Actin-capping protein is a key component of the actin cytoskeleton at sites of clathrin-mediated endocytosis. Farrell et al. show that a newly discovered component of the endocytic machinery belongs to the dynein light chain family and regulates the recruitment of actin-capping protein in a dynein motor–independent manner.
Abstract
Clathrin- and actin-mediated endocytosis is essential in eukaryotic cells. In this study, we demonstrate that Tda2 is a novel protein of the endocytic machinery necessary for normal internalization of native cargo in yeast. Tda2 has not been classified in any protein family. Unexpectedly, solving the crystal structure of Tda2 revealed it belongs to the dynein light chain family. However, Tda2 works independently of the dynein motor complex and microtubules. Tda2 forms a tight complex with the polyproline motif–rich protein Aim21, which interacts physically with the SH3 domain of the Arp2/3 complex regulator Bbc1. The Tda2–Aim21 complex localizes to endocytic sites in a Bbc1- and filamentous actin–dependent manner. Importantly, the Tda2–Aim21 complex interacts directly with and facilitates the recruitment of actin-capping protein, revealing barbed-end filament capping at endocytic sites to be a regulated event. Thus, we have uncovered a new layer of regulation of the actin cytoskeleton by a member of a conserved protein family that has not been previously associated with a function in endocytosis.
Introduction
Clathrin-mediated endocytosis (CME) is an essential process spanning all eukaryotes (Boettner et al., 2011; Traub and Bonifacino, 2013; Kirchhausen et al., 2014; Goode et al., 2015). CME has roles in the maintenance of membrane composition, signaling, protein trafficking, polarity, virus uptake, and nutrient and drug internalization (Brodsky et al., 2001; Brodsky, 2012). Applications of CME extend from basic eukaryotic cell biology to physiology and human disease (Robinson, 2015). CME is highly conserved between yeast and mammalian cells in protein composition, progression, and function. Close to 60 proteins have been identified with roles in CME, and live-cell fluorescent microscopy has revealed that they form patches on the plasma membrane, where they assemble in a precisely choreographed manner (Kaksonen et al., 2003, 2005; Weinberg and Drubin, 2012). Early in CME, a subset of coat proteins gathers at the membrane in an immobile but variable timed phase. Intermediate coat proteins and integral membrane protein cargoes then arrive at the endocytic site. Later, actin polymerization and actin-associated proteins are observed, concomitant with the inward movement of the membrane and most of the CME machinery. Lastly, scission and uncoating processes free the vesicle and allow the recycling of machinery components. Despite the description of the dynamics of many machinery proteins, several aspects of the endocytic process and its regulation are not well understood. For example, recently described putative endocytic factors are yet to be established as true components of the CME machinery and, if confirmed, the molecular function they may serve remains to be elucidated (Burston et al., 2009; Weinberg and Drubin, 2012; Farrell et al., 2015; Goode et al., 2015).
Actin polymerization produces force to overcome membrane tension and causes membrane bending during CME (Kukulski et al., 2012). When yeast cells are incubated with the actin monomer–sequestering agent latrunculin A, early endocytic coat proteins are recruited to the membrane, but invagination does not occur (Kaksonen et al., 2003; Newpher et al., 2005). Similarly, when the link between actin and the membrane is broken, such as in cells carrying a deletion of the SLA2 gene, large “comet tails” of actin are present, but the membrane does not internalize (Kaksonen et al., 2003; Newpher et al., 2005). Actin polymerization is a highly regulated process, and numerous actin-associated proteins are needed for the membrane invagination step of CME. For example, the α and β subunits of actin-capping protein localize to CME sites and are required for actin filament barbed-end capping function (Amatruda and Cooper, 1992; Amatruda et al., 1992). Thus, yeast cells carrying a deletion of the CAP1 or CAP2 genes encoding the actin-capping protein α and β subunits show a defect in CME (Kaksonen et al., 2005). The mechanisms and regulation of this CME network of actin-associated proteins are still being uncovered. For instance, it is not known whether capping protein simply binds in free fashion to newly formed barbed ends or if it is actively recruited via interactions with other proteins during CME.
In contrast to the fundamental functions of the actin cytoskeleton, microtubules and microtubule-associated motor proteins such as kinesin and dynein are not known to localize to CME sites or to have a role in endocytosis. Interestingly, some members of the dynein light chain protein family have functions independent of the dynein motor complex and microtubules (Chuang et al., 2005; Yeh et al., 2006, 2012; Conde et al., 2010; Li et al., 2011; Rapali et al., 2011). Nevertheless, no dynein light chain has been reported to work in endocytosis.
In this study, we show that Tda2 is a novel component of the CME machinery that associates with the actin cytoskeleton during the late stages of the endocytic process and is needed for the efficient uptake of native cargo. X-ray crystallography revealed Tda2 is an ancient dynein light chain. Tda2 is more closely related to the TcTex1 type than to the LC8 or LC7 branches of the dynein light chain family, thus representing the first TcTex1 dynein light chain found in yeast. Importantly, Tda2 has a dynein motor– and microtubule-independent role in CME. Tda2 forms a tight complex with another endocytic protein, Aim21, and together they are recruited to CME sites through interaction with Bbc1, a component of the WASp/Myo module and a regulator of actin polymerization during CME. The Tda2–Aim21 complex binds actin-capping protein directly and mediates its recruitment to CME sites. Therefore, we have discovered a new function for the dynein light chain working as a regulator of the actin cytoskeleton during CME.
Results
Tda2 is a novel late-stage component of the endocytic machinery
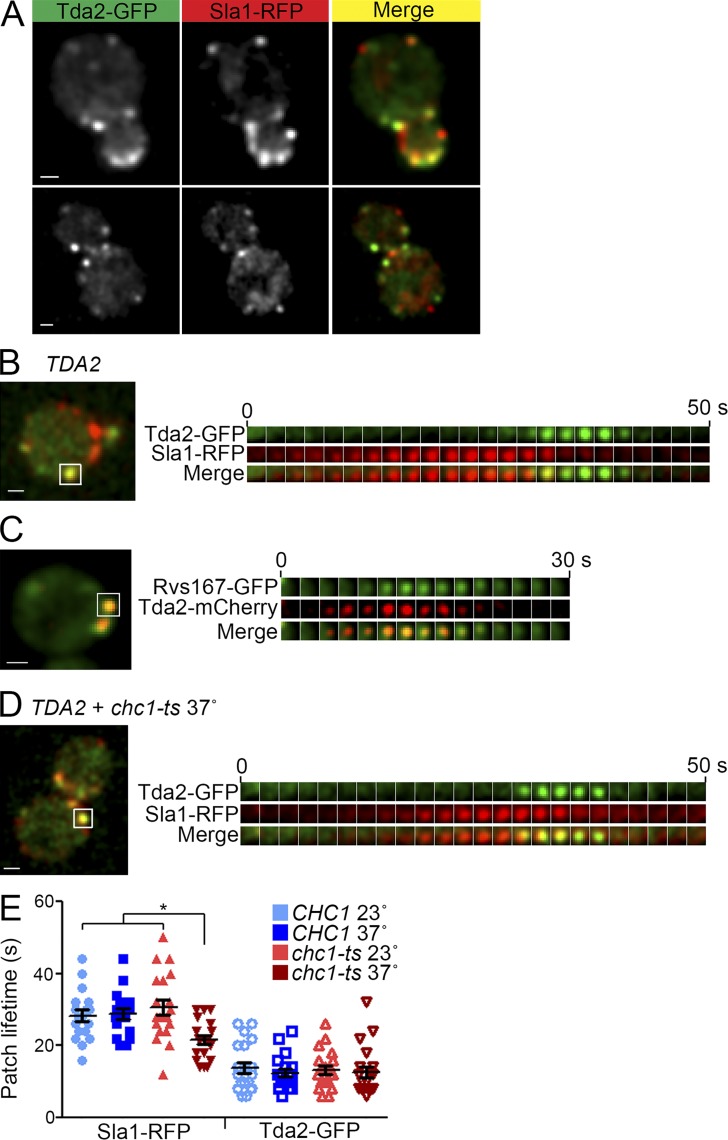
Tda2 was an uncharacterized protein of unknown function. In a screening for new candidate components of the CME machinery, we found that Tda2 localizes to endocytic sites, suggesting Tda2 may function in endocytosis (Farrell et al., 2015). In still images, Tda2-GFP colocalized with the well-established CME intermediate coat protein Sla1-RFP with a Pearson correlation coefficient of 0.49 ± 0.13 (Fig. 1 A; Farrell et al., 2015). In time-lapse imaging, Tda2-GFP appeared at 96% of Sla1-RFP patches and, conversely, Sla1-RFP appeared at 98% of Tda2-GFP patches. Tda2-GFP had a patch lifetime of 13 ± 4 s, appearing only at the late stages of Sla1-RFP fluorescence (Fig. 1 B and Video 1). Tda2-GFP overlapped Sla1-RFP fluorescence by several seconds and persisted after Sla1-RFP disappeared. Furthermore, when analyzed in cells with a temperature-sensitive clathrin heavy chain (chc1-ts), Tda2-GFP showed no patch lifetime change upon clathrin destabilization by high temperature, whereas coat proteins such as Sla1-RFP had a significantly decreased patch lifetime (Fig. 1, D and E). These results suggest that Tda2 may be a component of the CME machinery, in particular the actin module or the scission module, rather than a coat protein. Interestingly, Tda2-mCherry appeared at endocytic sites just before the scission protein Rvs167-GFP (Fig. 1 C). Localization, timing, dynamics, and independence from the clathrin coat suggest that Tda2 is part of the late-stage endocytic machinery, likely the actin cytoskeleton.
Figure 1.
Tda2 localizes to sites of CME at late stages of the endocytic process and independently of the clathrin coat. (A) Live-cell fluorescence microscopy showing strong colocalization between Tda2-GFP and Sla1-RFP. (B) Dynamics of Tda2-GFP and Sla1-RFP were compared. (Left) One frame of a video indicating with a white box the endocytic site used for constructing a kymograph. (Right) Kymograph illustrating how Tda2 is recruited to late stages of endocytosis. (C) Live-cell fluorescence microscopy showing strong colocalization between Tda2-mCherry and Rvs167-GFP. Notice Tda2-mCherry arrives a few seconds before Rvs167-GFP. (D and E) Patch lifetimes of Sla1-RFP and Tda2-GFP were measured in WT cells (CHC1) and cells carrying a temperature-sensitive allele of the clathrin heavy chain (chc1-ts) after incubating the cells at 23° or 37°. Clathrin disruption (37°) reduces patch lifetime in Sla1-RFP but not Tda2-GFP. Bars, 1 µm. Error bars indicate means ± SEM. *, P < 0.05.
