Phosphofructokinase-1 (PFK1) is an essential glycolysis enzyme as it catalyzes the step committing glucose to breakdown. Webb et al. show that the liver PFK1 isoform assembles into filaments in a tetramer- and substrate-dependent manner, providing insights into the spatial organization of isoform-specific glucose metabolism in cells.
Abstract
Despite abundant knowledge of the regulation and biochemistry of glycolytic enzymes, we have limited understanding on how they are spatially organized in the cell. Emerging evidence indicates that nonglycolytic metabolic enzymes regulating diverse pathways can assemble into polymers. We now show tetramer- and substrate-dependent filament assembly by phosphofructokinase-1 (PFK1), which is considered the “gatekeeper” of glycolysis because it catalyzes the step committing glucose to breakdown. Recombinant liver PFK1 (PFKL) isoform, but not platelet PFK1 (PFKP) or muscle PFK1 (PFKM) isoforms, assembles into filaments. Negative-stain electron micrographs reveal that filaments are apolar and made of stacked tetramers oriented with exposed catalytic sites positioned along the edge of the polymer. Electron micrographs and biochemical data with a PFKL/PFKP chimera indicate that the PFKL regulatory domain mediates filament assembly. Quantified live-cell imaging shows dynamic properties of localized PFKL puncta that are enriched at the plasma membrane. These findings reveal a new behavior of a key glycolytic enzyme with insights on spatial organization and isoform-specific glucose metabolism in cells.
Introduction
Intermediary metabolism, particularly catabolism of glucose, is central to normal cell function and is dysregulated in diseases such as cancer, diabetes, and neurodegenerative disorders. The cell biology of cytosolic metabolic enzymes, particularly their spatial organization, is critical for understanding normal and dysregulated metabolism but remains relatively understudied. Emerging evidence indicates that several cytosolic metabolic enzymes assemble into filamentous polymers for spatial and temporal organization. Filament assembly has been reported for several mammalian cytosolic metabolic enzymes, including acetyl-coA carboxylase (Meredith and Lane, 1978; Beaty and Lane, 1983), glutamine synthase (Frey et al., 1975), glutamate dehydrogenase (Eisenberg and Tomkins, 1968; Cohen et al., 1976), glutaminase (Olsen et al., 1970), β-glucosidase (Gunning, 1965; Kim et al., 2005), and cytidine triphosphate (CTP) synthase (Ingerson-Mahar et al., 2010; Liu, 2010; Noree et al., 2010; Habrian et al., 2016). Filament assembly by metabolic enzymes is not restricted to mammalian cells but is also seen in yeasts and bacteria (O’Connell et al., 2012; Shen et al., 2016), suggesting conserved biological processes. Several not necessarily exclusive functions for the evolution of metabolic enzyme filaments from bacteria to mammalian cells have been proposed (O’Connell et al., 2012). In some cases, assembly has a direct effect on enzyme activity and is believed to provide an additional layer of regulation in response to changing metabolic conditions; for example, bacterial CTP synthase is inactivated in the filament form (Barry et al., 2014). One function that is shared by filamentous assemblies of nonmetabolic enzymes is to maximize catalytic efficiency by channeling or funneling between active sites. An additional speculated function is to limit diffusion to enable a localized metabolic platform. Moreover, filaments of bacterial CTP synthase are also reported to have a structural role in cell shape (Ingerson-Mahar et al., 2010).
Glycolysis is an ancient metabolic pathway for processing glucose into pyruvate, generating energy in the form of ATP and NADH as well as anabolic building blocks for nucleotides, amino acids, and lipids. Glycolytic enzymes function as highly regulated molecular machines that fine-tune the rate of glucose utilization and are dysregulated as well as mutated in cancer. The committed step of glycolysis is catalyzed by phosphofructokinase-1 (PFK1). PFK1 was recently identified as a filament-forming enzyme in Saccharomyces cerevisiae (Shen et al., 2016). Filament formation by PFK1 may be evolutionarily conserved because purified liver PFK1 (PFKL) forms asymmetric oligomers in vitro, as determined by gel chromatography (Kemp, 1971), sedimentation with analytical ultracentrifugation (Trujillo and Deal, 1977), fluorescence polarization (Reinhart and Lardy, 1980), and fluorescence correlation spectroscopy (Ranjit et al., 2014). However, only one previous study showed filamentous structures by transmission EM (TEM; Foe and Trujillo, 1980), and this study did not address three important unknowns: whether filament assembly is regulated, the alignment of PFKL tetramers within the filaments, and whether filament formation occurs in cells. We now resolve these unknowns by showing the regulated assembly of PFKL filaments, their higher-order quaternary structure determined from negative-stain electron micrographs, and the dynamic behavior of punctate PFKL-EGFP particles in cells quantified from time-lapse imaging.
Results and discussion
We recently reported the first biologically relevant tetrameric mammalian PFK1 structure using recombinant platelet PFK1 (PFKP) produced using a baculovirus expression system (Webb et al., 2015). We used the same system to express and purify the liver and muscle isoforms of PFK1. While performing crystallization studies, we were unable to obtain highly concentrated PFKL, whereas PFKP was stable at a high concentration (>30 mg/ml). Because concentration-dependent aggregation is a hallmark of self-associating proteins, we sought to determine whether PFKL assembles into higher-order oligomers.
