A signaling module of NIMA-related kinases (Neks) regulates two kinesins, Mklp2 and Kif14, to spatiotemporally coordinate their subcellular localizations and activities. This is important for faithful completion of cytokinesis and reveals novel mechanisms by which Neks regulate late mitosis.
Abstract
In mitosis, cells undergo a precisely orchestrated series of spatiotemporal changes in cytoskeletal structure to divide their genetic material. These changes are coordinated by a sophisticated network of protein–protein interactions and posttranslational modifications. In this study, we report a bifurcation in a signaling cascade of the NIMA-related kinases (Neks) Nek6, Nek7, and Nek9 that is required for the localization and function of two kinesins essential for cytokinesis, Mklp2 and Kif14. We demonstrate that a Nek9, Nek6, and Mklp2 signaling module controls the timely localization and bundling activity of Mklp2 at the anaphase central spindle. We further show that a separate Nek9, Nek7, and Kif14 signaling module is required for the recruitment of the Rho-interacting kinase citron to the anaphase midzone. Our findings uncover an anaphase-specific function for these effector kinesins that is controlled by specific Nek kinase signaling modules to properly coordinate cytokinesis.
Introduction
Mitosis is a highly dynamic and tightly controlled phase of the cell cycle that spans diverse biological functions, including chromosome condensation, formation of a microtubule-based bipolar spindle, sister chromatid separation, and segregation, ultimately culminating in the formation of two genetically identical daughter cells (Musacchio and Salmon, 2007; Rieder, 2011). The spatiotemporal coordination of these processes is achieved by a complex array of signaling molecules, including kinases, phosphatases, and G proteins, among others. For example, the timely phosphorylation of key substrates by cyclin-dependent kinase 1 (Cdk1), the Aurora family (AurA and AurB), and Polo-like kinase 1 (Plk1) enzymes is crucial for successful completion of most aspects of cell division (Harper and Adams, 2001; Nigg, 2001; Carmena and Earnshaw, 2003). These upstream mediators of signal transduction in mitosis engage in protein–protein interactions with coactivators, inhibitors, and substrates to coordinate transient phosphorylation of their protein substrates and ensure proper mitotic progression. In addition to direct substrate phosphorylation to regulate their activity, localization, and abundance, some of their substrates are also kinases (Kettenbach et al., 2011), which in turn generate a complex network of signaling pathways that relay information and coordinate parallel mitotic functions.
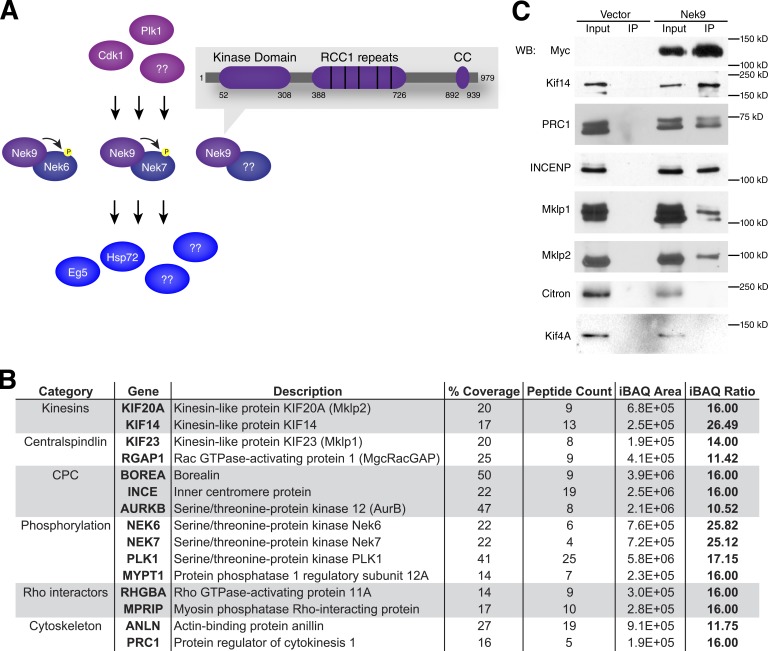
One such module of downstream kinases implicated in mitotic signal transduction consists of the three NIMA-related kinases (Neks) Nek9, Nek6, and Nek7, all of which are required for faithful cell division (O’Regan et al., 2007). In this signaling cascade, Nek9 is thought to lie upstream of Nek6 and Nek7 and activates them by both physical interaction (Richards et al., 2009) and phosphorylation of their respective activation loops in mitosis (Belham et al., 2003). In early mitosis, Nek9, Nek6, and the kinesin Eg5 form a signaling module downstream of Cdk1 and Plk1 that is required for centrosomes to separate and form a bipolar spindle (Rapley et al., 2008; Bertran et al., 2011). Nek9 also phosphorylates Nedd1 to recruit and retain γ-tubulin at centrosomes (Sdelci et al., 2012). Nek6 and Nek7 are thought to phosphorylate Nup98 and facilitate nuclear envelope permeabilization (Laurell et al., 2011). Nek6 has been shown to phosphorylate Hsp72, thereby stabilizing kinetochore–microtubule fibers (O’Regan et al., 2015). Finally, there is considerable evidence that Nek6, Nek7, and Nek9 contribute to faithful cytokinesis: Nek6, Nek7, and Nek9 localize to the midbody in cytokinesis (Roig et al., 2005; Kim et al., 2007; O’Regan and Fry, 2009), and depletion of Nek9 by siRNA (Kaneta and Ullrich, 2013), engineered knockout of Nek7 in mouse embryonic fibroblasts (Salem et al., 2010), or overexpression of kinase-dead Nek7 (Yissachar et al., 2006) leads to an increase in binucleated cells. Also, although overexpression of fully inactive Nek6 or Nek7 arrests cells in metaphase, overexpression of partially active Nek6 or Nek7 arrests cells in cytokinesis (O’Regan and Fry, 2009), indicating that higher amounts of Nek6 and Nek7 kinase activities are required to complete cytokinesis than to traverse metaphase. Although the mechanism by which Nek9 and Nek6 function in prometaphase has been investigated (Rapley et al., 2008; Bertran et al., 2011), there is currently no mechanistic insight into how Neks contribute to cytokinesis.
For successful completion of cytokinesis and abscission, a dramatic reorganization of the microtubule cytoskeleton is initiated in anaphase to form the central spindle at the midzone between the two poles (Glotzer, 2009; Green et al., 2012). The central spindle is a dynamic signaling platform composed of microtubule-associated proteins, kinesin motor proteins, mitotic kinases, and phosphatases. For instance, Mklp2 is a kinesin-6 family member that interacts with the chromosomal passenger complex (CPC) and targets it to the central spindle in anaphase in a manner regulated by Cdk1 (Gruneberg et al., 2004; Hümmer and Mayer, 2009; Kitagawa et al., 2014). In addition, Plk1 interacts with and phosphorylates Mklp2, which contributes to the localization of Plk1 to the central spindle and regulates the microtubule-bundling ability of Mklp2 (Neef et al., 2003). Kif14 is a kinesin-3 family member that is thought to recruit the effector kinase citron to the midzone (Gruneberg et al., 2006). Citron, in turn, is essential for proper midbody formation and abscission (Bassi et al., 2013). Both of these kinesins are required for accurate completion of cytokinesis.
In this study, we investigated the role of Neks in mitosis. We discovered that Nek9 regulates a bifurcated pathway in which its downstream effector kinases, Nek6 and Nek7, each control distinct kinesins that are required for faithful cytokinesis. We show that Nek9, Nek6, and the kinesin Mklp2 form a signaling module, which is required for Mklp2 to localize to the central spindle in anaphase. Nek6 also phosphorylates Mklp2 at Ser244, inhibiting its bundling activity until anaphase onset. Similarly, Nek9, Nek7, and Kif14 form a signaling module in which Nek7 phosphorylates Kif14, allowing the kinesin to localize citron kinase to the cleavage furrow in anaphase and contribute to proper midbody formation in cytokinesis. Although previously considered to be functionally equivocal in early phases of mitosis (Bertran et al., 2011), our data provide clear evidence for the participation of Nek6 and Nek7 in distinct arms of separate pathways that converge at cytokinesis. Collectively, these studies shed new light on the cellular and molecular mechanisms by which Neks contribute to faithful mitotic progression.
Results
Affinity purification mass spectrometry (AP-MS) identified proteins required for cytokinesis as interactors of Nek9 kinase
The physiological relevance of Nek-mediated regulation of cytokinesis is exhibited by a significant increase in cytokinesis failure upon depletion of Nek6, Nek7, or Nek9. Previous literature has shown that depletion of Nek9 or its downstream effectors Nek6 and Nek7 increases the number of binucleated cells, which are the result of failed cytokinesis (Salem et al., 2010; Kaneta and Ullrich, 2013), although the precise molecular mechanisms that underlie the relationship between these Nek kinases and faithful cell division have not been reported. In our hands, the cancer cell lines HeLa and U2OS display two- to fourfold increases in the percentage of binucleated cells after 2 d of Nek depletion when compared with control cells (Fig. 1, A and B). After 3 d, binucleation increased further such that a total of 10–18% of all cells were tetraploid, indicating a rate of cytokinesis failure that results in the accumulation of binucleated cells (Fig. 1, A and B). In the immortalized but nontransformed RPE1 cell line, the percentage of binucleated cells increased to a similar extent after 2 d of Nek depletion; however, tetraploid cells do not accumulate over time as in transformed cells (Fig. 1 C), consistent with previous literature that has shown p53-dependent senescence and apoptosis after cytokinesis failure (Minn et al., 1996; Lanni and Jacks, 1998; Andreassen et al., 2001; Castedo et al., 2006; Krzywicka-Racka and Sluder, 2011). These tetraploid cells are likely to result in higher rates of chromosome missegregation in subsequent cell divisions (Ganem et al., 2009), increasing chromosomal instability and promoting aneuploidy. Nek9 depletion also disrupted the structure of the midbody, as indicated by disorganized and asymmetrical staining of PRC1 and tubulin (Fig. 1 D). This observation is similar to the midbody defects previously observed upon depletion of essential midbody proteins such as citron kinase (Bassi et al., 2013).
