ER stress results in widespread aggregation of proteins that are not localized to the ER or are part of the secretory system. Hamdan et al. demonstrate that amorphous and amyloidogenic protein aggregation is an indirect consequence of perturbing ER homeostasis.
Abstract
Disturbances in endoplasmic reticulum (ER) homeostasis create a condition termed ER stress. This activates the unfolded protein response (UPR), which alters the expression of many genes involved in ER quality control. We show here that ER stress causes the aggregation of proteins, most of which are not ER or secretory pathway proteins. Proteomic analysis of the aggregated proteins revealed enrichment for intrinsically aggregation-prone proteins rather than proteins which are affected in a stress-specific manner. Aggregation does not arise because of overwhelming proteasome-mediated degradation but because of a general disruption of cellular protein homeostasis. We further show that overexpression of certain chaperones abrogates protein aggregation and protects a UPR mutant against ER stress conditions. The onset of ER stress is known to correlate with various disease processes, and our data indicate that widespread amorphous and amyloid protein aggregation is an unanticipated outcome of such stress.
Introduction
The ability to maintain the balance between protein biogenesis, folding, trafficking, and degradation in the face of changing conditions is essential for viability in all eukaryotic cells. This is illustrated by the plethora of human diseases influenced by alterations in protein homeostasis (Labbadia and Morimoto, 2015). The ER is the first organelle required for folding and trafficking of nascent membrane proteins and proteins that enter the secretory pathway. Proteins enter in an unfolded state, and ER-resident enzymes facilitate their oxidative folding, modification, and trafficking reactions (Walter and Ron, 2011). An ER stress occurs when there is a breakdown in ER protein homeostasis.
ER-associated degradation (ERAD) acts to remove proteins that do not fold properly from the ER. Proteins are retrotranslocated into the cytosol, where they are ubiquitinated and degraded by the proteasome (Nakatsukasa and Brodsky, 2008; Christianson and Ye, 2014; Zattas and Hochstrasser, 2015). Loss of ERAD results in an accumulation of misfolded ER proteins and promotes ER stress (Walter and Ron, 2011). Substrate recognition and membrane extraction is coordinated by ER-embedded ubiquitin ligase complexes; three distinct membrane protein complexes define different ERAD pathways (L-lumen, M-membrane, and C-cytosol), depending on the localization of the degradation signal (Carvalho et al., 2006; Nakatsukasa and Brodsky, 2008; Ruggiano et al., 2014). In yeast, ERAD-L and ERAD-M substrates are targeted for degradation by the Hrd1 complex, whereas, ERAD-C substrates are recognized by the Doa10 complex (Brodsky and Skach, 2011; Thibault and Ng, 2012; Christianson and Ye, 2014; Ruggiano et al., 2014; Zattas and Hochstrasser, 2015). The unfolded protein response (UPR) is a signaling pathway that is activated in response to ER stress by an ER-localized kinase, Ire1 (Travers et al., 2000). Ire1 senses the accumulation of unfolded proteins in the ER and acts as a specific endoribonuclease, splicing the HAC1 mRNA (Sidrauski and Walter, 1997). The translation product of spliced HAC1 mRNA is the transcriptional activator for genes affecting protein folding, degradation, and trafficking to restore ER homeostasis (Travers et al., 2000; Walter and Ron, 2011).
Although much is now known regarding the role of ER stress defense systems in maintaining ER protein homeostasis, little is known regarding the consequences of ER dysfunction on protein homeostasis in other cellular compartments. In the current study, we show that ER stress results in widespread cytoplasmic protein aggregation, including both amorphous and amyloidogenic aggregation.
Results and discussion
ER stress causes widespread protein aggregation
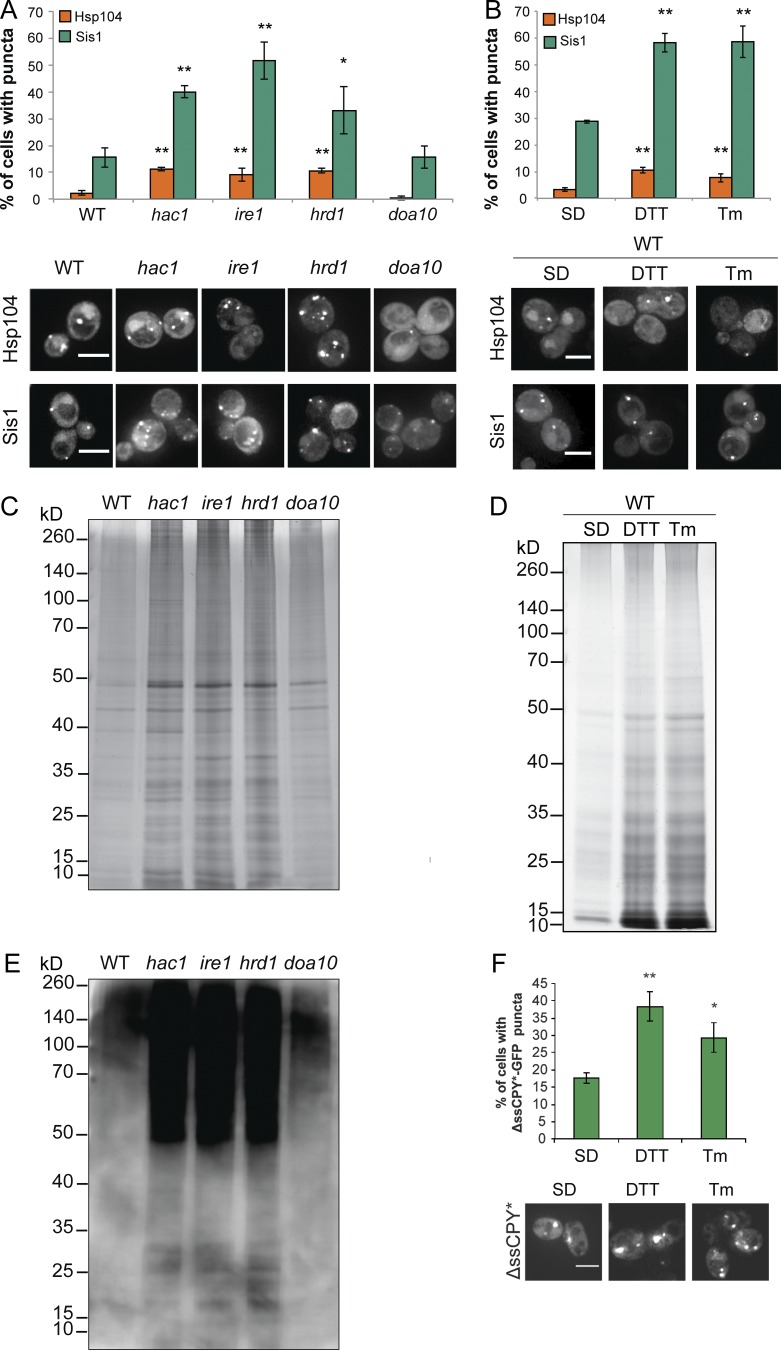
Protein aggregation was analyzed in mutants deficient in the UPR (HAC1 and IRE1) or ERAD (HRD1 and DOA10) and in cells exposed to tunicamycin (Tm) or DTT to promote ER stress (Cox et al., 1993; Kohno et al., 1993). Foci of protein aggregates can be detected using the fluorescently tagged Hsp104 disaggregase or Sis1 Hsp40 chaperone (Lum et al., 2004; Erjavec et al., 2007; Lee et al., 2010; Malinovska et al., 2012; Park et al., 2013). The number of cells containing Hsp104-RFP or Sis-GFP puncta was significantly increased in hac1, ire1, and hrd1 mutants (Fig. 1 A) and in wild-type cells exposed to DTT or Tm (Fig. 1 B). To confirm that protein aggregation occurs during ER stress, aggregates were purified using an established biochemical approach (Tomoyasu et al., 2001; Jang et al., 2004; Rand and Grant, 2006; Koplin et al., 2010). Elevated protein aggregation was observed in hac1, ire1, and hrd1 mutant strains (Fig. 1 C) and in wild-type cells exposed to DTT or Tm (Fig. 1 D). Ubiquitinated proteins were increased in the aggregate fractions isolated from the hac1, ire1, and hrd1 mutants, suggesting that aggregated proteins are targeted to the proteasome for degradation (Fig. 1 E). To further confirm that protein aggregates form during ER stress, a mutant version of the secretory protein carboxypeptidase Y lacking its signal sequence (ΔssCPY∗) was used, which is rapidly degraded by the ubiquitin proteasome system (UPS; Eisele and Wolf, 2008; Park et al., 2013). ΔssCPY∗-GFP aggregate formation was elevated in response to ER stress imposed by DTT or Tm treatment (Fig. 1 F).
