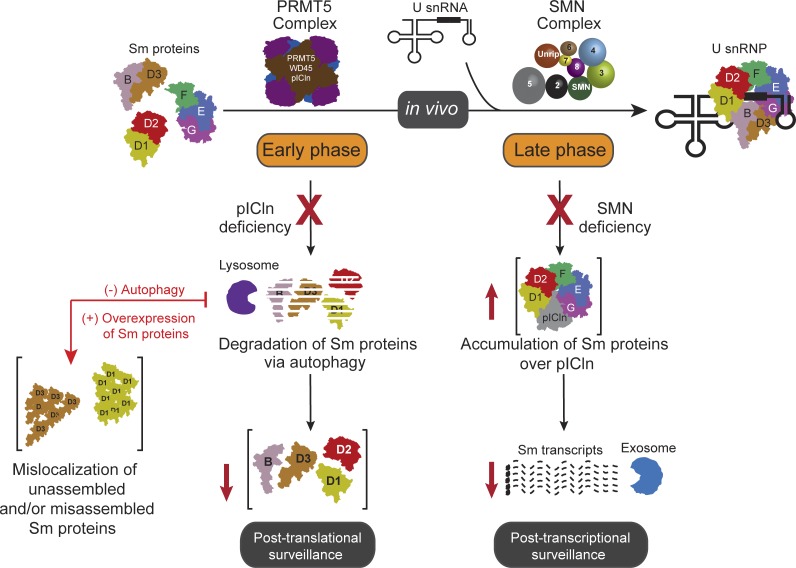
Cellular spliceosomal UsnRNP assembly is assisted by the PRMT5 and SMN complexes. Prusty et al. demonstrate that perturbations in the assembly machinery of UsnRNPs trigger complex cellular responses, using ribosomes, exosome-mediated RNA degradation, and autophagy to prevent Sm protein aggregation.
Abstract
Specialized assembly factors facilitate the formation of many macromolecular complexes in vivo. The formation of Sm core structures of spliceosomal U-rich small nuclear ribonucleoprotein particles (UsnRNPs) requires assembly factors united in protein arginine methyltransferase 5 (PRMT5) and survival motor neuron (SMN) complexes. We demonstrate that perturbations of this assembly machinery trigger complex cellular responses that prevent aggregation of unassembled Sm proteins. Inactivation of the SMN complex results in the initial tailback of Sm proteins on the PRMT5 complex, followed by down-regulation of their encoding mRNAs. In contrast, reduction of pICln, a PRMT5 complex subunit, leads to the retention of newly synthesized Sm proteins on ribosomes and their subsequent lysosomal degradation. Overexpression of Sm proteins under these conditions results in a surplus of Sm proteins over pICln, promoting their aggregation. Our studies identify an elaborate safeguarding system that prevents individual Sm proteins from aggregating, contributing to cellular UsnRNP homeostasis.
Introduction
The Sm class members of U-rich small nuclear ribonucleoprotein particles (UsnRNPs) constitute the central building blocks of major and minor spliceosomes. These macromolecular machines catalyze the removal of introns from pre-mRNAs, facilitating an essential step in the formation of a translatable mRNA (Matera and Wang, 2014). To cope with the task of splicing virtually all intron-containing pre-mRNAs, the steady-state level of cellular UsnRNPs is abundant in higher eukaryotes, reaching an estimated concentration of 10 µM per nucleus in HeLa cells (Montzka and Steitz, 1988). Therefore, efficient production of UsnRNPs and maintenance of their steady-state levels appear to be indispensable for eukaryotic cells.
UsnRNPs are assembled in cells in a segmented pathway that starts with the transcription of UsnRNA precursor molecules by either polymerase II (for U1, U2, U4, U5, U11, U12, and U4atac snRNAs) or polymerase III (for U6 and U6atac snRNAs). The polymerase II–transcribed snRNAs are transiently exported from the nucleus to the cytoplasm, where they associate with the seven common (Sm) proteins B/B′, D1, D2, D3, E, F, and G. This leads to the formation of a toroidal ring on the UsnRNA, termed the Sm core (Raker et al., 1996, 1999; Kambach et al., 1999b; Stark et al., 2001; Urlaub et al., 2001; Chari et al., 2008; Wahl et al., 2009; Zhang et al., 2011). The Sm core is a common structural denominator of these UsnRNPs that enables several subsequent steps in the biogenesis pathway. This includes the conversion of the m7G-cap of the UsnRNAs to its hypermethylated form (m2,2,73G-cap) and the nuclear import of the assembled UsnRNP particle (Fischer and Lührmann, 1990; Hamm et al., 1990; Mouaikel et al., 2002, 2003). The stage at which specific snRNP proteins join the respective particles appears to be variable. Some can bind in the cytosol to their respective snRNA target, whereas others join the UsnRNA in nuclear Cajal bodies where UsnRNA modifications also occur (Achsel et al., 1999; Sleeman and Lamond, 1999; Jády et al., 2003; Stanĕk and Neugebauer, 2004). Unlike the Sm class of UsnRNPs, formation of U6 and U6atac snRNPs occurs exclusively in the nucleus. These particles lack the Sm proteins but instead contain seven Sm-like proteins (LSm2–8) that associate with the 3′ end of the snRNA to form an RNP that is structurally similar to the Sm core domain (Pannone et al., 1998, 2001; Achsel et al., 1999).
The assembly of the Sm-class UsnRNPs involves several trans-acting factors. These are organized in two units, the protein arginine methyltransferase 5 (PRMT5) complex and the survival motor neuron (SMN) complex (Meister and Fischer, 2002; Paushkin et al., 2002; Fischer et al., 2011; Matera and Wang, 2014). The PRMT5 complex acts early in the assembly pathway and consists of the methyltransferase PRMT5, the assembly chaperone pICln, and WD45 (also termed MEP50). Its main task is to catalyze the symmetrical arginine-dimethylation in SmB/B′, SmD1, and SmD3 and the formation of higher-order Sm protein complexes (Friesen et al., 2001; Meister et al., 2001b; Gonsalvez et al., 2007; Barbarossa et al., 2014; Neuenkirchen et al., 2015). The latter activity is mediated predominantly by the assembly chaperone pICln, which recruits all newly synthesized Sm proteins to the PRMT5 complex. This leads to the formation of two kinetically trapped assembly intermediates: a ring-shaped 6S complex composed of pICln and the Sm proteins D1, D2, E, F, and G, as well as a pICln-SmB-D3 heterotrimer (Chari et al., 2008; Neuenkirchen et al., 2015). Association of pICln with Sm proteins inhibits, rather than enables, their binding onto UsnRNA, making the activity of the SMN complex in the late assembly phase pivotal (Fischer et al., 1997; Paushkin et al., 2002; Pellizzoni et al., 2002b; Shpargel and Matera, 2005; Wan et al., 2005; Winkler et al., 2005; Gabanella et al., 2007; Chari et al., 2008; Zhang et al., 2008, 2011; Boulisfane et al., 2011; Grimm et al., 2013). The SMN complex comprises the eponymous protein SMN, along with eight other proteins termed Gemins 2–8 and Unrip (Fischer et al., 1997; Hannus et al., 2000; Paushkin et al., 2000; Meister et al., 2001a; Gubitz et al., 2002; Pellizzoni et al., 2002a; Otter et al., 2007; Kroiss et al., 2008; Borg et al., 2015). Although SMN, Gemin2, and probably other components of the SMN complex engage with the Sm proteins and aid in the release of pICln (Grimm et al., 2013), Gemin5 has been reported to be the snRNA recruiter during UsnRNP assembly (Lau et al., 2009; Yong et al., 2010; Wahl and Fischer, 2016). All Sm-class UsnRNPs studied so far, including the U7snRNP particle containing a Sm/LSm hybrid core, require the SMN/PRMT5 system for assembly in vivo (Pillai et al., 2003). Although it is unclear whether U6 or other RNPs containing a pure LSm core likewise depend on this assembly machinery, SMN has been shown to be involved in the proper localization and assembly of mRNPs and telomerase RNP (Bachand et al., 2002; Donlin-Asp et al., 2016; Fallini et al., 2016). Nevertheless, consistent with its reported role in RNP biogenesis, several factors of the assembly machinery, including SMN, have been shown to be essential for viability (Schrank et al., 1997; Hsieh-Li et al., 2000; Paushkin et al., 2000; Shpargel et al., 2009; Praveen et al., 2012; Garcia et al., 2013). Furthermore, the reduced expression of functional SMN caused by genomic mutations has been linked to the debilitating human disorder spinal muscular atrophy (SMA; Lefebvre et al., 1995).
Although the assembly of UsnRNPs occurs spontaneously in vitro, the question arises why cells use a plethora of assembly factors to direct this reaction in vivo. The tendency of Sm proteins to bind nonspecifically to RNA under physiological conditions argues that the PRMT5/SMN system confers specificity to the assembly reaction. In accordance with this view, it was shown that binding of Sm proteins to the SMN complex prevents their association with nontarget RNAs (Pellizzoni et al., 2002b; Kroiss et al., 2008; Neuenkirchen et al., 2015). Whether the assembly machinery also serves other functions, however, remains elusive. We addressed in this study how cells respond to impaired assembly activity in vivo. Structural studies have shown that isolated Sm proteins possess two solvent-exposed hydrophobic interaction surfaces that are prone to nonspecific interactions (Kambach et al., 1999a,b; Zhang et al., 2011; Grimm et al., 2013). We therefore surmised that, apart from its safeguarding activity during UsnRNP formation, the assembly system could have evolved to prevent Sm protein aggregation. We further speculated that the shielding of these hydrophobic surfaces before assembly of the Sm proteins into the core might be the crucial task accomplished by the assembly factors. In line with these considerations, our studies uncovered an elaborate Sm protein protection system that acts on the posttranscriptional and posttranslational levels. We further show that these cellular measures prevent individual snRNP subunits from aggregation and help in maintaining the homeostasis of cellular snRNP levels.
Results
Knockdown of SMN causes Sm protein tailback on the assembly chaperone pICln
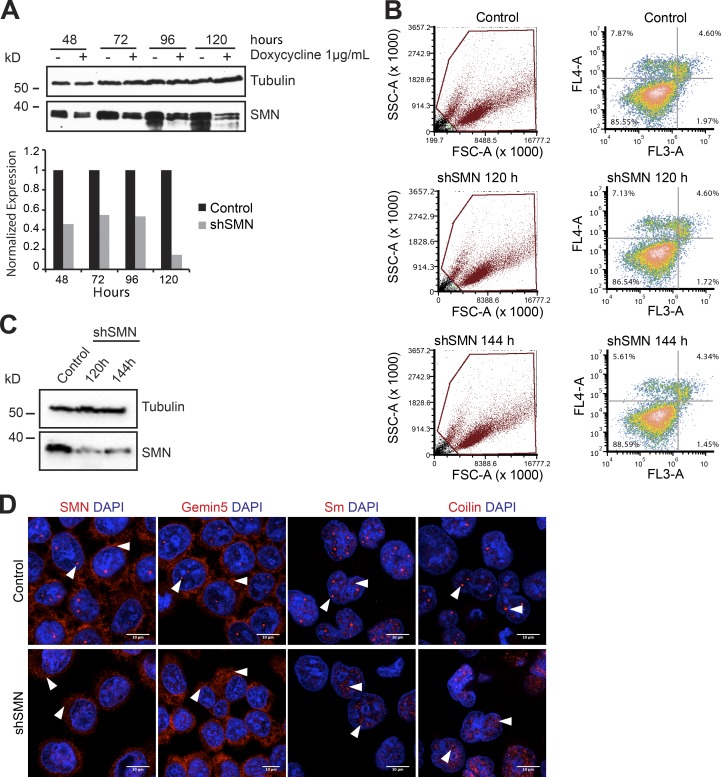
The SMN complex mediates the key steps of the late phase of UsnRNP assembly (Chari et al., 2008). Its core component SMN is crucial for these activities, and its inactivation has been shown to interfere with UsnRNP production in vivo (Wan et al., 2005; Winkler et al., 2005; Gabanella et al., 2007; Zhang et al., 2008; Boulisfane et al., 2011). Building on this observation, we investigated in the first set of experiments whether inhibition of the late assembly phase affects the level of newly synthesized Sm proteins. For this purpose, a stable cell line was generated that allowed the inducible expression of shRNAs against the SMN-encoding mRNAs (Table S1). The induction of SMN-shRNA caused near-complete suppression of SMN protein expression after 120 h (Fig. 1 A, shSMN) and, as a consequence, inactivation of the SMN complex. To test for viability, control and SMN shRNA–induced (shSMN) cells were analyzed by FACS after annexin V and propidium iodide staining at 120 and 144 h. These data revealed no significant changes in cell viability after SMN knockdown in comparison to control cells at the time points studied (Fig. 1, B and C, Western blot). Using indirect immunofluorescence analysis, we observed the absence of the SMN complex (SMN and Gemin5 as markers) from the nucleus, and in particular the Cajal bodies, upon SMN knockdown. Furthermore, although the preexisting UsnRNPs (Sm proteins) were still found in splicing speckles, we observed a clear disappearance of the Sm proteins from Cajal bodies (Fig. 1 D), suggesting interrupted UsnRNP biogenesis (Carvalho et al., 1999; Jády et al., 2003).