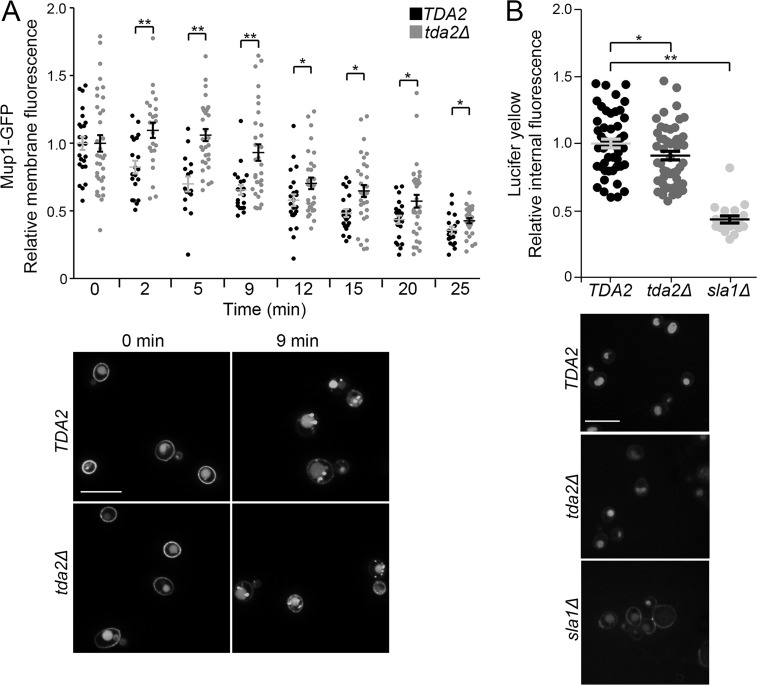
To analyze endocytic dependence on Tda2, endocytic assays were performed with cells carrying a deletion of the TDA2 gene (tda2Δ). First, we used Mup1-GFP to track cargo endocytosis. Mup1 is a methionine transporter that is localized to the plasma membrane in methionine-starved cells, but quickly internalized via CME after returning the cells to methionine-rich media. A delay in Mup1-GFP internalization was observed in tda2Δ cells relative to control cells (Fig. 2 A). Second, a small but significant defect in fluid-phase endocytosis was measured in tda2Δ cells using the fluorescent dye Lucifer yellow (Fig. 2 B). The Lucifer yellow internalization defect was less pronounced than the one observed with cells carrying a deletion of the SLA1 gene (sla1Δ), which is known to essentially stall CME (Fig. 2 B; Holtzman et al., 1993; Howard et al., 2002; Kaksonen et al., 2005; Di Pietro et al., 2010). The extent of the endocytic defect in tda2Δ cells was comparable to the one we observed for other endocytic machinery mutants including Las17–MP8-12, a mutant displaying misregulation of actin polymerization during CME (Feliciano and Di Pietro, 2012; Farrell et al., 2015). Collectively, these data indicate that Tda2 is a newly identified late-stage component of the CME machinery needed for optimal endocytosis.
Figure 2.
Tda2 is needed for optimal endocytosis of a native cargo. (A) Endocytosis of Mup1-GFP was analyzed in WT (TDA2) and tda2Δ cells as described in the Fluorescence microscopy and endocytosis assays section of Materials and methods. Internalization of Mup1-GFP was delayed in tda2Δ cells. Representative images at indicated time points are pictured below the plots. (B) WT (TDA2), tda2Δ, and sla1Δ cells were incubated with the fluorescent dye Lucifer yellow and analyzed by fluorescence microscopy. Lucifer yellow internalization was modestly reduced in tda2Δ cells, whereas sla1Δ cells portray a severe defect. Vacuoles containing Lucifer yellow are seen after 2 h incubation, pictured below the plots. Bars, 10 µm. Error bars indicate means ± SEM. *, P < 0.05; **, P < 0.01.
Tda2 is a dynein light chain
Tda2 has very low sequence similarity to any characterized protein in databases and therefore has not been classified into any protein family from which to infer function at a molecular level. To determine its 3D structure, recombinant 6-histidine–tagged Tda2 (His-Tda2) was expressed in Escherichia coli, purified, concentrated, and crystalized using the hanging drop vapor diffusion method. The crystal structure of Tda2 was determined by the multiwavelength anomalous diffraction method using data collected from a bromide-derivative crystal, where the anomalous signal extended to 2.5 Å. This model was refined against native data to a resolution of 2.3 Å and R/Rfree values of 21.2/26.5% (Table 1). The asymmetric unit was composed of two Tda2 molecules. The full-length protein chains, apart from the first four residues and some loops within the B chain monomer, were clearly observed in the electron density for both molecules.
Table 1. Tda2 data collection and refinement statistics.
| PDB Code | 5VKY |
|---|---|
| Data collection | |
| Space group | P21212 |
| Cell dimensions | |
| a, b, c (Å) | 50.39, 161.26, 33.26 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 53.75-2.30 (2.38-2.30) |
| Rmerge | 0.035 (0.195) |
| CC ½ | 1.00 (0.986) |
| Mean I/σI | 32.9 (8.4) |
| Completeness (%) | 100 (100) |
| Multiplicity | 6.8 (6.4) |
| Refinement | |
| Resolution (Å) | 40.31-2.3 (2.38-2.30) |
| Reflections | 12,714 (1,257) |
| Rwork/Rfree (%) | 21.1/26.5 (22.9/31.8) |
| Number of atoms | |
| Protein | 1,843 |
| Ligand/ion | 0 |
| Water | 87 |
| B-factors | |
| Protein | 45 |
| Water | 47.7 |
| Rmsd | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 0.87 |
| Ramachandran | |
| Favored (%) | 98.6 |
| Outliers (%) | 0 |
Values in parentheses are for highest-resolution shell.
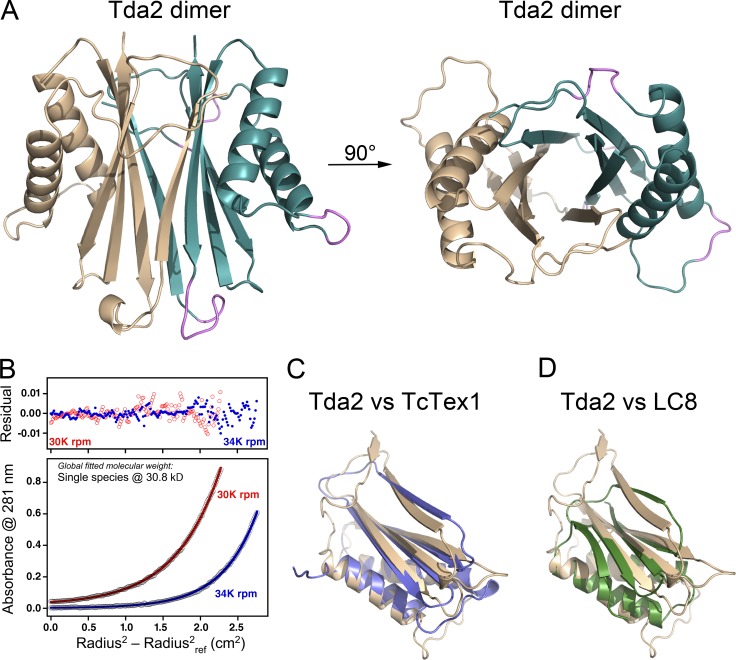
Tda2 forms a homodimer via a combined central β sandwich, with each β sheet composed of four strands arranged in an antiparallel fashion (β2’–β1–β4–β3), and the two helices from each Tda2 molecule flanked this central β sandwich (Fig. 3 A). The existence of the dimer in solution was verified by analytical ultracentrifugation that revealed a single 30.8-kD species, consistent with a dimer of 15.6-kD monomers (Fig. 3 B). The first strand of each β sheet was contributed by the homodimer partner, and the major interface between the two molecules was the antiparallel β2’–β1 strand interaction. The dimer interface buried a total of 3,138 Å2 solvent–accessible surface area (∼1,570 Å2 of each molecule).
Figure 3.
Tda2 belongs to the dynein light chain family. (A) Ribbon representation of the Tda2 crystal structure (PDB entry 5VKY) showing a homodimer via a combined central β sandwich, with each β sheet composed of four strands arranged in an antiparallel fashion and the two helices from each Tda2 molecule flanking the central β sandwich. One Tda2 molecule is shown in wheat color and the other in cyan, with some loops (magenta) on the cyan subunit being in poor density and omitted from the final model. Structure is rotated 90° for another view on the right. (B) Analytical ultracentrifugation data showing Tda2 to behave as a single species with a molecular weight of 30.8 kD, reflecting a dimer of 15.6-kD monomers in solution. (C and D) Cα-based superposition of Tda2 (wheat) and D. melanogaster TcTex1 (violet; PDB entry 1YGT, chain A) or S. cerevisiae LC8 (green; PDB entry 4HT6, chain E).
Surprisingly, the overall Tda2 fold was highly homologous to the dynein light chain of the TcTex1 type. The closest structural homologue is the Drosophila melanogaster TcTex1 (PDB: 1YGT chain A; root mean square deviation [rmsd] of ∼1.8 Å on 85/104 C pairs; Fig. 3 C; Williams et al., 2005, 2007; Hall et al., 2009). Tda2 also shared structural homology with the LC8-type dynein light chain. Among the LC8-type dynein light chains with known structure, the Saccharomyces cerevisiae LC8 is the closest structural homologue of Tda2 (PDB: 4HT6 chain E; rmsd of ∼2.1 Å on 71/86 C pairs; Fig. 3 D; Rao et al., 2013). This is consistent with the fact that TcTex1 and LC8 have a similar fold. However, Tda2 shares only 9% overall sequence identity with S. cerevisiae LC8 and 14–16% with TcTex1 from Homo sapiens, Mus musculus, D. melanogaster, and Caenorhabditis elegans, reinforcing the idea that Tda2 is a TcTex1-type dynein light chain. The third type of dynein light chain, LC7 (also known as Roadblock), has a more distantly related fold. For example, D. melanogaster LC7 (PDB: 3L7H chains C and D) has an rmsd of ∼3.6 Å on 71/90 C pairs. Also, Tda2 shares a mere 9–12% sequence identity with LC7 from H. sapiens, M. musculus, D. melanogaster, and C. elegans. The data therefore reveal that Tda2 is an ancient dynein light chain more closely related to the TcTex1 branch of the protein family.
Tda2 functions independently of the dynein motor complex and microtubules
The identification of Tda2 as a member of the dynein light chain family was unexpected as microtubules and the microtubule-associated dynein motor complex are not known to have roles in endocytosis. Although metazoan TcTex1 has been shown to have both dynein motor–dependent and –independent functions, it is not known to work in endocytosis (Chuang et al., 2005; Yeh et al., 2006, 2013; Conde et al., 2010; Li et al., 2011; Rapali et al., 2011). To investigate whether Tda2 is associated with the dynein motor complex and microtubules, we followed several approaches.
First, cells were treated with the microtubule-depolymerizing agent nocodazole. Previously known endocytic proteins (Sla1, Myo5, and Rvs167) did not show any change in recruitment or patch lifetimes (Fig. S1 A) under conditions in which microtubules were readily disrupted (Fig. S1 C). Similarly, Tda2 recruitment and lifetime was unaffected by microtubule depolymerization, suggesting that microtubules are not needed for Tda2 recruitment or normal progression of endocytosis (Fig. S1, A–C). Additionally, cells carrying a deletion of the TDA2 gene (tda2Δ) did not show any obvious defects in microtubules as observed with Tub1-GFP (Fig. S1 C).
Second, live-cell imaging analysis did not show any colocalization between Tda2-mCherry and Dyn2-GFP, the yeast LC8 dynein light chain component of the dynein motor complex (Pearson correlation coefficient = 0.04 ± 0.03; Fig. S1 D). Furthermore, Dyn2-GFP was not observed at endocytic sites (Fig. S1 E).