Figure 1.
ER stress causes protein aggregation. (A) Hsp104-RFP and Sis1-GFP were visualized in wild-type, hac1, ire1, hrd1, and doa10 mutant cells and in wild-type cells exposed to DTT or Tm. Bars, 4 µm. (B) The percentage of cells containing puncta is quantified for each strain from three independent biological repeat experiments ± SD. (C and D) Silver staining of protein aggregates isolated from the same strains as in A and B. (E) Protein ubiquitination was analyzed in the isolated protein aggregates by Western blot using an α-ubiquitin antibody. (F) Examples of cells containing ΔssCPY∗-GFP puncta in a wild-type strain after DTT or Tm treatment. *, P < 0.05; **, P < 0.01; ***, P < 0.005 (n = 3).
ER stress causes the aggregation of aggregation-prone proteins rather than specific ER targets
Aggregated proteins were identified using mass spectrometry. A large overlap in the proteins that aggregate in wild-type, hac1, and hrd1 mutants strains was observed, indicating that the majority of proteins do not aggregate in a mutant-specific manner (Fig. 2 A). Similar functional categories were enriched within aggregate fractions prepared from wild-type and ER stress mutants (Fig. S1), including common enrichment in major cellular processes such as metabolism, energy, protein synthesis, and protein fate. Strikingly, no enrichment for ER-related processes was observed in hac1 or hrd1 mutant strains. The majority of aggregated proteins were predicted to localize to the cytoplasm, and no enrichment for ER or secreted proteins was observed in hac1 or hrd1 mutant strains compared with the wild-type strain (Fig. 2 B).
Figure 2.
Aggregated proteins in wild-type and ER stress mutants have similar localization and physicochemical properties. (A) Venn diagram showing the overlaps between proteins aggregating in wild-type (green), hac1 (pink), and hrd1 (yellow) mutant strains. (B) Diagrams showing the localization of the proteins aggregating in wild-type, hac1, and hrd1 strains. (C–H) Box plots showing comparisons of physicochemical properties for the aggregated proteins common to the wild-type, hac1, and hrd1 mutant strains (common), present in the hac1 mutant, but not the wild-type, strain (hac1 only), and present in the hrd1 mutant, but not the wild-type, strain (hrd1 only). Aggregated proteins were compared with unaggregated proteins (MS). (C) Protein abundance. (D) Codon adaptation index (CAI). (E) Translation rates. (F) Grand mean of hydrophobicity (GRAVY). (G) Isoelectric points (pI). (H) Protein stability. Mann–Whitney U tests were used to assess the statistical significance of observed differences; *, P < 0.05; **, P < 0.01; ***, P < 0.005.
We assessed the physicochemical properties of aggregated proteins to determine whether they possess particular properties that make them aggregation prone. The 606 proteins that commonly aggregated in the wild-type, hac1, and hrd1 mutant strains (common set) were compared with the 189 and 190 proteins that aggregated in the hac1 and hrd1 mutants, but not in the wild-type strain. No functional classes were enriched in the hac1 or hrd1 mutants that were not also enriched in the common set (Fig. S2). The aggregated proteins were compared with a list of yeast proteins detectable by mass spectrometry (MS; MS set) to represent the properties of unaggregated proteins (Washburn et al., 2001). The common set was enriched for proteins with higher abundance (molecules per cell), higher expression levels (codon adaption index), and higher translation rate compared with the MS set (Fig. 2, C–E). In contrast, the proteins that specifically aggregated in hac1 or hrd1 mutants tended to be have lower abundances and translation rates compared with the MS set (Fig. 2, C–E). Hydrophobicity indicates a propensity to aggregate (Vabulas et al., 2010; Tamás et al., 2014), and we found that proteins in the common set showed a significant increase in hydrophobicity (GRAVY score) compared with the MS set, whereas the proteins which aggregate in hac1 or hrd1 mutants were similar to the MS set (Fig. 2 F). We found no differences in isoelectric points (pI) between the aggregated proteins and the MS set (Fig. 2 G). Using a global proteome turnover database (Christiano et al., 2014), we found that proteins in the common set and hrd1-only sets show, on average, a longer half-life than proteins in the MS proteome, suggesting that these proteins are normally stable in their native folded states (Fig. 2 H). Collectively, these data indicate that the proteins that commonly aggregate in the wild-type, hac1, and hrd1 mutants have common properties indicative of aggregation-prone proteins.
Protein aggregation does not arise because of inhibition of UPS-mediated protein degradation
We considered two mechanisms that might account for the high levels of protein aggregation in ER stress mutants. First, protein aggregation might arise because of a defect in proteasomal degradation that could result in the accumulation of misfolded proteins and subsequent aggregation rather than turnover. For example, an inhibition of UPS-mediated protein degradation might occur if ER stress generates more substrates than the UPS can cope with, effectively overwhelming the degradation machinery. We tested this possibility by examining the degradation kinetics of the ΔssCPY∗-GFP reporter as a model substrate for UPS-mediated degradation (Park et al., 2007, 2013). Rapid turnover of ΔssCPY∗-GFP was observed in the wild-type strain as well as in the hac1 or hrd1 mutants, suggesting that there is no defect in UPS degradation (Fig. 3 A). For comparison, we examined the turnover of Hmg2 (HMG-CoA reductase), a well-characterized ERAD substrate (Hampton et al., 1996), and we found that its turnover is slower in a hac1 mutant than in a wild-type strain (Fig. 3 B).