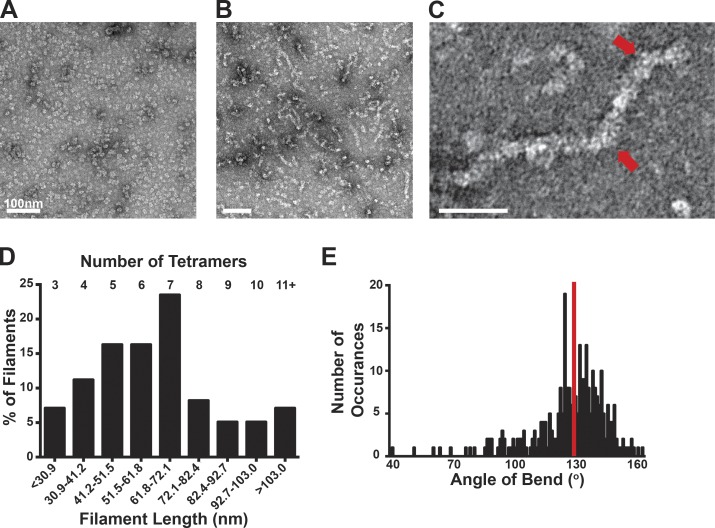
Baculovirus-expressed PFK1 isoforms were purified to homogeneity, as determined by Coomassie-stained SDS-PAGE gels (Fig. S1 A), and were tetrameric, as shown by TEM (Figs. 1 A and S1 B). All three recombinant enzymes showed isoform-specific activity and regulation, including sensitivity to inhibition by ATP and activation by the sugar substrate fructose 6-phosphate (F6P; Fig. S1, C–E). We visualized by TEM the quaternary structure of purified PFKL. In control buffer with 1 mM ATP, pH 7.4, PFKL was predominantly tetrameric and was occasionally seen as small aggregates (Fig. 1 A). However, in the presence of 2 mM F6P, PFKL assembled into filaments (Fig. 1, B and C). Filaments were heterogeneous in length, with a mean length of 65.4 ± 34.1 nm (mean ± SD; n = 98 filaments; Fig. 1 D). This corresponds with ∼6 tetramers similar in scale with previous biophysical measurements (Reinhart and Lardy, 1980). Some filaments displayed bends of a mean angle of 124 ± 18° (mean ± SD; n = 290 angles; Fig. 1, C and E). In contrast, F6P did not induce filament assembly of PFKP or muscle PFK1 (PFKM; Fig. S1 B), indicating an isoform-specific behavior. The presence of tetramers in control buffer suggests that tetramer formation precedes the formation of the elongated polymer. Consistent with this prediction, a catalytically inactive PFKL-Y638R mutant that does not form tetramers does not assemble into filaments in the presence of 2 mM F6P (Fig. S1, B and F).
Figure 1.
PFKL forms filaments of stacked tetramers. (A and B) TEM images of PFKL in control buffer (A) and in buffer containing 2 mM F6P (B). (C) Higher-magnification TEM image of a PFKL filament in the presence of 2 mM F6P. Red arrows highlight bends in the filament. (D) Chart of binned length determined relative to the percentage of filaments from 98 PFKL filaments measured from two separate protein preparations. The approximate number of tetramers that each bin represents is shown at the top of the chart. (E) Angle of bend determined relative to number of occurrences for a total of 290 bends measured from three separate protein preparations. The red line represents the mean of 124°.
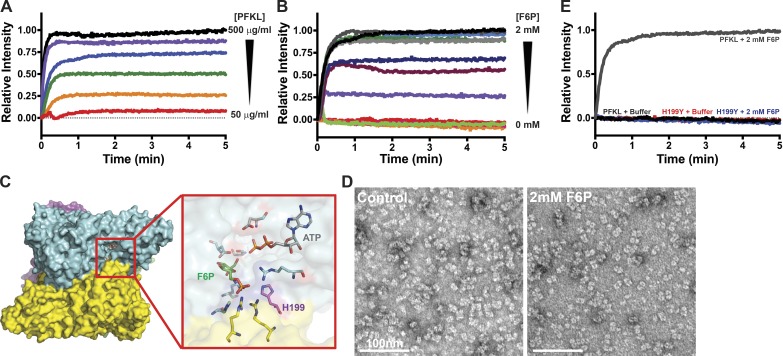
The assembly of PFKL into higher-order structures was also determined by 90° light scattering. Addition of 2 mM F6P but not a control buffer induced a rapid increase in the intensity of light scattering followed by a stable plateau (Fig. 2, A and B). Titrating the concentration of PFKL (50–500 µg/ml; 0.6–6.0 µM) showed a concentration-dependent increase in the rate and intensity of scattering at the plateau (Fig. 2 A). Filament formation as determined by TEM and oligomerization as determined by light scattering occurring at physiologically relevant concentrations of PFKL, which are estimated to be 20–50 µg/ml (Reinhart and Lardy, 1980). Titrating F6P from 0–2 mM at 200 µg/ml (2.4 µM) PFKL showed a concentration-dependent sigmoidal response to substrate. No increase in the intensity of scattering was observed at concentrations of <0.6 mM F6P (Fig. 2 B), an intermediate response occurred between 0.7 and 0.8 mM F6P, and a maximal response occurred at concentrations of >0.9 mM F6P. These concentrations were higher than the concentration of F6P in cells (∼100–350 µM; Albe et al., 1990), suggesting additional factors may be required for the formation of filaments in cells. Consistent with our TEM findings that PFKP and PFKM did not form filaments, 2 mM F6P did not increase light scattering with PFKP or PFKM (Fig. S1 G).
Figure 2.