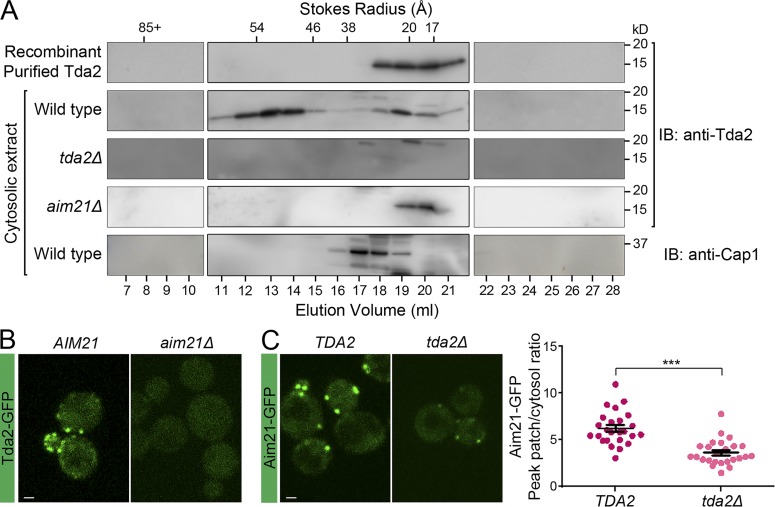
Third, biochemical experiments failed to detect Tda2 association with the dynein motor complex. To facilitate further investigation into endogenous Tda2, a rabbit affinity-purified anti-Tda2 antibody was generated and verified using recombinant His-Tda2 purified from bacteria as well as with cytosolic extracts from WT and tda2Δ cells (Fig. 4 A). Then, a TAP-tagged strain of the dynein heavy chain (DYN1-TAP) was used to purify the dynein motor complex under conditions that preserve motor integrity and activity (Markus and Lee, 2011; Markus et al., 2012; Lammers and Markus, 2015). Immunoblotting analysis showed that Tda2 was present in total cell extracts but did not copurify with the dynein heavy chain (Fig. S1 F). Size-exclusion chromatography on a Superpose-6 column was then performed to examine the behavior of Tda2 (Fig. 4 A). According to the column calibration with protein standards, recombinant purified Tda2 eluted at the size expected for the stable homodimer demonstrated in the crystal structure (Fig. 4 A; Stokes radius ∼20 Å and ∼29 kD). A cytosolic extract from tda2Δ cells showed no signal, confirming the specificity of our antibody (Fig. 4 A). In cytosolic extracts prepared from WT cells, Tda2 eluted as two peaks, the homodimer and a larger species with an estimated Stokes radius of ∼54 Å, corresponding with molecular weights of ∼250 kD for a globular complex (or less for an asymmetric complex; Fig. 4 A). This cytosolic Tda2 complex is much smaller than the large dynein motor protein complex of >1,000 kD, which eluted with the column exclusion volume (fractions 7–8), further supporting Tda2 as separate from the dynein motor complex.
Figure 4.
Tda2 exists both as a homodimer and in a larger complex with Aim21. (A) S. cerevisiae cytosol was fractionated by size-exclusion chromatography on a Superose 6 column, and the resulting fractions were analyzed by immunoblotting (IB) with the indicated antibodies. Recombinant Tda2 eluted at the expected position for the homodimer (peaking at fractions 19–20, Stokes radius ∼20 Å). Cytosolic cellular extracts prepared from WT cells showed endogenous Tda2 eluting as a homodimer (peaking at fractions 19–20) as well as a larger complex (peaking at fraction 13, Stokes radius ∼54 Å). Cytosolic cellular extracts prepared from tda2Δ cells portrayed no Tda2, confirming the specificity of our affinity-purified anti-Tda2 antibody. Cytosolic extracts prepared from aim21Δ cells showed the Tda2 homodimer species but no evidence of the larger Tda2 complex, indicating that Aim21 is essential for the larger Tda2 complex. In cytosolic extracts from WT cells, endogenous capping protein (Cap1) eluted in fractions 17–18. The Stokes radius of standard proteins is indicated at the top. (B) Live-cell fluorescence microscopy showing Tda2-GFP is no longer recruited to endocytic sites in aim21Δ cells. (C, left) Live-cell fluorescence microscopy showing Aim21-GFP recruitment to endocytic sites is severely reduced in tda2Δ cells. (Right) Quantification of the Aim21-GFP peak fluorescence intensity at endocytic patches in control (TDA2) and tda2Δ cells. Bars, 1 µm. Error bars indicate means ± SEM. ***, P < 0.001.
Fourth, the dynein motor pathway has a genetic interaction with the Kar9 pathway with dyn1Δkar9Δ double mutant cells showing a strong synthetic effect (Fig. S1 G; Lee et al., 2005). In contrast, tda2Δkar9Δ double mutant cells did not show a synthetic effect (Fig. S1 G). Therefore, the data indicate that Tda2 does not localize, function, associate, or interact genetically with the dynein motor complex and microtubules. However, the data suggest that Tda2 is part of a different complex.
Tda2 is in a stable complex with Aim21
Aim21 is a protein of unknown function that may be linked to Tda2 (Gavin et al., 2002). As determined by size-exclusion chromatography, the majority of cytosolic Tda2 exists in a complex of ∼54 Å (Fig. 4 A). However, in cytosolic fractions from aim21Δ cells analyzed in parallel, the ∼54-Å Tda2 species was no longer present, and only the ∼20-Å Tda2 homodimer was observed (Fig. 4 A), suggesting that Aim21 is an essential component of the ∼54-Å complex. To test for a direct physical association between Tda2 and Aim21, GST pulldown experiments with purified recombinant proteins were performed. GST alone and GST-Aim21 were immobilized on glutathione-Sepharose and subsequently incubated with His-Tda2. GST-Aim21 was able to specifically pull down His-Tda2, indicating a direct interaction between Tda2 and Aim21 (Fig. S2). Consistent with a Tda2–Aim21 complex, localization of Tda2-GFP was completely lost in aim21Δ cells, indicating that Tda2 recruitment to endocytic sites is strictly dependent on Aim21 (Fig. 4 B). Immunoblotting analysis of total cell extracts indicated that Tda2-GFP was not destabilized in aim21Δ cells (Fig. S2 B). Conversely, Aim21-GFP recruitment to endocytic sites was strongly dependent on Tda2, as determined with tda2Δ cells, although some residual Aim21-GFP was observed (Fig. 4 C). Quantification of the Aim21-GFP mislocalization in tda2Δ cells likely underestimated the defect as many patches had too little Aim21-GFP fluorescence to be detected and included in the analysis. Immunoblotting analysis of total cell extracts showed Aim21-GFP was not destabilized in tda2Δ cells (Fig. S2 B).
We further tested aim21Δ cells using the same endocytosis assays described in the first section of the Results, Mup1-GFP and Lucifer yellow (Fig. S3). In both assays, aim21Δ cells showed an endocytosis defect similar to the one observed with tda2Δ cells relative to WT cells. Finally, Tda2-RFP and Aim21-GFP colocalized tightly at CME sites (Fig. S4 D). Collectively, these data suggest that Tda2 interacts directly with Aim21, forming a stable complex of ∼54 Å that was present in the cytosol and recruited to CME sites to function during late stages of endocytosis.
The Tda2–Aim21 complex is recruited to the actin cytoskeleton at CME sites through direct interaction with Bbc1
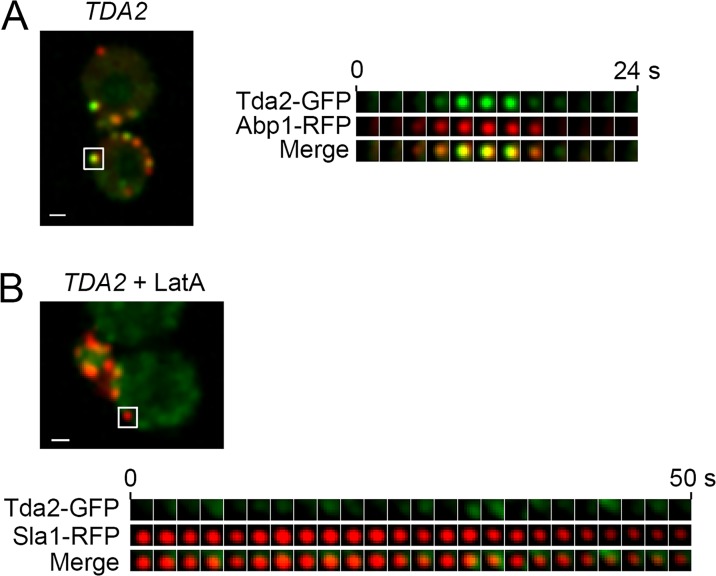
Because of the late recruitment of Tda2 to the endocytic site, its dynamics and lifetime are similar to filamentous actin-associated proteins (Fig. 1; Kaksonen et al., 2005). Indeed, live-cell imaging showed Tda2-GFP and the well-established marker of filamentous actin Abp1-RFP had very similar dynamics at endocytic sites (Fig. 5 A). More importantly, upon treatment of cells with the actin-depolymerizing agent latrunculin A, Tda2-GFP was no longer recruited to endocytic patches (Fig. 5 B). The behavior of Tda2 (and by extension Aim21) was typical of proteins associated with filamentous actin at CME sites. In latrunculin A–treated cells, coat proteins such as Sla1-RFP were recruited but remained immobile as the patch could not be internalized without actin polymerization (Fig. 5 B; Kaksonen et al., 2003; Newpher et al., 2005).
Figure 5.
Tda2 localizes to the actin cytoskeleton at sites of CME. (A) Live-cell fluorescence microscopy demonstrating strong colocalization between Tda2-GFP and Abp1-RFP. (B) When yeast cells were treated with latrunculin A (LatA), Tda2-GFP was not recruited to endocytic sites, whereas Sla1-RFP was recruited but remained largely immobile. White boxes indicate endocytic sites used for constructing kymographs on the right. Bars, 1 µm.
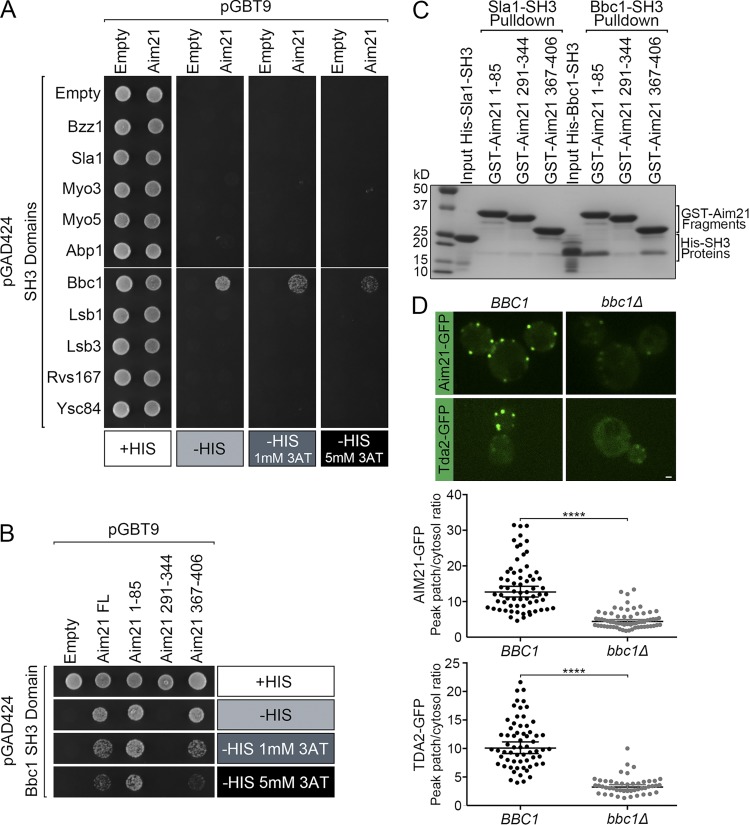
How is the Tda2–Aim21 complex recruited to the actin cytoskeleton at CME sites? Aim21 is a 679-residue protein predicted to be largely disordered. Interestingly, our analysis of its primary structure detected 10 sequences conforming to the polyproline motif consensus (PxxP, where x represents any amino acid). We reasoned that some of these sequences may bind to SH3 domains, which are prevalent among CME machinery proteins. Yeast two-hybrid analysis was performed to test for binding of full-length Aim21 to a panel of SH3 domains from known endocytic proteins (Tonikian et al., 2009). Only the SH3 domain of Bbc1 interacted with Aim21 as indicated by growth in selective plates lacking histidine (−HIS; Fig. 6 A). The Aim21 PxxP sequences were clustered in three separate segments of the protein corresponding with amino acids 1–85, 291–344, and 367–406. The ability of each of these fragments to interact with the Bbc1 SH3 domain was assessed by yeast two-hybrid analysis, and a comparison of the relative interaction strength was obtained by using selective plates with increasing concentrations of 3-amino-1,2,4-triazole (3-AT). The Bbc1 SH3 domain showed strong binding to Aim21 fragment 1–85, moderate binding to fragment 367–406, and no binding to fragment 291–344 (Fig. 6 B).