Figure 3.
Selected chaperones reduce protein aggregation in a hac1 mutant. (A) Turnover rate of ΔssCPY∗-GFP in wild-type, hac1, and hrd1 strains (n = 3) ± SD. (B) Turnover rate of Hmg2-myc in wild-type and hac1 strains (n = 3) ± SD. (C) The overlap between chaperones identified within the protein aggregate datasets of the wild-type, hac1, and hrd1 strains. The 21 chaperones listed were present in the aggregates of all strains. (D) Silver staining of protein aggregates isolated from the hac1 mutant containing galactose-regulatable expression plasmids for Ssa1, Ydj1, Sis1, Hsp104, Sse1, or Ssb1. Vector denotes an empty vector control. (E) Turnover rate of ΔssCPY∗-GFP in wild-type and hac1 strains containing galactose-regulatable expression plasmids for Sis1, Sse1, or Ssb1 or empty vector controls (n = 3) ± SD. *, P < 0.05; **, P < 0.01; ***, P < 0.005 (n = 3).
Overexpression of selected chaperones prevents ER stress–induced protein aggregation
The second possibility that we considered was that proteins that are protected from misfolding by the cellular protein quality control systems might accumulate and aggregate if these are disrupted or become overwhelmed. To counteract protein aggregation, cells contain many molecular chaperones (Flynn et al., 1991; Hartl et al., 2011; Verghese et al., 2012), but increased aggregation can arise because of insufficient availability of chaperones when ERAD substrates and aggregates accumulate. Increased cytosolic aggregation might therefore arise in ER stress mutants if protein aggregates or ERAD substrates sequester key chaperones, limiting their availability to maintain protein homeostasis. Of the 63 known chaperones in Saccharomyces cerevisiae (Gong et al., 2009), we identified 27 distributed between all the datasets (Fig. 3 C). These chaperones presumably localize to aggregates to mitigate any toxic consequences, so we examined whether the aggregates are enriched in proteins that have increased chaperone interactions. We found no enrichment for proteins with extensive chaperone interactions in the aggregated proteins compared with unaggregated proteins (Fig. S3 A).
We next tested whether overexpression of candidate chaperones identified in our aggregate fractions could prevent aggregate formation. Overexpression of Hsp104, Ssa1, or Ydj1 did not affect protein aggregation, whereas Sis1, Sse1, and Ssb1 dramatically reduced the levels of aggregation in a hac1 mutant (Fig. 3 D). Western blot analyses confirmed that chaperones were overexpressed (Fig. S3 B). The ΔssCPY∗-GFP reporter undergoes rapid UPS-mediated degradation dependent on cytosolic Hsp70 (Ssa1) and Hsp40’s (Ydj1 and Sis1; Park et al., 2007, 2013); thus, we tested whether chaperone overexpression affects UPS-mediated degradation (Fig. 3 E). Overexpression of Sis1 increased the rate of degradation of ΔssCPY∗-GFP in both the wild-type and hac1 mutant strains, in agreement with the idea that Sis1 is required for targeting misfolded proteins for degradation by the UPS system (Park et al., 2013; Shiber et al., 2013; Summers et al., 2013). In contrast, overexpression of Sse1 or Ssb1 did not affect the rate of ΔssCPY∗-GFP degradation. The finding that Sse1 and Ssb1 do not affect UPS-mediated degradation of ΔssCPY∗-GFP, while preventing protein aggregation, further confirms that a defect in UPS-mediated degradation does not account for the high levels of protein aggregation in a hac1 mutant. Together, these findings are in agreement with the idea that limitations in chaperone availability account for the increased protein aggregation in ER stress mutants.
ER stress increases the spontaneous frequency of prion formation
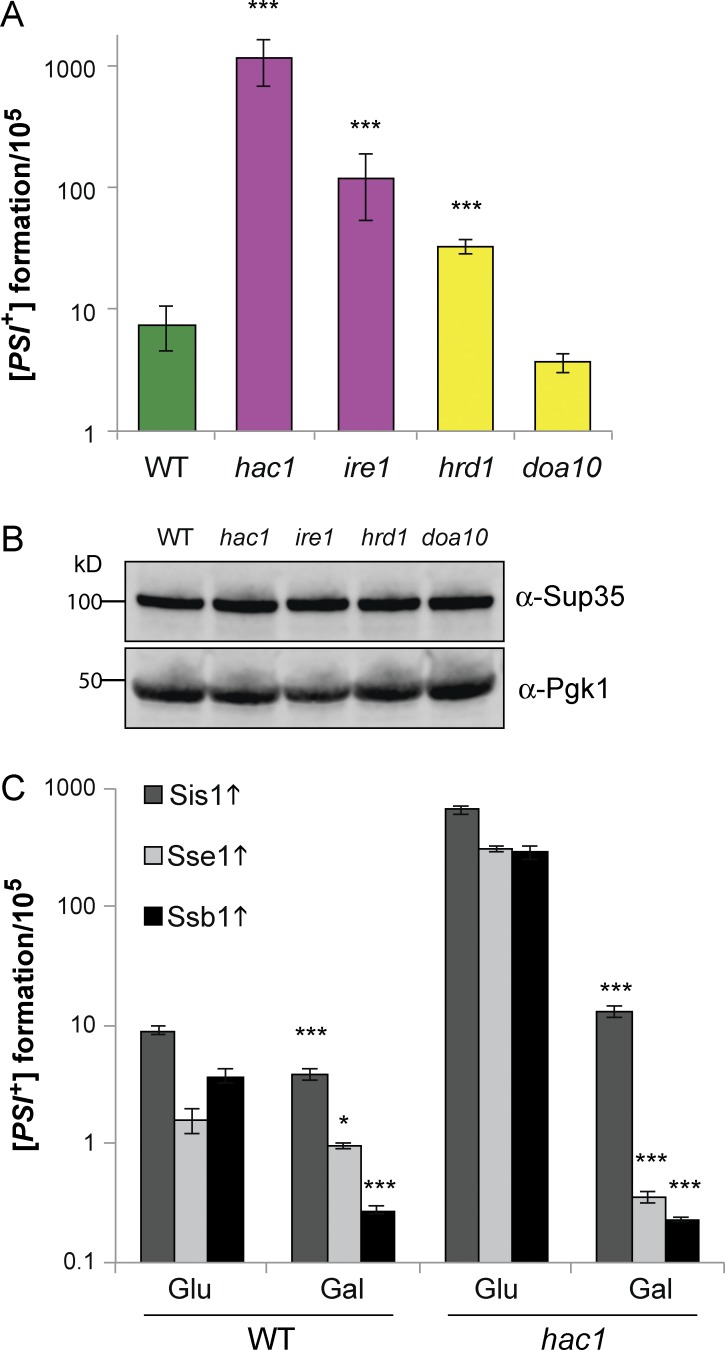
A genome-wide screen for factors that increase [PSI+] prion induction identified mutants in the UPR and ERAD pathways (Tyedmers et al., 2008), suggesting that prion formation may also be a consequence of ER stress. We examined prion formation in our mutants and found that the frequency of [PSI+] prion formation was elevated in hac1, ire1, and hrd1 mutants compared with wild-type and doa10 mutant strains (Fig. 4 A). This increase in [PSI+] prion formation did not arise because of differences in Sup35 protein levels (Fig. 4 B). We examined whether the chaperones that abrogate protein aggregation in a hac1 mutant could also prevent spontaneous prion formation and found that overexpression of Sis1, Sse1, and Ssb1 significantly reduced the frequency of [PSI+] prion formation in a hac1 mutant (Fig. 4 C).