Figure 1.
Nek depletion promotes cytokinesis failure. (A–C) HeLa (A), U2OS (B), and RPE1 (C) cells were treated with 10 nM of the indicated siRNAs for 48 and 72 h. Cells were fixed and stained for immunofluorescence, and the percentage of binucleated cells in >500 interphase cells was counted in three independent experiments. (D) Under conditions of Nek9 depletion, cytokinetic midbodies display disorganized PRC1 and tubulin staining that is consistent with depletion of citron kinase. Detergent-extracted HeLa cells stained with endogenous antibodies. n > 100 cytokinesis cells per condition in more than three independent experiments. *, P < 0.05 by Student’s t test; bar graphs show means ± SD. Bars: (main images) 20 µm; (insets) 5 µm.
Given that Nek9 contains several protein–protein interaction domains and has been shown to stably interact with Nek6 or Nek7 as well as other proteins (Fig. 2 A), we hypothesized that Nek9 may interact with proteins involved in cytokinesis and that knowledge of these proteins would provide additional insights into its function in this process. We generated HeLa cell lines stably expressing 6×Myc-tagged Nek9 or the tag alone and immunoprecipitated epitope-tagged Nek9 with anti-Myc antibody from mitotic cells. The resulting copurified proteins were separated by gel electrophoresis, digested to peptides, and identified by mass spectrometry using the tag-only sample as a negative control to distinguish specific from nonspecific interactors (Table S2). In addition to an array of protein interactors that participate in other mitotic functions, including KifC1, MYPT1, and NuMA1, we identified several proteins with known roles in cytokinesis as reproducible specific binding partners of Nek9 (Fig. 2 B). We confirmed these AP-MS results by Western blotting for the central spindle components Kif14, protein regulator of cytokinesis 1 (PRC1), inner centromere protein (INCENP), Mklp1, and Mklp2, demonstrating specific enrichment of these proteins in anti-Myc Nek9 versus anti-Myc control immunoprecipitations (Fig. 2 C). The absence of other midbody proteins by AP-MS (e.g., Kif4A, Ect2, citron, and Cep55; Table S2) and Western blot (Kif4A and citron; Fig. 2 C) supports the idea that the identified proteins are soluble interactors of Nek9 and not the result of precipitated midbodies. Collectively, Nek9 appears to interact with an array of regulatory proteins in mitosis, including select cytokinetic factors, raising the intriguing possibility that it may control their function in a way that promotes the formation of a stable midbody and successful completion of cytokinesis.
Figure 2.
Nek9 interacts with proteins involved in cytokinesis. (A) Nek9 signaling modules receive inputs from master mitotic regulators such as Cdk1 and Plk1 and either relay those signal to helper Nek6 and Nek7 kinases or signal directly to other targets. The integration of these signals is facilitated by protein–protein interaction domains on Nek9. CC, coiled-coil. (B) Proteins known to be involved in cytokinesis stably interact with Nek9. The coverage, peptide count, and intensity-based absolute quantification (iBAQ) area for proteins identified in Nek9 immunoprecipitates (IPs) and the iBAQ ratio (log2 ratio of iBAQ area in Nek9 immunoprecipitate over iBAQ area in control immunoprecipitates) represent the mean values from three independent AP-MS experiments. (C) Putative Nek9 interactors identified by mass spectrometry are confirmed by Western blot (WB). For the complete list of Nek9 interactors and corresponding quantification data, see Table S2.
Nek9 protein but not kinase activity is required for localizing the regulatory kinesins Mklp2 and Kif14 to the anaphase central spindle
To divide faithfully, cells begin the process of mitotic exit at anaphase, after chromosome segregation. The anaphase central spindle is organized as layers of signaling proteins on top of core structural components, the latter of which bind directly to and in some cases bundle central spindle microtubules for the recruitment of the former (Barr and Gruneberg, 2007). Although we observed Nek9 interacting with structural components of the central spindle, including PRC1, Mklp1, and INCENP (Fig. 1, B and C), depletion of Nek9 by siRNA (Fig. S1 A) did not affect the localization of these proteins by fixed-cell immunofluorescence microscopy (Fig. S1, B–D). In contrast, depletion of Nek9 abrogated the localization of two regulatory kinesins, Mklp2 and Kif14, to the central spindle in anaphase (Fig. 3, A and B). Surprisingly, however, both kinesins were observed at the midbody in telophase cells lacking Nek9, suggesting a phase-specific delay in their recruitment to the central spindle apparatus.
Figure 3.
Nek9 protein is required to localize Kif14 and Mklp2 to the anaphase midzone. (A and B) siRNA-mediated depletion of Nek9 results in mislocalization of Mklp2 (A) and Kif14 (B) in anaphase but not in telophase. Detergent-extracted HeLa cells stained with antibodies against endogenous proteins. (C) Quantification of localization behavior in A and B. n > 100 anaphase cells per condition in more than three independent experiments. (D) Live-cell imaging of a stable EGFP-Mklp2 HeLa cell line indicates that under conditions of Nek9 depletion, EGFP-Mklp2 localizes to the cell midzone 4–6 min later than in control cells. n ≥ 5 cells per condition. Bars: (A and B) 20 µm; (D) 10 µm. (E) Depletion of Mklp2 or Kif14 by siRNA significantly increases the proportion of binucleated cells observed after 48 h. Cells were fixed and stained for immunofluorescence, and the percentage of binucleated cells in >500 interphase cells was counted in three independent experiments. *, P < 0.05 by Student’s t test; points and bar graphs show means ± SD.
Prior studies have demonstrated persistent localization of Mklp2 at the central spindle and midbody from anaphase to cytokinesis. When mislocalized, for example in INCENP-depleted cells (Gruneberg et al., 2004; Hümmer and Mayer, 2009), Mklp2 was not observed at these structures at any point beyond metaphase. In our fixed-cell immunofluorescence microscopy experiments, we failed to observe endogenous Mklp2 localization in >90% of anaphases in cells transfected with Nek9 siRNA, whereas nearly all cells in telophase exhibited normal Mklp2 staining (Fig. 3 C). To confirm that we could observe both behaviors of Mklp2 in the same cell, we performed live-cell imaging at 2-min intervals from metaphase to mitotic exit of a HeLa cell line stably expressing EGFP-Mklp2 and transfected with Nek9 siRNA. As previously described (Gruneberg et al., 2004), EGFP-Mklp2 localized to the central spindle at the onset of anaphase and gradually became concentrated at the midbody in telophase (Fig. 3 D). However, upon depletion of Nek9, EGFP-Mklp2 accumulation at the central spindle was greatly reduced until telophase (Fig. 3 D). Comparison of midzone-localized EGFP-Mklp2 fluorescence intensity in control (n ≥ 6) and Nek9 siRNA (n ≥ 6)–transfected cells confirmed a statistically significant difference (*, P < 0.05; Student’s t test) between Mklp2 abundance at the central spindle in anaphase (Fig. 3 D, right). The decrease in Mklp2 abundance at the central spindle was likely the result of mislocalization to the cytosol rather than degradation, as treatment with siRNA to Nek9 or Nek6 did not decrease the total levels of endogenous or EGFP-Mklp2 protein, as measured by Western blot (Fig. S4 C).
As with Mklp2, Nek9 depletion displaced Kif14 from anaphase central spindles in >90% of siRNA-transfected cells in fixed-cell imaging experiments (Fig. 3 C). Consistent with previous literature (Maliga et al., 2013), repeated attempts to generate stable EGFP-Kif14 HeLa cells by transfection and by retroviral transduction were unsuccessful and uniformly resulted in cell death after a limited number of cell passages; this ultimately precluded us from performing live-cell imaging of Kif14. As an alternative strategy, however, we were able to synchronize mitotic HeLa cells by arresting WT or Nek9 siRNA–treated, thymidine-released cells with low-dose exposures to nocodazole to facilitate a transient mitotic arrest, followed by nocodazole washout and fixed-cell immunofluorescence microscopy at 5-min intervals after metaphase (Fig. S3 A). As was observed in asynchronous HeLa cells, >90% of synchronized cells exhibited a delay in recruitment of Kif14 to the midzone until telophase (Fig. S3 B).
Broadly speaking, we conclude that at least one aspect of Nek9 function is to ensure the timely localization of specific regulatory components to the central spindle such that these proteins can perform their biological roles at later points in cell division. Such roles are important for the faithful completion of cytokinesis, as depletion of Mklp2 or Kif14 by siRNA significantly increased the proportion of binucleated cells observed after 48 h (Fig. 3 E). Similar rates of cytokinesis failure have been shown to occur in Drosophila melanogaster S2 cells (Bassi et al., 2013) and HeLa cells (Gruneberg et al., 2006) upon Kif14 depletion, citron depletion (Gruneberg et al., 2006; Bassi et al., 2013), and Mklp2 depletion (Neef et al., 2003).
Having established that Nek9 is necessary for proper localization of Mklp2 and Kif14 in anaphase, we sought to determine whether Nek9 kinase activity is also required for this function. To do this, we generated two different kinase-dead mutants of Nek9 (K81M and D176A; Roig et al., 2002; Bertran et al., 2011). In our hands, HeLa cells transfected with the same amount of DNA expressed lower levels of 6×Myc-Nek9 K81M protein compared with 6×Myc-Nek9 WT and 6×Myc-Nek9 D176A (Fig. S1 E). Therefore, we used D176A as the kinase-dead mutant of Nek9. This mutant exhibited a minimal amount of kinase activity in vitro (Fig. 4 A), but still permitted observation of cellular effects that are highly sensitive to Nek9 kinase activity in cells.
Figure 4.