Figure 6.
Aim21 interacts physically with Bbc1, and recruitment of the Tda2–Aim21 complex to CME sites depends on Bbc1. (A) Yeast two-hybrid analysis: yeast cells were cotransformed with plasmids expressing the GAL4 DNA binding (pGBT9) and activation (pGAD424) domains fused to Aim21 and the SH3 domains from the indicated endocytic proteins, respectively. Cells were spotted onto plates containing histidine (+HIS; as control) or selective medium lacking histidine (−HIS) and containing various concentrations of 3-AT. Cell growth indicative of interaction was only detected within the Bbc1 SH3 domain. (B) Yeast two-hybrid analysis with polyproline motif–rich fragments of Aim21 (amino acids 1–85, 291–344, and 367–406) and the Bbc1 SH3 domain. Aim21 fragment 1–85 demonstrated the strongest interaction. (C) A GST pulldown assay was performed with GST fused to Aim21 fragments 1–85, 291–344, and 367–406. Each GST fusion protein was incubated with His-Sla1-SH3 and His-Bbc1-SH3. The bound proteins were analyzed by Coomassie staining demonstrating direct binding between the Bbc1 SH3 domain and Aim21. Fragment 1–85 showed the strongest binding. (D, top) Live-cell fluorescence microscopy showing recruitment of Aim21-GFP and Tda2-GFP to endocytic sites is dramatically reduced in bbc1Δ cells relative to WT cells (BBC1). (Bottom) Quantification of the Aim21-GFP and Tda2-GFP peak fluorescence intensity at endocytic patches in control (BBC1) and bbc1Δ cells. Bar, 1 µm. Error bars indicate means ± SEM. ****, P < 0.0001.
To corroborate a direct association between the Aim21 polyproline motif–rich fragments and the Bbc1 SH3 domain, we performed GST pulldown assays. GST fusions of the Aim21 fragments 1–85, 291–344, and 367–406 were immobilized on glutathione-Sepharose and subsequently incubated with purified 6-Histidine–tagged Bbc1 SH3 domain (His-Bbc1-SH3). Parallel experiments were performed with purified His-Sla1-SH3 as a negative control based on the yeast two-hybrid results. Coomassie blue staining showed that the Aim21 fragment 1–85 strongly bound His-Bbc1-SH3 but did not bind His-Sla1-SH3, demonstrating specificity (Fig. 6 C). Aim21 fragment 367–406 also bound His-Bbc1-SH3, although to a lesser extent, and the Aim21 fragment 291–344 showed no binding (Fig. 6 C). The relative level of binding paralleled the data obtained with the yeast two-hybrid system and confirmed a direct interaction between Aim21 and Bbc1. Given that Bbc1 is a component of the WASp/Myo module that regulates actin polymerization at CME sites, the physical interaction between Aim21 and Bbc1 is well positioned to mediate recruitment of the Tda2–Aim21 complex to CME sites. Confirming this idea, Aim21-GFP and Tda2-GFP localization to endocytic sites was drastically decreased in cells carrying a deletion of the BBC1 gene (bbc1Δ) compared with WT cells (Fig. 6 D). Collectively, these data suggest that the Tda2–Aim21 complex associates with the actin cytoskeleton at CME sites through interaction with Bbc1.
The Tda2–Aim21 complex binds actin-capping protein and regulates its recruitment to endocytic sites
As described in the previous section, Tda2-GFP and Abp1-RFP have very similar dynamics at endocytic sites (Fig. 5 A). Interestingly, upon close inspection, the fluorescent localization of Tda2-GFP appeared slightly offset from Abp1-RFP, with Tda2-GFP appearing closer to the membrane than Abp1-RFP fluorescence. Quantification showed the peak patch intensity of Tda2-GFP was 0.13 ± 0.06 µm (n = 20 patches and seven cells) closer to the plasma membrane than the peak patch intensity of Abp1-RFP (Fig. S4 A). A similar offset in localization had been previously demonstrated between actin-capping protein (Cap1/Cap2 dimer) and Abp1 in sla2Δ cells, with capping protein localizing closer to the plasma membrane (Michelot et al., 2013). When Tda2-GFP fluorescence was compared with Cap1-RFP, there was no shift in location of peak patch intensity (Fig. S4 B). This result was confirmed in cells with switched fluorescent tags (Fig. S5). Consistent with a tight Tda2–Aim21 complex, Aim21-GFP also showed no shift relative to Cap1-RFP (Fig. S4 C) and Tda2-RFP (Fig. S4 D). The analysis was further validated by determining the localization offset between Myo5-GFP and Abp1-mCherry, a pair of actin-associated proteins known to be spatially separated. Quantification showed the peak patch intensity of Myo5-GFP was 0.12 ± 0.01 µm (n = 29 patches and eight cells) closer to the plasma membrane than Abp1-RFP (Fig. S4 E). Thus, Tda2-Aim21 is an actin filament–associated protein complex with similar localization to actin-capping protein. Interestingly, a large-scale screening suggested actin-capping protein may be linked to Tda2 and Aim21, although no followup experiments were reported (Gavin et al., 2002).
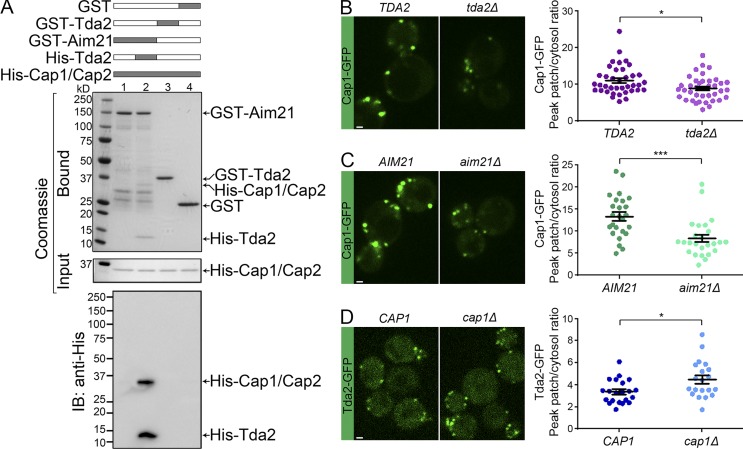
To test for a direct physical association between the Tda2–Aim21 complex and actin-capping protein, GST pulldown experiments with purified recombinant proteins were performed (Fig. 7 A). GST-Aim21 (lane 1), GST-Tda2 (lane 3), and GST alone (lane 4) were immobilized on glutathione-Sepharose and subsequently incubated with actin-capping protein carrying a 6His-tag in its Cap1 subunit (His-Cap1/Cap2 dimer). Neither GST-Aim21 nor GST-Tda2 were able to pull down actin-capping protein, as determined by Coomassie blue staining and anti–6His-tag immunoblotting analysis (Fig. 7 A). However, parallel experiments in which both 6His-tagged Tda2 and actin-capping protein were incubated with GST-Aim21 showed robust binding of both proteins (Fig. 7 A, lane 2). Anti–6His-tag immunoblotting analysis confirmed the identity of the bound protein bands (Fig. 7 A, bottom). Thus, a reconstituted Tda2–Aim21 complex, but not each protein separately, strongly and directly binds to actin-capping protein.
Figure 7.
The Tda2–Aim21 complex binds actin-capping protein and regulates its recruitment to CME sites. (A) GST pulldown assay was performed with GST-Aim21 (lanes 1 and 2), GST-Tda2 (lane 3), and GST alone (lane 4). Each GST fusion protein was incubated with purified actin-capping protein carrying a polyhistidine tag in its Cap1 subunit (His-Cap1/Cap2; lanes 1–4). One GST-Aim21 sample was incubated with His-Tda2 (to reconstitute the Tda2–Aim21 complex) in addition to His-Cap1/Cap2 (lane 2). The bound proteins were analyzed by Coomassie staining (top) and anti-His immunoblotting (IB; bottom). His-Cap1/Cap2 bound robustly to the GST-Aim21–His-Tda2 complex but not to GST-Aim21 alone or GST-Tda2 alone. (B, left) Live-cell fluorescence microscopy of actin-capping protein (Cap1-GFP) showing reduced recruitment to endocytic sites in tda2Δ cells relative to WT cells (TDA2). (Right) Quantification of the Cap1-GFP peak fluorescence intensity at endocytic patches in control (TDA2) and tda2Δ cells. (C, left) Live-cell fluorescence microscopy of actin-capping protein (Cap1-GFP) showing reduced recruitment to endocytic sites in aim21Δ cells relative to WT cells (AIM21). (Right) Quantification of the Cap1-GFP peak fluorescence intensity at endocytic patches in control (AIM21) and aim21Δ cells. (D, left) Live-cell fluorescence microscopy of Tda2-GFP showing increased recruitment to endocytic sites in cap1Δ cells relative to WT cells (CAP1). (Right) Quantification of the Tda2-GFP peak fluorescence intensity at endocytic patches in control (CAP1) and cap1Δ cells. Bars, 1 µm. Error bars indicate means ± SEM. *, P < 0.05; ***, P < 0.001.
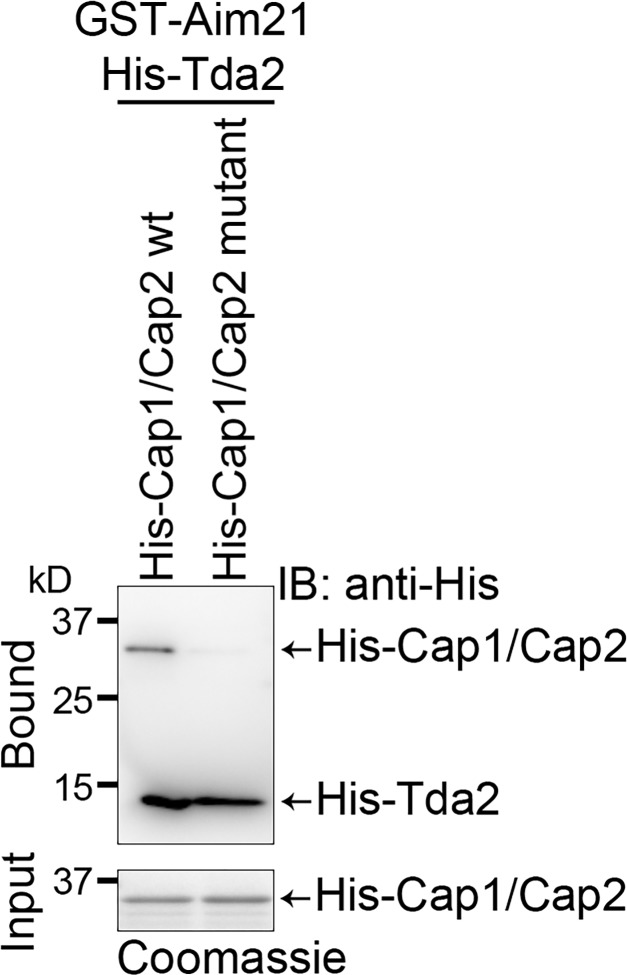
Mouse and chicken actin-capping proteins have been shown to interact physically with a diverse group of proteins containing a capping protein interaction motif (Hernandez-Valladares et al., 2010). Mutation of surface residues R15 and Y79 of the actin-capping protein β subunit abolished the interaction (Hernandez-Valladares et al., 2010; Edwards et al., 2015). In this study, we introduced homologous R15A and Y80A point mutations in yeast Cap2 and tested for binding to the Tda2–Aim21 complex using the GST pulldown assay with purified recombinant proteins described in Fig. 7 A. Binding of mutant actin-capping protein was significantly reduced compared with WT actin-capping protein, suggesting a conserved binding mechanism (Fig. 8).