Figure 1.
SMN knockdown results in mislocalization of SMN complex and UsnRNPs. (A) Lentivirus-mediated knockdown of SMN (shSMN). Western blot analysis of doxycycline-inducible shRNA knockdown of SMN, using tubulin as the normalization control. Quantification of the blot is shown in the bottom panel, with black and gray bars indicating control and SMN knockdown, respectively. (B) FACS analysis of control uninduced cells (top), shSMN at 120 h (middle) and shSMN at 144 h (bottom) after doxycycline induction of shRNA, after staining with annexin V (FL4-A channel) and propidium iodide (FL3-A channel). Left, gating of all cells (brown); right, distribution of necrotic (bottom right quadrant), early apoptotic (top right quadrant), and apoptotic (top left quadrant) cells along with the respective quantification. (C) Western blot using antibodies specific to SMN to validate knockdown in cells used for FACS analysis, with tubulin as loading control. (D) Confocal images of control cells (top) and shSMN cells (bottom) stained using antibodies against SMN, Gemin5, SmD1/B/B'/D3 (Y12 antibody), and coilin as well as DAPI for nuclear staining. White arrowheads in SMN and Gemin5 images indicate predominant staining pattern observed in cytoplasm (shSMN cells) and Cajal bodies (CBs) and cytoplasm (control cells). White arrowheads in Sm images indicate CBs, splicing speckles (control), and speckles (shSMN). White arrowheads in coilin images indicate intact CBs (control) and disintegrated CBs/mislocalized coilin (shSMN).
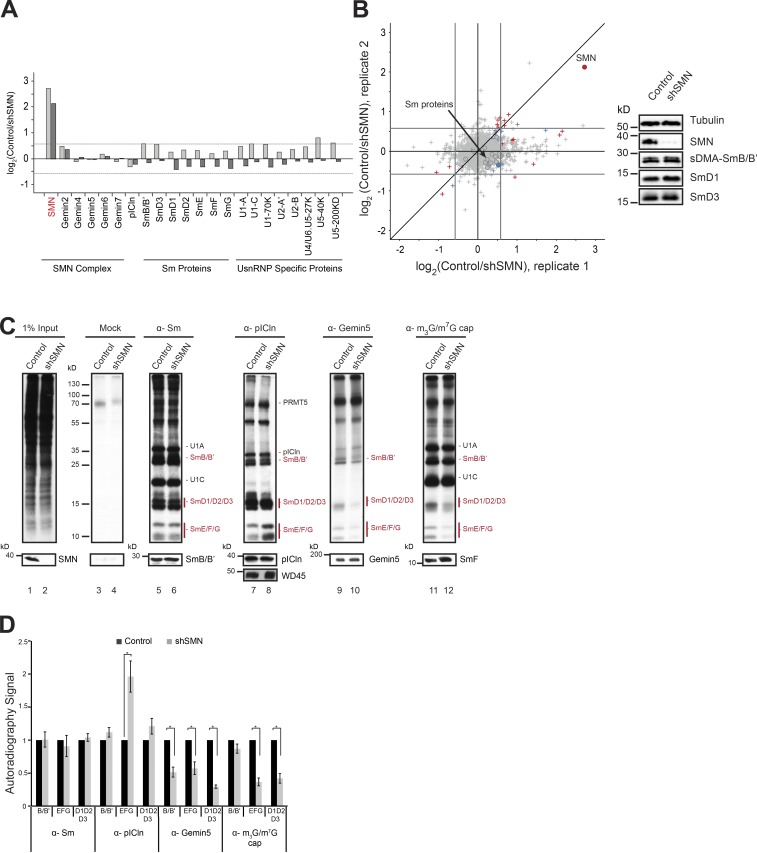
Initially, we investigated by pulsed stable isotope labeling by amino acids in cell culture (pSILAC; Schwanhäusser et al., 2009) whether the reduction of SMN affects the net translation of proteins and snRNP protein levels in particular. Uninduced and shSMN cells were grown in medium containing light amino acid (labeled [12C]lysine and [12C]arginine) for 120 h. This resulted in 80% reduction of SMN in the induced cell line compared with control (Fig. 1 A). Control and SMN knockdown cells were then cultivated for an additional 24 h in heavy amino acids ([13C] and [15N]lysine and [13C] and [15N]arginine) and medium heavy amino acids ([2H]lysine and [13C]arginine), respectively. The proteins from the respective cell lines were identified and quantified by mass spectrometry (MS). As expected, the SMN protein level was drastically reduced in the SMN knockdown cells in replicate experiments, and only a few proteins with no obvious direct connection to UsnRNP biogenesis were either up- or down-regulated compared to control (Fig. 2, A and B; and Table S3). In contrast, steady-state levels of the vast majority of cellular proteins, including Sm proteins translated during the pSILAC pulse, remained unchanged, indicating that the loss of SMN had no detectable influence on Sm protein homeostasis.
Figure 2.
Knockdown of SMN causes Sm protein tailback on pICln. (A) Bar graphs representing normalized log2 fold change in expression (control/shSMN) of UsnRNP-related proteins as well as those of SMN complex and pICln (circles in scatter plot; B) in pSILAC experiments. Dark- and light-gray bars represent two independent replicates. (B) Normalized values of fold change in protein expression during SMN depletion, from two independent pSILAC experiments. Circles indicate proteins involved in UsnRNP pathway that are listed in the bar graph in A. Arrow in the top right quadrant indicates SMN; arrow in the center indicates Sm proteins (gray area). Red, blue, and gray colors indicate high, moderate, or insignificant changes in expression, respectively. (Right) Western blot showing steady-state levels of Sm proteins (sDMA SmB/B', D1, D3) in control and shSMN samples. (C) [35S]methionine metabolic labeling and immunoprecipitation (IP) using antibodies against symmetrically dimethylated Sm proteins, pICln, Gemin5, and m3G/m7G-cap. (Top) Autoradiography of immunoprecipitated samples from control and SMN knockdown cells. (Bottom) Corresponding Western blot controls for immunoprecipitation with molecular weight (in kD) from the marker indicated on the left of the image. (D) Quantification of the autoradiography image shown in C, from three independent biological experiments for Sm, Gemin5, and m3G/m7G-cap IPs and five independent biological experiments for pICln IP. Black and gray bars represent control and SMN knockdown conditions, respectively. Error bars represent standard error (SE); *, P < 0.05 calculated using Student’s t test.
Considering that SMN deprivation results in the inhibition of snRNP assembly (Meister et al., 2001a; Wan et al., 2005; Winkler et al., 2005; So et al., 2016), we next determined where newly synthesized Sm proteins are deposited in the cell under these conditions. Metabolic labeling with [35S]methionine for 3.5 h in control and SMN knockdown cells allowed us to address this issue. Soluble cell extracts were prepared from both cell lines, and equal amounts were subjected to immunoprecipitation with antibodies that monitor different stages of the assembly reaction. The immunoprecipitates were then resolved by SDS-PAGE, and the newly synthesized proteins were visualized by autoradiography (Fig. 2 C).
Immunoprecipitation of the extracts with the monoclonal anti-Sm antibody Y12 specifically precipitated 35S-labeled Sm proteins (note that snRNP-specific proteins such as U1 A and C were also precipitated as part of assembled snRNPs; Fig. 2 C, lanes 5 and 6). The knockdown of SMN had no impact on the overall amount of newly synthesized (i.e., 35S-labeled) Sm proteins that are channeled into the UsnRNP assembly pathway. However, the association of Sm proteins with early assembly factors changed significantly, as determined by immunoprecipitation using an affinity-purified polyclonal anti-pICln antibody. Although this antibody precipitated equal amounts of 35S-labeled PRMT5 complex subunits (i.e., pICln, WD45, and PRMT5) from both the extracts, marked enrichment of Sm proteins over pICln was observed under the conditions of SMN deficiency. This effect was most pronounced for the Sm proteins E, F, and G but was less apparent for the remaining Sm proteins (Fig. 2 C, lanes 7 and 8; and Fig. 2 D, quantification of the data shown in Fig. 2 C). No clear autoradiography signal of WD45 was observed in immunoprecipitates, which is presumably a result of the slow turnover of this protein. Nevertheless, we confirmed the equal enrichment of WD45 in the anti-pICln immunoprecipitation by Western blot analysis using anti-WD45 antibody (Fig. 2 C, lanes 7 and 8).
We hypothesized that the tailback of Sm proteins on pICln might be a consequence of their incomplete handover to the late assembly machinery under SMN depletion. To analyze this possibility, antibodies against Gemin5 (a key component of the SMN complex) and the m3G/m7G-cap of the snRNA were used to monitor Sm protein transfer to the late-phase assembly machinery and subsequently onto the snRNA, respectively. Anti-Gemin5 antibodies precipitated equal amounts of Gemin5 protein from both extracts (Fig. 2 C, lanes 9 and 10; note that all components of the late assembly machinery were only extremely weakly 35S-labeled, indicating their very slow turnover; Meister and Fischer, 2002). However, the amount of coprecipitated Sm proteins was drastically reduced in extracts lacking SMN (Fig. 2 C, lane 10). As a consequence, fewer 35S-labeled Sm proteins were incorporated under SMN knockdown into snRNPs, as indicated by their reduced immunoprecipitation with antibodies recognizing the m3G/m7G-cap of the snRNA (Fig. 2 C, lanes 11 and 12). We note, however, that U1-specific proteins could assemble with snRNAs under these conditions. This is in line with previous observations that several UsnRNP-specific proteins directly interact with the UsnRNA through their RNA interaction motifs, in either the nucleus or the cytoplasm, independent of the Sm core assembly or functionality of the SMN complex (Kambach and Mattaj, 1992; McConnell et al., 2003; Law et al., 2006).
Collectively, these experiments confirm that Sm proteins flow through the assembly pathway from the PRMT5 complex via the SMN complex to UsnRNPs. Inhibition of this pathway in the late (i.e., SMN-driven) assembly phase induces a tailback of Sm proteins on the assembly chaperone pICln that acts upstream in the pathway. The level of total cellular Sm proteins, however, was not affected upon inhibition of the SMN system within a time frame of 24 h.
Destabilization of Sm transcripts after prolonged inhibition of the late assembly phase
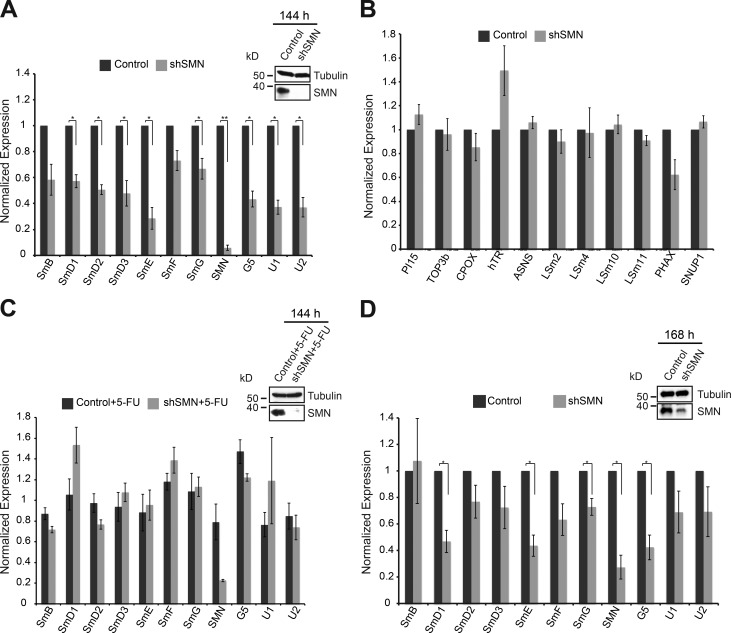
The capacity of pICln to sequester Sm proteins that cannot flow through the assembly system is likely to be limited. We therefore analyzed whether Sm protein levels are regulated upon SMN knockdown in an alternative manner when the system threatens to overflow. The absence of pICln results in the early accumulation of Sm D1 and D2 on the ribosomes near the exit tunnel (Paknia et al., 2016). We performed Western blot analysis for SmD1 and SmD3 using polysome gradient fractions. No accumulation of Sm proteins was observed on the ribosome upon inactivation of the SMN complex (i.e., the late phase of assembly; Fig. S1 A). We then compared transcript levels of individual Sm proteins by quantitative real-time PCR in control and SMN-deficient cells 144 h after shRNA induction. This resulted in the reduction not only of snRNA levels (Gabanella et al., 2007; Zhang et al., 2008) but also mRNAs encoding Sm proteins (Fig. 3 A). Of note, we observed no significant changes in the expression levels of several randomly selected transcripts that encode for either LSm proteins or proteins unrelated to UsnRNP biogenesis pathways (Fig. 3 B), demonstrating that snRNAs and Sm-encoding transcripts are specifically down-regulated. Surprisingly, however, we found down-regulation of the mRNA levels of PHAX, the adaptor protein for UsnRNA export to the cytoplasm (Fig. 3 B). This effect may be linked to the down-regulation of the UsnRNAs (Fig. 3 A) and UsnRNP assembly (Fig. 2, C and D).
