2007;71:413–451. doi: 10.1128/MMBR.00012-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gaddy JA, Arivett BA, McConnell MJ, Lopez-Rojas R, Pachon J, Actis LA. Infect Immun. 2012;80:1015–1024. doi: 10.1128/IAI.06279-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Fleming ID, Krezalek MA, Belogortseva N, Zaborin A, Defazio J, Chandrasekar L, Actis LA, Zaborina O, Alverdy JC. J Trauma Acute Care Surg. 2017;82:557–565. doi: 10.1097/TA.0000000000001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Proschak A, Lubuta P, Grun P, Lohr F, Wilharm G, De Berardinis V, Bode HB. Chembiochem. 2013;14:633–638. doi: 10.1002/cbic.201200764. [DOI] [PubMed] [Google Scholar]
- 204.Meneely KM, Lamb AL. Biochemistry. 2007;46:11930–11937. doi: 10.1021/bi700932q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Burrows LL. Annu Rev Microbiol. 2012;66:493–520. doi: 10.1146/annurev-micro-092611-150055. [DOI] [PubMed] [Google Scholar]
- 206.Mattick JS. Annu Rev Microbiol. 2002;56:289–314. doi: 10.1146/annurev.micro.56.012302.160938. [DOI] [PubMed] [Google Scholar]
- 207.Rumbo-Feal S, Gomez MJ, Gayoso C, Alvarez-Fraga L, Cabral MP, Aransay AM, Rodriguez-Ezpeleta N, Fullaondo A, Valle J, Tomas M, Bou G, Poza M. PLoS One. 2013;8:e72968. doi: 10.1371/journal.pone.0072968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Giles SK, Stroeher UH, Eijkelkamp BA, Brown MH. BMC Microbiol. 2015;15:116. doi: 10.1186/s12866-015-0440-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Allen CL, Gulick AM. Acta Crystallogr D Biol Crystallogr. 2014;70:1718–1725. doi: 10.1107/S1399004714008311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rumbo-Feal S, Perez A, Ramelot TA, Alvarez-Fraga L, Vallejo JA, Beceiro A, Ohneck EJ, Arivett BA, Merino M, Fiester SE, Kennedy MA, Actis LA, Bou G, Poza M. Front Cell Infect Microbiol. 2017;7:108. doi: 10.3389/fcimb.2017.00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Adams MD, Goglin K, Molyneaux N, Hujer KM, Lavender H, Jamison JJ, MacDonald IJ, Martin KM, Russo T, Campagnari AA, Hujer AM, Bonomo RA, Gill SR. J Bacteriol. 2008;190:8053–8064. doi: 10.1128/JB.00834-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Lyczak JB, Cannon CL, Pier GB. Microbes and infection / Institut Pasteur. 2000;2:1051–1060. doi: 10.1016/s1286-4579(00)01259-4. [DOI] [PubMed] [Google Scholar]
- 213.Winstanley C, O'Brien S, Brockhurst MA. Trends Microbiol. 2016;24:327–337. doi: 10.1016/j.tim.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Poole K. Front Microbiol. 2011;2:65. doi: 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Welsh MA, Blackwell HE. FEMS Microbiol Rev. 2016;40:774–794. doi: 10.1093/femsre/fuw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Schalk IJ, Guillon L. Environmental microbiology. 2013;15:1661–1673. doi: 10.1111/1462-2920.12013. [DOI] [PubMed] [Google Scholar]
- 217.Visca P, Imperi F, Lamont IL. Trends Microbiol. 2007;15:22–30. doi: 10.1016/j.tim.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 218.Cezard C, Farvacques N, Sonnet P. Curr Med Chem. 2015;22:165–186. doi: 10.2174/0929867321666141011194624. [DOI] [PubMed] [Google Scholar]
- 219.Vandenende CS, Vlasschaert M, Seah SY. J Bacteriol. 2004;186:5596–5602. doi: 10.1128/JB.186.17.5596-5602.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Visca P, Ciervo A, Orsi N. J Bacteriol. 1994;176:1128–1140. doi: 10.1128/jb.176.4.1128-1140.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ge L, Seah SY. J Bacteriol. 2006;188:7205–7210. doi: 10.1128/JB.00949-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.McMorran BJ, Kumara HM, Sullivan K, Lamont IL. Microbiology. 2001;147:1517–1524. doi: 10.1099/00221287-147-6-1517. [DOI] [PubMed] [Google Scholar]