Nek9 kinase activity is dispensable for Mklp2 and Kif14 localization. (A) Recombinant Nek9 WT and D176A (kinase-dead) were incubated with peptide substrate for 16 h at 28°C, and ATPase activity was measured. Graphs are normalized to mean WT kinase activity from three independent experiments. *, P < 0.05 by Student’s t test; bar graphs show means ± SD. (B and C) Stable HeLa cell lines containing either WT or D176A Nek9 rendered siRNA resistant by silent mutations were alternatively treated with Nek9 siRNA or transfection reagent alone, and then detergent was extracted, fixed, and stained for Mklp2 (B) or Kif14 (C) in anaphase and telophase cells. Note that both kinesins localize normally with or without Nek9 kinase activity in both mitotic phases. Bars, 20 µm.
We generated HeLa cell lines stably expressing either WT or D176A Nek9 that contained silent mutations to render them insensitive to Nek9 siRNAs (Fig. S1 F). We then compared the endogenous Mklp2 central spindle localization in HeLa cells expressing WT or D176A Nek9 in the presence or absence of endogenous Nek9, and we detected no difference in anaphase or telophase staining (Fig. 4 B). Mklp2 was readily recruited to the central spindle in anaphase in HeLa cells lacking Nek9 activity, but expressing Nek9 D176A protein. Similarly, endogenous Kif14 was also able to localize to the central spindle in anaphase in HeLa cells lacking endogenous Nek9 activity but expressing Nek9 D176A (Fig. 4 C). Collectively, these results imply that rather than being direct or indirect targets of Nek9 kinase activity, Mklp2 and Kif14 rely on Nek9 protein as a scaffold to promote their localization to the central spindle in anaphase.
Nek6 and Nek7 kinase activities are differentially required for Mklp2 and Kif14 anaphase localization
Nek9 is known to form mutually exclusive complexes with either Nek6 or Nek7 as well as to activate these kinases by direct binding and phosphorylation (Roig et al., 2002; Belham et al., 2003). To determine whether Nek9 impacts Mklp2 or Kif14 localization through the action of Nek6 or Nek7, we depleted the two kinases from HeLa cells using siRNA (Fig. S2, A–C). Interestingly, although Nek7 depletion had no effect on Mklp2 localization to the central spindle in anaphase, depletion of Nek6 mirrored the effects of Nek9 and significantly abolished Mklp2 central spindle localization in anaphase in >90% of HeLa cells (Fig. 5 A). We confirmed this result by live-cell imaging in HeLa cells expressing EGFP-Mklp2 in the presence and absence of Nek6 and found that Mlkp2 localization to the midzone in anaphase was strongly delayed and reduced upon Nek6 depletion (Fig. S2 D). This reduction in Mklp2 localization and timing at the central spindle upon Nek6 depletion recapitulated our prior observations for Nek9 depletion in live cells (Fig. 3 D). In contrast, depletion of Nek6 did not affect Kif14 localization (Fig. 5 B). However, Nek7 depletion abolished Kif14 at the central spindle in anaphase (Fig. 5 B), as was observed for Nek9 (Fig. 3 B).
Figure 5.
Nek6 or Nek7 kinase activity is required to localize Mklp2 or Kif14 to the central spindle. (A–D) siRNA-mediated depletion of Nek6 but not Nek7 results in mislocalization of Mklp2 in anaphase, whereas depletion of Nek7 but not Nek6 results in a similar phase-specific mislocalization of Kif14. Green indicates Mklp2 (A and C) or Kif14 (B and D); red indicates tubulin; blue indicates DNA. n > 100 anaphase cells per condition in more than three independent experiments. *, P < 0.05 by Student’s t test; bar graphs show means ± SD. (C) Mklp2 localization was analyzed in siRNA-resistant WT or kinase-dead 6×Myc-Nek6 cell lines plus or minus Nek6 siRNA to deplete endogenous protein. Central spindle localization is dependent on Nek6 kinase activity. (D) Kif14 localization was analyzed in siRNA-resistant WT or kinase-dead Nek7 stable cell lines with or without Nek7 siRNA to deplete endogenous protein. Note that Kif14 anaphase central spindle localization is dependent on Nek7 kinase activity. All panels depict detergent-extracted HeLa cells stained with antibodies against endogenous proteins. Bars, 20 µm. KD, knockdown.
We also asked whether Nek6 and Nek7 kinase activities were required for the localization of their respective kinesins. We generated HeLa cell lines stably expressing siRNA-resistant WT and D172A (kinase inactive) Nek6 as well as WT and D161A (kinase inactive) Nek7 (Fig. S2, E and F). When we investigated these cell lines for Mklp2 localization in the absence of endogenous Nek6, only WT and not D172A Nek6 was able to rescue localization of Mklp2 to the central spindle in anaphase (Fig. 5 C). Similarly, in the absence of endogenous Nek7, siRNA-resistant WT but not D161A Nek7 was able to rescue Kif14 localization to the central spindle in anaphase (Fig. 5 D). This indicates that in contrast to Nek9 (Fig. 4, B and C), Nek6 and Nek7 kinase activities are required for proper timing of Mklp2 and Kif14 localization to the central spindle, respectively, and suggests that these kinesins could be also be Nek substrates.
This observation raises the interesting question of how endogenous Nek6 and Nek7 were activated in our Nek9 D176A cell line such that Mklp2 and Kif14 were able to localize to the central spindle (Fig. 4, B and C). It is possible that the residual kinase activity of Nek9 D176A (Fig. 4 A) is sufficient to phosphorylate Nek6 and Nek7 over time in cells. In addition, it has been shown that Nek9 binding alone releases an autoinhibitory conformation to partially activate Nek7 without phosphorylation (Richards et al., 2009). Alternatively, there could exist additional signaling upstream of Nek6 and Nek7 that is parallel to but independent of Nek9. In any case, our data point to Mklp2 and Kif14 localization as more sensitive to Nek6 and Nek7 kinase activities, respectively, than to Nek9 activity.
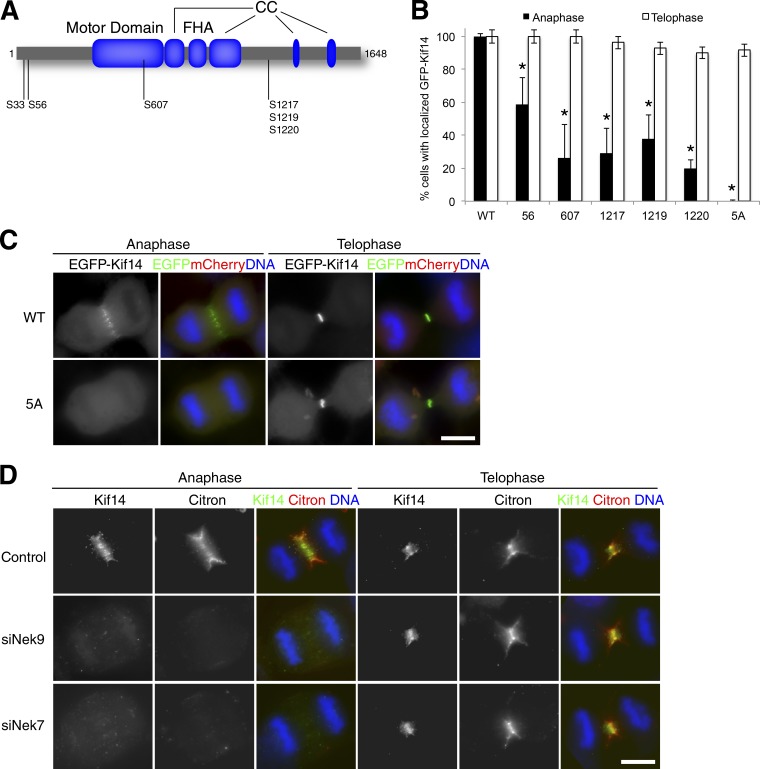
Nek7 direct phosphorylation is required for the anaphase localization of Kif14
To determine whether Kif14 is a direct substrate of Nek7, we performed in vitro kinase assays using recombinant active Nek7 and Kif14, followed by phosphorylation site identification by liquid chromatography–tandem mass spectrometry (LC-MS/MS). In these analyses, we identified five potential Nek7 phosphorylation sites on Kif14 that were present upon incubation of Kif14 with Nek7 but not in control reactions lacking Nek7: Ser33, Ser56, Ser607, Ser1217, and either Ser1219 or Ser1220, the last of which could not be confidently localized by mass spectrometry (Fig. 6 A). To determine the role of these phosphorylation sites in Kif14 central spindle localization, we generated six single-site Ser-to-Ala mutants by site-directed mutagenesis and tested for protein stability in HeLa cells by transient transfection. Except for EGFP-Kif14 S33A, protein expression of these phosphoablated mutant Kif14 constructs was detectable by Western blot analysis, and expression levels were comparable to WT EGFP-Kif14 protein levels (Fig. S3 C). Furthermore, immunofluorescence microscopy of the expressed Kif14 mutants showed different degrees of mislocalization of mutant Kif14 to the central spindle in anaphase cells (Fig. 6 B). Finally, to determine whether potential Nek7 phosphorylation sites had an additive effect on Kif14 localization, we generated an EGFP-Kif14-5A construct in which Ser56, Ser607, Ser1217, Ser1219, and Ser1220 were all mutated to Ala. When transfected into HeLa cells, EGFP-Kif14-5A was expressed to similar levels as WT Kif14 (Fig. S3 C), but its localization to the central spindle in anaphase cells was completely abolished (Fig. 6 C). These data suggest that phosphorylation of Kif14 by Nek7 may be required for accurate and timely central spindle localization of Kif14 in anaphase, but not in telophase.
Figure 6.