Figure 3.
Destabilization of Sm transcripts after extended inhibition of the late assembly phase. (A) Normalized fold change of transcripts encoding the indicated Sm proteins, SMN, and Gemin5, as well as U1 and U2 snRNAs in control cells (black bar) and SMN knockdown cells (gray bars), respectively, 144 h after doxycycline induction. (Top) Western blot analysis showing knockdown of SMN, with tubulin as loading control. (B) Normalized fold change of random transcripts (PI15, TOP3b, CPOX, hTR, and ASNS) that are unrelated to UsnRNP biogenesis as well as LSm encoding transcripts (LSm 2, 4,10, 11), PHAX and SNUP1. (C) qRT-PCR analysis of recovery of Sm encoding transcripts in control (black bars) and SMN knockdown (gray bars) cells upon treatment with 5-FU, an exosome inhibitor. Expression normalized to control cells treated with 5-FU. (Top) Western blot analysis showing knockdown of SMN, with tubulin as loading control. (D) Normalized fold change of transcripts encoding the indicated Sm proteins, SMN, and Gemin5, as well as U1 and U2 snRNAs in control cells (black bar) and SMN knockdown cells (gray bars) 168 h after doxycycline induction. (Top) Western blot analysis showing knockdown of SMN, with tubulin as loading control. Error bars indicate SE (n = 3, three independent biological experiments). **, P < 0.0005; *, P < 0.05, Student’s t test. GAPDH mRNA was used as a control for normalization.
The transcript levels of Sm-encoding mRNAs and UsnRNAs could be fully recovered in SMN knockdown cells treated with the antimetabolite 5-fluorouracil (5-FU; Kammler et al., 2008), suggesting that the exosome-mediated RNA degradation pathway is primarily responsible for the down-regulation (Fig. 3 C). To confirm this observation, we performed an RNA metabolic labeling experiment by feeding control and SMN knockdown cells with the uridine analog 5-ethynyluridine (5-EU) during exosome inhibition. We then used copper-catalyzed click chemistry (Jao and Salic, 2008) to covalently link the alkyne group on 5-EU to Alexa Fluor 488 azide, for labeling newly transcribed RNAs. Immunoprecipitation of the extracts with anti-Sm antibody Y12 and analysis of the labeled RNA confirmed decreased incorporation of the UsnRNAs into UsnRNPs, even after exosome inhibition (Fig. S1 B, quantification in Fig. S1 D). Finally, to address whether the recovery of UsnRNAs observed at 144 h of SMN knockdown represents free or assembled UsnRNAs, we analyzed the bulk UsnRNPs in control and SMN knockdown cells in the presence or absence of exosome inhibition with 5-FU (Fig. S1 C, quantification in Fig. S1 D). For this, we performed anti-Sm immunoprecipitations using Y12 antibody, end-labeled the coprecipitated RNA with [32P]pCp, and normalized the signal to Sm protein enrichment in the Western blot. We observed a nearly 50% decrease in UsnRNAs associated with Sm proteins (bulk UsnRNPs) in the absence of SMN (Fig. S1 C, lanes 2 and 3). This decrease in UsnRNA association with Sm proteins during SMN knockdown remained unchanged after exosome inhibition (Fig. S1 C, lanes 3 and 5). Therefore, the recovery of UsnRNAs observed upon exosome inhibition (Fig. S1 C, quantification in Fig. S1 D) reflects changes in the pool of free UsnRNAs rather than those of bulk UsnRNPs. The down-regulation of Sm-encoding mRNAs and UsnRNAs was apparent even at a later time point after SMN knockdown (Fig. 3 D). Because of a decrease in knockdown efficiency, however, the effect was not as strong as at the earlier time point.
Thus, the response to obstruction of the late assembly phase is biphasic. The sequestration of Sm proteins by pICln constitutes the immediate response, whereas the exosomal down-regulation of transcripts encoding Sm proteins and UsnRNAs accounts for the response at later time points.
Down-regulation of Sm proteins in the absence of the assembly chaperone pICln
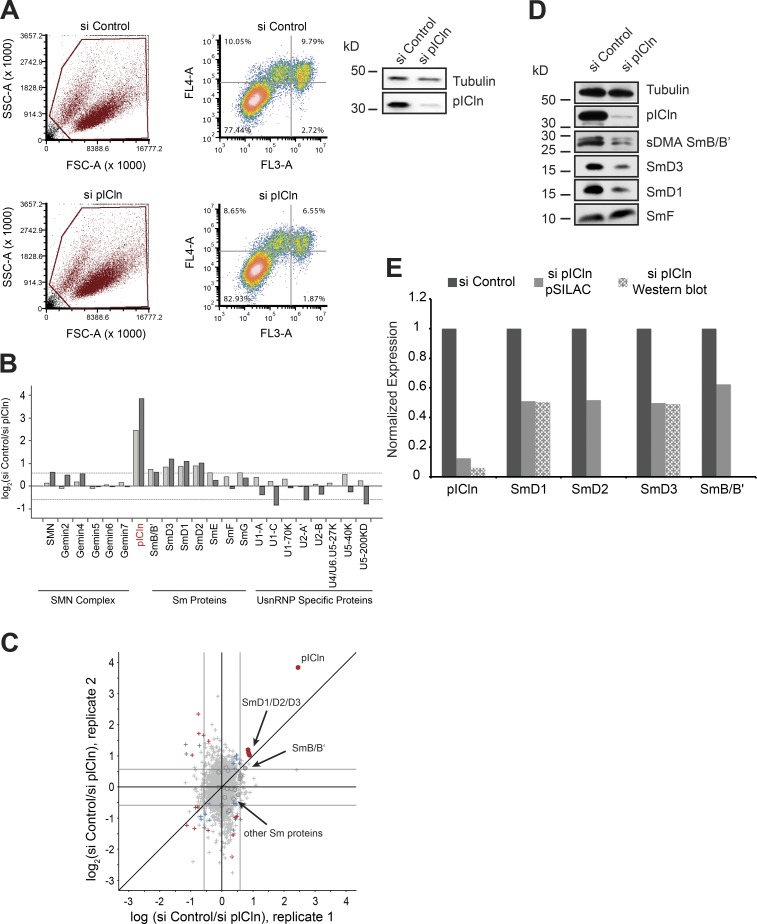
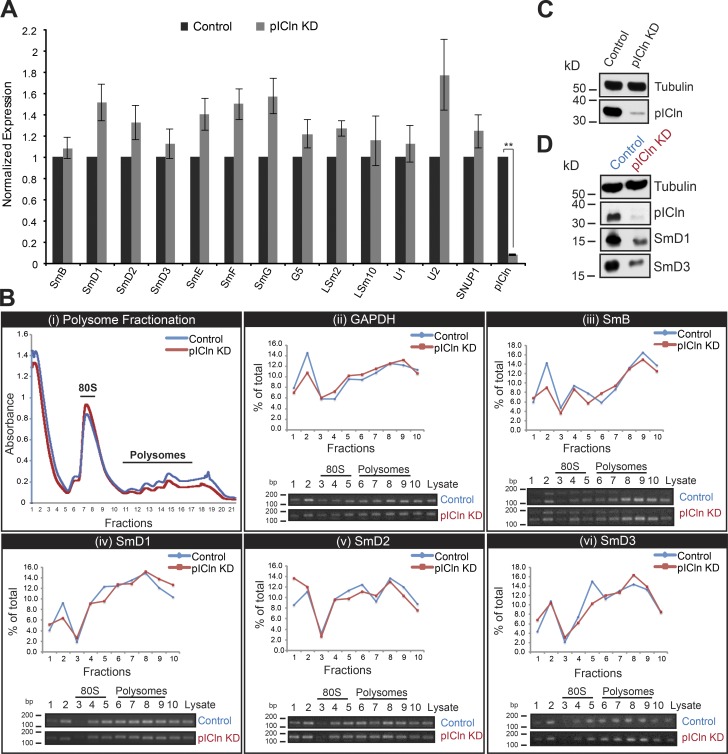
We next investigated how cells respond to perturbation of the early assembly phase. Our previous studies had already shown that the assembly chaperone pICln is required to guide newly synthesized Sm proteins from the ribosome into the assembly pathway (Paknia et al., 2016). Thus, the immediate response to loss of pICln is the accumulation of Sm proteins, in particular SmD1 and D2, at the ribosomal exit tunnel. Control siRNA or siRNA directed against the pICln mRNA was transfected into HeLa cells to suppress pICln expression, neither of which affected cell viability (Fig. 4 A). To assess the fate of Sm proteins during prolonged loss of pICln, we used the pSILAC approach to inspect changes in cellular protein synthesis by quantitative MS. Although the vast majority of cellular proteins remained unchanged, a small subset of proteins was specifically down-regulated (Fig. 4, B and C; and Table S4). These included pICln (i.e., the siRNA knockdown target) as well as SmB, SmD1, SmD2, and SmD3, i.e., those Sm proteins that interact with pICln immediately after their synthesis (Fig. 4, B, C, and E; Friesen et al., 2001; Meister et al., 2001b). Levels of the SmE, SmF, and SmG, however, were not significantly affected. Independent Western blot experiments using antibodies against the respective antigens showed a similar reduction in steady-state levels of Sm proteins (Fig. 4 D; quantification data in Fig. 4 E). Thus, limiting pICln levels caused the simultaneous down-regulation of a specific subset of Sm proteins. Importantly, the affected Sm proteins were those that directly interact with pICln after their translation and thereby are directed to the PRMT5 complex (Neuenkirchen et al., 2015; Paknia et al., 2016).
Figure 4.
Sm proteins are down-regulated in the absence of the pICln. (A) FACS analysis of control (top) and pICln knockdown cells (bottom) 120 h after siRNA transfection, after staining with annexin V (FL4-A channel) and propidium iodide (FL3-A channel). (Left) Gating of all cells (brown); right, distribution of necrotic (bottom right quadrant), early apoptotic (top right quadrant), and apoptotic (top left quadrant) cells along with the respective quantification. (Right) Western blot using antibodies specific to pICln to validate knockdown in cells used for FACS analysis, with tubulin as loading control. (B) Bar graphs representing normalized log2 fold change in expression of UsnRNP-related proteins and proteins of the SMN complex compared between control siRNA and sipICln cells during pSILAC experiment. Dark- and light-gray bars represent replicates 1 and 2, respectively. (C) Normalized values of fold change in protein expression during pICln depletion from two independent pSILAC experiments. Circles indicate proteins involved in UsnRNP pathway that are listed in the bar graph in B. Red dots in the top right quadrant indicate pICln and SmD1/D2/D3; arrow in the center indicates the other Sm proteins (gray area). Red, blue, and gray colors indicate high, moderate, or insignificant changes in expression, respectively. (D) Western blot analysis of soluble cell lysates prepared for pSILAC, from control and pICln knockdown cells, using anti-pICln, anti–sDMA-Sm (Y12), anti-SmD3, anti-SmD1, and anti-SmF antibodies, respectively. Anti-tubulin antibody was used to monitor sample loading. (E) Quantification of the Western blot images shown in D and pSILAC shown in B and C, from two independent biological replicates. Black, gray, and checkered gray bars indicate control, pSILAC, and Western blot quantifications, respectively.
