Abstract
We have Characterized overlapping cDNA clones encoding cGMP phosphodiesterase (PDE) α- and β-subunits of mouse retinal rod photoreceptors. The open reading frames predict an α-subunit of 100 kDa (856 residues), and a β-subunit of 99 kDa (853 residues). Sequence analysis of two of twelve β-subunit clones predicts the presence in the retina of an additional PDE, termed β′, which is generated by alternative splicing of the β-subunit gene. β′ differs from β only at the C-terminus being 55 residues shorter and lacking the Caax motif found at the C-termini of both the α- and β-subunits. A 300 residue segment thought to contain the active site is present in the C-terminal half of α, β and β′.
Keywords: Rod photoreceptor cGMP phosphodiesterase, cDNA cloning, Alternative splicing, Polymerase chain reaction (PCR), Mouse retina
1. INTRODUCTION
Phototransduction is a cascade of events that begins with absorption of light by rhodopsin, ultimately leading to the generation of a nerve impulse. In this cascade, rod photoreceptor cGMP phosphodiesterase (PDE) functions as a signal amplifier by rapidly hydrolyzing cGMP (reviewed in [7, 24]). PDE is thought to be a peripherally membrane-bound, heterotrimeric enzyme αβγ2 [3, 9, 10]. In dark adapted rods, the hydrolytic activity is greatly reduced due to an inhibitory constraint imposed by the PDE γ-subunit. Upon removal of γ, the turnover number for cGMP hydrolysis increases more than 20-fold. Depletion of cytoplasmic cGMP causes closure of cGMP-gated cation channels located in the plasma membrane and subsequent hyperpolarization of the rod photoreceptor cell.
Bovine and human cDNA clones encoding rod PDE α-subunits [18, 21, 22], and bovine PDE β-subunit cDNA clones [13, 14] have been characterized. The gene encoding the PDE β-subunit of mouse has recently been shown to be tightly linked to the rd locus on chromosome 5, and it has been suggested that a 300 bp insertion in the β-gene may be causative for the rd mutation [5, 8]. The α-subunit gene has been located on mouse chromosome 18 [8]. In this paper, we describe the primary structures of the α- and β-subunits of mouse rod PDE, and provide evidence by direct sequencing of PCR amplified genomic DNA that an additional isozyme, β′, is generated by alternative splicing of a large intron near the 3′-end of the coding region of the β-subunit.
2. MATERIALS AND METHODS
2.1. cDNA and PCR
Retinas of C57BL/6J mice were excised, and immediately dropped into liquid nitrogen. Poly(A) mRNA was isolated according to the Fasttrack procedure (Invitrogen) [2]. First strand cDNA suitable for PCR amplification was synthesized with AMV reverse transcriptase (Promega) using 1.5–2 μg RNA as a template and the 3′-T16·mcs primer as described [1]. The reaction product was diluted 10-fold with water and an aliquot used directly in a PCR reaction. PCR was performed according to standard protocols (Cetus) using Taq DNA polymerase (Promega).
2.2. Library screening and sequence analysis
A cDNA library in which first strand cDNA synthesis was initiated by both random hexanucleotide priming and oligo-dT priming was constructed in λzapII (Stratagene). Hybridizations were performed in 50% formamide containing 6 × SSC (0.9 M NaCl, 0.09 M Na citrate), 0.5% SDS, and 100 μg sheared salmon sperm DNA. Stringent hybridizations were carried out at 42°C (3 washings at 65°C in 2 M SSC/0.1% SDS, 1 M SSC/0.1% SDS, and 0.2 M SSC/0.1% SDS, 20 min each), and relaxed stringency hybridizations at 37°C with 3 washings at 50°C. Double stranded cDNA clones were sequenced with M13 universal and sequence specific oligonucleotide primers using the standard Sequenase protocol as described [21]. Oligonucleotides were synthesized on a PCR-MATE DNA synthesized (Applied Biosystems, Inc.) [1] and used without further purification. PCR products were purified by preparative agarose gel electrophoresis and sequenced directly after boiling 200–400 ng template and 100 ng primer for 3 min. Two μl of Sequenase 5 × buffer was added and standard sequenase reactions were performed. RNA sequencing was performed according to Geliebter [11].