A requirement for cellular Nek7 kinase activity and direct phosphorylation of Kif14 in anaphase is also linked to its cargo, citron kinase. (A) In vitro kinase assay reveals five Nek7-dependent phosphorylation sites on Kif14 by mass spectrometry. CC, coiled-coil; FHA, forkhead-associated domain. (B) Mutation of individual Nek7 phosphorylation sites to alanine partially disrupts the central spindle localization of Kif14 in HeLa cells transiently transfected with various EGFP-Kif14 WT or mutant constructs. n > 50 anaphase cells per condition in more than three independent experiments. *, P < 0.05 by Student’s t test; bar graphs show means ± SD. (C) Collective mutation of all Nek7 sites fully disrupts the central spindle localization of Kif14, but only in anaphase. HeLa cells transiently transfected with EGFP-Kif14 constructs and mCherry as a positive control for transfection. (D) Nek9- or Nek7-dependent Kif14 mislocalization results in a corresponding delay in the localization of a known Kif14 cargo protein, the Rho-dependent kinase citron. Detergent-extracted HeLa cells stained with antibodies against endogenous proteins. n > 100 anaphase cells per condition in more than three independent experiments. Bars, 20 µm. Green, EGFP-Kif14-5A (C) or Kif14 (D); red, mCherry (C) or citron (D); blue, DNA.
Little is known about the molecular and structural mechanisms through which Kif14 is required for cytokinesis. However, it was previously shown that Kif14 is essential for the recruitment of the Rho-interacting kinase citron to the central spindle (Gruneberg et al., 2006). Thus, we sought to determine whether Nek7 or Nek9 are also required for citron localization. Indeed, for HeLa cells transfected with Nek7 or Nek9 siRNA in which Kif14 was absent from the central spindle, citron kinase also failed to localize in anaphase, but both were present at the midbody in telophase (Fig. 6 D). A previous study of cells undergoing abscission while depleted of citron kinase (Bassi et al., 2013) revealed asymmetric, misshapen midbodies with structural defects similar to those we observed in cells depleted of Nek9 (Fig. 1 D).
Direct Nek6 phosphorylation is required for anaphase localization and function of Mklp2
To test whether Nek6 can directly phosphorylate Mklp2 and to identify potential Nek6-dependent phosphorylation sites on Mklp2, we performed in vitro kinase assays using recombinant active Nek6 and Mklp2 and analyzed them by LC-MS/MS. Using this approach, we identified five sites on Mklp2 that specifically increased in phosphorylation upon incubation with Nek6 versus control reactions: Ser240, Ser244, Ser683, Ser754, and Ser883, the first two of which are located in its motor domain (Fig. 7 A).
Figure 7.
Nek6 phosphorylates Mklp2 at Ser244, which affects both kinesin localization and anaphase progression. (A) In vitro kinase assay reveals five Nek6-dependent phosphorylation sites on Mklp2 by mass spectrometry. CC, coiled-coil. (B) Mutation of Ser244 to unphosphorylatable Ala disrupts the central spindle localization of Mklp2. Detergent-extracted HeLa cells transiently transfected with EGFP-Mklp2-WT or -S244A. n > 100 anaphase cells per condition in more than three independent experiments. (C) Live-cell imaging of a stable EGFP-Mklp2-S244A HeLa cell line reveals a decrease in the time to cleavage furrow ingression that is dependent on Nek6. n ≥ 5 cells per condition. (D) Quantification of furrow ingression relative to the equatorial cell diameter at metaphase. At 4–6 min after anaphase onset, EGFP-Mklp2-S244A has ingressed to a greater extent than EGFP-Mklp2-WT. (E) GFP immunoprecipitation of EGFP-Mklp2-WT or S244A expressed in stable or transiently transfected HeLa cells. EGFP-Mklp2-S244A forms a heterodimer with endogenous Mklp2 when expressed at low levels in the stable cell line and a homodimer when overexpressed by transient transfection. Black arrows indicate EGFP-Mklp2; gray arrows indicate endogenous Mklp2. WB, Western blot. (F) Depletion of endogenous Mklp2 results in a high proportion of binucleated cells. Simultaneous expression of EGFP-Mklp2-WT but not EGFP-Mklp2-S244A rescues this phenotype. Expression of EGFP-Mklp2-S244E can partially rescue. Cotransfected cells were fixed and stained for immunofluorescence, and the percentage of binucleated cells in >500 interphase cells was counted in three independent experiments. *, P < 0.05 by Student’s t test; bar graphs show means ± SD.
To determine the biological significance of these phosphorylation sites, we separately mutated each of these five Ser residues to a phosphoablated Ala and transfected the single-site mutants into HeLa cells. Western blot analysis of protein expression revealed that whereas EGFP-Mklp2-S240A protein could not be detected, EGFP-Mklp2-S883A expression was comparable to WT EGFP-Mklp2, and S244A, S683A, and S754A expression were slightly higher than WT EGFP-Mklp2 (Fig. S4 A). Immunofluorescence microscopy showed that transiently transfected EGFP-Mklp2-S683A, -S754A, and -S883A localized normally in anaphase and telophase (Fig. S4 B). However, transfected EGFP-Mklp2-S244A (Fig. 7 B) phenocopied the loss of endogenous Mklp2 from the central spindle in anaphase upon depletion of Nek6 and Nek9 protein (Figs. 3 A and 5 A) and Nek6 kinase activity (Fig. 5 C).
In addition to transient overexpression, we also made stable HeLa cell lines by retroviral transduction for live-cell imaging of EGFP-Mklp2 phosphomutants. Surprisingly, cells stably expressing EGFP-Mklp2-S244A displayed the correct localization of the S244A mutant in anaphase at the central spindle (Fig. 7 C, top). Given that Mklp2 is a dimer in cells, we surmised that the stable cell lines may exhibit lower levels of EGFP-Mklp2-S244A such that most of it would be in complex with endogenous Nek6-phosphorylated Mklp2, which could be sufficient for recruitment to the central spindle in anaphase. Conversely, we hypothesized that transient transfection of EGFP-S244A-Mklp2 would be likely to result in significant overexpression of the mutant compared with endogenous Mklp2, generating predominantly homodimeric EGFP-Mklp2-S244A, which is not capable of localizing to the anaphase midzone. To test this, we affinity-purified EGFP-Mklp2-WT and -S244A from transiently transfected or stably expressing HeLa cells and determined the ratio of endogenous Mklp2 bound to EGFP-Mklp2 proteins in the pulldowns by Western blot (Fig. 7 E). Indeed, although both EGFP-Mklp2-WT and -S244A coprecipitated endogenous Mklp2 in stable cell lines (Fig. 7 E, gray arrow), their transient transfection and purification resulted in mostly homomeric complexes that lacked endogenous Mklp2 (Fig. 7 E, black arrows).
Interestingly, live-cell imaging of EGFP-Mklp2-S244A cells progressing from metaphase to cytokinesis revealed a decreased time to cleavage furrow ingression by approximately one to two intervals (2–4 min; Fig. 7, C [top] and D). These data suggest that it is possible that phosphorylation of Mklp2 at Ser244 inhibits a post-anaphase function of Mklp2 while it is localized to the midzone in anaphase. Under this hypothesis, Mklp2 S244A might function prematurely, which would explain our observations of the decreased time to cleavage furrow ingression when it was localized to the central spindle via endogenous Mklp2 dimerization. Consistent with this, treatment of the EGFP-Mklp2-S244A cell line with siNek6 both disrupted the anaphase localization of the mutant and restored WT cell cycle timing (Fig. 7 C, bottom). Further supporting this hypothesis, in cells treated with Mklp2 siRNA, cotransfection of EGFP-Mklp2-WT rescued the binucleated cell phenotype, whereas cells expressing EGFP-Mklp2-S244A did not (Fig. 7 F). Interestingly, cells cotransfected with Mklp2 siRNA and EGFP-Mklp2-S244E showed an intermediate number of binucleated cells, suggesting that this phosphomimetic mutant was able to partially rescue the loss of endogenous Mklp2 (Fig. 7 F).
These observations are consistent with phosphorylation of Mklp2 S244 by Nek6 being necessary for both the anaphase localization of Mklp2 and also to prevent Mklp2 activation until an appropriate time. Persistent residence at the central spindle may then lead to dephosphorylation of S244, perhaps by the same as-yet-unidentified phosphatase implicated in dephosphorylating Cdk1 sites on Mklp2 (Kitagawa et al., 2014), which has been proposed to activate Mklp2 at anaphase onset so that cell division can progress.