- 223.Drake EJ, Cao J, Qu J, Shah MB, Straubinger RM, Gulick AM. J Biol Chem. 2007;282:20425–20434. doi: 10.1074/jbc.M611833200. [DOI] [PubMed] [Google Scholar]
- 224.Baltz RH. Journal of industrial microbiology & biotechnology. 2011;38:1747–1760. doi: 10.1007/s10295-011-1022-8. [DOI] [PubMed] [Google Scholar]
- 225.Felnagle EA, Barkei JJ, Park H, Podevels AM, McMahon MD, Drott DW, Thomas MG. Biochemistry. 2010;49:8815–8817. doi: 10.1021/bi1012854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhang W, Heemstra JR, Jr, Walsh CT, Imker HJ. Biochemistry. 2010;49:9946–9947. doi: 10.1021/bi101539b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Herbst DA, Boll B, Zocher G, Stehle T, Heide L. J Biol Chem. 2013;288:1991–2003. doi: 10.1074/jbc.M112.420182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Challis GL, Ravel J, Townsend CA. Chem Biol. 2000;7:211–224. doi: 10.1016/s1074-5521(00)00091-0. [DOI] [PubMed] [Google Scholar]
- 229.Stachelhaus T, Mootz HD, Marahiel MA. Chem Biol. 1999;6:493–505. doi: 10.1016/S1074-5521(99)80082-9. [DOI] [PubMed] [Google Scholar]
- 230.Kim Y, Hol WG. Chem Biol. 2001;8:1253–1264. doi: 10.1016/s1074-5521(01)00092-8. [DOI] [PubMed] [Google Scholar]
- 231.Drake EJ, Gulick AM. ACS Chem Biol. 2011;6:1277–1286. doi: 10.1021/cb2002973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Yeterian E, Martin LW, Guillon L, Journet L, Lamont IL, Schalk IJ. Amino Acids. 2010;38:1447–1459. doi: 10.1007/s00726-009-0358-0. [DOI] [PubMed] [Google Scholar]
- 233.Hannauer M, Schafer M, Hoegy F, Gizzi P, Wehrung P, Mislin GL, Budzikiewicz H, Schalk IJ. FEBS Lett. 2012;586:96–101. doi: 10.1016/j.febslet.2011.12.004. [DOI] [PubMed] [Google Scholar]
- 234.Clevenger KD, Mascarenhas R, Catlin D, Wu R, Kelleher NL, Drake EJ, Gulick AM, Liu D, Fast W. ACS Chem Biol. 2017;12:643–647. doi: 10.1021/acschembio.7b00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Dorrestein P, Begley TP. Bioorg Chem. 2005;33:136–148. doi: 10.1016/j.bioorg.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 236.Dorrestein PC, Poole K, Begley TP. Org Lett. 2003;5:2215–2217. doi: 10.1021/ol034531e. [DOI] [PubMed] [Google Scholar]
- 237.Nadal-Jimenez P, Koch G, Reis CR, Muntendam R, Raj H, Jeronimus-Stratingh CM, Cool RH, Quax WJ. J Bacteriol. 2014 doi: 10.1128/JB.01376-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Ringel MT, Drager G, Bruser T. J Biol Chem. 2016;291:23929–23938. doi: 10.1074/jbc.M116.755611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Drake EJ, Gulick AM. Acta Crystallogr F Struct Biol Commun. 2016;72:403–408. doi: 10.1107/S2053230X16006257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Yuan Z, Gao F, Bai G, Xia H, Gu L, Xu S. Biochem Biophys Res Commun. 2017;484:195–201. doi: 10.1016/j.bbrc.2016.12.181. [DOI] [PubMed] [Google Scholar]
- 241.Reid DW, Anderson GJ, Lamont IL. Am J Physiol Lung Cell Mol Physiol. 2009;297:L795–802. doi: 10.1152/ajplung.00132.2009. [DOI] [PubMed] [Google Scholar]
- 242.Konings AF, Martin LW, Sharples KJ, Roddam LF, Latham R, Reid DW, Lamont IL. Infect Immun. 2013;81:2697–2704. doi: 10.1128/IAI.00418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Granato ET, Harrison F, Kummerli R, Ross-Gillespie A. Front Microbiol. 2016;7:1952. doi: 10.3389/fmicb.2016.01952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Imperi F, Massai F, Facchini M, Frangipani E, Visaggio D, Leoni L, Bragonzi A, Visca P. Proc Natl Acad Sci U S A. 2013;110:7458–7463. doi: 10.1073/pnas.1222706110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Cornelis P, Dingemans J. Front Cell Infect Microbiol. 2013;3:75. doi: 10.3389/fcimb.2013.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Cornelis P. Appl Microbiol Biotechnol. 2010;86:1637–1645. doi: 10.1007/s00253-010-2550-2. [DOI] [PubMed] [Google Scholar]