3. RESULTS AND DISCUSSION
3.1. PDE cDNA clones
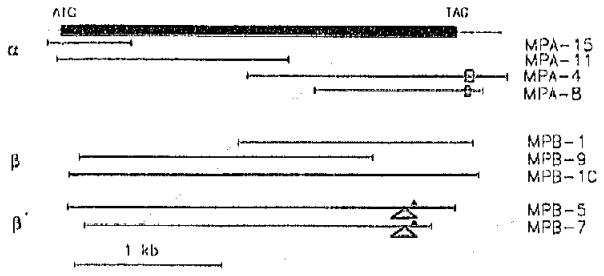
Screening a mouse retina cDNA library with a human PDE α subunit N-terminal probe (300 bp EcoRI/BamHI fragment of HPA1 [21]) and a bovine C-terminal probe (660 bp EcoRI fragment of the bovine sequence [21]) resulted in the isolation of overlapping α-subunit clones MPA-15, MPA-11, MPA-4, and MPA-8 (Fig. 1). The 3′-untranslated region of MPA-4 contains a variable number tandem repeats (VNTR) of (TTCTG)17, and MPA-8 of (TTCTG)9. The significance of this VNTR is unknown. Its conservation in human may prove useful for linkage studies as has been reported for (CA)n repeats [15].
Fig. 1.
Map of cDNA clones encoding the mouse PDE α-, β-and β′-polypeptides. The black box depicts the coding sequences of the α- and β-subunit. The extent of clones is indicated by bold-faced lines. The rectangles in the 3′ untranslated region of MPA-4 and MPA-8 represent (TTCTG)n repeats, where n = 17 in MPA-4 and n = 9 in MPA-8. The triangles in MPB-5 and MPB-7 symbolize a 10 bp insertion. The asterisk indicates the location of the first in-frame stop codon.
Using a combination of relaxed-stringency screening (sec Section 2) with the (C-terminal bovine α-subunit probe and stringent screening with PCR amplified full length bovine β-subunit [14] cDNA, we isolated a set of clones labeled MPB (Fig. 1). Sequence analysis allowed the β-subunit clones to be divided into two classes depending on the presence or absence of a 10 bp insertion near the 3′-end of the cDNA. The first class, comprising 12 clones (three of which, MPA-1, MPB-9, MPB-10 are shown in Fig, 1) represent putative PDE β-subunit clones as judged by their homology to the bovine β-subunit [14]. The second class contains two clones (MPB-5, MPB-7, Fig. 1) which predict a truncated version of the β-subunit, termed β′. Apart from the TTCTG VNTR and the 10 bp insertion, overlapping sequences of MPA, and MPB clones, respectively, are identical.