Figure 8.
Binding to the Tda2–Aim21 complex depends on conserved residues on the surface of actin-capping protein. GST-Aim21 pulldown assays were performed as in Fig. 7 A. GST-Aim21 was incubated with His-Tda2 and His-Cap1/Cap2 WT or a mutant in which the R15A and Y80A point mutations were introduced in the Cap2 subunit. Bound proteins were analyzed by anti-His immunoblotting (IB). Binding of His-Cap1/Cap2 mutant was significantly decreased compared with His-Cap1/Cap2 WT.
Consistent with a physical interaction between the Tda2–Aim21 complex and capping protein, Cap1-GFP localization to endocytic sites was decreased in cells carrying a deletion of the TDA2 gene (tda2Δ) or the AIM21 gene (aim21Δ) compared with WT cells (Fig. 7, B and C). However, Tda2-GFP recruitment to endocytic sites was not decreased but rather was increased in cells carrying a deletion of the CAP1 gene (cap1Δ; Fig. 7 D). These data suggest that the Tda2–Aim21 complex works upstream of capping protein and is needed for efficient capping protein recruitment to CME sites. Nevertheless, capping protein is not part of a stable complex with Tda2 and Aim21 because it does not cofractionate with the cytosolic ∼54-Å Tda2 peak in size-exclusion chromatography analysis (Fig. 4 A).
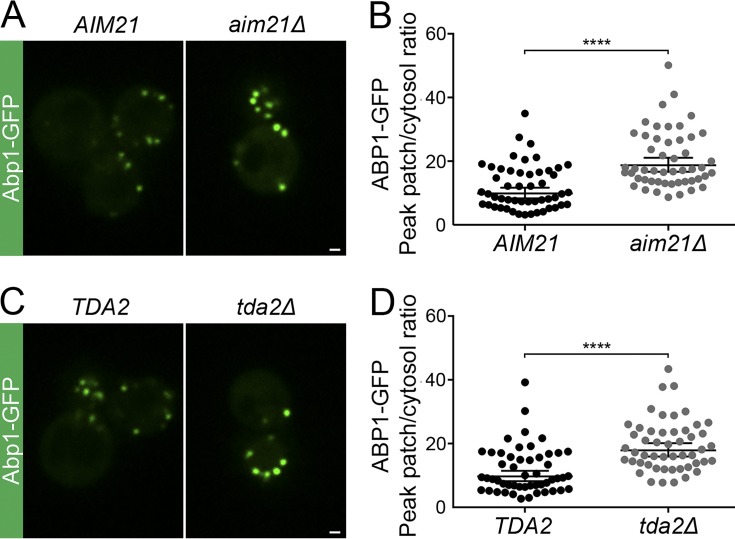
Deletion of actin-capping protein causes an endocytic phenotype characterized by increased Abp1 recruitment to endocytic sites presumably caused by the formation of longer, uncapped actin filaments (Kaksonen et al., 2005). We reasoned that if the Tda2–Aim21 complex functions in actin-capping protein recruitment to CME sites, deletion of TDA2 or AIM21 would cause a similar phenotype. Indeed, Abp1-GFP levels at endocytic sites were increased in cells carrying a deletion of the AIM21 gene (aim21Δ) or the TDA2 gene (tda2Δ) compared with WT cells (Fig. 9). Overall, the data suggest that the Tda2–Aim21 complex binds actin-capping protein directly and facilitates its recruitment to CME sites for efficient filament barbed-end capping.
Figure 9.
Deficiency in the Tda2–Aim21 complex elicits a similar phenotype as actin-capping protein deficiency. (A) Abp1-GFP was visualized by fluorescence microscopy using live WT cells (AIM21) or aim21Δ cells. (B) Quantification of the Abp1-GFP peak fluorescence intensity at endocytic patches in control (AIM21) and aim21Δ cells. (C) Abp1-GFP was visualized by fluorescence microscopy using live WT cells (TDA2) or tda2Δ cells. (D) Quantification of the Abp1-GFP peak fluorescence intensity at endocytic patches in control (TDA2) and tda2Δ cells. Bars, 1 µm. Error bars indicate means ± SEM. ****, P < 0.0001.
Discussion
Several uncharacterized proteins that may function in endocytosis still await establishment as true CME machinery components and, if confirmed, require elucidation of their function at a molecular level (Burston et al., 2009; Weinberg and Drubin, 2012; Farrell et al., 2015; Goode et al., 2015). In this study, we show how one such uncharacterized protein, Tda2, functions during late stages of the endocytic process and is needed for optimal internalization of native cargo, thereby defining Tda2 as a bona fide CME factor. Excitingly, Tda2 was revealed to be a member of the dynein light chain family that is associated with the actin cytoskeleton and required for proper recruitment of actin-capping protein to sites of CME. Thus, we have discovered a new regulatory mechanism of the actin cytoskeleton during CME by a member of a protein family that has not been previously associated with function in endocytosis or actin polymerization.
Despite not being previously classified in any protein family, the 3D structure of Tda2 demonstrated that it is a dynein light chain most closely related to the TcTex1 member of the family. This is the first identification of a TcTex1-type dynein light chain in yeast. In metazoans, TcTex1 corresponds with one of three dynein light chain subfamilies; the others are the well-studied LC8 as well as the LC7 (also known as Roadblock) light chains. TcTex1 copurifies with the dynein motor complex in metazoans, demonstrates binding to the dynein intermediate chain, and has been predicted to link cargo to the motor complex, though cargo linking is under debate (Williams et al., 2007; Hall et al., 2009). Importantly, the TcTex1 dynein light chain has also been observed in roles outside the dynein motor complex, but not in endocytosis or regulation of actin filament dynamics (Chuang et al., 2005; Yeh et al., 2006, 2013; Conde et al., 2010; Li et al., 2011; Rapali et al., 2011). Our results show Tda2 does not work associated with microtubules or the dynein motor complex. First, Tda2 localization was not affected by microtubule depolymerization. Furthermore, the CME machinery in general was unaffected by microtubule depolymerization. Second, Tda2 did not colocalize with the LC8 light chain, which does function with the dynein motor complex. Third, Tda2 failed to copurify with the dynein heavy chain. Fourth, in contrast with Dyn1, Tda2 did not interact genetically with Kar9. On the other hand, the data demonstrate that Tda2 exists in a distinct complex with Aim21 and that the Tda2–Aim21 complex is a component and regulator of the actin cytoskeleton at CME sites. This expands the repertoire of cellular processes performed by the dynein light chain family.
Aim21 is another uncharacterized protein with potential function in endocytosis (Burston et al., 2009). Our results directly link Tda2 with Aim21 both physically and functionally: (a) the ∼54-Å Tda2 complex detected by size-exclusion chromatography of yeast cytosol requires Aim21; (b) Tda2 and Aim21 interact directly in GST pulldown assays using purified recombinant proteins; (c) the recruitment of Tda2 to endocytic sites is absolutely dependent on Aim21 and, conversely, recruitment of Aim21 is largely dependent on Tda2; (d) Tda2 and Aim21 colocalize tightly at endocytic sites and have the same dynamics; (e) deletion of either gene elicits a similar defect in endocytosis of endogenous cargo Mup1; (f) localization of both Tda2 and Aim21 to CME sites depends on Bbc1; and (g) both Tda2 and Aim21 are needed for efficient actin-capping protein recruitment to CME sites. Collectively, these data suggest that Tda2 and Aim21 interact directly as part of a complex that is stable enough to withstand fractionation by size-exclusion chromatography. As TcTex1 and LC8 are strong dimers, these proteins have a “dimerization chaperone” activity, mediating dimerization of proteins both as part of and outside the dynein motor complex (Williams et al., 2007; Benison and Barbar, 2009; Hall et al., 2009; Rapali et al., 2011). The light chains typically bind disordered regions of their partners, promoting them to dimerize and form ordered structures, often coiled-coils (Williams et al., 2007; Benison and Barbar, 2009; Hall et al., 2009; Rapali et al., 2011). Interestingly, Aim21 is predicted to be a largely disordered protein with the exception of amino acids 85–110, which are predicted to form a coiled-coil. Thus, Tda2 likely performs a similar dimerization chaperone role with Aim21 (Fig. 10). The size-exclusion chromatography analysis is compatible with two Aim21 molecules per Tda2 dimer in the complex. The molecular weight of these four peptides (179 kD) is slightly below the estimated complex molecular weight (∼250 kD for a globular complex), which is consistent with an asymmetric complex (Fig. 10). We also cannot rule out the possibility of additional proteins in the complex that were not identified in the scope of this study.
Figure 10.
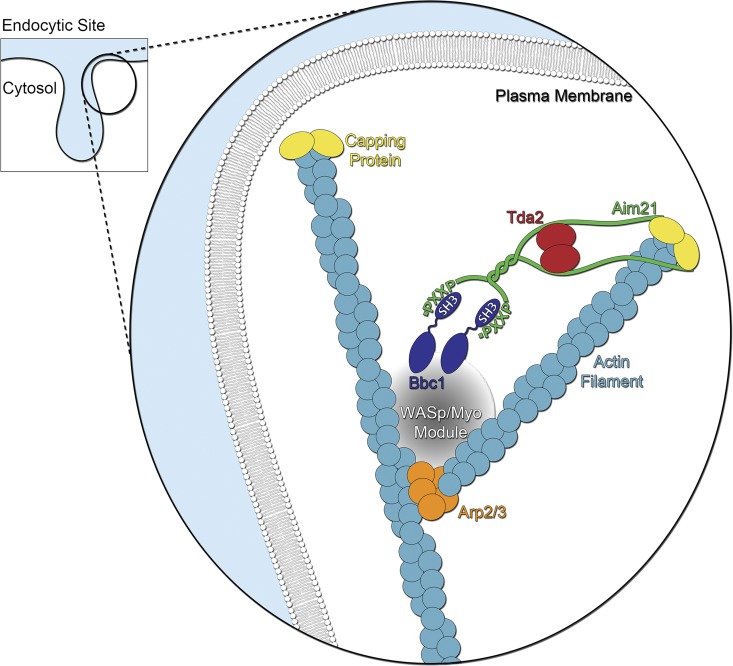
Model of Tda2–Aim21 complex function in CME. Tda2 is a TcTex1-type dynein light chain homologue and exists as a stable homodimer. Tda2 associates with Aim21 into a stable complex in which Tda2 likely acts as an Aim21 “dimerization chaperone.” The Tda2–Aim21 complex is recruited to endocytic sites via interaction with Bbc1, a component of the WASp/Myo module. The Tda2–Aim21 complex interacts physically with actin-capping protein and mediates its recruitment to the barbed end of actin filaments.