- 247.Cox CD, Rinehart KL, Jr, Moore ML, Cook JC., Jr Proc Natl Acad Sci U S A. 1981;78:4256–4260. doi: 10.1073/pnas.78.7.4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Cobessi D, Celia H, Pattus F. J Mol Biol. 2005;352:893–904. doi: 10.1016/j.jmb.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 249.Schlegel K, Lex J, Taraz K, Budzikiewicz H. Z Naturforsch C. 2006;61:263–266. doi: 10.1515/znc-2006-3-418. [DOI] [PubMed] [Google Scholar]
- 250.Serino L, Reimmann C, Visca P, Beyeler M, Chiesa VD, Haas D. J Bacteriol. 1997;179:248–257. doi: 10.1128/jb.179.1.248-257.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Reimmann C, Serino L, Beyeler M, Haas D. Microbiology. 1998;144(Pt 11):3135–3148. doi: 10.1099/00221287-144-11-3135. [DOI] [PubMed] [Google Scholar]
- 252.Reimmann C, Patel HM, Serino L, Barone M, Walsh CT, Haas D. J Bacteriol. 2001;183:813–820. doi: 10.1128/JB.183.3.813-820.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Reimmann C, Patel HM, Walsh CT, Haas D. J Bacteriol. 2004;186:6367–6373. doi: 10.1128/JB.186.19.6367-6373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Gaille C, Reimmann C, Haas D. J Biol Chem. 2003;278:16893–16898. doi: 10.1074/jbc.M212324200. [DOI] [PubMed] [Google Scholar]
- 255.Gaille C, Kast P, Haas D. J Biol Chem. 2002;277:21768–21775. doi: 10.1074/jbc.M202410200. [DOI] [PubMed] [Google Scholar]
- 256.Quadri LE, Keating TA, Patel HM, Walsh CT. Biochemistry. 1999;38:14941–14954. doi: 10.1021/bi991787c. [DOI] [PubMed] [Google Scholar]
- 257.Lee DG, Urbach JM, Wu G, Liberati NT, Feinbaum RL, Miyata S, Diggins LT, He J, Saucier M, Deziel E, Friedman L, Li L, Grills G, Montgomery K, Kucherlapati R, Rahme LG, Ausubel FM. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Maspoli A, Wenner N, Mislin GL, Reimmann C. Biometals. 2014;27:559–573. doi: 10.1007/s10534-014-9729-4. [DOI] [PubMed] [Google Scholar]
- 259.Harrison AJ, Yu M, Gardenborg T, Middleditch M, Ramsay RJ, Baker EN, Lott JS. J Bacteriol. 2006;188:6081–6091. doi: 10.1128/JB.00338-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Dumas Z, Ross-Gillespie A, Kummerli R. Proc Biol Sci. 2013;280:20131055. doi: 10.1098/rspb.2013.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Pruess DL, Scannell JP, Kellett M, Ax HA, Janecek J, Williams TH, Stempel A, Berger J. J Antibiot (Tokyo) 1974;27:229–233. doi: 10.7164/antibiotics.27.229. [DOI] [PubMed] [Google Scholar]
- 262.Scannel JP, Pruess DL, Demny TC, Sello LH, Williams T. J Antibiot (Tokyo) 1972;25:122–127. doi: 10.7164/antibiotics.25.122. [DOI] [PubMed] [Google Scholar]
- 263.Lee X, Azevedo MD, Armstrong DJ, Banowetz GM, Reimmann C. Environ Microbiol Rep. 2013;5:83–89. doi: 10.1111/j.1758-2229.2012.00395.x. [DOI] [PubMed] [Google Scholar]
- 264.Gehring H, Rando RR, Christen P. Biochemistry. 1977;16:4832–4836. doi: 10.1021/bi00641a012. [DOI] [PubMed] [Google Scholar]
- 265.Rando RR. Nature. 1974;250:586–587. doi: 10.1038/250586a0. [DOI] [PubMed] [Google Scholar]
- 266.Lee J, Wu J, Deng Y, Wang J, Wang C, Wang J, Chang C, Dong Y, Williams P, Zhang LH. Nat Chem Biol. 2013;9:339–343. doi: 10.1038/nchembio.1225. [DOI] [PubMed] [Google Scholar]