3.2. PDE α and β subunit sequences
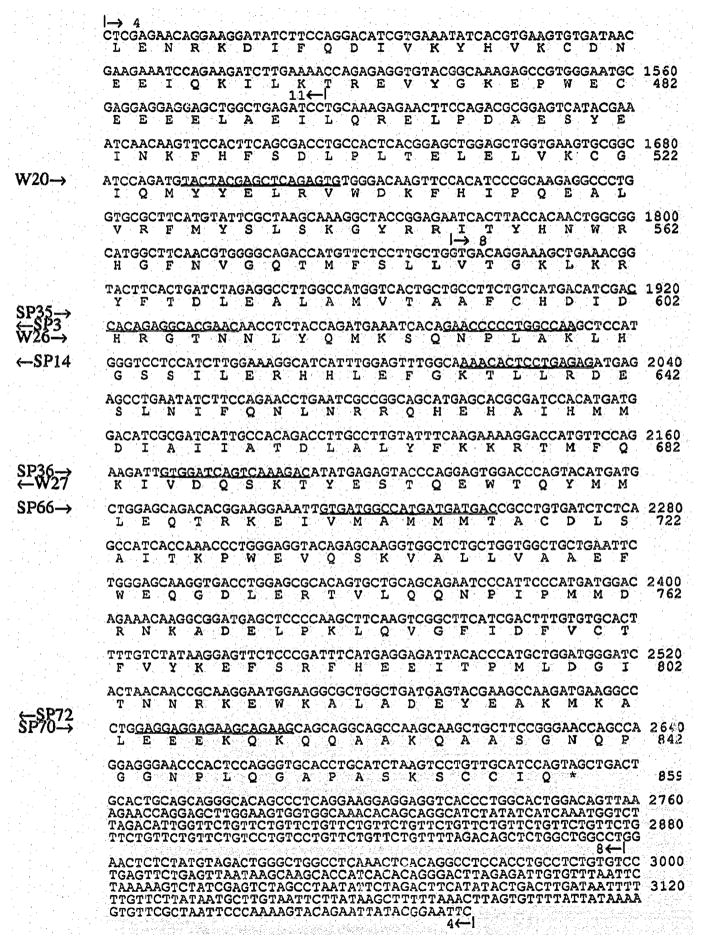
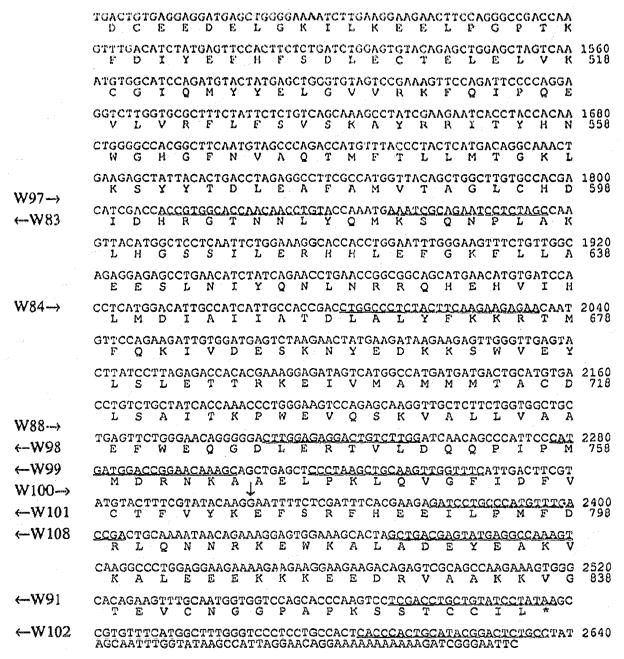
The α- and β-subunit cDNA sequences and their deduced amino acid sequences are shown in Fig 2 and 3A. The α-subunit sequence is a composite sequence of the 4 clones shown in Fig. 1, the β-subunit sequence is that of clone MPB-10 (Fig. 1). Clone MPB-10 starts at residue 21 lacking the first 14 nucleotides of the coding sequence. The sequence was completed by RNA sequencing extending an antisense oligonucleotide primer (W107) located at pos. 119–130 in Fig. 3.
Fig. 2.
cDNA and predicted amino acid sequence of the mouse PDE α-subunit. Primers used for DNA sequencing are underlined, and their sense or antisense direction is indicated on the left margin by arrows pointing to the right or left. Start and end points of overlapping clones (Fig. 1) are shown above the sequence.
Figure 3.
Fig. 3A. cDNA and predicted amino acid sequences of the mouse PDE β-subunit. The start point of clone MPB-10 is indicated above the sequence. Primers used for DNA and RNA sequencing, and for PCR amplification are underlined. The vertical arrow indicates the 10 bp insertion point.
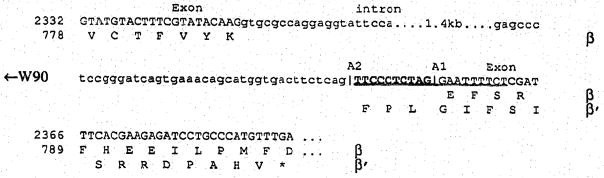
Fig. 3B. Junctions of the 1.4 kb intron that is alternatively spliced. Predicted amino acid sequences of the β-subunit and the β′-isozyme are shown below the sequence. The numbering on the left is according to Fig. 3A. W90 is an antisense, insertion specific primer. A1, junction one at the 3′ end of the intron used for generation of β; A2, junction two used for β′.