Figure 4.
The frequency of [PSI+] formation is increased in UPR and ERAD mutants. (A) The frequency of [PSI+] prion formation was quantified in wild-type, hac1, ire1, hrd1, and doa10 mutant strains. Data shown are the means of at least three independent biological repeat experiments ± SD. (B) Western blots probed with αSup35 or αPgk1. (C) The frequency of [PSI+] prion formation was quantified after the induction of the indicated chaperones (n = 3) ± SD. Significance is shown comparing strains grown on glucose media with strains grown on SGal media; *, P < 0.05; ***, P < 0.005 (n = 3).
Overexpression of Sis1, Sse1, and Ssb1 reduced both amorphous and amyloid aggregation. Sis1 is an Hsp40 chaperone that regulates the ATPase activity of Hsp70 chaperones. It has been implicated in targeting misfolded proteins for degradation, facilitating the transfer of misfolded proteins to the nucleus, where they are degraded by the UPS system (Park et al., 2013; Shiber et al., 2013; Summers et al., 2013). Sis1 therefore appears to be an essential but limiting factor for degrading misfolded and aggregated proteins. Ssb1 is a ribosome-associated chaperone required to prevent the aggregation of nascent polypeptides (Koplin et al., 2010; Preissler and Deuerling, 2012). Ssb1 reduced aggregation in a hac1 mutant, suggesting that proteins may misfold during translation, which would be particularly acute for highly abundant/translated proteins. Sse1 functions as a nucleotide exchange factor for Hsp70 chaperones (Easton et al., 2000; Shaner et al., 2005). Sse1 can bind unfolded peptides and suppress thermal aggregation by maintaining proteins in a folding-competent state (Oh et al., 1999). Sse1 is not thought to functionally refold proteins and hence may suppress protein aggregation in a hac1 mutant via its holdase function.
Chaperone overexpression rescues the sensitivity of a hac1 mutant to ER stress
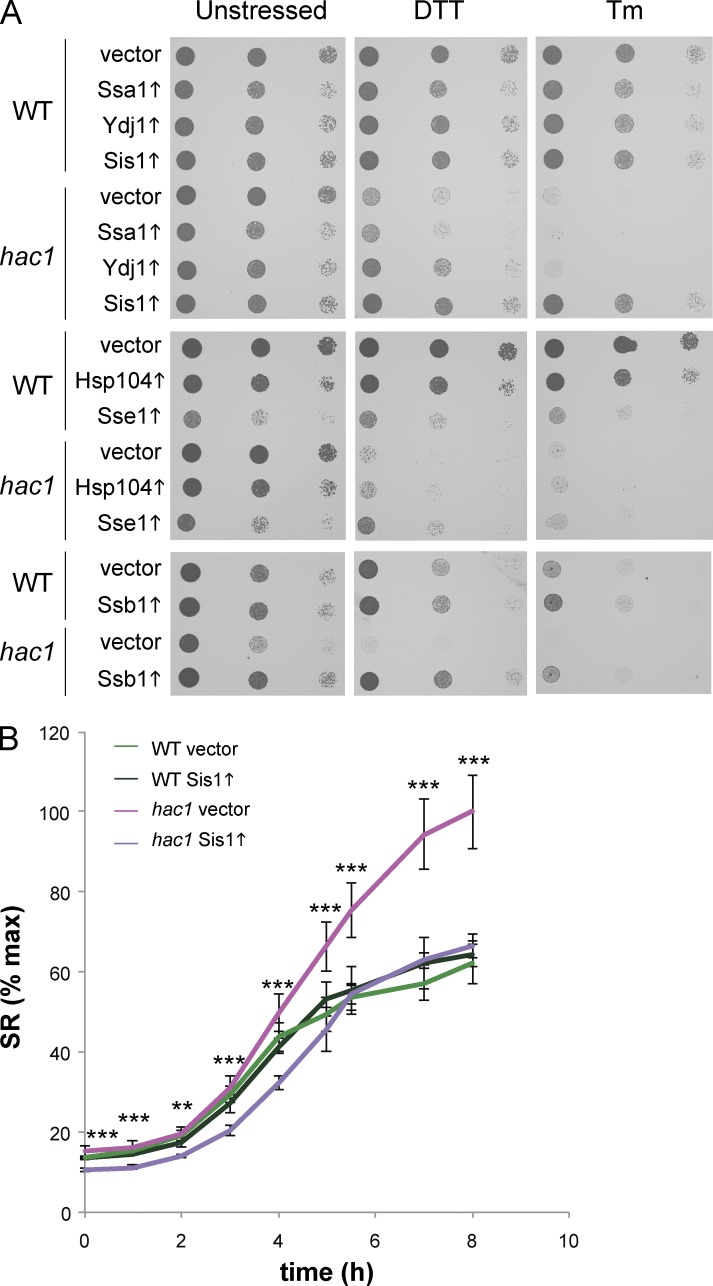
Because UPR mutants are sensitive to chemicals that promote ER stress, we tested whether the chaperones that abrogate protein aggregation influence the sensitivity of a hac1 mutant to chemically induced ER stress (Fig. 5 A). Overexpression of Ssa1 or Hsp104 did not affect the sensitivity of the hac1 mutant to DTT or Tm. Ydj1 minimally restored DTT tolerance but did not improve resistance to Tm stress. Sse1 was found to increase the sensitivity of the wild-type strain to DTT and Tm stress, whereas it increased the resistance of a hac1 mutant to DTT stress and did not alter its sensitivity to Tm. The strongest effects were seen with Sis1 and Ssb1 overexpression, which rescued the sensitivity of a hac1 mutant to both DTT and Tm; for Sis1, this resistance was comparable to wild-type levels (Fig. 5 A). We questioned whether chaperones rescue the hac1 mutant by enabling it to cope with the same amount of ER stress or by diminishing the level of ER stress. We tested this using a splicing reporter (SR) where the HAC1 open reading frame has been replaced with GFP (Pincus et al., 2010). This allows Ire1 activity to be monitored, because it splices the Hac1 intron to derepress GFP expression. We observed higher levels of SR activation in a hac1 mutant than in the wild-type upon Tm stress (Fig. 5 B). Overexpression of Sis1 dampened the increased SR induction in the hac1 mutant, indicating that Sis1 diminishes ER stress presumably by removing cytoplasmic protein aggregates.
Figure 5.