Sm proteins are degraded by autophagy
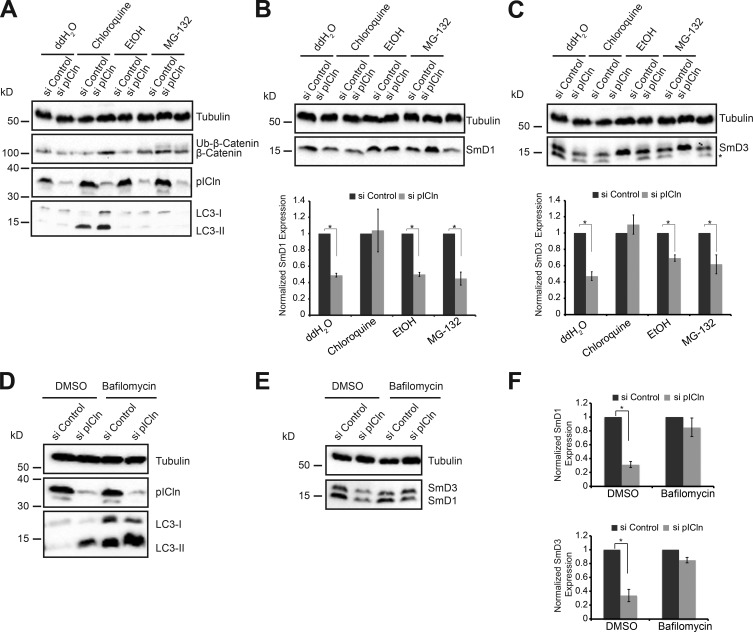
The decrease in Sm protein expression upon pICln knockdown could arise from down-regulation of transcripts (as observed under SMN deprivation), translational arrest, or protein degradation. We initially performed quantitative RT-PCR to analyze transcripts coding for Sm proteins in control and pICln knockdown cells. In contrast to our expectation, we found a noticeable up-regulation of Sm-encoding transcripts (Fig. 5 A; Western blot in Fig. 5 C). As there was also no apparent change in the association of Sm transcripts with monosomes and polysomes upon pICln depletion (Fig. 5 B; Western blot in Fig. 5 D), we reasoned that either proteasomal or autophagic pathways modulated Sm protein levels. To test for these possibilities, pICln-deprived cells were treated with either the proteasomal inhibitor MG-132 (Kisselev and Goldberg, 2001) or the autophagy inhibitor chloroquine (Barth et al., 2010) and analyzed 10 h later by Western blotting. To ascertain that the treatment was successful, we examined (a) the accumulation of microtubule-associated protein 1A/1B-light chain 3 I conjugated to phosphatidylethanolamine (LC3-II), which is indicative of reduced autophagosome turnover (Barth et al., 2010), and (b) the stabilization of β-catenin, which is selectively degraded by the proteasome (Aberle et al., 1997). We observed accumulation of LC3-II upon treatment with chloroquine and ubiquitinated β-catenin upon treatment with MG-132, confirming that either drug worked as predicted (Fig. 6 A). However, MG-132 treatment failed to cause accumulation of ubiquitinated Sm proteins upon reduction of pICln. In contrast, chloroquine efficiently prevented destabilization of Sm proteins (Fig. 6, B [SmD1] and C [SmD3]). To further support our findings, we used bafilomycin A (Yamamoto et al., 1998) as an alternate inhibitor of autophagy (Fig. 6, D–F). Inhibition of autophagy was confirmed by the accumulation of LC3-II as described for chloroquine treatment (Fig. 6 D). Of note, concomitant with the inhibition of autophagy, we observed a strong accumulation of SmD1 and D3 proteins (Fig. 6 E; quantification shown in Fig. 6 F). Together, these results link the autophagy degradation pathway to Sm protein degradation under conditions of pICln shortage.
Figure 5.
Absence of transcriptional or translational regulation of Sm transcripts during pICln knockdown. (A) Normalized fold change of Sm encoding transcripts, G5, LSm2, 10, U1, U2 snRNAs, and SNUP1 in control (black bar) and pICln depletion (gray bars), as analyzed using qRT-PCR experiments, with GAPDH as control for normalization. Error bars indicate SE for three biological replicates (n = 3); **, P < 0.0005. (B, i) Absorbance profile of polysome gradient fractions of control (blue) and pICln knockdown (red) lysates at 260 nm, harvested using Biocomp gradient fractionator. (B, ii) Quantification (top) of control GAPDH mRNA association with polysomes in control (blue) and pICln knockdown (red) cells by RT-PCR analysis, based on the ethidium bromide–stained agarose gel images shown at bottom. (B, iii–vi) RT-PCR analysis of polysome associated SmB/B′ (iii), SmD1 (iv), SmD2 (v), and SmD3 (vi) encoding mRNAs in control (blue) and pICln-deficient (red) cells, with quantification (top) of the ethidium bromide–stained agarose gel images shown at bottom. (C) Western blot analysis showing knockdown of pICln, with tubulin as loading control for the samples used in qRT-PCR experiments (A). (D) Western blot analysis of pICln, SmD1, and SmD3, with tubulin as loading control in input extracts used for polysome gradients (B).
Figure 6.
Sm proteins are degraded by autophagy upon pICln knockdown. (A) Western blot analysis of total cell lysates using antibodies against pICln (knockdown control), β-catenin (control for proteasome inhibition using MG-132), and LC3-II (control for autophagy inhibition using chloroquine). Anti-tubulin antibodies were used to monitor equal loading of all samples. (B and C, top) Western blot analysis of SmD1 (B) and SmD3 (C) using anti-tubulin antibodies to monitor equal loading, in control and pICln knockdown cells upon treatment with water (ddH2O), ethanol (EtOH)), chloroquine, or MG-132. (Bottom) Quantification of the Western blot from three independent biological experiments (n = 3). Error bars represent SE; *, P < 0.05. Black and gray bars represent control and pICln knockdown conditions, respectively. (D) Western blot analysis of total cell lysates using antibodies against LC3-II (control for autophagy inhibition using bafilomycin A) and pICln, with tubulin as loading control, in control and pICln knockdown conditions. (E) Western blot analysis of total cell lysates using antibodies against SmD1 and D3, with tubulin as loading control, in control and pICln knockdown conditions, upon bafilomycin A treatment, with DMSO as solvent control. (F) Quantification of the Western blot shown in E, from independent biological triplicates (n = 3). Error bars represent SE; *, P < 0.05. Black and gray bars represent control and pICln knockdown conditions, respectively.
Perturbation of Sm protein homeostasis leads to aggregation
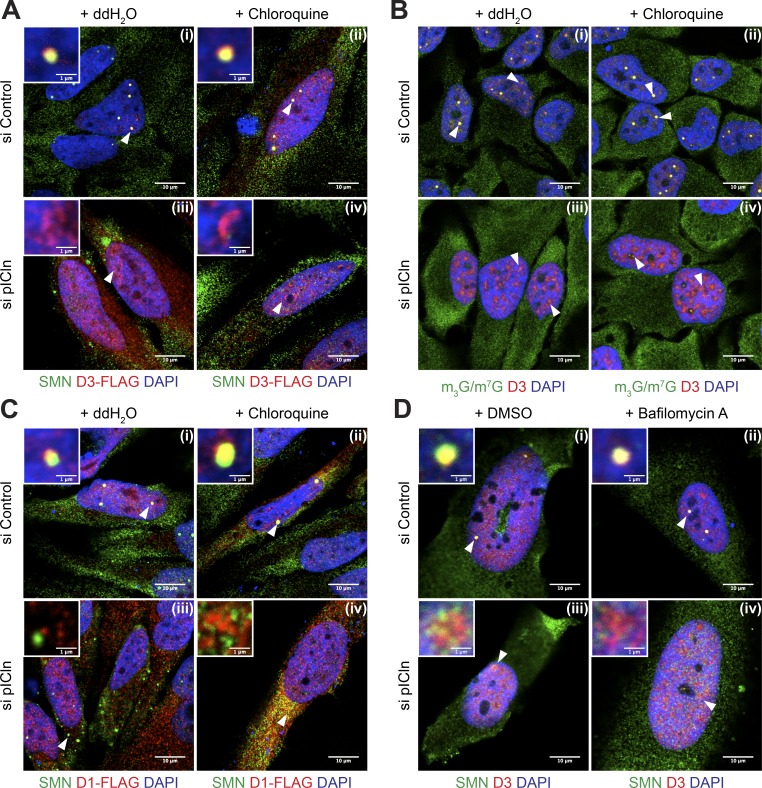
These data revealed an intricate posttranslational surveillance system that prevents accumulation of Sm proteins when their flow through the UsnRNP assembly pathway is blocked. We performed immunoprecipitation of the SMN complex using anti-SMN antibody, with and without bafilomycin-induced autophagy inhibition, to study the association of Sm proteins with the SMN complex upon pICln loss. Interestingly, we observed decreased transfer of Sm proteins to the SMN complex in pICln knockdown cells, even under conditions of autophagy inhibition (Fig. S2 A, quantification in Fig. S2 B). Taking into account these results and the hydrophobic nature of unassembled Sm proteins, we reasoned that one major function of this system is to prevent Sm protein aggregation. To test this experimentally, we initially devised an immunofluorescence-based strategy that allowed us to monitor Sm protein localization under conditions of pICln knockdown and blocked autophagy (Fig. 7; see also Fig. S3 [A–D] for individual fluorescence channel images and Fig. S4 [A–D] for color universal design [CUD] images). We designed FLAG-tagged SmD1 and SmD3 constructs for exogenous overexpression of SmD1 and D3 in HeLa cells. The efficient coimmunoprecipitation of UsnRNAs with the FLAG-tagged Sm proteins confirmed their proper incorporation into UsnRNPs (Fig. S2 C). Transfected FLAG-tagged SmD3 localized in Cajal bodies and nuclear speckles (sites of active mRNA transcription/processing and snRNP storage/recycling), a pattern characteristic of Sm proteins under control conditions (Fig. 7 A [i]). This localization was not altered upon treatment of cells with chloroquine (Fig. 7 A [ii]). However, knockdown of pICln leads to the formation of large nuclear structures containing SmD3, either within nucleoli or in ring-like areas at the nucleolar periphery, in nearly 80% of Sm-overexpressing pICln knockdown cells (Fig. 7, A [iii and iv]). In addition, severe mislocalization of SMN (Fig. 7 A [iii]) and other components of the SMN complex (Fig. S2 D, bottom, and Fig. S4 E, CUD images) became apparent in the pICln-deficient cells. A very similar reorganization was observed for endogenous SmD3 upon pICln knockdown with autophagy blockage using chloroquine or an alternate inhibitor, bafilomycin A (Yamamoto et al., 1998; Figs. 7 D [iv] and Fig. S3 D [iv]).
Figure 7.
Perturbation of Sm protein homeostasis leads to aggregation in the absence of pICln. (A) Overexpression of SmD3-FLAG in control and pICln-deficient cells analyzed by confocal microscopy by indirect immunofluorescence using antibodies against SMN (green), FLAG (exogenous D3-FLAG; red), or DAPI (blue) upon water (ddH2O) or chloroquine treatment. White arrowheads indicate regions shown in insets at the top left corner of each image. (B) Confocal microscopy of indirect immunofluorescence of m3G/m7G capped RNA (H20 antibody; green), endogenous SmD3 (red), and DAPI (blue) during control or pICln knockdown, upon chloroquine or water treatment. White arrowheads indicate predominant colocalization/staining pattern observed in splicing speckles or Cajal bodies (CBs; control) or disperse higher-order structures (pICln knockdown). (C) Overexpression of SmD1-FLAG in control and pICln-deficient cells analyzed by confocal microscopy. Indirect immunofluorescence using antibodies targeting SMN (green), FLAG (exogenous D1-FLAG; red), and DAPI (blue) upon water (ddH2O) or chloroquine treatment. White arrowheads indicate regions shown in insets at the top left corner of each image. (D) Indirect immunofluorescence of endogenous SMN (green) and SmD3 (red) upon treatment of control (i and ii) and pICln-deficient (iii and iv) cells with DMSO solvent or the lysosome inhibitor bafilomycin A. DNA is visualized using DAPI. White arrowheads indicate regions shown in insets at the top left corner of each image. Bars: (A–D, main) 10 µm; (A, C, and D, insets) 1 µm.
Importantly, the SmD3-containing regions appear largely devoid of UsnRNAs, as we failed to stain them with m7G/m3G-specific monoclonal antibody H20 (Fig. 7 B [iv]). To confirm our observation, we calculated the Pearson’s colocalization coefficient (PCC) for the extent of overlap of UsnRNA with SmD3, using ImarisColoc software. Whereas the PCC reached 0.68 in control cells and hence indicated a high degree of colocalization, the value dropped to 0.18 in pICln knockdown cells. The tendency of Sm proteins to reorganize in cells under pICln deficiency was not restricted to SmD3 but was also readily apparent for SmD1. Of note, however, SmD1 accumulated in cytoplasmic rather than nuclear structures as observed for SmD3 (Fig. 7 C [iv]).
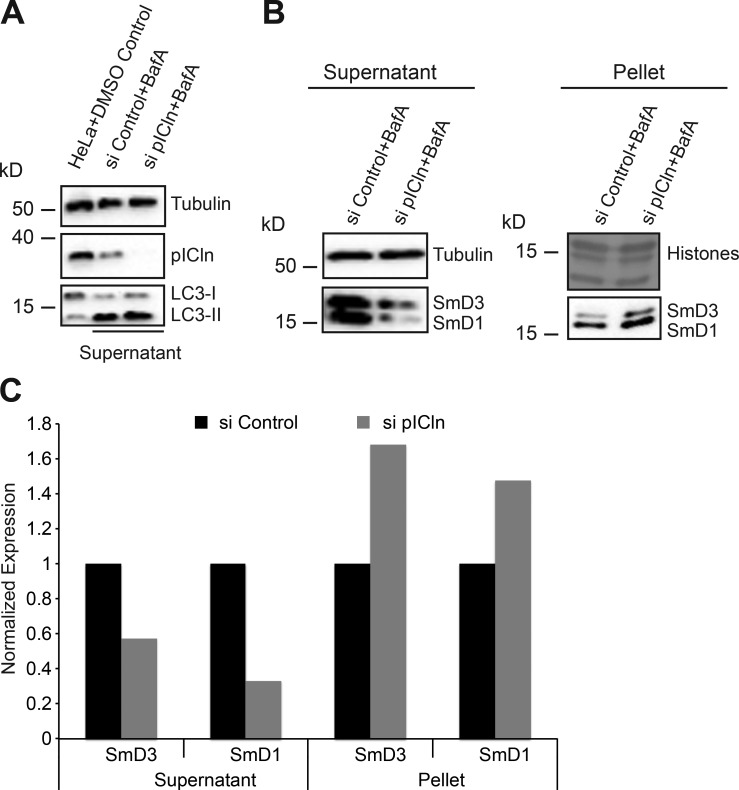
Next, we analyzed whether under pICln shortage the Sm proteins could still engage with the late assembly machinery. In this case, one would expect their colocalization with SMN, but that was not the case (Fig. 7, A [iii and iv], C [iii and iv], and D [iii and iv]). In fact, the calculated PCC for colocalization of SmD1-FLAG with SMN (Fig. 7 C [iv]) was nearly zero. These experiments suggest that during pICln shortage, the bulk of the forcibly expressed Sm proteins bypass the canonical UsnRNP assembly pathway and hence form aberrant structures. This finding raised the possibility that the Sm proteins in the absence of pICln form insoluble aggregates. To test this possibility, we adapted an established protocol for yeast cells to determine the fraction of insoluble proteins by Western blotting (Koplin et al., 2010; Nedialkova and Leidel, 2015). These studies revealed a shift of Sm proteins from the soluble into the insoluble fraction, suggesting that they form aggregates in the absence of pICln (Fig. 8, A and B; quantification in Fig. 8 C). Collectively, these data suggest that pICln acts not only as a topological organizer in the UsnRNP pathway but also as a factor that shields unassembled Sm proteins from the engaging in unwanted interactions or aggregation.