The α-subunit cDNA sequence predicts a polypeptide of 859 residues, exactly the same length as for bovine and human [21]. The predicted β-subunit is 856 residues in length, 3 residues longer than its bovine counterpart. Sequence similarity between mouse and bovine PDE subunits [13, 14, 18, 21] is at least 90%. The calculated molecular weights are 100000 and 99000, considerably larger than estimated by SDS-PAGE mobility [3]. Both subunits are acidic polypeptides with C-termini that conform to the Caax motif (C = cysteine, a = aliphatic, x = any residue) [16] signaling multistep posttranslational processing [12, 17].
C-terminal processing involving lipidation, proteolysis and carboxymethylation has been shown to occur on the bovine PDE α-subunit, but not on the β-subunit [25]. Since a Caax tetrapeptide is sufficient to signal processing [26], it appears likely that the β-subunit will also be processed. Lamin A undergoes Caax processing which is then followed by cleavage of 19 residues from the C-terminus [23]. Such proteolysis may follow β-subunit Caax processing which could account in part for the size discrepancy observed in the α- and β-subunits.
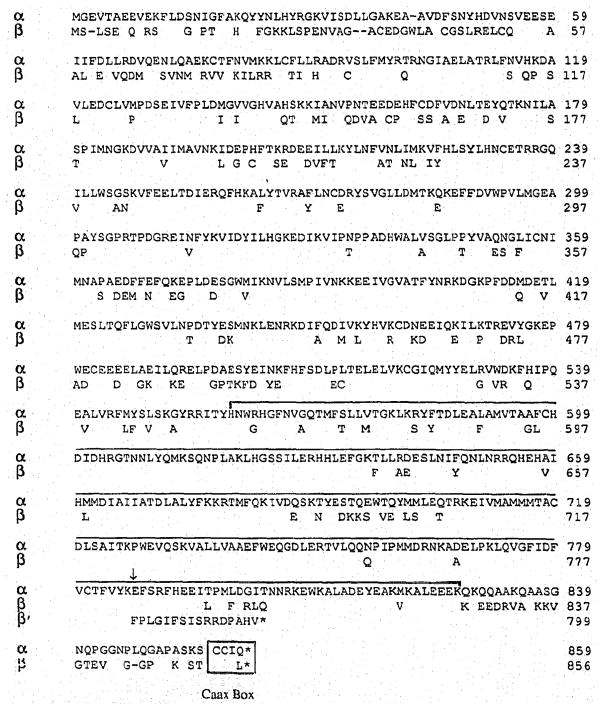
Alignment of the two amino acid sequences (Fig. 4) shows that the overall sequence similarity between the mouse PDE α- and β-subunits is 73%. The domain with greatest sequence similarity is located near the C-terminus (bracketed in Fig. 4), consisting of approximately 250 amino acids. This domain is thought to contain the catalytic site of several cNMP PDEs from yeast to man [4, 20]. The domains that are most dissimilar (Fig. 4) are at the N-terminus (first 100 residues) and at the C-terminus (last 30 residues).
Fig. 4.
Alignment of mouse PDE α- and β-subunit amino acid sequences. Gaps are indicated by a hyphen, identical residues by blanks. The C-terminal 16 residues of β′ are shown from the point (arrow) where the frame shift occurs in the cDNA sequence. An asterisk depicts a stop codon. The conserved domain thought to be involved in catalysis is bracketed. The Caax motif at the C-terminus is boxed.
3.3. The β′-isozyme is generated by alternative splicing
Two β cDNA clones (MPB-5 and MPB-7 in Fig. 1) have a 10 bp insertion (TTCCCTCTAG) near the C-terminus. This insertion conforms to the consensus sequence (Y)6XAG (Y = pyrimidine, X = any nucleotide) of the 3′ end of an intron [19]. The insertion causes a shift in the reading frame, and a stop codon is encountered 16 codons after the insertion (Fig. 3B). The predicted amino acid sequence of β′ thus would have a C-terminus different from β (Fig. 3A), be 55 residues shorter (Fig. 4), lack the Caax motif, but contain most of the catalytic domain. The existence of an mRNA in the retina poly (A) mRNA pool with the 10 bp insertion was verified by PCR amplification of normal adult mouse retina cDNA using an N-terminal primer common to both β and β′ (W89 in Fig. 3), and an insertion specific primer (W90 in Fig. 3B).