Our results show that the Tda2–Aim21 complex defines a new player of the endocytic actin cytoskeleton network. Localization of the Tda2–Aim21 complex to CME sites depends on actin filaments, and its recruitment time tightly matches that of Abp1 and other filamentous actin-associated proteins. Direct physical interaction between polyproline motif–rich regions of Aim21 and the SH3 domain of Bbc1 provide a mechanistic link between the Tda2–Aim21 complex and a well-established component of the actin cytoskeleton at CME sites (Fig. 10). Accordingly, the drastic reduction in Tda2 and Aim21 localization to CME sites in cells carrying a deletion of the BBC1 gene suggests that the interaction between Aim21 and Bbc1 mediates recruitment of the Tda2–Aim21 complex to the actin cytoskeleton. Further linking the Tda2–Aim21 complex with the actin cytoskeleton, we also found it interacts directly with actin-capping protein in in vitro assays using purified recombinant proteins. Actin-capping protein is likely not stably associated with the Tda2–Aim21 complex in vivo because it is not detected in the cytosolic gel filtration fractions corresponding with ∼54 Å. Instead, the data support a model in which actin-capping protein and the Tda2–Aim21 complex associate in a transient manner at CME sites where Tda2–Aim21 facilitates actin-capping protein recruitment (Fig. 10). This model is supported by the fact that Cap1-GFP recruitment to CME sites was decreased in tda2Δ and aim21Δ cells. Conversely, in cap1Δ cells, Tda2-GFP recruitment to CME sites was not decreased but slightly increased, consistent with the model that the Tda2–Aim21 complex functions upstream of actin-capping protein. These results have several important conceptual implications.
The first implication is that actin-capping protein has critical roles in actin assembly in vitro as well as in live cells by binding to the barbed end of the filament and blocking addition and loss of subunits (Kim et al., 2004). Consequently, yeast cells carrying a deletion of the CAP1 and CAP2 genes display defective CMEs (Kaksonen et al., 2005). It was previously assumed that capping protein simply diffuses in free fashion to bind and cap newly formed barbed ends. However, it was recently demonstrated that in human cells, capping protein localization and function at the leading edge of the cell is mediated by interaction with other proteins (Edwards et al., 2015). Our findings suggest capping protein recruitment and function at yeast CME sites is facilitated by interaction with the Tda2–Aim21 component of the actin network. Furthermore, our results show the surface residues of actin-capping protein involved in binding to recruitment partners are conserved between yeast, chicken, and mouse, suggesting a conserved mechanism. Therefore, the concept that actin-capping protein is actively recruited to specific cellular locations is broader and more ancient than previously appreciated.
The second implication is that mathematical modeling of endocytic actin patch kinetics in fission yeast concluded that in vivo binding of capping protein to the filament barbed end is 10 times faster than determined in vitro with purified capping protein and actin (Berro et al., 2010). Binding to the Tda2–Aim21 complex should increase the local concentration of actin-capping protein at CME sites, thus increasing the rate of filament capping and providing an elegant explanation to this apparent discrepancy.
The third implication is that Bbc1 is a well-established factor of the WASp/Myo module that works with the Arp2/3 complex in the formation of a branched actin filament network at CME sites (Fig. 10). Bbc1 regulates the WASp-mediated activation of the Arp2/3 complex in vitro, Bbc1 polyproline motifs interact physically with the SH3 domains of Myo3 and Myo5, and deletion of the BBC1 gene contributes to an exaggerated network of actin filaments at CME sites in live cells. Because of its ability to bind both Bbc1 and capping protein, the Tda2–Aim21 complex is well positioned to physically link the pointed and barbed filament ends, thus limiting the length of actin filaments (Fig. 10). Such architectural constraint would explain the highly dendritic actin network observed at CME sites, which is composed of short actin filaments of only 20 actin subunits and ideally suited to effectively exert the force needed for membrane bending (Young et al., 2004). The observation of excessive actin polymerization (higher Abp1-GFP levels) and deficient internalization in cap1Δ cells (Kaksonen et al., 2005) and Tda2–Aim21 complex–deficient cells described in this study is likely a result of uncapped, long, and wobbly filaments that are less effective at applying membrane-bending force.
In conclusion, we have firmly established two new components of the endocytic machinery and uncovered a mechanism for recruitment of actin-capping protein showing that barbed-end filament capping at endocytic sites is a regulated event. This newly discovered layer of regulation of the actin cytoskeleton is mediated by a member of the conserved dynein light chain family, which was not known to function in endocytosis or the regulation of actin polymerization.
Materials and methods
Plasmids, yeast strains, and yeast two-hybrid assay
Plasmid for bacterial expression of recombinant His-Tda2 was created by PCR amplification of full-length ORF TDA2 from S. cerevisiae genomic DNA and was cloned into pET30a(+) using NdeI and SalI restriction sites. The 6-histidine tag was encoded in the forward primer used in the PCR so that the protein contained the sequence MHHHHHH directly attached to the amino terminus of Tda2. Plasmid for bacterial expression of recombinant GST-Aim21 was created by PCR amplification of full-length ORF AIM21 from S. cerevisiae genomic DNA and cloned in-frame into pGEX-5X-1 using EcoRI and SalI sites. Bicistronic plasmid for simultaneous expression in bacteria and of both yeast actin-capping protein subunits was created by PCR amplification of full-length ORF CAP1 and CAP2 from S. cerevisiae genomic DNA and cloned into pETDuet using restriction sites EcoRI and SalI for CAP1 (multiple cloning site I) and FseI and XhoI for CAP2 (multiple cloning site II). The pETDuet plasmid encodes for a 6-histidine tag attached to Cap1 for protein purification. The R15A and Y80A point mutations were introduced in CAP2 using the In-Fusion system (Takara Bio Inc.).
Plasmids for yeast two-hybrid analysis of the SH3 domains of BZZ1, Sla1, Myo3, Myo5, Abp1, Bbc1, Lsb1, Lsb3, Rvs167, and Ysc84 were generated by PCR amplification of the corresponding DNA fragment from S. cerevisiae genomic DNA and cloning into pGAD424 using the In-Fusion system. Plasmids for yeast two-hybrid analysis of full-length Aim21 and fragments encoding Aim21 amino acids 1–85, 291–344, and 367–406 were created by PCR amplification of the corresponding DNA sequence from S. cerevisiae genomic DNA and cloned into pGBT9 using the In-Fusion system. Plasmids for bacterial expression of recombinant GST–Aim21 fusions encoding Aim21 amino acids 1–85, 291–344, and 367–406 were created by PCR amplification of the corresponding DNA fragment from S. cerevisiae genomic DNA and cloned into pGEX-5X-1 using the In-Fusion system. Plasmids for bacterial expression of recombinant His-Bbc1 SH3 domain was created by PCR amplification of the corresponding DNA fragment from S. cerevisiae genomic DNA and cloned into pET30(+) using the In-Fusion system. Plasmids for expression of His-Sla1 SH3 domains 1 and 2 (His-Sla1-SH3) was previously reported (Feliciano and Di Pietro, 2012). Plasmid for expression of GFP-Tub1 was a gift from S. Markus (Colorado State University, Fort Collins, CO). Constructs were verified by DNA sequencing.
Background strains of S. cerevisiae BY4741 and BY4742 were used for this study. WT GFP-expressing strains (Tda2-GFP, Mup1-GFP, Sla1-GFP, Dyn2-GFP, Cap1-GFP, Aim21-GFP, Abp1-GFP, and Rvs167-GFP) were obtained from the yeast GFP library (Invitrogen). The library Tda2-GFP strain (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, and TDA2-GFP::HIS3) was mated with (MATα his3Δ1, leu2Δ0, met15-Δ0, and ura3Δ0), and the resulting diploid strain was sporulated and subjected to tetrad dissection. From this cross, we obtained SDY425 (MATα his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, and TDA2-GFP::HIS3). SDY425 was subsequently crossed with the knockout library strains cap1Δ and am21Δ, provided by L. Stargell (Colorado State University, Fort Collins, CO), to obtain SDY741 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, cap1Δ::KanMX4, and TDA2-GFP::HIS3) and SDY754 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, aim21Δ::KanMX4, and TDA2-GFP::HIS3). SDY425 was also crossed with SDY360 to create SDY710 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, ABP1-RFP::TRP1, and TDA2-GFP::HIS3). The TDA2 and CAP1 ORFs were tagged with RFP or mCherry by PCR amplification of pFA6a-mRFP-KanMX6 or pFA6a-mCherry-KanMX6 and homologous recombination (Ito et al., 1983; Longtine et al., 1998) to create SDY740 (MATα his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, and TDA2-RFP::KanMX6), SDY820 (MATα his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, and TDA2-mCherry::KanMX6), and SDY756 (MATα his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, and CAP1-RFP::KanMX6). SDY740 was then crossed with GFP library strains to obtain SDY751 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, CAP1-GFP::HIS3, and TDA2-RFP::KanMX6) and SDY781 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, AIM21-GFP::HIS3, and TDA2-RFP::KanMX6). SDY756 was crossed with Tda2-GFP or Aim21-GFP library strains to create SDY776 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, CAP1-RFP::KanMX6, and TDA2-GFP::HIS3) and SDY778 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, CAP1-RFP::KanMX6, and AIM21-GFP::HIS3). SDY820 was crossed with Dyn2-GFP and Rvs167-GFP library strains to create SDY827 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, TDA2-mCherry::KanMX6, and DYN2-GFP::HIS3) and SDY828 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, TDA2-mCherry::KanMX6, and RVS167-GFP::HIS3). Deletion strains (tda2Δ, sla1Δ, cap1Δ, aim21Δ, and kar9Δ) were obtained from the yeast knockout library (GE Healthcare). The tda2Δ strain was mated, sporulated, and dissected to create SDY722 (MATα his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, and tda2Δ::KanMX4). SDY722 was then crossed with GFP library strains to create SDY656 (MATα his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, tda2Δ::KanMX4, and MUP1-GFP::HIS3), SDY725 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, tda2Δ::KanMX4, and CAP1-GFP::HIS3), and SDY727 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, tda2Δ::KanMX4, and AIM21-GFP::HIS3). SDY647 (MATα his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, and MUP1-GFP::HIS3) was mated with aim21Δ strain from the library, sporulated, and dissected to create SDY729 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, aim21Δ::KanMX4, and MUP1-GFP::HIS3). SDY423 (MATα ura3-52, leu2-3,112, his3-Δ200, trp1-Δ901, lys2-801, suc2-Δ9, GAL-MEL chc1-521::URA3, TDA2-GFP::HIS3, and SLA1-RFP::KanMX6) was used for experiments in Fig. 1 (A, B, D, and E). Deletion strains bbc1Δ, aim21Δ, and tda2Δ from the yeast knockout library were subjected to GFP tagging of AIM21, TDA2, and ABP1 ORFs by PCR amplification of pFA6a-GFP-HIS3MX6 and homologous recombination (Ito et al., 1983; Longtine et al., 1998) to create SDY1011 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, bbc1Δ::KanMX4, and AIM21-GFP::HIS3), SDY1012 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, bbc1Δ::KanMX4, and TDA2-GFP::HIS3), SDY1013 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, aim21Δ::KanMX4, and ABP1-GFP::HIS3), and SDY1030 (MATa his3Δ1, leu2Δ0, met15-Δ0, ura3Δ0, tda2Δ::KanMX4, and ABP1-GFP::HIS3). DYN1-TAP and deletion strains kar9Δ (SMY193) and dyn1Δ (SMY16) were gifts from S. Markus (Markus et al., 2009, 2012; Markus and Lee, 2011; Lammers and Markus, 2015). GPY1805 was subjected to GFP tagging of MYO5 and mCherry tagging of ABP1 by PCR amplification of pFA6a-GFP-TRP1 and pFA6a-mCherry-HIS3MX6 and homologous recombination to obtain SDY871 (MATa, ura3Δ-52, leu2-3,112, his3-Δ200, trp1-Δ901, lys2-801, suc2-Δ9, sst1Δ::LYS2, ste2Δ::LEU2, MYO5-GFP::TRP1, and ABP1-mCherry::HIS3).