Overexpression of selected chaperones rescues the sensitivity of a hac1 mutant to ER stress. (A) Strains as shown in Fig. 4 D were spotting onto SGal media containing DTT or Tm. (B) Wild-type and hac1 mutant cells expressing GFP splicing reporter (SR) and containing a galactose-regulatable expression plasmid for Sis1 or empty vector (pRS413) were treated with 0.2 µg/ml Tm for 8 h. Fluorescence was measured by flow cytometry and is expressed relative to 100% maximum ± SD. Significance is shown comparing hac1 vector with hac1 SIS1 (n = 3). **, P < 0.01; ***, P < 0.005.
In summary, our results demonstrate that ER stress causes defects in cytosolic protein homeostasis and the formation of protein aggregates. Protein aggregation occurs when proteins adopt aberrant conformations or misfold resulting in their abnormal association into larger, often insoluble structures (Hartl et al., 2011; Hipp et al., 2014). ER stress appears to lead to the aggregation of aggregation-prone proteins rather than the aggregation of ER or secretory pathway proteins. Most of the commonly aggregated proteins tended to be abundant, highly translated, stable proteins, in agreement with the idea that protein abundance is a good indicator of protein aggregation in vivo (Weids et al., 2016). It is generally thought that highly expressed and abundant proteins have evolved to be highly soluble and resistant to aggregation (Tartaglia et al., 2007; Castillo et al., 2011; Gsponer and Babu, 2012; Bednarska et al., 2013). However, there is relatively little flexibility in this equilibrium, and any conditions that alter protein solubility or concentration can promote aggregation (Tartaglia et al., 2007).
Mutants deleted for HAC1 are sensitive to conditions that cause unfolded proteins to accumulate in the ER (Cox et al., 1993; Mori et al., 1993). Induction of gene expression by Hac1 promotes tolerance to ER stress by increasing ER chaperone concentrations. It is therefore surprising that overexpression of cytoplasmic chaperones rescued the sensitivity of a hac1 mutant to conditions which cause unfolded protein accumulation in the ER. This suggests that in the absence of active UPR, ER stress causes cellular toxicity via cytoplasmic protein aggregation. There are established links between cytosolic chaperones and prevention of ERAD substrates from aggregation before degradation (Nishikawa et al., 2005). Hence, clearance or prevention of cytosolic aggregates may explain this stress rescue. In agreement, the Hac1 SR indicated that overexpression of Sis1 reduces ER stress caused by Tm exposure, and restored wild-type levels of ER stress tolerance to a UPR mutant. This new finding links the toxicity of ER stress with cytosolic protein aggregation and emphasizes the importance of interorganelle cross talk.
Materials and methods
Yeast strains and plasmids
The wild-type yeast strain 74D-694 (MATa ade1-14 ura3-52 leu2-3,112 trp1-289 his3-200 [PIN+][psi−]) was used for all experiments. Strains were deleted for HRD1 (hrd1::HIS3, hrd1::TRP1), DOA10 (doa10::HIS3), IRE1 (ire1::LEU2), and HAC1 (hac1::HIS3, hac1::TRP1) using standard yeast methodology. Plasmids expressing fluorescently tagged proteins, including Hsp104-RFP, Sis1-GFP, and ΔssCPY∗-GFP (Lee et al., 2010; Malinovska et al., 2012; Park et al., 2013) and Hmg2-Myc (Hampton and Rine, 1994), have been described previously. Chaperones were overexpressed under the control of the GAL1 promoter, including pESC-Leu-SSA1 (Park et al., 2013), pESC-Leu-YDJ1 (Park et al., 2013), pRS415GAL-HA-SIS1 (Park et al., 2013), p425GAL1-HSP104 (O’Driscoll et al., 2015), p425GAL1-SSE1 (O’Driscoll et al., 2015), and pAG416GAL-SSB1 (Malinovska et al., 2012). The GFP SR used to monitor Ire1 activity has been described previously (Pincus et al., 2010).
Growth and stress conditions
Yeast strains were grown at 30°C with shaking at 180 rpm in rich YEPD medium (2% wt/vol glucose, 2% wt/vol bactopeptone, and 1% wt/vol yeast extract) or minimal SD (0.67% wt/vol yeast nitrogen base without amino acids and 2% wt/vol glucose) supplemented with appropriate amino acids and bases. SRaf media contained 2% wt/vol raffinose, and SGal media contained 1% wt/vol galactose/1% wt/vol raffinose in place of glucose. Media were solidified by the addition of 2% (wt/vol) agar. For chaperone expression, cultures were initially grown in SRaf media to exponential phase before switching to SGal media for a further 24 h to induce GAL1 expression. For ER stress conditions, cells were treated with 2 mM DTT or 0.2 µg/ml Tm for 2 h. Stress sensitivity was determined by growing cells to stationery phase in SRaf media and spotting diluted cultures (A600 = 1.0, 0.1, 0.01) onto SGal agar plates containing various concentrations of DTT or Tm.
Analysis of insoluble protein aggregates
Insoluble protein aggregates were isolated by separating insoluble proteins from soluble proteins by differential centrifugation and removing any contaminating membrane proteins using detergent washes (Weids and Grant, 2014). In brief, 20 ODs of cells were harvested by centrifugation at 4,000 rpm, 4°C, for 10 min. Cells were washed in 1 ml aggregate lysis buffer (ALB; 50 mM potassium phosphate, 1 mM EDTA, 5% (vol/vol) glycerol, 1 mM PMSF, and 1× complete mini-protease cocktail; Roche). Cells were resuspended in 300 µl ALB and spheroplasts were generated after treatment with 1 mg/ml lyticase for 30 min at 30°C. Cell breakage was achieved by sonication (Sonifier 150; Branson; 8 × 5 s, level 4), and samples were adjusted to equal protein concentrations before isolation of protein aggregates by centrifugation at 13,000 rpm, 4°C for 20 min. Insoluble fractions were resuspended in a buffer containing ALB buffer containing 2% (vol/vol) Igepal CA-630 (Sigma-Aldrich) through sonication (4 × 5 s, level 4). Samples were centrifuged at 13,000 rpm for 20 min at 4°C and the detergent wash repeated. Residual detergent was removed by two washes with ALB and the pellet resuspended by sonication. The final insoluble fraction was resuspended in 80 µl ALB and 20 µl reducing 4× protein loading buffer, separated by reducing SDS/PAGE (10% gels), and visualized by silver staining using the silver stain plus kit (Bio-Rad).