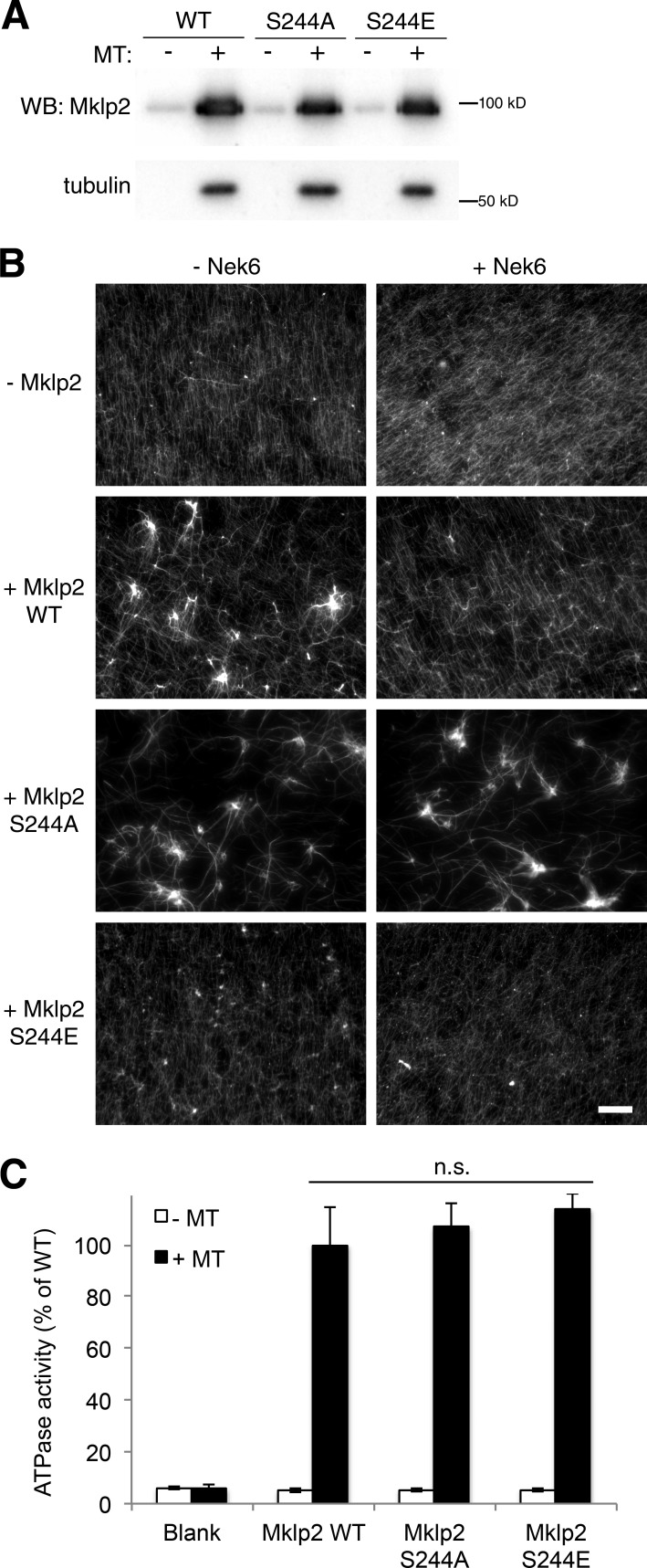
Nek6-dependent Mklp2 Ser244 phosphorylation inhibits microtubule bundling activity but not microtubule binding or ATP hydrolysis in vitro
Mklp2 has been shown to bundle microtubules in vitro in a manner that is negatively regulated by Plk1 and Cdk1 activity (Neef et al., 2003; Kitagawa et al., 2014). We considered that bundling may be an anaphase function of Mklp2 also inhibited by Nek6 phosphorylation. Such inhibition may occur by reducing the capacity for Mklp2 to interact with microtubules or, more directly, by impacting its kinesin activity. Incubation of recombinant WT, S244A, and S244E-Mklp2 with taxol-stabilized microtubules resulted in a similar degree of microtubule-dependent pelleting (Fig. 8 A), indicating that Nek6 phosphorylation does not reduce Mklp2 interaction with microtubules in vitro. However, preincubation of recombinant WT Mklp2 with Nek6 and ATP strongly inhibited microtubule bundling, whereas S244A-Mklp2 exhibited robust microtubule bundling in the presence or absence of Nek6 (Fig. 8 B). In contrast, the phosphomimetic S244E-Mklp2 mutant did not efficiently bundle microtubules in this assay under any condition (Fig. 8 B). In this study, bundling activity appeared to be distinct from ATPase activity, as results from endpoint assays for the extent of microtubule-dependent hydrolysis of ATP are indistinguishable between Mklp2 WT, S244A, and S244E (Fig. 8 C).
Figure 8.
Ser244 phosphorylation inhibits microtubule bundling but not microtubule binding or ATPase activity. (A) Western blot (WB) of pellets from microtubule (MT) pelleting assays performed in 25 mM KCl. Ser244 mutation to Ala or Glu does not affect the ability of Mklp2 to bind microtubules. (B) Mklp2 WT, S244A, or S244E was incubated with or without Nek6 followed by addition of taxol-stabilized microtubules and centrifugation onto coverslips. Microtubule bundling was observed by immunofluorescence. Nek6 inhibits bundling of Mklp2 WT, but Mklp2 S244A is refractory to Nek6 inhibition, whereas Mklp2 S244E is significantly attenuated in bundling activity in both conditions. Representative images of three independent experiments are shown. Bar, 20 µm. (C) Ser244 mutation to Ala or Glu does not affect the microtubule-dependent ATPase activity of Mklp2. Recombinant Mklp2 WT, S244A, and S244E were incubated with or without taxol-stabilized microtubules for 30 min at 28°C, and then ATPase activity was measured. Graphs are normalized to mean WT ATPase activity from three independent experiments. *, P < 0.05 by Student’s t test; bar graphs show means ± SD.
Discussion
In this study, we describe a new cellular mechanism by which bifurcation in the Nek signaling cascade downstream of Nek9 affects the fidelity of cell division by regulating the localization and activity of essential kinesins in anaphase. Collectively, these data demonstrate distinct, noninterchangeable roles of Nek6 and Nek7 kinase activity downstream of Nek9 in mitosis and support a model in which Nek9 forms at least two separate signaling modules: one composed of Nek6 and Mklp2, and the other of Nek7 and Kif14. In these modules, Nek9 activates Nek6 and Nek7 by releasing autoinhibitory conformations through direct binding (Richards et al., 2009) and by phosphorylating their activation loops (Belham et al., 2003; Richards et al., 2009) and facilitates their recruitment to the respective substrate kinesins, enabling specific phosphorylation of Mklp2 by Nek6 and Kif14 by Nek7 in prometaphase (Fig. 9).
Figure 9.
Model for Nek kinase-dependent signaling modules that specify anaphase behavior of Mklp2 and Kif14. For Mklp2, preanaphase cells contain high Cdk1 and Nek activity, rendering Mklp2 cytosolic and inactive. Dephosphorylation at anaphase onset by loss of Cdk1 activity and/or gain of phosphatase activity while maintaining Nek-dependent phosphorylation status licenses Mklp2 for deposition to the anaphase central spindle. Residence at the anaphase central spindle then registers it via an unknown mechanism for functions in telophase and beyond; analogous mechanisms are proposed to exist for Kif14. Pptase, phosphatase.
Our findings suggest that the Nek9–Nek6–Mklp2 signaling module that results in Ser244 phosphorylation before chromosome separation promotes timely recruitment of Mklp2 to the central spindle in anaphase while keeping the bundling activity of the kinesin in check until dephosphorylation by a phosphatase later in anaphase, which could activate its bundling activity (Fig. 9). Consistent with this model, we found that localization of nonphosphorylatable, bundling-competent Mklp2-S244A to the central spindle in anaphase by heterodimerization with WT Mklp2 (Fig. 7 C, top) results in decreased time to cleavage furrow ingression (Fig. 7 D). Similarly, participation in a Nek9–Nek7–Kif14 complex to facilitate phosphorylation at multiple sites by Nek7 promotes localization of Kif14 to the central spindle, where it traffics citron kinase, an event necessary for proper cleavage furrow ingression and abscission (Bassi et al., 2013).
Interestingly, siRNA-mediated depletion of Nek kinases resulted in mislocalization of these kinesins in anaphase but not telophase; although their central spindle localization was abrogated, Mklp2 and Kif14 still localized to the midbody in telophase. This indicates that temporally distinct mechanisms are responsible for the localization of Mklp2 and Kif14 to the central spindle and midbody. Although Mklp2 and Kif14 were eventually able to localize to the telophase midbody under conditions of Nek depletion, evidence suggests that these midbodies were not fully functional. Both tubulin and PRC1 (Fig. 1 D) appeared disorganized, and Nek-depleted cells have high incidences of cytokinesis failure that result in binucleated cells as observed in this study and in others (Yissachar et al., 2006; Salem et al., 2010; Kaneta and Ullrich, 2013). Additionally, expression of EGFP-Mklp2-S244A was unable to rescue depletion of endogenous Mklp2 (Fig. 7 F). Even though the mutant protein localized to the telophase midbody, these cells failed to complete cytokinesis efficiently, suggesting that localization of this mutant in telophase and beyond is insufficient to rescue the entirety of its post-anaphase function or functions. Although Mklp2 may be responsible for additional functions at the anaphase central spindle, our data suggest that it also receives signals, possibly in the form of additional posttranslational modifications, that are required to modulate its function after anaphase. Alternatively, soluble Mklp2 may perform as of yet underappreciated functions in a Nek activity–dependent manner. Our data are consistent with the hypothesis that Nek-dependent residence of Mklp2 and Kif14 at the central spindle in anaphase is important for their later functions to coordinate faithful completion of cytokinesis, although the precise mechanisms by which this occurs remain to be elucidated.
Intriguingly, recent evidence suggests that Cdk1-dependent phosphorylation of Mklp2 inhibits both microtubule binding as well as bundling activity of Mklp2 (Kitagawa et al., 2014). In that study, the authors concluded that Cdk1 phosphorylation of Mklp2 maintains its solubility in the cytosol in early mitosis; rapid turnover of Cdk1 phosphorylation sites by a phosphatase at anaphase onset was posited as a requirement for loading of Mklp2 onto chromosomes, where it subsequently relocalizes to the central spindle. Additionally, transit of the CPC from chromosomes to the central spindle immediately at anaphase onset has been shown to depend on the Cdk1 phosphorylation status of Mklp2 (Hümmer and Mayer, 2009; Kitagawa et al., 2014). Our data in which the CPC member INCENP localizes at the anaphase central spindle whereas Mklp2 does not in Nek9 protein– and Nek6 activity–depleted cells suggest that additional layers of regulation beyond Cdk1 phosphorylation are required for faithful deposition of the CPC to the midzone and that Cdk1-dependent physical interactions between INCENP and Mklp2 alone may not be sufficient to explain the mechanism by which Mklp2 promotes this event. Finally, we suggest that the removal of the Nek6 phosphorylation site or sites on Mklp2 is likely to occur later, after anaphase, in light of our observations of reduced time to furrow ingression for S244A-Mklp2–containing cells. The identity of the phosphatase (or phosphatases) responsible for the phase- and site-specific activation of Mklp2 will likely shed additional light on how these events are coordinated to ensure faithful exit from mitosis.
Nek6 and Nek7 share 87% sequence identity and potentially redundant functions (Bertran et al., 2011). Although the presence of separate Nek9–Nek6 and Nek9–Nek7 complexes has been known for some time (Roig et al., 2002; Belham et al., 2003), the strict dependence of Mklp2 localization on Nek6 and Kif14 localization on Nek7 represents a concrete, functional distinction between these two very similar kinases. In the context of kinesin localization in anaphase, depletion of either Nek6 or Nek7 results in unique defects that cannot be compensated for by the other kinase, providing strong evidence for their unique cellular roles in cytokinesis. Analogous to AurA and AurB kinases, which are highly similar enzymes with very distinct cellular functions, the divergent N termini of Nek6 and Nek7 may provide clues into the mechanisms by which they carry out their separate roles (O’Regan et al., 2015) as well as how they may be differentially regulated by Nek9 or other upstream signals.