Figure 8.
Sm proteins rescued from autophagy form insoluble aggregates. (A) Western blot analysis of lysates from HeLa cells treated with DMSO, compared with soluble extracts prepared from control and pICln siRNA–transfected cells, treated with autophagy inhibitor bafilomycin A, using antibodies specific to pICln and LC3 to monitor pICln knockdown and autophagy inhibition, with tubulin as loading control. (B) Western blot analysis of soluble (supernatant) and insoluble (pellet) fractions of control and pICln knockdown cells upon bafilomycin A treatment. Right top, amido black staining of histones after PVDF membrane transfer as loading control for the insoluble fraction; left top, tubulin (loading control for soluble fraction). (C) Quantification of the Western blots shown in B from n = 1 replicate.
Discussion
The production of macromolecular machines in vivo imposes a major challenge on cells, as unassembled individual protein components often expose hydrophobic residues to the solvent and, as a consequence, are prone to aggregation. It is hence an indispensable task for cells to tightly control the production, assembly, and turnover of all components participating in the formation of the macromolecular assembly. Here, we analyzed how such a system works for the production of the Sm class of splicing snRNPs, which constitute one of the most abundant RNP classes in eukaryotes.
The active role of SMN and PRMT5 complexes in UsnRNP assembly has been proven and confirmed by many laboratories (Fischer et al., 1997; Meister et al., 2001a; Meister and Fischer, 2002; Paushkin et al., 2002; Pellizzoni et al., 2002b; Shpargel and Matera, 2005; Gonsalvez et al., 2007; Chari et al., 2008; Zhang et al., 2011; Neuenkirchen et al., 2015). For the late assembly factors (i.e., the SMN complex and its subunit Gemin5 in particular), a proofreading activity has been described (Pellizzoni et al., 2002b; Kroiss et al., 2008; Wahl and Fischer, 2016). This activity ensures the discrimination of nontarget RNAs during UsnRNP assembly, providing a plausible explanation for the requirement of the SMN complex for assembly. Recent studies further demonstrated cellular UsnRNA surveillance activity in which mutant UsnRNAs that fail to incorporate into UsnRNPs are targeted to P-bodies for exosomal Rrp6- or Xrn1-dependent degradation (Ishikawa et al., 2014; Shukla and Parker, 2014). A similar fate has been observed for UsnRNAs in tissues of a mouse model of SMA that lack sufficient amounts of SMN to ensure proper UsnRNP assembly (Gabanella et al., 2007; Zhang et al., 2008). However, a surveillance system for the potentially harmful Sm proteins has not yet been described.
Early assembly factors preorganize Sm proteins into higher-order structures to allow their concerted transfer onto the SMN complex (Neuenkirchen et al., 2015). Similar to the assembly reaction by itself, loading of the SMN complex with Sm proteins can be faithfully recapitulated in vitro without pICln, suggesting additional functions for the early UsnRNP assembly factors. In keeping with this notion, we uncovered a crucial role of pICln in shielding newly synthesized Sm proteins from unwanted interactions. The induced depletion of SMN leads to a biphasic cellular response, starting with an initial accumulation of Sm proteins in their pICln-bound and kinetically trapped state. When the expression of Gemin5, an essential SMN-complex component, was reduced by inducible shRNA-mediated knockdown (Fig. S5 A), the expression of newly synthesized Sm proteins likewise remained unchanged (Fig. S5 B). Of note, a similar tailback of the Sm proteins on pICln (Fig. S5 C, lanes 7 and 8; quantification in Fig. S5 D) decreased loading of Sm proteins on the SMN complex (Fig. S5 C, lanes 9 and 10; quantification in Fig. S5 D); reduced UsnRNP assembly was also observed and was comparable to that of SMN knockdown conditions (Fig. S5 C, lanes 11 and 12; quantification in Fig. S5 D). The uncontrolled release of newly synthesized proteins and the risk of UsnRNP misassembly are thereby prevented. The amount of available pICln is likely to determine how many Sm proteins can be stored. When this buffering system becomes saturated, mRNAs encoding Sm proteins are destabilized, which is reminiscent of UsnRNA degradation in SMA mice or in cell culture that mimics the SMA situation (Gabanella et al., 2007; Zhang et al., 2008). Currently, how these transcripts are selectively degraded by the exosome awaits further investigation.
The inhibition of the early UsnRNP assembly phase by pICln depletion causes a different response. We had shown previously that under these conditions, the Sm proteins D1 and D2 (and the structural counterparts LSm10 and 11 of U7snRNP) are retained at the ribosomal exit tunnel to prevent their unassisted release into the cytosol (Paknia et al., 2016). During prolonged pICln limitation, however, Sm proteins are degraded by autophagy. The most severe effect on stability was observed for those Sm proteins that interact with pICln immediately after their synthesis (i.e., D1, D2, D3, and B/B′) whereas E, F, and G were less affected. These latter proteins, in the absence of UsnRNA, can form a closed hexameric ring in which all hydrophobic surfaces are protected alike in the assembled Sm core (Raker et al., 1996). This “self-protecting” oligomerization, which cannot be seen for the other Sm proteins, may explain their stability even during pICln shortage. The majority of cellular Sm proteins exist as part of UsnRNPs. In our pSILAC and Western blot experiments performed during pICln depletion, we observed nearly 50% down-regulation of both newly translated and steady-state levels of Sm proteins (Fig. 3). This observation suggests that in addition to decreased levels of newly translated Sm proteins, UsnRNP turnover is strongly affected as a consequence of pICln deficiency.
The protection of cells from protein aggregation is a likely reason for the selective degradation of Sm proteins upon perturbation of the early assembly phase. Our finding of massive Sm protein mislocalization and aggregation upon forced overproduction supports this view. The biochemical nature of Sm protein aggregates is currently unknown, but their obvious separation from the SMN complex and snRNA suggests that they are nonfunctional deposits that fail to proceed in the assembly reaction. It is quite likely that these nonfunctional Sm proteins associate with the chromatin or other noncognate RNAs based on simple interaction of countercharged residues and nucleotides. However, large-scale RNA metabolic labeling accompanied by Sm immunoprecipitation experiments will be necessary to analyze such nonspecific interactions, as the large excess of bulk cellular UsnRNPs mask these minority signals in simple end-radiolabeling experiments. Alternatively, large-scale pSILAC experiments performed under autophagy inhibition conditions with significant incorporation of isotope amino acids in the insoluble cellular fraction will have to be analyzed under pICln knockdown conditions to differentiate the behavior of bulk Sm proteins and newly synthesized proteins. However, irrespective of their nature and composition, these deposits can be seen only upon pICln knockdown. Therefore, our data support the view that pICln acts not only as an assembly chaperone but also a classic chaperone that shields Sm proteins from the environment, thereby preventing their aggregation.
Cellular protein aggregation has been linked to several human diseases, including Alzheimer’s, Huntington’s, and type II diabetes (Ross and Poirier, 2004). To limit their toxicity, protein aggregates are often processed in stress granules and cleared by cells through several mechanisms, including autophagy (Wolozin, 2012). Whether SMN deficiency, as observed in SMA, or mutations in pICln can be linked to toxic (Sm) protein aggregation is currently unclear. In favor of such a scenario, SMN has been shown to play a role in stress granule assembly and cellular stress regulation under different conditions (Hua and Zhou, 2004; Zou et al., 2011). Furthermore, we observed cytoplasmic mislocalization of SMN complex in distinct cytosolic foci during pICln knockdown and their disappearance upon autophagy inhibition. This indicates that the SMN-containing foci are indeed stress granules (Seguin et al., 2014) that are induced upon cellular stress during pICln depletion. No diseases have so far been linked to mutations in the pICln gene, but knockout mice are not viable (Pu et al., 2000). Based on the findings presented in this article, it will be interesting to analyze whether disturbed Sm proteostasis (or Sm protein aggregation) can be observed in these animals and whether this is causative for cell lethality.
Autophagy has been shown to be mostly nonspecific bulk degradation machinery for long-lived proteins and a preferred pathway for aggregates of proteins in neurodegenerative pathways (He and Klionsky, 2009; Kroemer et al., 2010). Furthermore, the core spliceosomal proteins have been linked to the mTOR pathway and autophagy (Quidville et al., 2013). It is currently unclear, however, how the Sm proteins are selected for autophagy, as they lack common sequence motifs that might be identified by the known chaperones of lysosomal proteolysis (Dice, 1990).
Our results, summarized in Fig. 9, demonstrate an intricate, multilayered regulation that streamlines the assembly of UsnRNPs, beginning with the acceptance of the Sm proteins immediately posttranslation by the assembly chaperone pICln. The controlled transfer of the Sm proteins to the SMN complex and eventually their loading onto snRNA completes the assembly pathway. In case of exhaustion or improper functioning of the assembly pathway, the cell activates fail-safe measures, including targeted autophagosome-mediated Sm protein degradation and exosome-processed Sm-encoding transcript degradation. These measures refine cellular quality control mechanisms to prevent proteotoxicity during imbalances in UsnRNP assembly.
Figure 9.
Elaborate regulation of Sm proteins during UsnRNP assembly prevents cellular proteotoxicity. Schematic representation of the regulatory events ensuring homeostasis of UsnRNPs. For details, see Discussion.
Materials and methods
Cell culture, RNAi-mediated knockdown, and antibodies
HeLa cells and stable lentivirus-mediated inducible shRNA knockdown HeLa cell lines were cultured in DMEM supplemented with 10% FCS (tetracycline negative, US origin), penicillin, and streptomycin (Gibco). HeLa cells expressing doxycycline-inducible shRNAs against SMN (shSMN) and Gemin5 (shGemin5) was generated using a lentivirus-based system (Wiznerowicz and Trono, 2003) with a SMARTpool of shRNAs (Table S1), and single-cell clones positive for EGFP (lentiviral vector encoded) were analyzed for knockdown of SMN and Gemin5, respectively. Cells were induced for shRNA expression using 1 µg/ml doxycycline for 120 or 144 h for knockdown of SMN and 96 h for Gemin5. Control cells were not treated with doxycycline. Knockdown of pICln was performed using On-TargetPlus SmartPool CLNS1A siRNA (L-012571-00-0005; GE Healthcare; Table S1) for 120 h. Control cells were transfected with siRNA against firefly luciferase (custom synthesized by Eurofins; Table S1). siRNA transfections were performed using Lipofectamine RNAiMAX (13778-150; Thermo Fisher Scientific) per the manufacturer’s protocol.