MS and statistical analysis
Aggregated proteins were identified by MS (performed by the Biomolecular Analysis Core Facility, The University of Manchester) in triplicate for each condition. For protein identification, protein samples were run a short distance into SDS-PAGE gels and stained using colloidal Coomassie blue (Sigma-Aldrich). Total proteins were excised; trypsin was digested and identified using liquid chromatography MS. Proteins were identified using the Mascot mass fingerprinting program (http://www.matrixscience.com) to search the NCBInr and Swissprot databases. Final datasets for each condition were determined by selecting proteins that were identified in at least two of the three replicates. We identified 688, 820, and 818 proteins that aggregate in the wild-type, hac1, and hrd1 mutants strains, respectively. Venn diagrams and analysis of the distribution of protein hits between different strains was performed using Venny (http://bioinfogp.cnb.csic.es/tools/venny/). Significantly enriched (5% false discovery rate) functional categories were identified using the MIPS Functional Catalogue (Ruepp et al., 2004). Datasets for each condition were assessed for functional enrichment (P < 0.01) of functional categories (MIPS database) using FunCat (available at http://www.helmholtz-muenchen.de/en/ibis). Mann–Whiney U tests were used to assess the statistical significance of observed differences in protein abundance (molecules per cell; Ghaemmaghami et al., 2003), estimated translation rates per protein (Arava et al., 2003), CAI, GRAVY score, pI, and protein stability (Christiano et al., 2014).
Western blot analysis
The turnover of ΔssCPY∗-GFP and Hmg2-myc was assessed in cells by inhibiting protein synthesis with cycloheximide. Cells were collected at the indicated time points and ΔssCPY∗-GFP and Hmg2-myc levels analyzed by immunoblotting and quantified by densitometry. Values shown are percentages relative to zero time points from three independent biological repeats. Protein extracts were electrophoresed under reducing conditions on NuPAGE minigels (Thermo Fisher Scientific) and electroblotted onto polyvinylidene fluoride membrane (GE Healthcare). Primary antibodies used were rabbit α-Sup35 (Ness et al., 2002), ubiquitin (sc-8017; Santa Cruz Biotechnology, Inc.), Ydj1 (ab74442; Abcam), Sis1 (COP-080051; Operon Biotechnologies), Ssa1 (ADI-SPA-822; Enzo Life Sciences), GFP (A6465, Invitrogen), Pgk1 (459250; Thermo Fisher Scientific), Hsp104 (ab2924; Abcam), rabbit α-Sse1 (Chiabudini et al., 2012), and Myc 4A6 (05–724; EMD Millipore).
Fluorescence microscopy
Sites of protein aggregation were detected using fluorescently tagged chaperones. Strains transformed with Hsp104-RFP or Sis1-GFP plasmids were harvested at the indicated time points and resuspended in water. The cells were immediately placed on poly-l-lysine–coated slides (Sigma-Aldrich) and visualized at room temperature using a IX71 (Olympus) Delta Vision (Applied Precision Ltd.) microscope with a 100×/NA.140 UPlan SAPO (oil) objective and FITC (BP490/20, BP531/28) and TRITC (BP555/28, BP617/63) band-pass filters from the Sedat QUAD filter set (Chroma Technology Corp.). Images were acquired using a Coolsnap HQ2 (Photometrics) camera using a Z optical spacing of 0.2 to 0.5 µm with Softworx 5.5.1 (Applied Precision Ltd.) and were then deconvolved with measured point spread function and maximum intensity quick projections were generated using the same software. For display, images were processed and analyzed using ImageJ.
Analyses of prion formation
Yeast strain 74D-694 (MATa ade1-14 trp1-289 his3-200 ura3-52 leu2-3,112) contains an assayable nonsense (UGA) mutation in the ADE1 gene. The induction of [PSI+] prion formation was quantified using the ade1-14 mutant allele, which confers adenine auxotrophy and prions differentiated from nuclear gene mutations by their irreversible elimination in guanidine hydrochloride (GdnHCl). GdnHCl blocks the propagation of yeast prions by inhibiting the key ATPase activity of Hsp104, a molecular chaperone that is absolutely required for yeast prion propagation (Ferreira et al., 2001; Jung and Masison, 2001). The frequency of spontaneous [PSI+] prion formation was scored by growth in the absence of adenine. Diluted cell cultures were plated onto SD plates lacking adenine (SD-Ade) and incubated for 7–10 d. Colonies which grew on SD-Ade plates were counted and then picked onto new SD-Ade plates before replica-printing onto SD-Ade and SD-Ade containing 4 mM GdnHCl. Colonies that grew on SD-Ade, but not on SD-Ade with GdnHCl, were scored as [PSI+]. [PSI+] colonies were also scored by visual differentiation of red/white colony formation on YEPD plates and by the conversion of pink/white [PSI+] colonies to red [psi−] colonies on YEPD plates containing GdnHCl. Data shown are the means of at least three independent biological repeat experiments expressed as the number of colonies per 105 viable cells.
SR assay
Cells were grown in SGal media and treated with 0.2 μg/ml Tm for 8 h. Samples were collected hourly, and 10,000 cells were analyzed in an LSR Fortessa Flow cytometer (BD) with excitation at 488 nm and emission at 530/30 nm. The mean median value of the intensity of fluorescence of three independent biological repeats was calculated. The mean median value of the intensity of the hac1 vector strain at the 8-h time point was considered the maximum and set to 100%. All other values were expressed relative to that.
Statistical analyses
Data are presented as mean values ± SD. Statistical analysis for multiple groups was performed using one-way ANOVA with pairwise comparisons of sample means via the Turkey HSD test, and results were considered statistically significant with a p-value <0.05. The physicochemical, translation, and degradation rates of proteins in aggregates were evaluated with pairwise Mann–Whitney–Wilcoxon U tests.
Online supplemental material
Fig. S1 presents MIPS functional categorization of aggregated proteins identified in the wild type and hac1 and hrd1 mutant strains. Fig. S2 presents MIPS functional categorization of aggregated proteins identified in the common, hac1-only, and hrd1-only sets. Fig. S3 A presents the proportion of proteins in each aggregate set that interact with specific chaperones, and Fig. S3 B presents immunoblot analysis to confirm overexpression of selected chaperones.
Supplementary Material
Acknowledgments
We thank Lynn Megeney, Helen Saibil, Ulrich Hartl, Martin Poole, Simon Alberti, Mick Tuite, Peter Walter, David Pincus, and Sabine Rospert for supplying resources used in this study. We thank Lisa Swanton and Alan Weids for critical reading of the manuscript.
This work was supported by a Malaysian Government Agency studentship (People’s Trust Council-MARA; N. Hamdan), a Wellcome Trust PhD studentship (096604/Z/11/Z; P. Kritsiligkou), and a Biotechnology and Biological Sciences Research Council (BBSRC) project grant (BB/M020770/1; C.M. Grant). We thank the Bioimaging (microscopes used in this study were purchased with grants from the BBSRC, Wellcome Trust, and the University of Manchester Strategic Fund) and biological mass spectrometry facilities at the University of Manchester.
The authors declare no competing financial interests.
Author contributions: N. Hamdan and P. Kritsiligkou performed the experiments and analyzed the data together with C.M. Grant. C.M. Grant, P. Kritsiligkou, and N. Hamdan wrote the manuscript. C.M. Grant designed and supervised the project.