Our AP-MS experiment for Nek9-interacting proteins revealed a large number of proteins associated with Nek9 during nocodazole arrest, of which a subset were already known to play roles in cell division and cytokinesis. Although this study has implicated two of them, Kif14 and Mklp2, as downstream targets of Nek9 signaling, it is also very likely that our AP-MS data contain additional protein effectors of Nek biology in cell division, cytokinesis, or other aspects of mitosis that were not in this study. Indeed, the presence of cytoskeletal proteins and other motors (NuMA1, dynein, and anillin) raises the likelihood that the net impact of Nek9 loss on cytokinesis is a mixture of the direct effects on Mklp2 and Kif14, as described in this study, and potentially indirect effects on other as of yet unstudied Nek9 effectors. As a kinase family, Neks are broadly involved in microtubule organization, either through the formation of cilia (Nek1 and Nek8; Mahjoub et al., 2004; Evangelista et al., 2008; Shalom et al., 2008; Zalli et al., 2012) or more explicitly during mitotic functions (Nek2, Nek6, Nek7, and Nek9; Fry et al., 2012). With this study, we provide evidence that the kinesins Mklp2 and Kif14 are regulated by Nek6, Nek7, and Nek9 in anaphase, and previous work has shown Nek regulation of Eg5 (Rapley et al., 2008; Bertran et al., 2011). Perhaps coordination of kinesin activity is a general characteristic of Neks. Future work focused on whether other Nek family members perform their microtubule-organizing functions via kinesins or other mechanisms will further clarify the biological roles for these essential enzymes.
Materials and methods
Cell lines
pBMN-IRES-Neo(6×Myc-Nek9), pBMN-IRES-Neo(6×Myc-Nek6), pBMN-IRES-Neo(6×Myc-Nek7), or pBMN-IRES-Neo(EGFP-Mklp2) was transfected into Phoenix Ampho cells, and the resulting retrovirus was used to infect HeLa cells; this was followed by neomycin selection to isolate a polyclonal population of cells. QuikChange Lightning Multi (Agilent Technologies) site-directed mutagenesis was used to generate siRNA-resistant silent mutations in pBMN-IRES-Neo(6×Myc-Nek9), pBMN-IRES-Neo(6×Myc-Nek6), and pBMN-IRES-Neo(6×Myc-Nek7), and then these constructs were used to create stable cell lines by neomycin selection. QuikChange Lightning (Agilent Technologies) was used to generate kinase-dead pBMN-IRES-Neo(6×Myc-Nek9 siRNA D176A), pBMN-IRES-Neo(6×Myc-Nek6 siRNA D172A), and pBMN-IRES-Neo(6×Myc-Nek7 siRNA D161A) as well as pBMN-IRES-Neo(EGFP-Mklp2-S244A), and then these constructs were used to create stable cell lines by neomycin selection.
Antibodies
Nek9, Nek6, and Nek7 antibodies were generated in rabbits by Covance against three Nek9 antigens, one Nek6 antigen, and one Nek7 antigen. DNA corresponding with Nek9 amino acids 1–58, 301–349, and 360–388, Nek6 amino acids 1–39, and Nek7 amino acids 1–35 was cloned into pGEX-6p1, expressed in BL21DE3 pLys Escherichia coli, and purified by affinity chromatography using glutathione-Sepharose resin (GE Healthcare). GST was cleaved, antigen solutions were lyophilized, and the three Nek9 antigens were combined and submitted to Covance; the resulting sera was affinity-purified using antigens coupled to N-hydroxysuccinimide ester–Sepharose per Covance protocols.
Lamin A/C antibody was a gift from F. McKeon (University of Houston, Houston, TX). All other antibodies were commercially available as follows: PRC1 and INCENP (Cell Signaling Technology), Mklp1 and GFP (Genetex), Mklp2 (Abcam), Kif14 (Bethyl Laboratories, Inc.), α-tubulin (Sigma-Aldrich), myc (clone 9E10), and citron (BD).
Coimmunoprecipitation
Four 15-cm-diameter dishes of HeLa cells were arrested in 2-mM thymidine for 22 h followed by 100 ng/ml nocodazole for 15 h. Cells were harvested by mitotic shakeoff, washed in PBS, and resuspended in immunoprecipitate lysis buffer (150 mM NaCl, 50 mM Tris, pH 7.5, 1 mM MgCl2, 1 mM EDTA, 0.5% Triton X-100, 1 mM β-glycerophosphate, 1 mM NaMoO4, 1 mM NaF, 1 mM Na tartrate, 1 mM DTT, and protease inhibitor [Roche]) to an approximate protein concentration of 2 mg/ml. Cells were lysed by sonication and then centrifuged in a refrigerated benchtop centrifuge at max speed at 4°C for 15 min. Supernatants were cleared by rotating at 4°C for 4–6 h and then centrifuging. Anti-myc antibody clone 9E10 was cross-linked to protein G Agarose (Thermo Fisher Scientific) using 20 mM dimethyl pimelimidate (Sigma-Aldrich). A volume of bead slurry equivalent to 30 µg antibody was added to the cleared lysate and incubated at 4°C for 16 h with rotation. Beads were washed with immunoprecipitate lysis buffer, and bound proteins were eluted in SDS-PAGE sample buffer without DTT.
Eluates were reduced with 5 mM DTT, alkylated with 15 mM iodoacetamide (Sigma-Aldrich), and separated by SDS-PAGE. Gel regions were excised and digested with trypsin (Promega). Proteins were identified by LC-MS/MS on an Orbitrap Fusion (Thermo Fisher Scientific) exactly as was described previously (Lyons et al., 2009; Choi et al., 2010; Yore et al., 2011; Hood et al., 2012; Zhang et al., 2012).
siRNA and plasmid transfections
siGENOME SMARTpool siRNAs (GE Healthcare) for Nek9, Nek6, and Nek7 were transfected at 10 nM for 48 h using RNAiMax (Thermo Fisher Scientific). siRNAs targeting Mklp2 and Kif14 were synthesized according to previously published sequences (Gruneberg et al., 2006) and transfected at 40 nM for 48 h using RNAiMax. All siRNA sequences are listed in Table S1. pEGFP-C1(Mklp2) and pEGFP-C1(Kif14) were transfected at 1 µg/ml for 24 h using jetPRIME (Polyplus). QuikChange Lightning (Agilent Technologies) was used to generate phosphorylation site mutants of pEGFP-C1(Mklp2) and pEGFP-C1(Kif14), which were then transfected (see Fig. 6, B and C; Fig. 7, B, E, and F; Fig. S3 C, and Fig. S4, A and B). For rescue experiments, 250 ng pEGFP-C1(Mklp2) and 40 nM siMklp2 were cotransfected for 48 h using jetPRIME.
Immunofluorescence microscopy
Cells were extracted with MTSB (100 mM Pipes, pH 6.8, 5 mM MgCl2, 1 mM EGTA, 30% glycerol, and 0.5% Triton X-100) for 2 min and fixed in 3.5% paraformaldehyde in PBS. Primary antibodies were diluted in TBS-BSA (10 mM Tris, pH 7.5, 150 mM NaCl, 10 mg/ml BSA, 0.1% NaN3, and 0.1% Triton X-100) and incubated 1–24 h at 4°C. Secondary antibodies (goat anti–rabbit Alexa Fluor 488 or goat anti–mouse Alexa Fluor 568; Thermo Fisher Scientific) were diluted 1:1,000 in TBS-BSA and 1 µg/ml Hoechst 33258 (Sigma-Aldrich) and then incubated 1 h at room temperature. Coverslips were mounted on slides using ProLong Diamond (Molecular Probes). Images were acquired with a Clara cooled charge-coupled device camera (Andor Technology) mounted on an Axioplan2 microscope (ZEISS) with an Apochromat 100× 1.4 NA oil immersion objective. Exposure times were determined for each protein under WT conditions, and then the same time was applied to any mutants and siRNA-treated cells. Image channels were merged, contrast-adjusted (histogram stretching according to Cromey [2010]), and assembled into figure panels in Photoshop (CS6 v13; Adobe).
Live-cell imaging
EGFP-Mklp2-WT and -S244A HeLa cell lines were plated on glass-bottomed dishes (MatTek Corporation) and transfected with Nek6 or Nek9 siRNA as described in Figs. 3 D, 7 (C and D), and S2 D. During imaging, cells were maintained at 37°C with 5% CO2 in an incubator surrounding the stage on an IX83 inverted microscope (Olympus) with a Zyla sCMOS camera (Andor Technology). Metaphase cells with bipolar spindles were identified using differential interference contrast microscopy. 10 1-µm optical sections were collected along the z axis at 2-min intervals with an Apochromat 63× 1.4 NA oil immersion lens for a total of 20 min. The exposure time was determined for EGFP-Mklp2-WT cells untreated with siRNA, and this time was used consistently for all cells and conditions. Images were obtained using MetaMorph Advanced software (v7.8.3.0). To quantify fluorescence, we measured pixel intensities within a rectangular area surrounding the metaphase plate and subsequent spindle midzones and midbodies as identified by differential interference contrast microscopy. A cytosolic area of the same size was measured and subtracted from the midzone intensity. Maximum projections were used for all quantification and figure displays. To quantify furrow ingression, the distance across the cell equator from membrane to membrane as identified by differential interference contrast microscopy was measured at each time point and normalized to the metaphase value to generate the equatorial cell diameter as a percentage of the metaphase cell width. Images were quantified using ImageJ (National Institutes of Health). Images were contrast-adjusted and assembled into figure panels in Photoshop.