PFKL filament formation is concentration- and F6P-dependent. (A) 90° light scattering of 50–500 µg/ml PFKL upon the addition of 2 mM F6P at time 0. Red, 50 µg/ml; orange, 100 µg/ml; green, 150 µg/ml; blue, 200 µg/ml; purple, 400 µg/ml; black, 500 µg/ml. (B) 90° light scattering at the 200 µg/ml PFKL upon the addition of the indicated concentration of F6P at time 0. Red, 0 mM; orange, 0.25 mM; pink, 0.5 mM; green, 0.6 mM; purple, 0.7 mM; maroon, 0.75 mM; dark blue, 0.8 mM; light blue, 0.9 mM; dark green, 1.0 mM; light gray, 1.25 mM; dark gray, 1.5 mM; black, 2.0 mM. (C) Location of His199 in the F6P binding pocket of PFK1. (D) TEM images of PFKL-H199Y in control buffer (left) and in buffer containing 2 mM F6P (right). (E) 90° light scattering at 200 µg/ml PFKL or PFKL-H199Y upon the addition of 2 mM F6P at time 0. Data are representative of three determinations from two separate protein preparations.
To confirm F6P-dependent assembly of PFKL filaments, we generated a PFKL mutant that was unable to bind F6P. Using our recently resolved PFKP crystal structure (Webb et al., 2015) as a guide, we identified His199 (His208 of PFKP) in the substrate-binding pocket as a critical residue for binding F6P (Fig. 2 C). We predicted that mutation of His199 to Tyr would occlude F6P from the binding pocket but not inhibit tetramer formation. Consistent with an inability to bind to F6P, PFKL-H199Y was catalytically inactive (Fig. S1 F). TEM analysis showed that PFKL-H199Y was mainly tetrameric in control buffer and did not assemble into filaments with 2 mM F6P (Fig. 2 D). Additionally, no increase in light scattering was observed with F6P (Fig. 2 E). Collectively, these data confirm that substrate binding is essential for the formation of PFKL filaments in vitro.
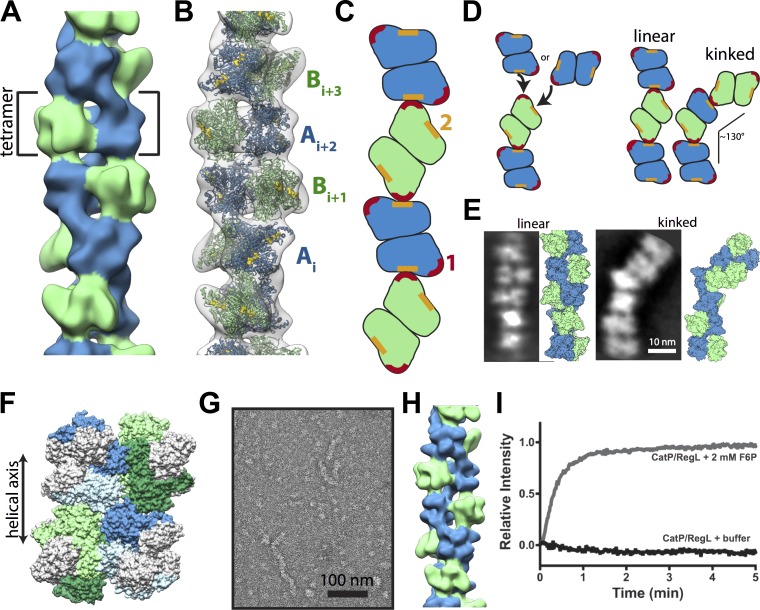
To better understand the PFKL filament architecture, we determined filament structure by negative-stain EM and iterative helical real-space reconstruction at 25-Å resolution (Fig. S2 A). The filament was formed from stacked tetramers that were related by a rotation of 221° and a translation of 83 Å between subunits (Fig. 3, A and B). Although we did not directly determine the hand of the PFKL filament by EM, the PFK crystal structure only fit well in the right-handed helix. The PFKL tetramer had D2 symmetry, with three perpendicular twofold symmetry axes; one twofold axis was perpendicular to the helical symmetry axis, whereas the other two were tilted ∼30° from the helical axis (Fig. S2, B and C). This arrangement resulted in different assembly interactions for each of the two dimers that make up the PFKL tetramer, which we refer to as dimers A and B. Although the overall approximate D2 symmetry of the PFKL tetramer remained intact, the resolution of our structure was insufficient to visualize small conformational differences in dimers A and B that may arise from their unique packing environments in the polymer; such asymmetry, however, would be consistent with asymmetry observed in PFKP crystal structures (Webb et al., 2015). The asymmetry in assembly interfaces between the A and B dimers of PFKL was unusual among helical filaments with multimeric helical asymmetric units. Typically, each protomer makes identical assembly contacts, although asymmetries like the one seen in this study have been seen previously (Egelman et al., 2015).
Figure 3.
Architecture of PFKL filament. (A) Negative-stain 3D reconstruction of the PFKL at 25-Å resolution. The PFKL tetramer is the repeating helical unit. Dimers on each half of the tetramer, colored blue or green, are engaged in different assembly contacts. (B) Fit of the PFKP crystal structure into the EM structure, with distinct A and B dimers labeled. (C) Diagram of longitudinal interactions between A and B dimers. (D) PFKL tetramers can add to the end of a filament through interfaces 1 or 2, resulting in either a continued linear polymer or a kink of ∼130°. (E) Examples of linear and straight filaments from reference-free means of PFKL segments with corresponding molecular models. (F) Model of CatP/RegL chimera with the catalytic domain of PFKP (gray) and the regulatory domain of PFKL (colored blue for dimer A and green for dimer B) interacting in the filament. (G) Negative-stain micrograph of the CatP/RegL in the presence of F6P. (H) Reconstruction of CatP/RegL at 25-Å resolution. (I) 90° light scattering at the 200 µg/ml CatP/RegL upon the addition of buffer alone (black line) or 2 mM F6P (gray line) at time 0. Data are representative of three determinations from two separate protein preparations.