In this study, the following primary antibodies were used: mouse anti-SMN (clone 7B10; affinity purified from hybridoma supernatant; Meister et al., 2001a; 0176-01; ImmunoGlobe), mouse anti-Gemin5 (clone 10G11, 05-1535; EMD Millipore), rabbit anti-pICln (Chari et al., 2008), mouse anti-m3G/m7G-cap (Bochnig et al., 1987; H-20, gift from R. Lührmann, MPI for Biophysical Chemistry, Goettingen, Germany), mouse anti-Sm (Lerner et al., 1981; Y12, gift from J.A. Steitz, Yale School of Medicine, New Haven, CT), rabbit anti-coilin (H-300, sc-32860; Santa Cruz Biotechnology, Inc.), rat anti-Gemin3 (Grundhoff et al., 1999; gift from F. Grässer, Universitätsklinkum des Saarlandes, Homburg, Germany), mouse anti–β-catenin (610153; BD), rabbit anti-LC3 (L7543; Sigma-Aldrich), rabbit anti-SmD3 (PA5-26288; Thermo Fisher Scientific), rabbit anti-SmD1 (PA5-12459; Thermo Fisher Scientific), rabbit anti-SmF (ab66895; Abcam), rabbit anti-FLAG (F7425; Sigma-Aldrich), mouse anti–β-actin (A5316; Sigma-Aldrich), and mouse anti–α-tubulin (T5168; Sigma-Aldrich). For Western blotting, we used secondary goat antibodies conjugated with HRP anti–mouse (A4416; Sigma-Aldrich) and anti–rabbit (A6154; Sigma-Aldrich). For indirect immunostaining, we used Cy5-conjugated goat secondary antibodies (red channel) from Jackson ImmunoResearch Laboratories, Inc., anti–rat IgG (712-175-150), anti–mouse IgG (115-175-146), and anti–rabbit IgG (111-175-144), and Alexa Fluor 488–conjugated goat secondary antibodies (green channel) from Thermo Fisher Scientific, anti–rabbit (A11070) and anti–mouse (A11017).
pSILAC labeling
Control, shSMN, and shGemin5 cells were grown in DMEM containing 10% dialyzed FCS and light amino acids L-8662 ([12C]lysine; Sigma-Aldrich) and A-6969 ([12C]arginine; Sigma-Aldrich). After efficient knockdown using doxycycline induction, the control cells were grown in DMEM containing heavy isotope amino acids CNLM-291-H ([13C], [15N]lysine; Cambridge Isotope Laboratories) and CNLM-539-H ([13C], [15N]arginine; Cambridge Isotope Laboratories), whereas the SMN or Gemin5 knockdown cells were grown in medium heavy amino acids DLM-2640 ([2H]lysine; Cambridge Isotope Laboratories) and CLM-2265-H ([13C]arginine; Cambridge Isotope Laboratories) for 24 h. Next, cells were lysed in lysis buffer (50 mM Hepes-NaOH, pH 7.5, 150 mM NaCl, 1% NP-40, 2.5 mM MgCl2, and protease inhibitor cocktail). After incubation on ice for 10 min, the cells were lysed using 26G needle followed by mild water bath sonication to ensure complete nuclear lysis of the cells and centrifugation at 13,200 rpm for 20 min to remove cell debris. Heavy and medium pulse-labeled cell lysates were mixed in a 1:1 ratio, based on whole protein content estimated by Bradford assay (500-0006; Bio-Rad Laboratories) before processing for MS. siRNA-mediated control and pICln knockdown cells were processed similarly after the transfections.
Mass spectrometry and MS data analysis
For reduction, samples were incubated in NuPAGE LDS sample buffer (Thermo Fisher Scientific) supplemented with 50 mM DTT, incubated for 10 min at 70°C, and alkylated by incubation with iodoacetamide (final concentration 120 mM) for 20 min at RT. Reduced and alkylated samples were loaded on NuPAGE Novex Bis-Tris 4–12% gradient gels (NP0321BOX; Thermo Fisher Scientific) and stained with Coomassie (Simply Blue, LC6060; Thermo Fisher Scientific). Whole lanes were cut into 15 bands. The bands were destained with 30% acetonitrile, shrunk with 100% acetonitrile, and dried in a vacuum concentrator. Digestion 0.1 µg trypsin (V5280; Promega) per gel band was performed overnight at 37°C in 50 mM ammonium bicarbonate buffer. Peptides were extracted from the gel slices with 5% formic acid.
Nano–liquid chromatography/tandem MS analyses were performed on an LTQ-Orbitrap Velos Pro mass spectrometer equipped with an Easy-Spray ion source and coupled to an Easy-nLC 1000 UHPLC system (Thermo Fisher Scientific). Peptides were loaded on a trapping column (2 cm × 75 µm ID PepMap C18 3 µm particles, 100 Å pore size) and separated on an Easy-Spray column (25 cm × 75 µm ID, PepMap C18 2 µm particles, 100 Å pore size). pSILAC samples were analyzed with a 120-min linear gradient from 3% to 30% acetonitrile, 0.1% formic acid, and 200 or 400 nl/min flow rate. MS scans were acquired in the Orbitrap analyzer with a resolution of 30,000 at m/z 400. MS/MS scans were acquired in the LTQ Velos analyzer using collision-induced dissociation fragmentation with a TOP15 data-dependent MS/MS method. The minimum signal threshold for precursor selection was set to 10,000. Predictive automatic gain control (AGC) was used with an AGC target value of 1e6 for MS scans and 1e4 for MS/MS scans (e = exponent). Lock mass option was applied for internal calibration using background ions from protonated decamethylcyclopentasiloxane (m/z 371.10124). For all experiments, a dynamic exclusion was applied with a repeat count of 1 and exclusion duration of 60 s. Singly charged precursors were excluded from the selection.
For protein identification and quantification, MS raw data files were analyzed with MaxQuant version 1.5.2.8, and database searches were performed with the integrated search engine Andromeda. The UniProt human reference proteome database was used in combination with a database containing common contaminants as a reverse-concatenated target-decoy database. Protein identification was under the control of the false-discovery rate (FDR; <1% FDR on protein and peptide level). In addition to MaxQuant default settings (e.g., at least one razor/unique peptide for identification, two allowed miscleavages), the search was performed against the following variable modifications: protein N-terminal acetylation, Gln to pyro-Glu formation, and oxidation (on Met). For quantification of pSILAC-labeled proteins, the median was calculated of the log2-transformed normalized peptide ratios heavy to median (H/M) for each protein. At least two ratio counts were required for protein quantification. Protein ratios were normalized for each experiment in intensity bins (at least 300 proteins per bin), and outliers were identified by boxplot statistics as significantly altered, if their values were outside a 1.5× or 3× interquartile range (extreme outliers).
35S metabolic labeling, immunoprecipitation, and autoradiography
Cells were grown in DMEM supplemented with 10% FCS to 90–95% confluence (control without doxycycline and knockdown cells with 1 µg/ml doxycycline). After starvation in serum- and methionine-free medium for 30 min, the cells were labeled using [35S]methionine for 3.5 h at 25 µCi/ml in methionine-free medium supplemented with 10% dialyzed FCS and 20 mM Hepes, pH 7.4. The cells were then washed extensively using 1× PBS and lysed in lysis buffer (50 mM Hepes-NaOH, pH 7.5, 150 mM NaCl, 1% NP-40, 2.5 mM MgCl2, 0.8 U/µl murine RNase inhibitor [M0314S; New England Biolabs, Inc.], and protease inhibitor cocktail) as described in pSILAC labeling. Lysates were precleared using Sepharose–protein G beads (GE Healthcare) for 1 h at 4°C on a head-over-tail rotor. In parallel, the antibody against the protein of interest was incubated with protein G–Sepharose beads on a head-over-tail rotor for 1 h at RT and washed once with 1× PBS and twice with lysis buffer. The cleared lysates were incubated with primary antibody–bound protein G–Sepharose beads for 3 h at 4°C on a head-over-tail rotor for immunoprecipitation. After three washes using wash buffer (50 mM Hepes-NaOH, pH 7.5, 250 mM NaCl, 2.5 mM MgCl2, and 0.01% NP-40) and one wash with 1× PBS, the beads were boiled in 1× SDS sample loading buffer for 10 min at 95°C. Part of the samples were run on 13% high-tetramethylethylenediamine Bis-Tris SDS-PAGE gels, fixed, and incubated with Amplify fluorographic reagent (NAMP100; GE Healthcare). The gels were dried before being exposed to hypersensitive autoradiography films. The autoradiography signals were quantified using Fiji ImageJ software, and P values were calculated using Student’s t test. Part of the immunoprecipitated samples were used for Western blot analysis.
RNA isolation and quantitative RT-PCR
RNA and protein was isolated from HeLa cells lines using TRIzol reagent (15596018; Thermo Fisher Scientific) according to the manufacturer’s protocol. The precipitated RNA was treated with DNase using Turbo DNA-free kit (AM1907; Thermo Fisher Scientific) and reverse transcribed using SuperScript III reverse transcription kit (18080-093; Thermo Fisher Scientific) using random hexamer primers. After RNase H treatment, the cDNA was used in quantitative PCR using iTaq Universal SYBR Green Supermix (172-5122; Bio-Rad Laboratories) and analyzed using Bio-Rad CFX 2.0. Protein pellets were resuspended in 1× SDS sample Lämmli loading dye and analyzed by immunoblotting. Table S2 lists qPCR primers.
Polysome gradients
Control, SMN, and pICln knockdown cells were treated for 30 min at 37°C with 50 µg/ml cycloheximide to arrest translation and then washed with cold PBS. Cell extracts were prepared using gradient lysis buffer (100 mM KCl, 20 mM Tris HCl, pH 7.5, 5 mM MgCl2, 0.01 U/µl murine RNase inhibitor, 0.5% NP-40, 0.1 mg/ml cycloheximide, 1 mM DTT, and protease inhibitor cocktail). Cells were incubated on ice for 10 min in lysis buffer and centrifuged at 10,000 rpm for 10 min to clear the lysate. Cell extracts were separated by ultracentrifugation on a 5–45% (wt/vol) sucrose gradient (SW40Ti; 38,000 rpm, 90 min, 4°C), and the gradient was harvested using Biocomp piston gradient harvester connected to a Bio-Rad 2110 fraction collector, using Biocomp gradient profiler software. The gradient fractions were analyzed by Western blot or RNA was isolated from gradient fractions by phenol extraction and reverse-transcribed using Superscript III (Thermo Fisher Scientific) as described in RNA isolation and qRT-PCR. cDNA was amplified by PCR and analyzed by agarose gel electrophoresis.
Sm overexpression, FLAG immunoprecipitation, RNA end-labeling, and inhibition of exosome, lysosome, and proteasome
For overexpression of SmD3 and SmD1, C-terminal FLAG-tagged constructs were cloned in pcDNA3 vector. The tagged constructs were transfected at 0.5 µg vector per ∼0.15 × 106 cells using polyethylenimine, 48 h before fixing the cells for immunofluorescence. For anti-FLAG immunoprecipitation, 8 µg vector was transfected per 6 × 106 HeLa cells for 48 h. Cell were lysed using the lysis conditions described in pSILAC labeling and incubated with anti-FLAG M2 affinity gel (A2220; Sigma-Aldrich) for 3 h at 4°C on a head-over-tail rotor for immunoprecipitation. After washes using wash buffer (see 35S metabolic labeling, immunoprecipitation, and autoradiography), the immunoprecipitates were eluted twice using 1% SDS in 1× PBS buffer at 37°C on a thermomixer. The eluates were phenolized and processed for RNA precipitation and end-labeling. For exosome inhibition, cells were treated with 40 µM 5-FU for 24 h before cell lysis and RNA extraction. The cells were treated with 10 µM MG-132 (BML-PI102-0005; Enzo Life Sciences) for 10 h to induce proteasomal block using an equal volume of ethanol solvent as control. For inhibition of autophagy, cells were treated with 50 µM chloroquine (C6628; Sigma-Aldrich) for 10–12 h. Bafilomycin A1 (B1793; Sigma-Aldrich), diluted in 0.5% DMSO, was used at 200 nM for 24 h to induce block in autophagy, with an equal volume of 0.5% DMSO as solvent control. Soluble cell lysates were prepared as described in 35S metabolic labeling, immunoprecipitation, and autoradiography. For analysis of Sm aggregates, after removal of the supernatant containing the soluble lysates, the pellets were washed twice with 1% NP40 lysis buffer and dissolved by boiling in 1× SDS-Laemmli loading dye.
RNA 3′ end-labeling and metabolic labeling
The RNA samples were 3′ end-labeled using [32P]pCp with T4 RNA ligase (EL0021; Thermo Fisher Scientific) overnight at 4°C. After phenolization and RNA precipitation, the samples were run on a 10% denaturing urea-PAGE gel and exposed for autoradiography.
For RNA metabolic labeling, cells were grown in medium containing 10% FCS, antibiotics, and 5-EU (ab146642; Abcam) at 0.1 mM for 24 h in the presence of 5-FU. Cells were washed once with 1× PBS, lysed, and subjected to anti-Sm immunoprecipitation using Y12 antibody as described above for 35S metabolic labeling and immunoprecipitation. After washes, the beads were incubated with 5 µM Alexa Fluor 488 picolyl azide containing Click-iT Plus reaction cocktail (C10641; Thermo Fisher Scientific) with low CuSO4 concentration for 30 min at RT and protected from light. The beads were then washed with 1× PBS containing 3% BSA. The immunoprecipitated protein and RNA were extracted using TRIzol reagent per the manufacturer’s protocol. The extracted RNA was run on a 10% denaturing urea-PAGE gel and transferred onto a positively charged nylon membrane by semidry transfer at 1 mA/cm2 for 1 h with 1× TBE buffer. The membrane was then subjected to Alexa Fluor 488 fluorescence scan with the Bio-Rad Laboratories XRS+ Chemidoc system using Image Lab software.