Protein purification
pFastBac1(GST-Nek9), pFastBac1(6×HN-Nek6), pFastBac1(6×HN-Nek7), pFastBac1(6×His-Mklp2), and pFastBac1(GST-Kif14) were transformed into DH10Bac E. coli. Bacmids were purified, and recombination was confirmed by PCR according to the manufacturer’s protocol (Thermo Fisher Scientific). Bacmids were transfected into Sf9 cells using Cellfectin (Thermo Fisher Scientific). 5–7 d after transfection, p0 viral supernatants were harvested and further amplified. Sf9 cells were infected with amplified virus and incubated at 28°C for 60–72 h. Infected cells were treated with 100 nM okadaic acid 2 h before harvest. Cells were pipetted off the flask, pelleted, and washed in PBS.
For 6×His-Mklp2, cells were lysed via sonication in pulldown buffer (10 mM Hepes, pH 7.7, 20 mM β-glycerophosphate, 5 mM EGTA, 5 mM β-mercaptoethanol, 150 mM NaCl, 1% Chaps, 20 mM imidazole, and protease inhibitors), clarified at 8,000 g and 4°C for 30 min, and bound to Ni-NTA beads (QIAGEN) for 2 h at 4°C while rotating. Beads were washed in wash buffer (10 mM Hepes, pH 7.7, 20 mM β-glycerophosphate, 5 mM EGTA, 5 mM β-mercaptoethanol, 500 mM NaCl, and 20 mM imidazole) and eluted in elution buffer (10 mM Hepes, pH 7.7, 20 mM β-glycerophosphate, 5 mM EGTA, 5 mM β-mercaptoethanol, 25 mM NaCl, 400 mM imidazole, 1 mM DTT, 1 mM EDTA, and protease inhibitors). Protein was dialyzed overnight at 4°C against dialysis buffer (10 mM Hepes, pH 7.7, 100 mM NaCl, 1 mM DTT, 0.1 mM EDTA, and 10% glycerol). 6×HN-tagged proteins were purified as in Figs. 4 A, 6 A, 7 A, and 8 B, except with 10, 10, and 250 mM imidazole in the pulldown, wash, and elution buffers, respectively.
For GST-tagged proteins, cells were lysed via sonication in GST lysis buffer (PBS to 350 mM NaCl, 0.5% Triton X-100, 1 mM EDTA, 0.5 mM DTT, and protease inhibitors), clarified at 8,000 g and 4°C for 30 min, and bound to glutathione-Sepharose (GE Healthcare) for 1 h at 4°C while rotating. Beads were washed in PBS and eluted in GST elution buffer (50 mM Tris, pH 8.7, 150 mM NaCl, 50 mM reduced glutathione, 0.5 mM DTT, and 0.1% Chaps). Protein was dialyzed overnight at 4°C against dialysis buffer.
In vitro kinase assays
To measure kinase activity, 600 ng recombinant kinase and 100 µM dephosphorylated tryptic peptide substrate were incubated in kinase buffer (20 mM Hepes, pH 7.7, 1 mM DTT, 20 mM MgCl2, 150 mM NaCl, and 100 µM ATP) at 28°C for 2 h. Negative controls contained 100 µM substrate and buffer only. The ATP consumed in each reaction was measured using the ADP-Glo Kinase Assay kit (Promega) according to the manufacturer’s instructions.
To determine in vitro phosphorylation sites, 1 µg of recombinant kinesin substrate and 100 ng of recombinant kinase were incubated in kinase buffer at 28°C for 16 h. Negative controls contained 1 µg substrate and buffer only. Reactions were reduced with 5 mM DTT, alkylated with 15 mM iodoacetamide (Sigma-Aldrich), and separated by SDS-PAGE. Substrate bands were excised and digested with trypsin (Promega), desalted on a micro solid phase extraction (μSPE) plate (Oasis), and dried. Phosphorylation sites were identified by LC-MS/MS on a QExactive+ (Thermo Fisher Scientific) exactly as described previously (Baker et al., 2009; Hood et al., 2012).
Microtubule bundling, pelleting, and ATP hydrolysis assays
Bundling and pelleting assays were generally performed as described previously (Neef et al., 2003) with the following specifications: taxol-stabilized microtubules were polymerized in vitro from tubulin monomers (Cytoskeleton) by incubation in BRB80 (80 mM Pipes, pH 6.8, 1 mM MgCl2, and 1 mM EGTA) + 1 mM DTT + 1 mM GTP at 37°C with increasing additions of taxol (1 µM, 10 µM, and 100 µM). Microtubules were centrifuged through a 40% glycerol cushion at 70,000 rpm for 12 min in a TLA100 rotor (Beckman Coulter). Microtubule pellet was washed once then resuspended in BRB80 + 1 mM DTT + 25 µM taxol to a final concentration of 1 mg/ml.
500 ng 6×His-Mklp2 WT or S244A was incubated with 25 ng 6×HN-Nek6 or buffer alone in BRB80 + 1 mM DTT + 1 mM ATP + 25 mM KCl at 28°C for 30 min. For bundling assays, 1 µl taxol-stabilized microtubules was added, and the reaction was incubated at 25°C for 15 min. Microtubules were cross-linked with 1% glutaraldehyde and then centrifuged through a 25% glycerol cushion onto acid-washed, poly-lysine–coated coverslips at 15,000 rpm for 1 h in a Sorvall SureSpin rotor. Coverslips were fixed with cold methanol and stained with antitubulin antibody.
For pelleting assays, kinase reactions were performed as in Fig. 8 A, 5 µl taxol-stabilized microtubules or buffer was added, and then reactions were incubated at 25°C for 15 min. Reactions were centrifuged through a 40% glycerol cushion at 55,000 rpm for 20 min in a TLA100 rotor (Beckman Coulter). Microtubule pellets were washed once and then resuspended in SDS-PAGE buffer.
To measure ATP hydrolysis, 250 ng recombinant Mklp2 was incubated with or without 500 ng taxol-stabilized microtubules in BRB80 + 1 mM DTT + 500 µM ATP + 20 mM KCl at 28°C for 30 min. The ATP consumed in each reaction was measured using the ADP-Glo Kinase Assay kit (Promega) according to the manufacturer’s instructions.
Online supplemental material
Fig. S1 shows controls for the Nek9 siRNA efficiency, siRNA-resistant cell lines, and coimmunoprecipitations as well as immunofluorescence images of central spindle proteins not mislocalized by Nek9 depletion. Fig. S2 shows controls for the Nek6 and Nek7 siRNA efficiency and siRNA-resistant cell lines as well as live-cell imaging of EGFP-Mklp2 in siNek6. Fig. S3 includes an experiment analyzing the localization of Kif14 in a synchronized cell population and controls for the production of Kif14-phosphoablated mutants. Fig. S4 shows controls for the production of Mklp2-phosphoablated mutants and EGFP-Mklp2 cell lines, additional immunofluorescence images of Mklp2 in early mitosis, and images of INCENP localization when Mklp2 and Nek9 are depleted concurrently. Table S1 lists the sequences and sources of siRNAs used in this study. Table S2 contains the complete list of Nek9 interactors and quantitative data acquired by LC-MS/MS.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. Duane Compton, Bernardo Orr, Kristina Godek, Dean Madden, and Arminja N. Kettenbach for helpful discussions.
This work was supported by National Institutes of Health grants P20-GM103413 [Project 1], S10-OD016212, and R01-CA155260 to S.A. Gerber and R35-GM119455 to A.N. Kettenbach, and the Dartmouth College Molecular and Cellular Biology training grant T32-GM008704 to support S.N. Cullati.
The authors declare no competing financial interests.
Author contributions: S.A. Gerber and S.N. Cullati conceived the project, designed the experiments, analyzed the data, and wrote the manuscript. L. Kabeche performed the live-cell imaging experiments. A.N. Kettenbach performed AP-MS experiments. All other experiments were performed by S.N. Cullati.