Two different surfaces of the C-terminal regulatory domain mediate the interaction of PFKL tetramers in filaments. Although the resolution of the structure prevents a precise description of the assembly interface, the approximate regions are residues (PFKL numbering) 396–401, 476–490, 510–519, and 693–705 (interface 1; red) and residues 327–354, 372–377, and 712–718 (interface 2; orange; Fig. S2, D and E; and Fig. S3 A). Longitudinal contacts between PFKL tetramers alternate along the filament, with interface 2 of A dimers interacting with interface 1 of B dimers. In B dimers, interface 2 was exposed, whereas in A dimers, interface 1 made contact with interface 1 of the A dimer in the next subunit along the filament (Figs. 3 C and S2, D and E). This unusual helical symmetry meant that the last subunit in a polymer presented two different but mutually exclusive assembly interfaces for the addition of another subunit; addition at one site produced a linear filament, whereas addition at the other introduced a kink of ∼130° into the filament (Fig. 3 D). This closely matched the bends we observed in filaments (Fig. 1, C and E). 2D reference-free averages of PFKL filament segments revealed well-ordered classes with defined kinks (Fig. 3 E), suggesting that the kinked assembly interactions happen frequently and that the bends observed in PFKL filaments are not the result of inherent flexibility but rather reflect alternate assembly contacts. The formation of polymers with irregular long-range structure may reflect a lack of constraints on PFKL architecture and suggests that assembled filaments may be more important for aggregating PFKL subunits than for organization into a specific configuration.
Based on the observed contact sites between tetramers, we tested whether the regulatory domain of PFKL is sufficient for filament formation. We generated a chimera containing the catalytic region of PFKP (amino acids 1–368) and the regulatory region of PFKL (amino acids 360–780; CatP/RegL; Figs. 3 F and S3 A). Recombinant CatP/RegL was catalytically active (Fig. S3, B and C) but less stable than the wild-type protein, and it required addition of 5 mM of the allosteric activator ammonium sulfate to maintain protein stability and activity (unpublished data). In the presence but not the absence of F6P, CatP/RegL formed elongated filaments as visualized by TEM, and the 3D reconstruction was essentially identical to the PFKL filament structure (Fig. 3, G and H). Additionally, 2 mM F6P but not control buffer induced an increase in light scattering (Fig. 3 I). Collectively, these results are consistent with our TEM data and confirm that the residues required for filament formation are in the regulatory region of PFKL.
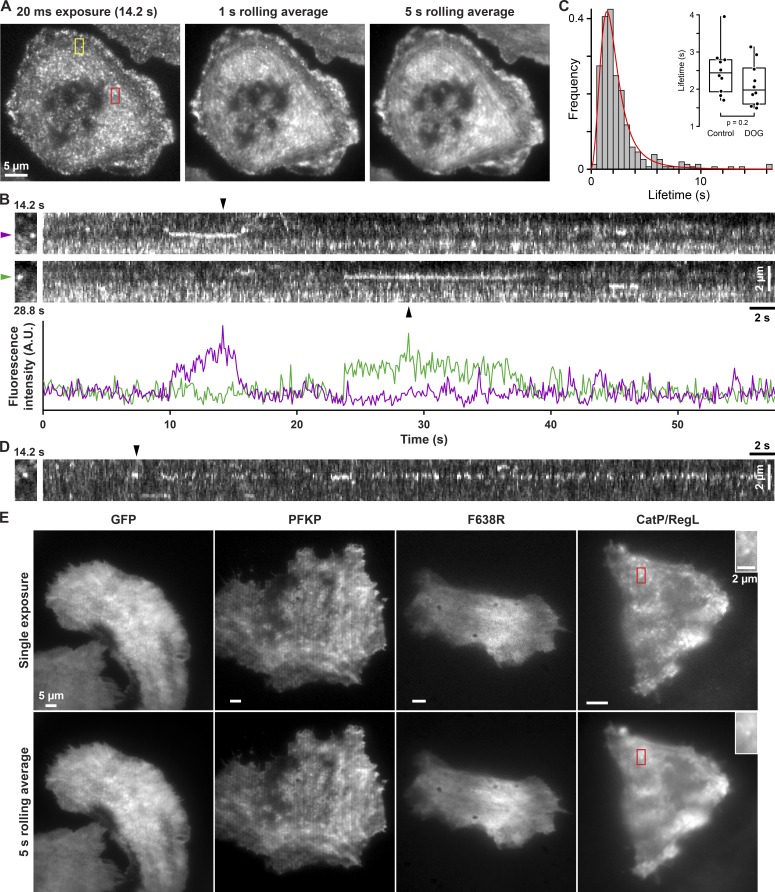
We also asked whether PFKL forms higher-order structures in cells by expressing EGFP-tagged PFK1 isoforms in rat carcinoma MTLn3 cells. High-speed total internal reflection fluorescence (TIRF) microscopy at 5–10-frames-per-second acquisition rates revealed that PFKL has a punctate localization in cells (Fig. 4 A and Video 1). Because of the high density of the particle field and movement of PFKL-EGFP in and out of the TIRF plane, it was not possible to track individual particles, but the majority of PFKL-EGFP particles appeared to move around the cytoplasm in a nondirected fashion, likely by Brownian diffusion. However, further analysis of PFKL-EGFP time-lapse sequences by applying rolling averages of 1–5-s temporal windows to average highly mobile particles identified a smaller population of more stable PFKL-EGFP that remained stationary for several seconds and were clearly visible on kymographs of nonaveraged data (Fig. 4, B and C). Because the TIRF excitation plane is only a few hundred nanometers thick, these stationary PFKL-EGFP particles likely represent temporary docking events at or near the ventral plasma membrane. Furthermore, some PFKL-EGFP particles were seen to pause at the same location proximal to the membrane (Fig. 4 D), suggesting that the docking site can remain stationary. The identity of the docking site or sites is currently unknown, but we speculate that PFKL-EGFP particles are interacting with transmembrane ion transport proteins.