To verify that the insertion is due to an alternative acceptor site usage and not the product of another gene, we amplified a 1.4 kb intron with two exon primers W98 and W108 (Fig. 3A). Direct sequencing of the amplified DNA showed that the 3′ end of the intron has two intron/exon junctions, A1 and A2, that are preceded by (Y)6XAG consensus sequences (Fig. 3B). Junction A1 is preferentially used to produce the β-subunit mRNA, junction A2 to produce β′-mRNA. Although alternative splicing as a means to produce protein diversity has been shown to be a ubiquitous phenomenon from Drosophila to man [6], this is the first example of such a pathway in a gene involved in mammalian phototransduction. The cellular origin, and function of the truncated PDE is not known. To determine the cellular location, an antipeptide antibody specific for the C-terminus of β′ is currently being prepared.
Acknowledgments
This research was supported by grants from the National Eye Institute (EY08123), the National Retinitis Pigmentosa Foundation/Gund Foundation, the Retina Research Foundation (Houston) to W.B., and a NRSA fellowship (EY06172) and a grant from the Knight’s Templar Eye Foundation, Inc. to S.J.P. W.B. is a Jules and Doris Stein Research to Prevent Blindness Professor.
Footnotes
NOTE ADDED IN PROOF
Since submission of this manuscript, a paper describing a partial sequence of the mouse PDE β-subunit has been published [Bowes et al.(1990) Nature 347, 677–680]. Comparison of their sequence with Fig. 3A shows 8 discrepancies E (Fig. 3A) vs G (Bowes et al.) at position 5. A vs S at pos. 19, E vs D at pos. 49, L vs V at pos. 50, P vs T at pos. 158, L vs C at pos. 176, G vs S at pos. 236, and D vs Q at pos. 752. All deviations are within the N-terminal 236 residues (except D vs Q pos. 752). Our sequence has been further verified with two full length clones which were isolated since submission of this manuscript. Moreover, residues at positions 5, 19, 49, 158, and 236 of our sequence are identical with residues in a bovine β-Subunit sequence published by Lipkin et al. [Proc. Natl. Acad. Sci. USA (1990) 265, 12955–12959]. Since the N-terminal 236 residues of the Bowes et al. sequence were determined by analysis of only one cloned PCR copy of MPB1 and MPB61-12, the difference may be the result of PCR artifacts (e.g. sec Ennis et al., ‘Rapid cloning of HLA-A, B cDNA by using the polymerase chain reaction: frequency and nature of errors produced in amplification’, Proc. Natl. Acad. Sci. USA (1990) 87, 2833–2837).