- 267.Rojas Murcia N, Lee X, Waridel P, Maspoli A, Imker HJ, Chai T, Walsh CT, Reimmann C. Front Microbiol. 2015;6:170. doi: 10.3389/fmicb.2015.00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Lee X, Fox A, Sufrin J, Henry H, Majcherczyk P, Haas D, Reimmann C. J Bacteriol. 2010;192:4251–4255. doi: 10.1128/JB.00492-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Hausinger RP. Crit Rev Biochem Mol Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 270.Schuster M, Lostroh CP, Ogi T, Greenberg EP. J Bacteriol. 2003;185:2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Wagner VE, Bushnell D, Passador L, Brooks AI, Iglewski BH. J Bacteriol. 2003;185:2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Chugani S, Kim BS, Phattarasukol S, Brittnacher MJ, Choi SH, Harwood CS, Greenberg EP. Proc Natl Acad Sci U S A. 2012;109:E2823–2831. doi: 10.1073/pnas.1214128109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Bijtenhoorn P, Mayerhofer H, Muller-Dieckmann J, Utpatel C, Schipper C, Hornung C, Szesny M, Grond S, Thurmer A, Brzuszkiewicz E, Daniel R, Dierking K, Schulenburg H, Streit WR. PLoS One. 2011;6:e26278. doi: 10.1371/journal.pone.0026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Yan Q, Philmus B, Chang JH, Loper JE. Elife. 2017;6 doi: 10.7554/eLife.22835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Salunkhe P, Smart CH, Morgan JA, Panagea S, Walshaw MJ, Hart CA, Geffers R, Tummler B, Winstanley C. J Bacteriol. 2005;187:4908–4920. doi: 10.1128/JB.187.14.4908-4920.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Kukavica-Ibrulj I, Bragonzi A, Paroni M, Winstanley C, Sanschagrin F, O'Toole GA, Levesque RC. J Bacteriol. 2008;190:2804–2813. doi: 10.1128/JB.01572-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Huang X, Yan A, Zhang X, Xu Y. Gene. 2006;376:68–78. doi: 10.1016/j.gene.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 278.Nowak-Thompson B, Chaney N, Wing JS, Gould SJ, Loper JE. J Bacteriol. 1999;181:2166–2174. doi: 10.1128/jb.181.7.2166-2174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Thomas MG, Burkart MD, Walsh CT. Chem Biol. 2002;9:171–184. doi: 10.1016/s1074-5521(02)00100-x. [DOI] [PubMed] [Google Scholar]
- 280.Dorrestein PC, Yeh E, Garneau-Tsodikova S, Kelleher NL, Walsh CT. Proc Natl Acad Sci U S A. 2005;102:13843–13848. doi: 10.1073/pnas.0506964102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Jaremko MJ, Lee DJ, Opella SJ, Burkart MD. J Am Chem Soc. 2015;137:11546–11549. doi: 10.1021/jacs.5b04525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.de Bentzmann S, Giraud C, Bernard CS, Calderon V, Ewald F, Plesiat P, Nguyen C, Grunwald D, Attree I, Jeannot K, Fauvarque MO, Bordi C. PLoS pathogens. 2012;8:e1003052. doi: 10.1371/journal.ppat.1003052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Zaborin A, Gerdes S, Holbrook C, Liu DC, Zaborina OY, Alverdy JC. PLoS One. 2012;7:e34883. doi: 10.1371/journal.pone.0034883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Liu WY, Wong CF, Chung KM, Jiang JW, Leung FC. PLoS One. 2013;8:e74487. doi: 10.1371/journal.pone.0074487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Paauw A, Caspers MP, Schuren FH, Leverstein-van Hall MA, Deletoile A, Montijn RC, Verhoef J, Fluit AC. PLoS One. 2008;3:e3018. doi: 10.1371/journal.pone.0003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Ohad S, Block C, Kravitz V, Farber A, Pilo S, Breuer R, Rorman E. Journal of applied microbiology. 2014;116:1315–1321. doi: 10.1111/jam.12439. [DOI] [PubMed] [Google Scholar]
- 287.Ren Y, Ren Y, Zhou Z, Guo X, Li Y, Feng L, Wang L. J Bacteriol. 2010;192:2463–2464. doi: 10.1128/JB.00067-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Hoffmann H, Stindl S, Ludwig W, Stumpf A, Mehlen A, Monget D, Pierard D, Ziesing S, Heesemann J, Roggenkamp A, Schleifer KH. Journal of clinical microbiology. 2005;43:3297–3303. doi: 10.1128/JCM.43.7.3297-3303.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]