Figure 4.
PFKL forms dynamic particles in MTLn3 rat breast cancer cells. (A) TIRF image of PFKL-EGFP expressing MTLn3 rat mammary adenocarcinoma cell from a time-lapse sequence acquired at 10 frames per second and 1-s (10 frames) and 5-s (50 frames) rolling averages highlighting docked PFKL-EGFP particles. (B) Kymographs showing docked PFKL-EGFP particles in the yellow box indicated in A. Black arrowheads correspond with the individual time points shown on the left. The plot shows fluorescence intensity of the two example particles highlighted by colored arrowheads as a function of time. (C) Docked PFKL-EGFP lifetime distribution from control cells (n = 306 particles from 10 cells). Histogram bin width was set according to the Freedman–Diaconis rule. The red line is a log-logistic fit of the lifetime distribution. The inset is a box plot of the mean particle docking time per cell for control cells or cells treated with 2-deoxyglucose (DOG; n = 298 particles from 10 cells) for 23 h. A t test was used to determine statistical significance. (D) Kymograph of the area in the red box indicated in A showing PFKL-EGFP particles docking at the similar location over the course of the video. (E) Single exposure and 5-s rolling averages for MTLn3 cells expressing EGFP or various EGFP-PFK1 constructs as labeled. The frame rate of time-lapse sequences was: EGFP, 2.6 frames per second; PFKP-EGFP, 1.5 frames per second; PFKL-F638R-EGFP, 2.8 frames per second; CatP/RegL-EGFP, 2.9 frames per second. Higher-magnification images of the areas of the red boxes in the CatP/RegL-EGFP cells highlight a docked particle. Data are representative of three independent cell preparations.
Although we cannot unambiguously confirm that these intracellular PFKL particles are filaments, several experimental observations strongly support this idea. First, PFKP-EGFP, which does not form filaments in vitro, did not form particles in cells but in contrast remained completely diffuse throughout the cytoplasm and indistinguishable from free EGFP (Fig. 4 E and Video 2). Second, consistent with our in vitro data showing that tetramers are necessary for filament assembly, we did not observe intracellular particles of PFKL-F638R-EGFP (Fig. 4 E), which does not form tetramers. Third, the CatP/RegL chimeric PFK1 that forms filaments in vitro was also seen as particles in cells (Fig. 4 E and Video 3). Hence, there was a strong experimental correlation between intracellular particle formation and in vitro filament assembly. Lastly, PFKL-EGFP particles did not appear elongated by resolution-limited microscopy, indicating that if the punctate appearance indeed represents PFKL filaments, their length must be well below the Abbe resolution limit of the light microscope of ∼200 nm; i.e., intracellular PFKL filaments must have fewer than ∼15 tetramer subunits, in agreement with the length of filaments formed in vitro (Fig. 1 D). The fluorescence intensity of PFKL-EGFP particles was also relatively homogenous, further indicating a narrow filament length distribution.
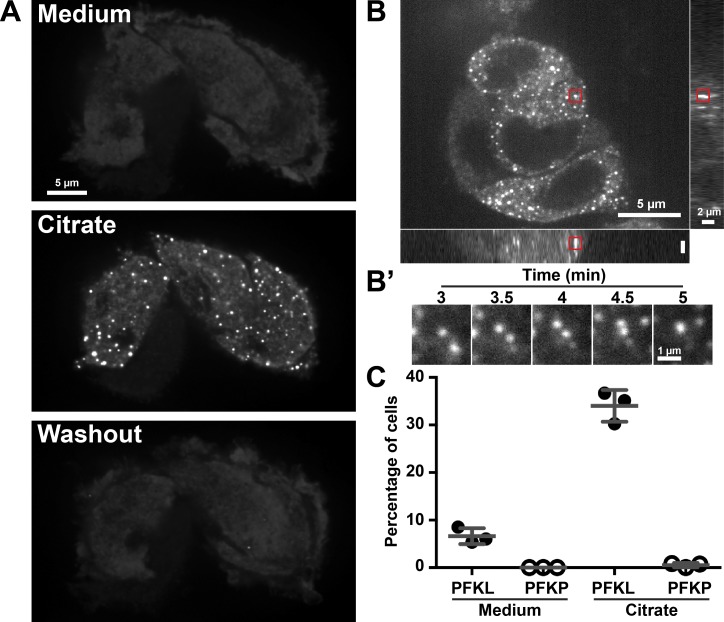
Finally, we sought to determine whether PFKL-EGFP particle size and/or dynamics are regulated by glucose availability. Neither glucose starvation (not depicted) nor incubation of cells for 23 h with 2-deoxyglucose, a glucose analogue that cannot be metabolized, had a discernable impact on punctae size, dynamics, or membrane docking (Fig. 4 C). While examining the impact of metabolic ligands of PFK1 on the dynamics of PFKL-EGFP particles, we found that the addition of citrate, an allosteric inhibitor of PFK1, to the media induced the formation of large PFKL-EGFP–positive structures that we termed citrate-induced punctae (CIP). By confocal microscopy, PFKL-EGFP appeared diffuse in the cytoplasm of MTLn3 cells grown in control medium (Fig. 5 A and Video 4). Addition of 10 mM citrate induced the formation of μm-scale PFKL-EGFP punctae with a mean ± SD diameter of 0.39 ± 0.15 µm (n = 24 total cells from three independent preparations; Fig. S3 D) within 4 min. The formation of these structures was reversible, as CIP disappeared within minutes of returning cells to normal growth medium. Fusion of smaller particles to form larger particles was occasionally observed (Fig. 5, B and B′). CIP were specific to cells expressing PFKL-EGFP as they did not form in MTLn3 expressing PFKP-EGFP (Fig. 5 C). CIP appeared similar in size and morphology to stress granules, P-bodies, and aggresomes (Mahboubi and Stochaj, 2017), suggesting that PFKL-EGFP can undergo a phase transition upon citrate challenge (Li et al., 2012). The significance and function of CIP are currently unknown but are important to resolve in future studies.