References
- 1.Al-Ubaidi MR, Pittler SJ, Champagne MS, Triantafyllos JT, McGinnis JF, Baehr W. J Biol Chem. 1990:265. in press. [PubMed] [Google Scholar]
- 2.Badley JE, Bishop GA, St John T, Frelinger JA. BioFeedback. 1988;6:114–116. [PubMed] [Google Scholar]
- 3.Baehr W, Devlin MJ, Applebury ML. J Biol Chem. 1979;254:11669–11677. [PubMed] [Google Scholar]
- 4.Beavo JA. Adv Second Messenger Phosphoprotein Res. 1988;22:1–38. [PubMed] [Google Scholar]
- 5.Bowes C, Danciger M, Kozak CA, Farber DB. Proc Natl Acad Sci USA. 1989;86:9722–9726. doi: 10.1073/pnas.86.24.9722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breitbart RE, Andreadis A, Nadal-Ginard B. Annu Rev Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- 7.Chabre M, Deterre P. Eur J Biochem. 1989;179:255–265. doi: 10.1111/j.1432-1033.1989.tb14549.x. [DOI] [PubMed] [Google Scholar]
- 8.Danciger M, Kozak CA, Li T, Applebury ML, Farber DB. Exp Eye Res. 1990;51:185–189. doi: 10.1016/0014-4835(90)90071-2. [DOI] [PubMed] [Google Scholar]
- 9.Deterre P, Bigay J, Forquet F, Robert M, Chabre M. Proc Natl Acad Sci USA. 1988;85:2424–2428. doi: 10.1073/pnas.85.8.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fung BKK, Young JH, Yamane HK, Griswold-Prenner I. Biochemistry. 1990;29:2657–2664. doi: 10.1021/bi00463a006. [DOI] [PubMed] [Google Scholar]
- 11.Geliebter J. Focus. 1987;9:5–8. [Google Scholar]
- 12.Hancock JF, Magee AI, Childs JE, Marshall CJ. Cell. 1989;57:1167–1177. doi: 10.1016/0092-8674(89)90054-8. [DOI] [PubMed] [Google Scholar]
- 13.Lipkin VM, Gubanov VV, Khramtsov NV, Vasilevskaya IA, Atabckova NV, Muradov KG, Shuvaeva TM, Surina EA, Zagranichny VE, Li T. Bioorg Khimia. 1990;16:118–120. [PubMed] [Google Scholar]
- 14.Lipkin VM, Khramtsov NV, Vasilevskaya IA, Atabekova NV, Muradov KG, Gubanov VV, Li T, Johnston JP, Volpp KJ, Applebury ML. J Biol Chem. 1990;265:12955–12959. [PubMed] [Google Scholar]
- 15.Litt M, Luty JA. Am J Hum Genet. 1989;44:397–401. [PMC free article] [PubMed] [Google Scholar]
- 16.Magee T, Henley M. Nature. 1988;335:114–115. doi: 10.1038/335114a0. [DOI] [PubMed] [Google Scholar]
- 17.Ong OC, Ota IM, Clarke S, Fung BKK. Proc Natl Acad Sci USA. 1989;86:9238–9242. doi: 10.1073/pnas.86.23.9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ovchinnikov YA, Gubanov VV, Khramtsov NV, Ischenko KA, Zagranichny VE, Muradov KG, Shuvaeva TM, Lipkin VM. FEBS Lett. 1987;223:169–173. doi: 10.1016/0014-5793(87)80530-6. [DOI] [PubMed] [Google Scholar]
- 19.Padgett RA, Grabowski PJ, Konarska MM, Seiler S, Sharp PA. Annu Rev Biochem. 1986;55:1119–1150. doi: 10.1146/annurev.bi.55.070186.005351. [DOI] [PubMed] [Google Scholar]
- 20.Pittler SJ, Baehr W. In: Degenerative Retinal Disorders: Clinical and Laboratory Investigation. Chader GJ, Farber DB, editors. Alan R. Liss; New York: 1990. in press. [Google Scholar]
- 21.Pittler SJ, Baehr W, Wasmuth JJ, McConnel DG, Champagne MS, VanTuinen P, Ledbetter D, Davis RL. Genomies. 1990;6:272–283. doi: 10.1016/0888-7543(90)90567-e. [DOI] [PubMed] [Google Scholar]
- 22.Pittler SJ, McConnel DG, Davis RL. Am J Hum Genet. 1987;41:A234. (Abstract) [Google Scholar]
- 23.Reiss Y, Goldstein JL, Seabra MC, Casey P, Brown MS. Cell. 1990;62:81–88. doi: 10.1016/0092-8674(90)90242-7. [DOI] [PubMed] [Google Scholar]
- 24.Stryer L. Cold Spring Harbor Symp Quant Biol IIII. 1998:283–294. doi: 10.1101/sqb.1988.053.01.035. [DOI] [PubMed] [Google Scholar]
- 25.Swanson RJ, Applebury ML. J Biol Chem. 1983;258:10509–10605. [PubMed] [Google Scholar]
- 26.Vorburger K, Kitten GT, Nigg EA. EMBO J. 1989;8:4007–4013. doi: 10.1002/j.1460-2075.1989.tb08583.x. [DOI] [PMC free article] [PubMed] [Google Scholar]