Footnotes
Abbreviations used:
- ERAD
- ER-associated degradation
- MS
- mass spectrometry
- SR
- splicing reporter
- Tm
- tunicamycin
- UPR
- unfolded protein response
- UPS
- ubiquitin proteasome system
References
- Arava Y., Wang Y., Storey J.D., Liu C.L., Brown P.O., and Herschlag D.. 2003. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 100:3889–3894. 10.1073/pnas.0635171100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarska N.G., Schymkowitz J., Rousseau F., and Van Eldere J.. 2013. Protein aggregation in bacteria: the thin boundary between functionality and toxicity. Microbiology. 159:1795–1806. 10.1099/mic.0.069575-0 [DOI] [PubMed] [Google Scholar]
- Brodsky J.L., and Skach W.R.. 2011. Protein folding and quality control in the endoplasmic reticulum: Recent lessons from yeast and mammalian cell systems. Curr. Opin. Cell Biol. 23:464–475. 10.1016/j.ceb.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho P., Goder V., and Rapoport T.A.. 2006. Distinct ubiquitin-ligase complexes define convergent pathways for the degradation of ER proteins. Cell. 126:361–373. 10.1016/j.cell.2006.05.043 [DOI] [PubMed] [Google Scholar]
- Castillo V., Graña-Montes R., and Ventura S.. 2011. The aggregation properties of Escherichia coli proteins associated with their cellular abundance. Biotechnol. J. 6:752–760. 10.1002/biot.201100014 [DOI] [PubMed] [Google Scholar]
- Chiabudini M., Conz C., Reckmann F., and Rospert S.. 2012. Ribosome-associated complex and Ssb are required for translational repression induced by polylysine segments within nascent chains. Mol. Cell. Biol. 32:4769–4779. 10.1128/MCB.00809-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christiano R., Nagaraj N., Fröhlich F., and Walther T.C.. 2014. Global proteome turnover analyses of the yeasts S. cerevisiae and S. pombe. Cell Reports. 9:1959–1965. 10.1016/j.celrep.2014.10.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson J.C., and Ye Y.. 2014. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nat. Struct. Mol. Biol. 21:325–335. 10.1038/nsmb.2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.S., Shamu C.E., and Walter P.. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 73:1197–1206. 10.1016/0092-8674(93)90648-A [DOI] [PubMed] [Google Scholar]
- Easton D.P., Kaneko Y., and Subjeck J.R.. 2000. The hsp110 and Grp1 70 stress proteins: Newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 5:276–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisele F., and Wolf D.H.. 2008. Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett. 582:4143–4146. 10.1016/j.febslet.2008.11.015 [DOI] [PubMed] [Google Scholar]
- Erjavec N., Larsson L., Grantham J., and Nyström T.. 2007. Accelerated aging and failure to segregate damaged proteins in Sir2 mutants can be suppressed by overproducing the protein aggregation-remodeling factor Hsp104p. Genes Dev. 21:2410–2421. 10.1101/gad.439307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira P.C., Ness F., Edwards S.R., Cox B.S., and Tuite M.F.. 2001. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol. Microbiol. 40:1357–1369. 10.1046/j.1365-2958.2001.02478.x [DOI] [PubMed] [Google Scholar]
- Flynn G.C., Pohl J., Flocco M.T., and Rothman J.E.. 1991. Peptide-binding specificity of the molecular chaperone BiP. Nature. 353:726–730. 10.1038/353726a0 [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S., Huh W.K., Bower K., Howson R.W., Belle A., Dephoure N., O’Shea E.K., and Weissman J.S.. 2003. Global analysis of protein expression in yeast. Nature. 425:737–741. 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Gong Y., Kakihara Y., Krogan N., Greenblatt J., Emili A., Zhang Z., and Houry W.A.. 2009. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: implications to protein folding pathways in the cell. Mol. Syst. Biol. 5:275 10.1038/msb.2009.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsponer J., and Babu M.M.. 2012. Cellular strategies for regulating functional and nonfunctional protein aggregation. Cell Reports. 2:1425–1437. 10.1016/j.celrep.2012.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R.Y., and Rine J.. 1994. Regulated degradation of HMG-CoA reductase, an integral membrane protein of the endoplasmic reticulum, in yeast. J. Cell Biol. 125:299–312. 10.1083/jcb.125.2.299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton R.Y., Gardner R.G., and Rine J.. 1996. Role of 26S proteasome and HRD genes in the degradation of 3-hydroxy-3-methylglutaryl-CoA reductase, an integral endoplasmic reticulum membrane protein. Mol. Biol. Cell. 7:2029–2044. 10.1091/mbc.7.12.2029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl F.U., Bracher A., and Hayer-Hartl M.. 2011. Molecular chaperones in protein folding and proteostasis. Nature. 475:324–332. 10.1038/nature10317 [DOI] [PubMed] [Google Scholar]
- Hipp M.S., Park S.H., and Hartl F.U.. 2014. Proteostasis impairment in protein-misfolding and -aggregation diseases. Trends Cell Biol. 24:506–514. 10.1016/j.tcb.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Jang H.H., Lee K.O., Chi Y.H., Jung B.G., Park S.K., Park J.H., Lee J.R., Lee S.S., Moon J.C., Yun J.W., et al. 2004. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 117:625–635. 10.1016/j.cell.2004.05.002 [DOI] [PubMed] [Google Scholar]
- Jung G., and Masison D.C.. 2001. Guanidine hydrochloride inhibits Hsp104 activity in vivo: A possible explanation for its effect in curing yeast prions. Curr. Microbiol. 43:7–10. 10.1007/s002840010251 [DOI] [PubMed] [Google Scholar]
- Kohno K., Normington K., Sambrook J., Gething M.-J., and Mori K.. 1993. The promoter region of the yeast KAR2 (BiP) gene contains a regulatory domain that responds to the presence of unfolded proteins in the endoplasmic reticulum. Mol. Cell. Biol. 13:877–890. 10.1128/MCB.13.2.877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koplin A., Preissler S., Ilina Y., Koch M., Scior A., Erhardt M., and Deuerling E.. 2010. A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J. Cell Biol. 189:57–68. 10.1083/jcb.200910074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J., and Morimoto R.I.. 2015. The biology of proteostasis in aging and disease. Annu. Rev. Biochem. 84:435–464. 10.1146/annurev-biochem-060614-033955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.E., Brunette S., Puente L.G., and Megeney L.A.. 2010. Metacaspase Yca1 is required for clearance of insoluble protein aggregates. Proc. Natl. Acad. Sci. USA. 107:13348–13353. 10.1073/pnas.1006610107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum R., Tkach J.M., Vierling E., and Glover J.R.. 2004. Evidence for an unfolding/threading mechanism for protein disaggregation by Saccharomyces cerevisiae Hsp104. J. Biol. Chem. 279:29139–29146. 10.1074/jbc.M403777200 [DOI] [PubMed] [Google Scholar]
- Malinovska L., Kroschwald S., Munder M.C., Richter D., and Alberti S.. 2012. Molecular chaperones and stress-inducible protein-sorting factors coordinate the spatiotemporal distribution of protein aggregates. Mol. Biol. Cell. 23:3041–3056. 10.1091/mbc.E12-03-0194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Ma W., Gething M.J., and Sambrook J.. 1993. A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell. 74:743–756. 10.1016/0092-8674(93)90521-Q [DOI] [PubMed] [Google Scholar]