Figure 5.
Citrate reversibly induces the formation of large PFKL-EGFP punctae. (A) Spinning-disk confocal image of PFKL-EGFP–expressing MTLn3 cells from a time-lapse sequence acquired at two frames per minute. Cells were imaged in growth medium for 5 min before the addition of 10 mM citrate. After imaging for 5 min, the citrate-containing medium was replaced with growth medium, and cells were imaged for an additional 5 min. Medium, image directly before the addition of 10 mM citrate; citrate, image 4 min after the addition of citrate; washout, image 5 min after removal of citrate-containing buffer. (B) Spinning-disk confocal z-stack of PFKL-EGFP–expressing cells 4 min after the addition of 10 mM citrate. (B′) Area of the red box in B at the time indicated after the addition of citrate. (C) Percentage of MTLn3 cells expressing either PFKL-EGFP or PFKP-EGFP showing the formation of large punctae in growth medium or 4 min after the addition of 10 mM citrate. Error bars represent means ± SD. Data are representative of three independent cell preparations.
To our knowledge, this is the first study showing regulated assembly of a glycolytic enzyme into filamentous structures. The substrate-dependent filament assembly of PFKL and the alignment of tetramers in filaments with an exposed catalytic site suggest that the enzyme is an active state in assembled polymers. When assembled into filaments, acetyl-CoA carboxylase (Meredith and Lane, 1978; Beaty and Lane, 1983) and β-glucosidase (Gunning, 1965; Kim et al., 2005) are enzymatically active; however, bacterial CTP synthase is enzymatically inactive in polymers, likely because of conformational constraints imposed by the filament assembly interactions (Barry et al., 2014).
Homomeric filament formation by PFKL could serve multiple, not necessarily exclusive functions. Filaments of other metabolic enzymes are proposed, but some are not conclusively determined, to function in scaffolding, regulated activity, and efficient substrate transfer. Filament formation may enable allosteric regulation of activity, perhaps for selective roles of the liver isoform in gluconeogenesis. Finally, PFKL filaments could allow for localized ATP production in cells. A subset of PFKL-EGFP particles localize near the plasma membrane (Fig. 4, A–D), where localized ATP production by glycolysis could support processes including protrusion, endocytosis, and the activity of ATP-dependent ion transporters and channels. Proteomics analyses indicate that several glycolytic enzymes, including pyruvate kinase, aldolase, and lactate dehydrogenase, localize in pseudopodia or invadopodia (Beckner et al., 1990; Jia et al., 2005; Attanasio et al., 2011), and a plasma membrane–localized glycolytic metabolon was reported to enable vessel sprouting of endothelial cells (De Bock et al., 2013) and to generate localized energy in neurons for synaptic transmission (Jang et al., 2016). Several glycolytic enzymes, such as PFKM and aldolase, associate with the cytoskeleton, where this interaction regulates enzyme localization and activity (Clarke and Masters, 1975; Wang et al., 1996). Thus, enzymes that do not form filaments may bind to filamentous cytoskeletal structures instead. Further studies are needed to resolve the function of PFKL filaments, with the distinct role of glycolysis in ATP production perhaps providing an important determinant. Moreover, our findings open a clinically relevant new direction for determining whether PFKL filaments regulate metabolic programming in diseases such as cancer.