Immunoblotting
Protein samples were run on an SDS-PAGE gel and transferred onto 0.22-µm pore size PVDF membranes using 1× towbin buffer containing 20% methanol. The membrane was blocked using 10% skim milk for 1 h at RT, incubated in primary antibody overnight at 4°C, and washed three times with 1× PBS-T (1× PBS, 0.05% [vol/vol] Tween-20, and 0.2% [vol/vol] Triton X-100) before incubation in HRP secondary antibody for 2 h at RT. The membrane was washed three times with 1× PBS-T and developed using luminol reagent or chemiluminescence substrate (170-5060; Bio-Rad Laboratories). The blots were quantified using Fiji software, and P values were calculated using Student’s t test.
Indirect immunofluorescence and image acquisition
Cells were grown on coverslips, washed, and fixed with 4% PFA for 20 min. After washes with 1× PBS, the cells were permeabilized for 20 min using 0.2% Triton X-100. After washes with 1× PBS, the cells were blocked using 10% FCS for 1 h. Primary and secondary antibodies were diluted in 2% FCS. Primary antibody incubation, followed by DAPI and secondary antibody incubation, was performed in a humidified chamber at RT for 1 h each and mounted using 2.5% Mowiol 4-88-DABCO mounting medium (0713, 0718; Carl Roth). Confocal images were acquired using a TCS SP5 confocal microscope (Leica Biosystems) with a photomultiplier, using a 63× oil-immersion UV objective, with numerical aperture of 1.4. All imaging was performed at RT using the Leica Biosystems Application Suite software package for image acquisition with 1,024 × 1,024-pixel resolution. The images were analyzed using Fiji software. CUD images were generated using Fiji software. ImarisColoc software was used to calculate PCC.
Online supplemental material
Table S1 lists RNAi sequences, Table S2 lists qPCR primers, Table S3 lists significantly regulated targets during SMN knockdown as identified by pSILAC, and Table S4 lists significantly regulated targets during pICln knockdown as identified by pSILAC. Fig. S1 shows that SMN knockdown does not result in the retention of Sm proteins on the polysomes and that UsnRNP biogenesis is inhibited upon SMN knockdown. Fig. S2 shows that pICln knockdown results in decreased transfer of Sm proteins to the SMN complex and formation of large cytoplasmic foci containing the SMN complex subunits. Fig. S3 shows individual channel images: perturbation of Sm protein homeostasis leads to aggregation in the absence of pICln. Fig. S4 shows individual channel images (CUD): perturbation of Sm protein homeostasis leads to aggregation in the absence of pICln. Fig. S5 shows that knockdown of Gemin5 results in the tailback of Sm proteins on pICln.
Supplementary Material
Acknowledgments
We thank R. Lührmann, F. Grässer, and J.A. Steitz for gifts of antibodies. We thank Suvagata Roy Chowdhury for guidance with confocal microscopy, Nitish Gulve for help with FACS, and Dr. Timothy Krüger for help with Imaris image analysis.
This work was supported by grants of the Deutsche Forschungsgemeinschaft (DFG; Fi573/8-2; U. Fischer), DFG Excellence Initiative at the University of Würzburg (Postdoc Plus program; A.B. Prusty), and a Graduate School of Life Sciences PhD fellowship (R. Meduri).
The authors declare no competing financial interests.
Author contributions: A.B. Prusty, R. Meduri, and U. Fischer conceived and designed the experiments. A.B. Prusty and R. Meduri carried out all the experiments. B.K. Prusty established the lentiviral-mediated shRNA knockdown cell lines and single cell clones. J. Vanselow and A. Schlosser performed MS of pSILAC samples and analyzed MS data. A.B. Prusty, R. Meduri, and U. Fischer analyzed the data and wrote the manuscript.
Footnotes
Abbreviations used:
- 5-EU
- 5-ethynyluridine
- 5-FU
- 5-fluorouracil
- CUD
- color universal design
- MS
- mass spectrometry
- PCC
- Pearson’s colocalization coefficient
- PRMT5
- protein arginine methyltransferase 5
- pSILAC
- pulsed stable isotope labeling by amino acids in cell culture
- SE
- standard error
- SMA
- spinal muscular atrophy
- SMN
- survival motor neuron
- UsnRNP
- U-rich small nuclear ribonucleoprotein particle
References
- Aberle H., Bauer A., Stappert J., Kispert A., and Kemler R.. 1997. β-Catenin is a target for the ubiquitin-proteasome pathway. EMBO J. 16:3797–3804. 10.1093/emboj/16.13.3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achsel T., Brahms H., Kastner B., Bachi A., Wilm M., and Lührmann R.. 1999. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 18:5789–5802. 10.1093/emboj/18.20.5789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachand F., Boisvert F.M., Côté J., Richard S., and Autexier C.. 2002. The product of the survival of motor neuron (SMN) gene is a human telomerase-associated protein. Mol. Biol. Cell. 13:3192–3202. 10.1091/mbc.E02-04-0216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbarossa A., Antoine E., Neel H., Gostan T., Soret J., and Bordonné R.. 2014. Characterization and in vivo functional analysis of the Schizosaccharomyces pombe ICLN gene. Mol. Cell. Biol. 34:595–605. 10.1128/MCB.01407-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S., Glick D., and Macleod K.F.. 2010. Autophagy: Assays and artifacts. J. Pathol. 221:117–124. 10.1002/path.2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochnig P., Reuter R., Bringmann P., and Lührmann R.. 1987. A monoclonal antibody against 2,2,7-trimethylguanosine that reacts with intact, class U, small nuclear ribonucleoproteins as well as with 7-methylguanosine-capped RNAs. Eur. J. Biochem. 168:461–467. 10.1111/j.1432-1033.1987.tb13439.x [DOI] [PubMed] [Google Scholar]
- Borg R.M., Bordonne R., Vassallo N., and Cauchi R.J.. 2015. Genetic interactions between the members of the SMN-gemins complex in Drosophila. PLoS One. 10:e0130974 10.1371/journal.pone.0130974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulisfane N., Choleza M., Rage F., Neel H., Soret J., and Bordonné R.. 2011. Impaired minor tri-snRNP assembly generates differential splicing defects of U12-type introns in lymphoblasts derived from a type I SMA patient. Hum. Mol. Genet. 20:641–648. 10.1093/hmg/ddq508 [DOI] [PubMed] [Google Scholar]
- Carvalho T., Almeida F., Calapez A., Lafarga M., Berciano M.T., and Carmo-Fonseca M.. 1999. The spinal muscular atrophy disease gene product, SMN: A link between snRNP biogenesis and the Cajal (coiled) body. J. Cell Biol. 147:715–728. 10.1083/jcb.147.4.715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari A., Golas M.M., Klingenhäger M., Neuenkirchen N., Sander B., Englbrecht C., Sickmann A., Stark H., and Fischer U.. 2008. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 135:497–509. 10.1016/j.cell.2008.09.020 [DOI] [PubMed] [Google Scholar]
- Dice J.F. 1990. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem. Sci. 15:305–309. 10.1016/0968-0004(90)90019-8 [DOI] [PubMed] [Google Scholar]
- Donlin-Asp P.G., Bassell G.J., and Rossoll W.. 2016. A role for the survival of motor neuron protein in mRNP assembly and transport. Curr. Opin. Neurobiol. 39:53–61. 10.1016/j.conb.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Fallini C., Donlin-Asp P.G., Rouanet J.P., Bassell G.J., and Rossoll W.. 2016. Deficiency of the survival of motor neuron protein impairs mRNA localization and local translation in the growth cone of motor neurons. J. Neurosci. 36:3811–3820. 10.1523/JNEUROSCI.2396-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., and Lührmann R.. 1990. An essential signaling role for the m3G cap in the transport of U1 snRNP to the nucleus. Science. 249:786–790. 10.1126/science.2143847 [DOI] [PubMed] [Google Scholar]
- Fischer U., Liu Q., and Dreyfuss G.. 1997. The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 90:1023–1029. 10.1016/S0092-8674(00)80368-2 [DOI] [PubMed] [Google Scholar]
- Fischer U., Englbrecht C., and Chari A.. 2011. Biogenesis of spliceosomal small nuclear ribonucleoproteins. Wiley Interdiscip. Rev. RNA. 2:718–731. 10.1002/wrna.87 [DOI] [PubMed] [Google Scholar]
- Friesen W.J., Paushkin S., Wyce A., Massenet S., Pesiridis G.S., Van Duyne G., Rappsilber J., Mann M., and Dreyfuss G.. 2001. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 21:8289–8300. 10.1128/MCB.21.24.8289-8300.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabanella F., Butchbach M.E., Saieva L., Carissimi C., Burghes A.H., and Pellizzoni L.. 2007. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS One. 2:e921 10.1371/journal.pone.0000921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia E.L., Lu Z., Meers M.P., Praveen K., and Matera A.G.. 2013. Developmental arrest of Drosophila survival motor neuron (Smn) mutants accounts for differences in expression of minor intron-containing genes. RNA. 19:1510–1516. 10.1261/rna.038919.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez G.B., Tian L., Ospina J.K., Boisvert F.M., Lamond A.I., and Matera A.G.. 2007. Two distinct arginine methyltransferases are required for biogenesis of Sm-class ribonucleoproteins. J. Cell Biol. 178:733–740. 10.1083/jcb.200702147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm C., Chari A., Pelz J.P., Kuper J., Kisker C., Diederichs K., Stark H., Schindelin H., and Fischer U.. 2013. Structural basis of assembly chaperone-mediated snRNP formation. Mol. Cell. 49:692–703. 10.1016/j.molcel.2012.12.009 [DOI] [PubMed] [Google Scholar]
- Grundhoff A.T., Kremmer E., Türeci O., Glieden A., Gindorf C., Atz J., Mueller-Lantzsch N., Schubach W.H., and Grässer F.A.. 1999. Characterization of DP103, a novel DEAD box protein that binds to the Epstein-Barr virus nuclear proteins EBNA2 and EBNA3C. J. Biol. Chem. 274:19136–19144. 10.1074/jbc.274.27.19136 [DOI] [PubMed] [Google Scholar]
- Gubitz A.K., Mourelatos Z., Abel L., Rappsilber J., Mann M., and Dreyfuss G.. 2002. Gemin5, a novel WD repeat protein component of the SMN complex that binds Sm proteins. J. Biol. Chem. 277:5631–5636. 10.1074/jbc.M109448200 [DOI] [PubMed] [Google Scholar]
- Hamm J., Darzynkiewicz E., Tahara S.M., and Mattaj I.W.. 1990. The trimethylguanosine cap structure of U1 snRNA is a component of a bipartite nuclear targeting signal. Cell. 62:569–577. 10.1016/0092-8674(90)90021-6 [DOI] [PubMed] [Google Scholar]
- Hannus S., Bühler D., Romano M., Seraphin B., and Fischer U.. 2000. The Schizosaccharomyces pombe protein Yab8p and a novel factor, Yip1p, share structural and functional similarity with the spinal muscular atrophy-associated proteins SMN and SIP1. Hum. Mol. Genet. 9:663–674. 10.1093/hmg/9.5.663 [DOI] [PubMed] [Google Scholar]
- He C., and Klionsky D.J.. 2009. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43:67–93. 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., and Li H.. 2000. A mouse model for spinal muscular atrophy. Nat. Genet. 24:66–70. 10.1038/71709 [DOI] [PubMed] [Google Scholar]
- Hua Y., and Zhou J.. 2004. Survival motor neuron protein facilitates assembly of stress granules. FEBS Lett. 572:69–74. 10.1016/j.febslet.2004.07.010 [DOI] [PubMed] [Google Scholar]
- Ishikawa H., Nobe Y., Izumikawa K., Yoshikawa H., Miyazawa N., Terukina G., Kurokawa N., Taoka M., Yamauchi Y., Nakayama H., et al. 2014. Identification of truncated forms of U1 snRNA reveals a novel RNA degradation pathway during snRNP biogenesis. Nucleic Acids Res. 42:2708–2724. 10.1093/nar/gkt1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády B.E., Darzacq X., Tucker K.E., Matera A.G., Bertrand E., and Kiss T.. 2003. Modification of Sm small nuclear RNAs occurs in the nucleoplasmic Cajal body following import from the cytoplasm. EMBO J. 22:1878–1888. 10.1093/emboj/cdg187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao C.Y., and Salic A.. 2008. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl. Acad. Sci. USA. 105:15779–15784. 10.1073/pnas.0808480105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C., and Mattaj I.W.. 1992. Intracellular distribution of the U1A protein depends on active transport and nuclear binding to U1 snRNA. J. Cell Biol. 118:11–21. 10.1083/jcb.118.1.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambach C., Walke S., and Nagai K.. 1999a Structure and assembly of the spliceosomal small nuclear ribonucleoprotein particles. Curr. Opin. Struct. Biol. 9:222–230. 10.1016/S0959-440X(99)80032-3 [DOI] [PubMed] [Google Scholar]
- Kambach C., Walke S., Young R., Avis J.M., de la Fortelle E., Raker V.A., Lührmann R., Li J., and Nagai K.. 1999b Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 96:375–387. 10.1016/S0092-8674(00)80550-4 [DOI] [PubMed] [Google Scholar]
- Kammler S., Lykke-Andersen S., and Jensen T.H.. 2008. The RNA exosome component hRrp6 is a target for 5-fluorouracil in human cells. Mol. Cancer Res. 6:990–995. 10.1158/1541-7786.MCR-07-2217 [DOI] [PubMed] [Google Scholar]
- Kisselev A.F., and Goldberg A.L.. 2001. Proteasome inhibitors: From research tools to drug candidates. Chem. Biol. 8:739–758. 10.1016/S1074-5521(01)00056-4 [DOI] [PubMed] [Google Scholar]