Footnotes
Abbreviations used:
- AP-MS
- affinity purification mass spectrometry
- CPC
- chromosomal passenger complex
- iBAQ
- intensity-based absolute quantification
- INCENP
- inner centromere protein
- LC-MS/MS
- liquid chromatography–tandem mass spectrometry
- Nek
- NIMA-related kinase
References
- Andreassen P.R., Lohez O.D., Lacroix F.B., and Margolis R.L.. 2001. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol. Biol. Cell. 12:1315–1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker C.L., Kettenbach A.N., Loros J.J., Gerber S.A., and Dunlap J.C.. 2009. Quantitative proteomics reveals a dynamic interactome and phase-specific phosphorylation in the Neurospora circadian clock. Mol. Cell. 34:354–363. 10.1016/j.molcel.2009.04.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr F.A., and Gruneberg U.. 2007. Cytokinesis: Placing and making the final cut. Cell. 131:847–860. 10.1016/j.cell.2007.11.011 [DOI] [PubMed] [Google Scholar]
- Bassi Z.I., Audusseau M., Riparbelli M.G., Callaini G., and D’Avino P.P.. 2013. Citron kinase controls a molecular network required for midbody formation in cytokinesis. Proc. Natl. Acad. Sci. USA. 110:9782–9787. 10.1073/pnas.1301328110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belham C., Roig J., Caldwell J.A., Aoyama Y., Kemp B.E., Comb M., and Avruch J.. 2003. A mitotic cascade of NIMA family kinases: Nercc1/Nek9 activates the Nek6 and Nek7 kinases. J. Biol. Chem. 278:34897–34909. 10.1074/jbc.M303663200 [DOI] [PubMed] [Google Scholar]
- Bertran M.T., Sdelci S., Regué L., Avruch J., Caelles C., and Roig J.. 2011. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 30:2634–2647. 10.1038/emboj.2011.179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M., and Earnshaw W.C.. 2003. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 4:842–854. 10.1038/nrm1245 [DOI] [PubMed] [Google Scholar]
- Castedo M., Coquelle A., Vitale I., Vivet S., Mouhamad S., Viaud S., Zitvogel L., and Kroemer G.. 2006. Selective resistance of tetraploid cancer cells against DNA damage-induced apoptosis. Ann. N. Y. Acad. Sci. 1090:35–49. 10.1196/annals.1378.004 [DOI] [PubMed] [Google Scholar]
- Choi S.H., Wright J.B., Gerber S.A., and Cole M.D.. 2010. Myc protein is stabilized by suppression of a novel E3 ligase complex in cancer cells. Genes Dev. 24:1236–1241. 10.1101/gad.1920310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromey D.W. 2010. Avoiding twisted pixels: Ethical guidelines for the appropriate use and manipulation of scientific digital images. Sci. Eng. Ethics. 16:639–667. 10.1007/s11948-010-9201-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evangelista M., Lim T.Y., Lee J., Parker L., Ashique A., Peterson A.S., Ye W., Davis D.P., and de Sauvage F.J.. 2008. Kinome siRNA screen identifies regulators of ciliogenesis and hedgehog signal transduction. Sci. Signal. 1:ra7 10.1126/scisignal.1162925 [DOI] [PubMed] [Google Scholar]
- Fry A.M., O’Regan L., Sabir S.R., and Bayliss R.. 2012. Cell cycle regulation by the NEK family of protein kinases. J. Cell Sci. 125:4423–4433. 10.1242/jcs.111195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., and Pellman D.. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282. 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. 2009. The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat. Rev. Mol. Cell Biol. 10:9–20. 10.1038/nrm2609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green R.A., Paluch E., and Oegema K.. 2012. Cytokinesis in animal cells. Annu. Rev. Cell Dev. Biol. 28:29–58. 10.1146/annurev-cellbio-101011-155718 [DOI] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Honda R., Nigg E.A., and Barr F.A.. 2004. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 166:167–172. 10.1083/jcb.200403084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Li X., Chan E.H., Chalamalasetty R.B., Nigg E.A., and Barr F.A.. 2006. KIF14 and citron kinase act together to promote efficient cytokinesis. J. Cell Biol. 172:363–372. 10.1083/jcb.200511061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J.W., and Adams P.D.. 2001. Cyclin-dependent kinases. Chem. Rev. 101:2511–2526. 10.1021/cr0001030 [DOI] [PubMed] [Google Scholar]
- Hood E.A., Kettenbach A.N., Gerber S.A., and Compton D.A.. 2012. Plk1 regulates the kinesin-13 protein Kif2b to promote faithful chromosome segregation. Mol. Biol. Cell. 23:2264–2274. 10.1091/mbc.E11-12-1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hümmer S., and Mayer T.U.. 2009. Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Curr. Biol. 19:607–612. 10.1016/j.cub.2009.02.046 [DOI] [PubMed] [Google Scholar]
- Kaneta Y., and Ullrich A.. 2013. NEK9 depletion induces catastrophic mitosis by impairment of mitotic checkpoint control and spindle dynamics. Biochem. Biophys. Res. Commun. 442:139–146. 10.1016/j.bbrc.2013.04.105 [DOI] [PubMed] [Google Scholar]
- Kettenbach A.N., Schweppe D.K., Faherty B.K., Pechenick D., Pletnev A.A., and Gerber S.A.. 2011. Quantitative phosphoproteomics identifies substrates and functional modules of Aurora and Polo-like kinase activities in mitotic cells. Sci. Signal. 4:rs5 10.1126/scisignal.2001497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Lee K., and Rhee K.. 2007. NEK7 is a centrosomal kinase critical for microtubule nucleation. Biochem. Biophys. Res. Commun. 360:56–62. 10.1016/j.bbrc.2007.05.206 [DOI] [PubMed] [Google Scholar]
- Kitagawa M., Fung S.Y., Hameed U.F., Goto H., Inagaki M., and Lee S.H.. 2014. Cdk1 coordinates timely activation of MKlp2 kinesin with relocation of the chromosome passenger complex for cytokinesis. Cell Reports. 7:166–179. 10.1016/j.celrep.2014.02.034 [DOI] [PubMed] [Google Scholar]
- Krzywicka-Racka A., and Sluder G.. 2011. Repeated cleavage failure does not establish centrosome amplification in untransformed human cells. J. Cell Biol. 194:199–207. 10.1083/jcb.201101073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni J.S., and Jacks T.. 1998. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol. Cell. Biol. 18:1055–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurell E., Beck K., Krupina K., Theerthagiri G., Bodenmiller B., Horvath P., Aebersold R., Antonin W., and Kutay U.. 2011. Phosphorylation of Nup98 by multiple kinases is crucial for NPC disassembly during mitotic entry. Cell. 144:539–550. 10.1016/j.cell.2011.01.012 [DOI] [PubMed] [Google Scholar]
- Lyons P.D., Peck G.R., Kettenbach A.N., Gerber S.A., Roudaia L., and Lienhard G.E.. 2009. Insulin stimulates the phosphorylation of the exocyst protein Sec8 in adipocytes. Biosci. Rep. 29:229–235. 10.1042/BSR20080162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoub M.R., Qasim Rasi M., and Quarmby L.M.. 2004. A NIMA-related kinase, Fa2p, localizes to a novel site in the proximal cilia of Chlamydomonas and mouse kidney cells. Mol. Biol. Cell. 15:5172–5186. 10.1091/mbc.E04-07-0571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliga Z., Junqueira M., Toyoda Y., Ettinger A., Mora-Bermúdez F., Klemm R.W., Vasilj A., Guhr E., Ibarlucea-Benitez I., Poser I., et al. . 2013. A genomic toolkit to investigate kinesin and myosin motor function in cells. Nat. Cell Biol. 15:325–334. 10.1038/ncb2689 [DOI] [PubMed] [Google Scholar]
- Minn A.J., Boise L.H., and Thompson C.B.. 1996. Experssion of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 10:2621–2631. 10.1101/gad.10.20.2621 [DOI] [PubMed] [Google Scholar]
- Musacchio A., and Salmon E.D.. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393. 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Neef R., Preisinger C., Sutcliffe J., Kopajtich R., Nigg E.A., Mayer T.U., and Barr F.A.. 2003. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J. Cell Biol. 162:863–875. 10.1083/jcb.200306009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A. 2001. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2:21–32. 10.1038/35048096 [DOI] [PubMed] [Google Scholar]
- O’Regan L., and Fry A.M.. 2009. The Nek6 and Nek7 protein kinases are required for robust mitotic spindle formation and cytokinesis. Mol. Cell. Biol. 29:3975–3990. 10.1128/MCB.01867-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan L., Blot J., and Fry A.M.. 2007. Mitotic regulation by NIMA-related kinases. Cell Div. 2:25 10.1186/1747-1028-2-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan L., Sampson J., Richards M.W., Knebel A., Roth D., Hood F.E., Straube A., Royle S.J., Bayliss R., and Fry A.M.. 2015. Hsp72 is targeted to the mitotic spindle by Nek6 to promote K-fiber assembly and mitotic progression. J. Cell Biol. 209:349–358. 10.1083/jcb.201409151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapley J., Nicolàs M., Groen A., Regué L., Bertran M.T., Caelles C., Avruch J., and Roig J.. 2008. The NIMA-family kinase Nek6 phosphorylates the kinesin Eg5 at a novel site necessary for mitotic spindle formation. J. Cell Sci. 121:3912–3921. 10.1242/jcs.035360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards M.W., O’Regan L., Mas-Droux C., Blot J.M., Cheung J., Hoelder S., Fry A.M., and Bayliss R.. 2009. An autoinhibitory tyrosine motif in the cell-cycle-regulated Nek7 kinase is released through binding of Nek9. Mol. Cell. 36:560–570. 10.1016/j.molcel.2009.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L. 2011. Mitosis in vertebrates: the G2/M and M/A transitions and their associated checkpoints. Chromosome Res. 19:291–306. 10.1007/s10577-010-9178-z [DOI] [PubMed] [Google Scholar]
- Roig J., Mikhailov A., Belham C., and Avruch J.. 2002. Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev. 16:1640–1658. 10.1101/gad.972202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roig J., Groen A., Caldwell J., and Avruch J.. 2005. Active Nercc1 protein kinase concentrates at centrosomes early in mitosis and is necessary for proper spindle assembly. Mol. Biol. Cell. 16:4827–4840. 10.1091/mbc.E05-04-0315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem H., Rachmin I., Yissachar N., Cohen S., Amiel A., Haffner R., Lavi L., and Motro B.. 2010. Nek7 kinase targeting leads to early mortality, cytokinesis disturbance and polyploidy. Oncogene. 29:4046–4057. 10.1038/onc.2010.162 [DOI] [PubMed] [Google Scholar]
- Sdelci S., Schütz M., Pinyol R., Bertran M.T., Regué L., Caelles C., Vernos I., and Roig J.. 2012. Nek9 phosphorylation of NEDD1/GCP-WD contributes to Plk1 control of γ-tubulin recruitment to the mitotic centrosome. Curr. Biol. 22:1516–1523. 10.1016/j.cub.2012.06.027 [DOI] [PubMed] [Google Scholar]
- Shalom O., Shalva N., Altschuler Y., and Motro B.. 2008. The mammalian Nek1 kinase is involved in primary cilium formation. FEBS Lett. 582:1465–1470. 10.1016/j.febslet.2008.03.036 [DOI] [PubMed] [Google Scholar]
- Yissachar N., Salem H., Tennenbaum T., and Motro B.. 2006. Nek7 kinase is enriched at the centrosome, and is required for proper spindle assembly and mitotic progression. FEBS Lett. 580:6489–6495. 10.1016/j.febslet.2006.10.069 [DOI] [PubMed] [Google Scholar]
- Yore M.M., Kettenbach A.N., Sporn M.B., Gerber S.A., and Liby K.T.. 2011. Proteomic analysis shows synthetic oleanane triterpenoid binds to mTOR. PLoS One. 6:e22862 10.1371/journal.pone.0022862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalli D., Bayliss R., and Fry A.M.. 2012. The Nek8 protein kinase, mutated in the human cystic kidney disease nephronophthisis, is both activated and degraded during ciliogenesis. Hum. Mol. Genet. 21:1155–1171. 10.1093/hmg/ddr544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A., Petrov K.O., Hyun E.R., Liu Z., Gerber S.A., and Myers L.C.. 2012. The Tlo proteins are stoichiometric components of Candida albicans mediator anchored via the Med3 subunit. Eukaryot. Cell. 11:874–884. 10.1128/EC.00095-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.