- Nakatsukasa K., and Brodsky J.L.. 2008. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 9:861–870. 10.1111/j.1600-0854.2008.00729.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ness F., Ferreira P., Cox B.S., and Tuite M.F.. 2002. Guanidine hydrochloride inhibits the generation of prion “seeds” but not prion protein aggregation in yeast. Mol. Cell. Biol. 22:5593–5605. 10.1128/MCB.22.15.5593-5605.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S., Brodsky J.L., and Nakatsukasa K.. 2005. Roles of molecular chaperones in endoplasmic reticulum (ER) quality control and ER-associated degradation (ERAD). J. Biochem. 137:551–555. 10.1093/jb/mvi068 [DOI] [PubMed] [Google Scholar]
- O’Driscoll J., Clare D., and Saibil H.. 2015. Prion aggregate structure in yeast cells is determined by the Hsp104-Hsp110 disaggregase machinery. J. Cell Biol. 211:145–158. 10.1083/jcb.201505104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh H.J., Easton D., Murawski M., Kaneko Y., and Subjeck J.R.. 1999. The chaperoning activity of hsp110. Identification of functional domains by use of targeted deletions. J. Biol. Chem. 274:15712–15718. 10.1074/jbc.274.22.15712 [DOI] [PubMed] [Google Scholar]
- Park S.H., Bolender N., Eisele F., Kostova Z., Takeuchi J., Coffino P., and Wolf D.H.. 2007. The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol. Biol. Cell. 18:153–165. 10.1091/mbc.E06-04-0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.H., Kukushkin Y., Gupta R., Chen T., Konagai A., Hipp M.S., Hayer-Hartl M., and Hartl F.U.. 2013. PolyQ proteins interfere with nuclear degradation of cytosolic proteins by sequestering the Sis1p chaperone. Cell. 154:134–145. 10.1016/j.cell.2013.06.003 [DOI] [PubMed] [Google Scholar]
- Pincus D., Chevalier M.W., Aragón T., van Anken E., Vidal S.E., El-Samad H., and Walter P.. 2010. BiP binding to the ER-stress sensor Ire1 tunes the homeostatic behavior of the unfolded protein response. PLoS Biol. 8:e1000415 10.1371/journal.pbio.1000415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S., and Deuerling E.. 2012. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem. Sci. 37:274–283. 10.1016/j.tibs.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Rand J.D., and Grant C.M.. 2006. The thioredoxin system protects ribosomes against stress-induced aggregation. Mol. Biol. Cell. 17:387–401. 10.1091/mbc.E05-06-0520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruepp A., Zollner A., Maier D., Albermann K., Hani J., Mokrejs M., Tetko I., Güldener U., Mannhaupt G., Münsterkötter M., and Mewes H.W.. 2004. The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res. 32:5539–5545. 10.1093/nar/gkh894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiano A., Foresti O., and Carvalho P.. 2014. Quality control: ER-associated degradation: Protein quality control and beyond. J. Cell Biol. 204:869–879. 10.1083/jcb.201312042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L., Wegele H., Buchner J., and Morano K.A.. 2005. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J. Biol. Chem. 280:41262–41269. 10.1074/jbc.M503614200 [DOI] [PubMed] [Google Scholar]
- Shiber A., Breuer W., Brandeis M., and Ravid T.. 2013. Ubiquitin conjugation triggers misfolded protein sequestration into quality control foci when Hsp70 chaperone levels are limiting. Mol. Biol. Cell. 24:2076–2087. 10.1091/mbc.E13-01-0010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidrauski C., and Walter P.. 1997. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 90:1031–1039. 10.1016/S0092-8674(00)80369-4 [DOI] [PubMed] [Google Scholar]
- Summers D.W., Wolfe K.J., Ren H.Y., and Cyr D.M.. 2013. The Type II Hsp40 Sis1 cooperates with Hsp70 and the E3 ligase Ubr1 to promote degradation of terminally misfolded cytosolic protein. PLoS One. 8:e52099 10.1371/journal.pone.0052099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamás M.J., Sharma S.K., Ibstedt S., Jacobson T., and Christen P.. 2014. Heavy metals and metalloids as a cause for protein misfolding and aggregation. Biomolecules. 4:252–267. 10.3390/biom4010252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia G.G., Pechmann S., Dobson C.M., and Vendruscolo M.. 2007. Life on the edge: A link between gene expression levels and aggregation rates of human proteins. Trends Biochem. Sci. 32:204–206. 10.1016/j.tibs.2007.03.005 [DOI] [PubMed] [Google Scholar]
- Thibault G., and Ng D.T.. 2012. The endoplasmic reticulum-associated degradation pathways of budding yeast. Cold Spring Harb. Perspect. Biol. 4:a013193 10.1101/cshperspect.a013193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T., Mogk A., Langen H., Goloubinoff P., and Bukau B.. 2001. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 40:397–413. 10.1046/j.1365-2958.2001.02383.x [DOI] [PubMed] [Google Scholar]
- Travers K.J., Patil C.K., Wodicka L., Lockhart D.J., Weissman J.S., and Walter P.. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 101:249–258. 10.1016/S0092-8674(00)80835-1 [DOI] [PubMed] [Google Scholar]
- Tyedmers J., Madariaga M.L., and Lindquist S.. 2008. Prion switching in response to environmental stress. PLoS Biol. 6:e294 10.1371/journal.pbio.0060294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas R.M., Raychaudhuri S., Hayer-Hartl M., and Hartl F.U.. 2010. Protein folding in the cytoplasm and the heat shock response. Cold Spring Harb. Perspect. Biol. 2:a004390 10.1101/cshperspect.a004390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verghese J., Abrams J., Wang Y., and Morano K.A.. 2012. Biology of the heat shock response and protein chaperones: Budding yeast (Saccharomyces cerevisiae) as a model system. Microbiol. Mol. Biol. Rev. 76:115–158. 10.1128/MMBR.05018-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., and Ron D.. 2011. The unfolded protein response: from stress pathway to homeostatic regulation. Science. 334:1081–1086. 10.1126/science.1209038 [DOI] [PubMed] [Google Scholar]
- Washburn M.P., Wolters D., and Yates J.R. III. 2001. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 19:242–247. 10.1038/85686 [DOI] [PubMed] [Google Scholar]
- Weids A.J., and Grant C.M.. 2014. The yeast peroxiredoxin Tsa1 protects against protein-aggregate-induced oxidative stress. J. Cell Sci. 127:1327–1335. 10.1242/jcs.144022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weids A.J., Ibstedt S., Tamás M.J., and Grant C.M.. 2016. Distinct stress conditions result in aggregation of proteins with similar properties. Sci. Rep. 6:24554 10.1038/srep24554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zattas D., and Hochstrasser M.. 2015. Ubiquitin-dependent protein degradation at the yeast endoplasmic reticulum and nuclear envelope. Crit. Rev. Biochem. Mol. Biol. 50:1–17. 10.3109/10409238.2014.959889 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.