- Koplin A., Preissler S., Ilina Y., Koch M., Scior A., Erhardt M., and Deuerling E.. 2010. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 189:57–68. 10.1083/jcb.200910074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G., Mariño G., and Levine B.. 2010. Autophagy and the integrated stress response. Mol. Cell. 40:280–293. 10.1016/j.molcel.2010.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroiss M., Schultz J., Wiesner J., Chari A., Sickmann A., and Fischer U.. 2008. Evolution of an RNP assembly system: A minimal SMN complex facilitates formation of UsnRNPs in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA. 105:10045–10050. 10.1073/pnas.0802287105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau C.K., Bachorik J.L., and Dreyfuss G.. 2009. Gemin5-snRNA interaction reveals an RNA binding function for WD repeat domains. Nat. Struct. Mol. Biol. 16:486–491. 10.1038/nsmb.1584 [DOI] [PubMed] [Google Scholar]
- Law M.J., Rice A.J., Lin P., and Laird-Offringa I.A.. 2006. The role of RNA structure in the interaction of U1A protein with U1 hairpin II RNA. RNA. 12:1168–1178. 10.1261/rna.75206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S., Bürglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. 1995. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 80:155–165. 10.1016/0092-8674(95)90460-3 [DOI] [PubMed] [Google Scholar]
- Lerner E.A., Lerner M.R., Janeway C.A. Jr., and Steitz J.A.. 1981. Monoclonal antibodies to nucleic acid-containing cellular constituents: Probes for molecular biology and autoimmune disease. Proc. Natl. Acad. Sci. USA. 78:2737–2741. 10.1073/pnas.78.5.2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera A.G., and Wang Z.. 2014. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15:108–121. 10.1038/nrm3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell T.S., Lokken R.P., and Steitz J.A.. 2003. Assembly of the U1 snRNP involves interactions with the backbone of the terminal stem of U1 snRNA. RNA. 9:193–201. 10.1261/rna.2136103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., and Fischer U.. 2002. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 21:5853–5863. 10.1093/emboj/cdf585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meister G., Bühler D., Pillai R., Lottspeich F., and Fischer U.. 2001a A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 3:945–949. 10.1038/ncb1101-945 [DOI] [PubMed] [Google Scholar]
- Meister G., Eggert C., Bühler D., Brahms H., Kambach C., and Fischer U.. 2001b Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr. Biol. 11:1990–1994. 10.1016/S0960-9822(01)00592-9 [DOI] [PubMed] [Google Scholar]
- Montzka K.A., and Steitz J.A.. 1988. Additional low-abundance human small nuclear ribonucleoproteins: U11, U12, etc. Proc. Natl. Acad. Sci. USA. 85:8885–8889. 10.1073/pnas.85.23.8885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouaikel J., Verheggen C., Bertrand E., Tazi J., and Bordonné R.. 2002. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell. 9:891–901. 10.1016/S1097-2765(02)00484-7 [DOI] [PubMed] [Google Scholar]
- Mouaikel J., Narayanan U., Verheggen C., Matera A.G., Bertrand E., Tazi J., and Bordonné R.. 2003. Interaction between the small-nuclear-RNA cap hypermethylase and the spinal muscular atrophy protein, survival of motor neuron. EMBO Rep. 4:616–622. 10.1038/sj.embor.embor863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedialkova D.D., and Leidel S.A.. 2015. Optimization of codon translation rates via tRNA modifications maintains proteome integrity. Cell. 161:1606–1618. 10.1016/j.cell.2015.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuenkirchen N., Englbrecht C., Ohmer J., Ziegenhals T., Chari A., and Fischer U.. 2015. Reconstitution of the human U snRNP assembly machinery reveals stepwise Sm protein organization. EMBO J. 34:1925–1941. 10.15252/embj.201490350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otter S., Grimmler M., Neuenkirchen N., Chari A., Sickmann A., and Fischer U.. 2007. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J. Biol. Chem. 282:5825–5833. 10.1074/jbc.M608528200 [DOI] [PubMed] [Google Scholar]
- Paknia E., Chari A., Stark H., and Fischer U.. 2016. The ribosome cooperates with the assembly chaperone pICln to initiate formation of snRNPs. Cell Reports. 16:3103–3112. 10.1016/j.celrep.2016.08.047 [DOI] [PubMed] [Google Scholar]
- Pannone B.K., Xue D., and Wolin S.L.. 1998. A role for the yeast La protein in U6 snRNP assembly: Evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 17:7442–7453. 10.1093/emboj/17.24.7442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone B.K., Kim S.D., Noe D.A., and Wolin S.L.. 2001. Multiple functional interactions between components of the Lsm2-Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics. 158:187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paushkin S., Charroux B., Abel L., Perkinson R.A., Pellizzoni L., and Dreyfuss G.. 2000. The survival motor neuron protein of Schizosacharomyces pombe. Conservation of survival motor neuron interaction domains in divergent organisms. J. Biol. Chem. 275:23841–23846. 10.1074/jbc.M001441200 [DOI] [PubMed] [Google Scholar]
- Paushkin S., Gubitz A.K., Massenet S., and Dreyfuss G.. 2002. The SMN complex, an assemblyosome of ribonucleoproteins. Curr. Opin. Cell Biol. 14:305–312. 10.1016/S0955-0674(02)00332-0 [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Baccon J., Rappsilber J., Mann M., and Dreyfuss G.. 2002a Purification of native survival of motor neurons complexes and identification of Gemin6 as a novel component. J. Biol. Chem. 277:7540–7545. 10.1074/jbc.M110141200 [DOI] [PubMed] [Google Scholar]
- Pellizzoni L., Yong J., and Dreyfuss G.. 2002b Essential role for the SMN complex in the specificity of snRNP assembly. Science. 298:1775–1779. 10.1126/science.1074962 [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Grimmler M., Meister G., Will C.L., Lührmann R., Fischer U., and Schümperli D.. 2003. Unique Sm core structure of U7 snRNPs: Assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 17:2321–2333. 10.1101/gad.274403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praveen K., Wen Y., and Matera A.G.. 2012. A Drosophila model of spinal muscular atrophy uncouples snRNP biogenesis functions of survival motor neuron from locomotion and viability defects. Cell Reports. 1:624–631. 10.1016/j.celrep.2012.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu W.T., Wickman K., and Clapham D.E.. 2000. ICln is essential for cellular and early embryonic viability. J. Biol. Chem. 275:12363–12366. 10.1074/jbc.275.17.12363 [DOI] [PubMed] [Google Scholar]
- Quidville V., Alsafadi S., Goubar A., Commo F., Scott V., Pioche-Durieu C., Girault I., Baconnais S., Le Cam E., Lazar V., et al. 2013. Targeting the deregulated spliceosome core machinery in cancer cells triggers mTOR blockade and autophagy. Cancer Res. 73:2247–2258. 10.1158/0008-5472.CAN-12-2501 [DOI] [PubMed] [Google Scholar]
- Raker V.A., Plessel G., and Lührmann R.. 1996. The snRNP core assembly pathway: Identification of stable core protein heteromeric complexes and an snRNP subcore particle in vitro. EMBO J. 15:2256–2269. [PMC free article] [PubMed] [Google Scholar]
- Raker V.A., Hartmuth K., Kastner B., and Lührmann R.. 1999. Spliceosomal U snRNP core assembly: Sm proteins assemble onto an Sm site RNA nonanucleotide in a specific and thermodynamically stable manner. Mol. Cell. Biol. 19:6554–6565. 10.1128/MCB.19.10.6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross C.A., and Poirier M.A.. 2004. Protein aggregation and neurodegenerative disease. Nat. Med. 10(7, Suppl):S10–S17. 10.1038/nm1066 [DOI] [PubMed] [Google Scholar]
- Schrank B., Götz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., and Sendtner M.. 1997. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl. Acad. Sci. USA. 94:9920–9925. 10.1073/pnas.94.18.9920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhäusser B., Gossen M., Dittmar G., and Selbach M.. 2009. Global analysis of cellular protein translation by pulsed SILAC. Proteomics. 9:205–209. 10.1002/pmic.200800275 [DOI] [PubMed] [Google Scholar]
- Seguin S.J., Morelli F.F., Vinet J., Amore D., De Biasi S., Poletti A., Rubinsztein D.C., and Carra S.. 2014. Inhibition of autophagy, lysosome and VCP function impairs stress granule assembly. Cell Death Differ. 21:1838–1851. 10.1038/cdd.2014.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel K.B., and Matera A.G.. 2005. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl. Acad. Sci. USA. 102:17372–17377. 10.1073/pnas.0508947102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpargel K.B., Praveen K., Rajendra T.K., and Matera A.G.. 2009. Gemin3 is an essential gene required for larval motor function and pupation in Drosophila. Mol. Biol. Cell. 20:90–101. 10.1091/mbc.E08-01-0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla S., and Parker R.. 2014. Quality control of assembly-defective U1 snRNAs by decapping and 5′-to-3′ exonucleolytic digestion. Proc. Natl. Acad. Sci. USA. 111:E3277–E3286. 10.1073/pnas.1412614111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeman J.E., and Lamond A.I.. 1999. Newly assembled snRNPs associate with coiled bodies before speckles, suggesting a nuclear snRNP maturation pathway. Curr. Biol. 9:1065–1074. 10.1016/S0960-9822(99)80475-8 [DOI] [PubMed] [Google Scholar]
- So B.R., Wan L., Zhang Z., Li P., Babiash E., Duan J., Younis I., and Dreyfuss G.. 2016. A U1 snRNP-specific assembly pathway reveals the SMN complex as a versatile hub for RNP exchange. Nat. Struct. Mol. Biol. 23:225–230. 10.1038/nsmb.3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanĕk D., and Neugebauer K.M.. 2004. Detection of snRNP assembly intermediates in Cajal bodies by fluorescence resonance energy transfer. J. Cell Biol. 166:1015–1025. 10.1083/jcb.200405160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark H., Dube P., Lührmann R., and Kastner B.. 2001. Arrangement of RNA and proteins in the spliceosomal U1 small nuclear ribonucleoprotein particle. Nature. 409:539–542. 10.1038/35054102 [DOI] [PubMed] [Google Scholar]
- Urlaub H., Raker V.A., Kostka S., and Lührmann R.. 2001. Sm protein-Sm site RNA interactions within the inner ring of the spliceosomal snRNP core structure. EMBO J. 20:187–196. 10.1093/emboj/20.1.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M.C., and Fischer U.. 2016. The right pick: Structural basis of snRNA selection by Gemin5. Genes Dev. 30:2341–2344. 10.1101/gad.293084.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl M.C., Will C.L., and Lührmann R.. 2009. The spliceosome: Design principles of a dynamic RNP machine. Cell. 136:701–718. 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- Wan L., Battle D.J., Yong J., Gubitz A.K., Kolb S.J., Wang J., and Dreyfuss G.. 2005. The survival of motor neurons protein determines the capacity for snRNP assembly: Biochemical deficiency in spinal muscular atrophy. Mol. Cell. Biol. 25:5543–5551. 10.1128/MCB.25.13.5543-5551.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler C., Eggert C., Gradl D., Meister G., Giegerich M., Wedlich D., Laggerbauer B., and Fischer U.. 2005. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 19:2320–2330. 10.1101/gad.342005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiznerowicz M., and Trono D.. 2003. Conditional suppression of cellular genes: Lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 77:8957–8961. 10.1128/JVI.77.16.8957-8951.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolozin B. 2012. Regulated protein aggregation: Stress granules and neurodegeneration. Mol. Neurodegener. 7:56 10.1186/1750-1326-7-56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., and Tashiro Y.. 1998. Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23:33–42. 10.1247/csf.23.33 [DOI] [PubMed] [Google Scholar]
- Yong J., Kasim M., Bachorik J.L., Wan L., and Dreyfuss G.. 2010. Gemin5 delivers snRNA precursors to the SMN complex for snRNP biogenesis. Mol. Cell. 38:551–562. 10.1016/j.molcel.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., So B.R., Li P., Yong J., Glisovic T., Wan L., and Dreyfuss G.. 2011. Structure of a key intermediate of the SMN complex reveals Gemin2’s crucial function in snRNP assembly. Cell. 146:384–395. 10.1016/j.cell.2011.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., and Dreyfuss G.. 2008. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 133:585–600. 10.1016/j.cell.2008.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou T., Yang X., Pan D., Huang J., Sahin M., and Zhou J.. 2011. SMN deficiency reduces cellular ability to form stress granules, sensitizing cells to stress. Cell. Mol. Neurobiol. 31:541–550. 10.1007/s10571-011-9647-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.