Yeast two-hybrid experiments were performed using pGBT9 and pGAD424 vectors (Takara Bio Inc.), and AH109 S. cerevisiae cells were grown on synthetic dropout media lacking leucine and tryptophan as selection markers as previously described (Starcevic and Dell’Angelica, 2004). 3-AT was used to test for higher binding stringency.
Biochemical methods
Cytosolic yeast extracts were obtained as follows: liquid nitrogen frozen yeast pellets were ground in a blender, resuspended in PBS (12 mM phosphate, 147 mM NaCl, and 3 mM KCl, pH 7.35) supplemented with protease inhibitor cocktail (Sigma-Aldrich), and were subjected to ultracentrifugation at 4°C for 15 min at 400,000 g as previously described (Feliciano et al., 2011; Feliciano and Di Pietro, 2012).
Size-exclusion chromatography fractionation of cytosolic extracts (∼2 mg/ml) was performed on a Superose 6 10/300 GL column (GE Healthcare) connected to a fast protein liquid chromatography system (GE Healthcare) equilibrated with PBS. Elution was performed at a flow rate of 0.4 ml/min at 4°C with fractions collected every 1 ml and analyzed by immunoblotting. The column was calibrated using blue dextran and the standard proteins cytochrome C, carbonic anhydrase, BSA, alcohol dehydrogenase, β-amylase, apoferritin, and thyroglobulin.
Recombinant fusion proteins His-Tda2, His-Bbc1-SH3, His-Sla1-SH3, the His-Cap1/Cap2 capping protein dimer, GST-Aim21, GST-Aim21 1–85, GST-Aim21 291–344, and GST-Aim21 367–406 were expressed in BL21 codon plus E. coli and purified using the TALON cobalt affinity resin or glutathione–Sepharose-4B (Takara Bio Inc.; BD) as described previously (Di Pietro et al., 2004, 2010).
The generation and characterization of antibodies against Tda2 was performed as described previously (Di Pietro et al., 2004; Bultema et al., 2012). Polyclonal antibodies were raised in rabbit by immunizing with recombinant His-Tda2 protein. Subsequently, these antibodies were affinity-purified using as a ligand the same recombinant His-Tda2 protein that had been covalently coupled to Affi-Gel 15 activated media (Bio-Rad Laboratories). The antibodies were validated using yeast total cell extracts from WT, Tda2-GFP, and tda2Δ strains. Antibodies against yeast actin-capping protein were provided by J. Cooper (Washington University School of Medicine, St. Louis, MO).
GST-pulldown assays were performed as previously described (Farrell et al., 2015; Feliciano et al., 2015). In brief, recombinant polyhistidine-tagged proteins (10 µg) were incubated with GST and GST fusion proteins (10 µg) bound to glutathione-Sepharose beads in 1 ml of PBS containing 0.1% Triton X-100 for 1 h at 4°C. Beads were washed with PBS containing 0.1% Triton X-100 four times, boiled in Laemmli sample buffer, and analyzed by SDS-PAGE and Coomassie blue staining and/or immunoblotting.
Fluorescence microscopy and endocytosis assays
All fluorescently tagged proteins were expressed from the corresponding endogenous locus and visualized by confocal fluorescence microscopy using live cells grown to early logarithmic phase. Fluorescence microscopy was performed as previously described using an IX81 spinning-disk confocal microscope (Olympus) with a Cascade II camera (Photometrics) and a 100× 1.40 NA objective (Di Pietro et al., 2010; Feliciano and Di Pietro, 2012; Farrell et al., 2015). Time-lapse videos were generated by collecting 60 images every 2 s at room temperature using cells grown until early log phase. Slidebook 6 software (Intelligent Imaging Innovations) was used for quantifying colocalization (Pearson correlation coefficient), patch lifetimes, patch intensities, and intensities of Mup1-GFP cargo and Lucifer yellow (Ambrosio et al., 2012, 2015; Farrell et al., 2015). A mask was generated covering endocytic spots, the entire plasma membrane, or regions inside the cell, depending on the assay. Mup1-GFP cells were grown in minimal media with methionine to early log phase, moved to minimal media lacking methionine for 2 h, and imaged at each time point after return to methionine-rich media. Fluorescence in the membrane was measured using a mask drawn on the cell periphery and normalized to the background.
Lucifer yellow uptake experiments were performed as described previously (Duncan et al., 2001), incubating cells in dye (Invitrogen) for 2 h at room temperature (excepting heat shock strains at 37°C). Fluorescence was measured with a mask drawn on the vacuole and normalized to the background.
Localization displacement between Tda2-GFP and Abp1-RFP (and other pairs of endocytic proteins) was determined using Slidebook 6 software. For each endocytic patch, a single frame of the video in which Tda2-GFP fluorescence intensity was maximal was used for analysis. A line scan analysis was performed across the endocytic patch, perpendicular to the plasma membrane, and the distance between the fluorescence intensity peaks of the two channels was measured in pixels (1 pixel = 0.11 µm).
Statistical significance was determined using an unpaired Student’s t test (GraphPad Software) comparing mean, SEM, and n (cells or patches). Nocodazole and latrunculin A treatment were performed as described previously (Newpher et al., 2005; Aravamudhan et al., 2015).
Crystallization and structure determination
Native crystals of full-length Tda2 were grown at 16°C by hanging drop vapor diffusion. Crystals grew in drops containing one part 15 mg/ml protein (in 10 mM Hepes, pH 7.0, 100 mM NaCl, and 5% glycerol) and one part well solution (0.1 M Bis-Tris, pH 4.9–5.1, 1.5–1.7 M (NH4)2SO4, and 15% xylitol). All crystals were frozen in liquid nitrogen after soaking in 10-µl drops of well solution made with 20% xylitol for 1–5 min. Bromide derivative crystals were obtained by soaking native crystals in 10-µl drops consisting of well solution made with 20% xylitol and 1 M NaBr for 30 s.
Native diffraction data were collected on the Molecular Biology Consortium beamline 4.2.2 at the Advanced Light Source using shutterless data collection on a CMOS-based Taurus detector (RDI, Inc.) with 0.1° frames covering 180° in 180 s at a wavelength of 1 Å. Bromide-derivative multiwavelength anomalous diffraction data were collected with 0.2° frames over 180° total at wavelengths corresponding with the peak (0.9193 Å), inflection (0.9196 Å), and high remote (0.90502 Å) bromide absorption edge.
All data were processed with XDS, and initial phases were determined from the bromide multiwavelength anomalous diffraction data using Phenix (phenix.autosol), which located two bromide sites and computed a density-modified map into which an initial model was built. This model was used as a molecular replacement model for a native crystal dataset and was refined to 2.3 Å resolution. Model building, refinement, and validation were performed with Coot, Phenix, and MolProbity, respectively, as implemented in the SBgrid suite (Morin et al., 2013). Loops composed of residues 10–14 and 36–39 showed weak but traceable density within the A chain but were omitted altogether from the final B chain model. The residue 76–83 loop appeared to be flexible and was omitted from the B chain because of a lack of density, but the A chain was stabilized into one conformation by a crystal packing contact. The coordinates and molecular structure factors of the Tda2 dimer have been deposited in the PDB under accession code 5VKY.
Analytical ultracentrifugation
The solution molecular weight of Tda2 was determined by sedimentation equilibrium ultracentrifugation in an XL-I centrifuge (Beckman Coulter) with an AnTi60 rotor and six-sector centerpieces at 20°C in a buffer composed of 2 mM KH2PO4, 10 mM Na2HPO4, 137 mM NaCl, and 2.7 mM KCl. Data obtained at rotor speeds of 26,000, 30,000, and 34,000 rpm were globally fit using Ultrascan II v9.9 (http:/www.ultrascan.uthscsa.edu) with a protein partial specific volume of 0.73066 cm3/g and a buffer density of 1.0057 g/cm3, revealing an excellent fit to a single-species model with a 30.8 kD molecular weight that corresponded with a Tda2 dimer.
Online supplemental material
Fig. S1 contains supporting data showing that Tda2 does not work as part of the dynein motor complex or associate with microtubules. Fig. S2 contains supporting data showing that Aim21 and Tda2 interact physically and that they are not destabilized in the absence of the partner protein. Fig. S3 contains data supporting a role for Aim21 in endocytosis. Fig. S4 contains supporting data demonstrating that Tda2 and Aim21 localize closer to the plasma membrane than Abp1. Fig. S5 contains supporting data showing Tda2 has the same localization as Cap1 regardless of the fluorescent protein tag used for visualization. Video 1 shows the dynamics of Tda2 and Sla1 at endocytic sites in live cells.
Supplementary Material
Acknowledgments
We thank John Cooper, Steven Markus, and Laurie Stargell for generous gift of reagents; Dustin Steele, Al Aradi, and Lena Cuevas for assistance with pulldown assays; Charles McDonald for assistance with ultracentrifugation; Jay Nix at ALS beamline 4.2.2; and Andrea Ambrosio for help with figure preparation.
This work was supported by National Science Foundation grant 1616775. K.B. Farrell acknowledges an American Heart Association predoctoral fellowship.
The authors declare no competing financial interests.
Author contributions: conceptualization, S.M. Di Pietro; investigation, K.B. Farrell, S. McDonald, A.K. Lamb, C. Worcester, O.B. Peersen, and S.M. Di Pietro; formal analysis, K.B. Farrell, S. McDonald, A.K. Lamb, O.B. Peersen, and S.M. Di Pietro; writing (original draft), K.B. Farrell, S. McDonald, O.B. Peersen, and S.M. Di Pietro; writing (review and editing), O.B. Peersen and S.M. Di Pietro; visualization, K.B. Farrell, O.B. Peersen, and S.M. Di Pietro; funding acquisition, O.B. Peersen and S.M. Di Pietro; resources, O.B. Peersen and S.M. Di Pietro; supervision, O.B. Peersen and S.M. Di Pietro.