Materials and methods
Cloning, expression, and purification of recombinant human PFK1 isoforms
The Homo sapiens cDNA encoding the 780-amino-acid isoform b of PFKL (NP_002617) and the 780-amino-acid isoform 2 of the muscle isoform PFKM (NP_000280) were cloned into pFastBac HTa or pFastBac HTb vectors, respectively. PFKL point mutants at the sugar substrate binding site H199Y and at the tetramer interface F638R were generated by using a commercially available site-directed mutagenesis kit (QuikChange Lightning; Agilent Technologies). DNA primers were designed using an online primer design tool (Agilent Technologies) and were purchased from Elim Biopharmaceuticals. Cloning of human PFKP was previously described (Webb et al., 2015). The chimera of PFKP and PFKL was generated by Gibson Assembly (New England Biolabs, Inc.). Baculovirus expression and PFK1 purification were performed as previously described (Webb et al., 2015). In brief, 200–500 million sf21 cells were used to express PFK1 at a multiplicity of infection of 2 for 48 h. Cells were pelleted by low-speed centrifugation (1,000 g), then media was discarded, and pellets were stored at −80°C until the time of purification. Cell pellets were defrosted on ice, resuspended in 50 ml lysis buffer per 200 million cells (20 mM Tris-HCl, pH 7.5, 50 mM potassium phosphate, 1 mM 2-mercaptoethanol, 10% glycerol, 10 mM imidazole, and cOmplete Protease Inhibitor Cocktail [Roche]). Cells were lysed with ∼15 passes of a Dounce homogenizer, and debris was removed by centrifugation. The supernatant was incubated with Talon resin (Takara Bio Inc.) and then washed with 20 bed volumes of lysis buffer, and PFK1 was eluted with a minimal volume of elution buffer (lysis buffer with 100 mM imidazole). Fractions containing protein were pooled and dialyzed into a buffer containing 20 mM Hepes, pH 7.5, 100 mM potassium chloride, 1 mM dithiothreitol, 1 mM ATP, 1 mM magnesium chloride, and 5% glycerol. Protein concentration was determined using a Bradford Protein Assay kit (Thermo Fisher Scientific). After purification, PFK1 was stored on ice in a 4°C fridge. The sequences of PFKL, PFKP, and CatP/RegL were aligned using Clustal Omega (Sievers et al., 2011).
PFK1 activity assays
PFK1 activity was determined using an auxiliary enzyme assay (Brüser et al., 2012) with kinetic analysis using 200-µl reactions containing 50 mM Tris-HCl, pH 7.4, 100 mM KCl, 10 mM MgCl2, 0.15 mM NADH, 0.675 U/ml aldolase, 5 U/ml triosephosphate isomerase, and 2 U/ml glycerol phosphate dehydrogenase. ATP and F6P were used as indicated. Auxiliary enzymes were desalted using an Amicon Ultracel-10K Centrifugal Filter Unit (EMD Millipore) before use. The temperature was equilibrated to 25°C for 10 min before initiating the reaction with the addition of magnesium chloride. The absorbance at 340 nm was measured using a SpectraMax M5 microplate reader (Molecular Devices). Kinetic parameters were generated by linear regression analysis of the Michalas-Menton or Hill equations using Prism (GraphPad Software) and are the mean of a minimum of three measurements from two independent preparations of protein. One unit of activity was defined as the amount of enzyme that catalyzed the formation of 1 mmol of fructose 1,6-bisphosphate per minute at 25°C.
TEM
20 µl of 50 µg/ml PFK1 was applied to glow-discharged carbon-coated grids and stained with 2% (wt/vol) uranyl acetate. Grids were examined and photographed with a 100CX II transmission electron microscope (JEOL). For estimation of size of PFK1 particles and filaments, the length and width of individual particles from TEM images were measured using FIJI software (ImageJ; National Institutes of Health; Schindelin et al., 2012). The mean length and width ± SD are reported.
3D EM
For both PFKL and the PFKP/PFKL chimera, purified protein was concentrated to 0.5 mg/ml. Final concentrations of 2 mM F6P and 20 mM (NH4)SO4 were then added to the protein, and the assembly reaction was left to proceed for 10 min at room temperature. 3.5 µl of this reaction mixture was then spotted onto a 400 mesh glow-discharged copper-coated grid using a 0.7% uranyl formate solution. Electron micrographs were acquired on a Tecnai T12 microscope (FEI) operating at 120 kV at 52,000× magnification on a Gatan US4000 charge-coupled device camera with a 2.07 Å/px.
For both reconstructions, images were collected with Leginon (Suloway et al., 2005). Contrast transfer function (CTF) parameters were estimated using CTFFIND3 (Rohou and Grigorieff, 2015) with a defocus range of 0.6–1.4 µm. Polymer assemblies were manually picked in Appion (Lander et al., 2009). Particle boxing was performed in Appion using a 240-px box with an 80-Å helical step for both constructs, and CTF correction was performed by phase-flipping. For PFKL, 297 images were acquired, resulting in 25,364 segments corresponding with ∼31,700 phosphofructokinase tetramers. For CatP/RegL, 96 images were acquired, resulting in 5,206 segments corresponding with ∼6,500 PFK tetramers. 2D classification was performed in Relion (Scheres, 2012), and poorly behaved classes (poorly defined features or obvious kinks in the filament) were excluded from 3D reconstructions. The final datasets consisted of 9,324 segments for PFKL and 5,206 segments for CatP/RegL.
The Iterative Helical Real Space Reconstruction (IHRSR) method (Egelman, 2010) in Spider (Shaikh et al., 2008) was used for helical reconstruction. We modified the standard IHRSR protocol for gold-standard refinement, with all overlapping segments from a given filament assigned to the same half group to prevent overfitting. For the CatP/RegL reconstruction, a featureless cylinder was subject to 26 rounds of gold-standard refinement (Scheres and Chen, 2012). This resulted in a 26-Å converged map, with 221.6° azimuthal rotation and 83 Å helical rise as the final helical parameters. For the PFKL reconstruction, the final PFKP/PFKL map filtered to 50 Å was used as an initial model for gold-standard refinement. After 15 rounds, this resulted in a 25-Å converged map, with 221.5° azimuthal rotation and 82.8 Å helical rise as the final helical parameters.