Footnotes
Abbreviations used:
- 3-AT
- 3-amino-1,2,4-triazole
- CME
- clathrin-mediated endocytosis
- rmsd
- root mean square deviation
References
- Amatruda J.F., and Cooper J.A.. 1992. Purification, characterization, and immunofluorescence localization of Saccharomyces cerevisiae capping protein. J. Cell Biol. 117:1067–1076. 10.1083/jcb.117.5.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda J.F., Gattermeir D.J., Karpova T.S., and Cooper J.A.. 1992. Effects of null mutations and overexpression of capping protein on morphogenesis, actin distribution and polarized secretion in yeast. J. Cell Biol. 119:1151–1162. 10.1083/jcb.119.5.1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio A.L., Boyle J.A., and Di Pietro S.M.. 2012. Mechanism of platelet dense granule biogenesis: study of cargo transport and function of Rab32 and Rab38 in a model system. Blood. 120:4072–4081. 10.1182/blood-2012-04-420745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrosio A.L., Boyle J.A., and Di Pietro S.M.. 2015. TPC2 mediates new mechanisms of platelet dense granule membrane dynamics through regulation of Ca2+ release. Mol. Biol. Cell. 26:3263–3274. 10.1091/mbc.E15-01-0058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravamudhan P., Goldfarb A.A., and Joglekar A.P.. 2015. The kinetochore encodes a mechanical switch to disrupt spindle assembly checkpoint signalling. Nat. Cell Biol. 17:868–879. 10.1038/ncb3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benison G., and Barbar E.. 2009. NMR analysis of dynein light chain dimerization and interactions with diverse ligands. Methods Enzymol. 455:237–258. 10.1016/S0076-6879(08)04209-2 [DOI] [PubMed] [Google Scholar]
- Berro J., Sirotkin V., and Pollard T.D.. 2010. Mathematical modeling of endocytic actin patch kinetics in fission yeast: Disassembly requires release of actin filament fragments. Mol. Biol. Cell. 21:2905–2915. 10.1091/mbc.E10-06-0494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner D.R., Chi R.J., and Lemmon S.K.. 2011. Lessons from yeast for clathrin-mediated endocytosis. Nat. Cell Biol. 14:2–10. 10.1038/ncb2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky F.M. 2012. Diversity of clathrin function: New tricks for an old protein. Annu. Rev. Cell Dev. Biol. 28:309–336. 10.1146/annurev-cellbio-101011-155716 [DOI] [PubMed] [Google Scholar]
- Brodsky F.M., Chen C.Y., Knuehl C., Towler M.C., and Wakeham D.E.. 2001. Biological basket weaving: Formation and function of clathrin-coated vesicles. Annu. Rev. Cell Dev. Biol. 17:517–568. 10.1146/annurev.cellbio.17.1.517 [DOI] [PubMed] [Google Scholar]
- Bultema J.J., Ambrosio A.L., Burek C.L., and Di Pietro S.M.. 2012. BLOC-2, AP-3, and AP-1 proteins function in concert with Rab38 and Rab32 proteins to mediate protein trafficking to lysosome-related organelles. J. Biol. Chem. 287:19550–19563. 10.1074/jbc.M112.351908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burston H.E., Maldonado-Báez L., Davey M., Montpetit B., Schluter C., Wendland B., and Conibear E.. 2009. Regulators of yeast endocytosis identified by systematic quantitative analysis. J. Cell Biol. 185:1097–1110. 10.1083/jcb.200811116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang J.Z., Yeh T.Y., Bollati F., Conde C., Canavosio F., Caceres A., and Sung C.H.. 2005. The dynein light chain Tctex-1 has a dynein-independent role in actin remodeling during neurite outgrowth. Dev. Cell. 9:75–86. 10.1016/j.devcel.2005.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conde C., Arias C., Robin M., Li A., Saito M., Chuang J.Z., Nairn A.C., Sung C.H., and Cáceres A.. 2010. Evidence for the involvement of Lfc and Tctex-1 in axon formation. J. Neurosci. 30:6793–6800. 10.1523/JNEUROSCI.5420-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro S.M., Falcón-Pérez J.M., and Dell’Angelica E.C.. 2004. Characterization of BLOC-2, a complex containing the Hermansky–Pudlak syndrome proteins HPS3, HPS5 and HPS6. Traffic. 5:276–283. 10.1111/j.1600-0854.2004.0171.x [DOI] [PubMed] [Google Scholar]
- Di Pietro S.M., Cascio D., Feliciano D., Bowie J.U., and Payne G.S.. 2010. Regulation of clathrin adaptor function in endocytosis: novel role for the SAM domain. EMBO J. 29:1033–1044. 10.1038/emboj.2010.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan M.C., Cope M.J., Goode B.L., Wendland B., and Drubin D.G.. 2001. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat. Cell Biol. 3:687–690. 10.1038/35083087 [DOI] [PubMed] [Google Scholar]
- Edwards M., McConnell P., Schafer D.A., and Cooper J.A.. 2015. CPI motif interaction is necessary for capping protein function in cells. Nat. Commun. 6:8415 10.1038/ncomms9415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell K.B., Grossman C., and Di Pietro S.M.. 2015. New regulators of clathrin-mediated endocytosis identified in Saccharomyces cerevisiae by systematic quantitative fluorescence microscopy. Genetics. 201:1061–1070. 10.1534/genetics.115.180729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciano D., and Di Pietro S.M.. 2012. SLAC, a complex between Sla1 and Las17, regulates actin polymerization during clathrin-mediated endocytosis. Mol. Biol. Cell. 23:4256–4272. 10.1091/mbc.E11-12-1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciano D., Bultema J.J., Ambrosio A.L., and Di Pietro S.M.. 2011. In vivo andin vitro studies of adaptor-clathrin interaction. J. Vis. Exp. 47:2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliciano D., Tolsma T.O., Farrell K.B., Aradi A., and Di Pietro S.M.. 2015. A second Las17 monomeric actin-binding motif functions in Arp2/3-dependent actin polymerization during endocytosis. Traffic. 16:379–397. 10.1111/tra.12259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. . 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 415:141–147. 10.1038/415141a [DOI] [PubMed] [Google Scholar]
- Goode B.L., Eskin J.A., and Wendland B.. 2015. Actin and endocytosis in budding yeast. Genetics. 199:315–358. 10.1534/genetics.112.145540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J., Karplus P.A., and Barbar E.. 2009. Multivalency in the assembly of intrinsically disordered Dynein intermediate chain. J. Biol. Chem. 284:33115–33121. 10.1074/jbc.M109.048587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Valladares M., Kim T., Kannan B., Tung A., Aguda A.H., Larsson M., Cooper J.A., and Robinson R.C.. 2010. Structural characterization of a capping protein interaction motif defines a family of actin filament regulators. Nat. Struct. Mol. Biol. 17:497–503. 10.1038/nsmb.1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman D.A., Yang S., and Drubin D.G.. 1993. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J. Cell Biol. 122:635–644. 10.1083/jcb.122.3.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J.P., Hutton J.L., Olson J.M., and Payne G.S.. 2002. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J. Cell Biol. 157:315–326. 10.1083/jcb.200110027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., and Kimura A.. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaksonen M., Sun Y., and Drubin D.G.. 2003. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 115:475–487. 10.1016/S0092-8674(03)00883-3 [DOI] [PubMed] [Google Scholar]
- Kaksonen M., Toret C.P., and Drubin D.G.. 2005. A modular design for the clathrin- and actin-mediated endocytosis machinery. Cell. 123:305–320. 10.1016/j.cell.2005.09.024 [DOI] [PubMed] [Google Scholar]
- Kim K., Yamashita A., Wear M.A., Maéda Y., and Cooper J.A.. 2004. Capping protein binding to actin in yeast. J. Cell Biol. 164:567–580. 10.1083/jcb.200308061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T., Owen D., and Harrison S.C.. 2014. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb. Perspect. Biol. 6:a016725 10.1101/cshperspect.a016725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kukulski W., Schorb M., Kaksonen M., and Briggs J.A.. 2012. Plasma membrane reshaping during endocytosis is revealed by time-resolved electron tomography. Cell. 150:508–520. 10.1016/j.cell.2012.05.046 [DOI] [PubMed] [Google Scholar]
- Lammers L.G., and Markus S.M.. 2015. The dynein cortical anchor Num1 activates dynein motility by relieving Pac1/LIS1-mediated inhibition. J. Cell Biol. 211:309–322. 10.1083/jcb.201506119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W.L., Kaiser M.A., and Cooper J.A.. 2005. The offloading model for dynein function. J. Cell Biol. 168:201–207. 10.1083/jcb.200407036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., Saito M., Chuang J.Z., Tseng Y.Y., Dedesma C., Tomizawa K., Kaitsuka T., and Sung C.H.. 2011. Ciliary transition zone activation of phosphorylated Tctex-1 controls ciliary resorption, S-phase entry and fate of neural progenitors. Nat. Cell Biol. 13:402–411. 10.1038/ncb2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Markus S.M., and Lee W.L.. 2011. Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev. Cell. 20:639–651. 10.1016/j.devcel.2011.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus S.M., Punch J.J., and Lee W.L.. 2009. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr. Biol. 19:196–205. 10.1016/j.cub.2008.12.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus S.M., Kalutkiewicz K.A., and Lee W.L.. 2012. She1-mediated inhibition of dynein motility along astral microtubules promotes polarized spindle movements. Curr. Biol. 22:2221–2230. 10.1016/j.cub.2012.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A., Grassart A., Okreglak V., Costanzo M., Boone C., and Drubin D.G.. 2013. Actin filament elongation in Arp2/3-derived networks is controlled by three distinct mechanisms. Dev. Cell. 24:182–195. 10.1016/j.devcel.2012.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin A., Eisenbraun B., Key J., Sanschagrin P.C., Timony M.A., Ottaviano M., and Sliz P.. 2013. Collaboration gets the most out of software. eLife. 2:e01456 10.7554/eLife.01456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newpher T.M., Smith R.P., Lemmon V., and Lemmon S.K.. 2005. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev. Cell. 9:87–98. 10.1016/j.devcel.2005.04.014 [DOI] [PubMed] [Google Scholar]
- Rao L., Romes E.M., Nicholas M.P., Brenner S., Tripathy A., Gennerich A., and Slep K.C.. 2013. The yeast dynein Dyn2-Pac11 complex is a dynein dimerization/processivity factor: structural and single-molecule characterization. Mol. Biol. Cell. 24:2362–2377. 10.1091/mbc.E13-03-0166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rapali P., Szenes Á., Radnai L., Bakos A., Pál G., and Nyitray L.. 2011. DYNLL/LC8: a light chain subunit of the dynein motor complex and beyond. FEBS J. 278:2980–2996. 10.1111/j.1742-4658.2011.08254.x [DOI] [PubMed] [Google Scholar]
- Robinson M.S. 2015. Forty years of clathrin-coated vesicles. Traffic. 16:1210–1238. 10.1111/tra.12335 [DOI] [PubMed] [Google Scholar]
- Starcevic M., and Dell’Angelica E.C.. 2004. Identification of snapin and three novel proteins (BLOS1, BLOS2, and BLOS3/reduced pigmentation) as subunits of biogenesis of lysosome-related organelles complex-1 (BLOC-1). J. Biol. Chem. 279:28393–28401. 10.1074/jbc.M402513200 [DOI] [PubMed] [Google Scholar]
- Tonikian R., Xin X., Toret C.P., Gfeller D., Landgraf C., Panni S., Paoluzi S., Castagnoli L., Currell B., Seshagiri S., et al. . 2009. Bayesian modeling of the yeast SH3 domain interactome predicts spatiotemporal dynamics of endocytosis proteins. PLoS Biol. 7:e1000218 10.1371/journal.pbio.1000218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub L.M., and Bonifacino J.S.. 2013. Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 5:a016790 10.1101/cshperspect.a016790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg J., and Drubin D.G.. 2012. Clathrin-mediated endocytosis in budding yeast. Trends Cell Biol. 22:1–13. 10.1016/j.tcb.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J.C., Xie H., and Hendrickson W.A.. 2005. Crystal structure of dynein light chain TcTex-1. J. Biol. Chem. 280:21981–21986. 10.1074/jbc.M414643200 [DOI] [PubMed] [Google Scholar]
- Williams J.C., Roulhac P.L., Roy A.G., Vallee R.B., Fitzgerald M.C., and Hendrickson W.A.. 2007. Structural and thermodynamic characterization of a cytoplasmic dynein light chain–intermediate chain complex. Proc. Natl. Acad. Sci. USA. 104:10028–10033. 10.1073/pnas.0703614104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh C., Li A., Chuang J.Z., Saito M., Cáceres A., and Sung C.H.. 2013. IGF-1 activates a cilium-localized noncanonical Gβγ signaling pathway that regulates cell-cycle progression. Dev. Cell. 26:358–368. 10.1016/j.devcel.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh T.Y., Peretti D., Chuang J.Z., Rodriguez-Boulan E., and Sung C.H.. 2006. Regulatory dissociation of Tctex-1 light chain from dynein complex is essential for the apical delivery of rhodopsin. Traffic. 7:1495–1502. 10.1111/j.1600-0854.2006.00482.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young M.E., Cooper J.A., and Bridgman P.C.. 2004. Yeast actin patches are networks of branched actin filaments. J. Cell Biol. 166:629–635. 10.1083/jcb.200404159 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.