90° light scattering
PFK1 polymerization was measured by 90°-angle light scattering in a Shimadzu RF-5301PC spectrofluorophotometer with 350-nm excitation and emission wavelengths and 1.5-nm slit width at 25°C. PFK1 was added at a final concentration up to 500 µg/ml (6 µM) as specified. Data were collected for ∼2 min to establish a baseline. Buffer or F6P was added to the cuvette from a 50× stock and gently mixed by pipetting. The elapsed time for the addition of substrate was ∼20 s. The reading at time 0 was the first reading after the addition of buffer or F6P with data collected every second. The change in light scattering relative to the maximal signal was plotted as a function of time.
Plasmid generation, transfection, cell culture, and microscopy
To generate PFK1 constructs with a carboxy-terminal EGFP tag, cDNA for wild-type and mutant PFKL and wild-type PFKP were cloned into pEGFP N2 and N1 vectors, respectively. The integrity of EGFP-tagged PFK1 was confirmed by Western blot analysis of total cell lysates of 293FT cells using rabbit anti-GFP (A-11122; 1DB-001-0000868907; Invitrogen) and mouse anti-actin clone C4 (MAB1501; 1DB-001-0000850281; EMD Millipore) antibodies (Fig. S3 E). MTLn3 rat mammary adenocarcinoma cells (Segall et al., 1996) were maintained in high-glucose MEM α media supplemented with 10% heat-inactivated fetal bovine serum and 1× penicillin/streptomycin at 37°C and 5% CO2. Cells were tested for mycoplasma contamination before use (DNA Diagnostics Center). Fluorescently tagged PFK1 constructs were expressed by transfecting plasmid DNA into cells using FuGENE HD (Promega) as previously described (Webb et al., 2015). Cells were treated with 1 mM 2-deoxyglucose (Sigma-Aldrich) in cOmplete growth medium for 23 h. For acute treatment of cells with 10 mM citrate (Sigma-Aldrich), citrate was added from a 1 M, pH 7.4, stock directly to growth medium.
For live-cell microscopy, cells were grown in 35-mm glass-bottom dishes (MatTek Corporation). TIRF images were acquired on a 37°C environmentally controlled inverted microscope stand (Ti; Nikon) equipped with a motorized TIRF illuminator (Nikon), an iXon electron-multiplying charge-coupled device camera (Andor Technology), and a 100× 1.49 NA CFI Apochromat TIRF objective (Nikon) using 1.5× intermediate magnification optimized for live-cell imaging. Confocal images were acquired with a 60× Plan Apochromat TIRF objective (Nikon) 1.49 NA oil immersion objective on an inverted microscope system (Eclipse Ti Perfect Focus System; Nikon) equipped with a spinning-disk confocal scanner unit (Borealis-modified CSU-X1; Yokogawa Electric Corporation; Spectral Applied Research), a multipoint stage (MS-2000; Applied Scientific Instruments), and a cMyo cooled charge-coupled device camera (Photometrics; Stehbens et al., 2012). All microscope hardware was controlled by NIS Elements software 4.5 (Nikon), and image processing and analysis was performed in NIS Elements. Stationary PFKL-EGFP particles were identified by 1- or 5-s rolling averages generated by the “ND Image Average” function of NIS Elements. The lifetime of docked particles was estimated to be the time between the half-maximum intensities in 2 × 2–pixel regions of interest in which fluorescence intensity was measured as a function of time. The histogram and the log-logistic curve fit of the lifetime distribution was calculated in MATLAB (MathWorks). Kymographs were generated from the “Show Slices View” in NIS Elements by building a stack of y-t views and displaying a maximum-intensity projection of 3–4 y-t views to suppress image noise essentially as described (Stehbens et al., 2014). Final figures were assembled in Illustrator CS5 (Adobe).
Online supplemental material
Fig. S1 shows the activity, TEM, and light scattering of purified recombinant wild-type and mutant PFK1. Fig. S2 shows the determination of the PFKL filament structure. Fig. S3 shows the sequence, purification, and activity of CatP/RegL, size distribution of CIP, and immunoblot of PFK1-EGFP constructs. Video 1 shows how PFKL-EGFP forms dynamic particles with punctate localization in cells and corresponds with Fig. 4 (A–D). Video 2 shows how PFKP-EGFP is diffuse in cells and corresponds with Fig. 4 E. Video 3 shows how CatP/RegL-EGFP forms dynamic particles with punctate localization in cells and corresponds with Fig. 4 E. Video 4 shows how citrate reversibly induces the formation of large PFKL-EGFP punctae and corresponds with Fig. 5 A.
Supplementary Material
Acknowledgments
This work was supported by a Canadian Institutes of Health Research postdoctoral fellowship and a Pilot/Feasibility grant from the University of California, San Francisco Liver Center (P30 DK026743) to B.A. Webb, National Institutes of Health (R01 GM079139 and S10 RR26758) to T. Wittmann, National Institutes of Health (R01 GM118396) to J.M. Kollman, and National Institutes of Health (R01 CA197855) to D.L. Barber.
The authors declare no competing financial interests.
Author contributions: B.A. Webb conceived initial studies and generated all constructs. B.A. Webb expressed and purified recombinant PFK1, performed biochemical and TEM analysis of recombinant protein, and performed live-cell imaging. A.M. Dosey and J.M. Kollman performed EM, 3D reconstruction, and structural analysis of PFKL filaments. T. Wittmann performed analysis of live-cell imaging. T. Wittmann, B.A. Webb, J.M. Kollman, and D.L. Barber contributed ideas to the project and experimental design and also contributed to writing the manuscript.
Footnotes
Abbreviations used:
- CIP
- citrate-induced punctate
- CTP
- cytidine triphosphate
- F6P
- fructose 6-phosphate
- PFKL
- liver PFK1
- PFKM
- muscle PFK1
- PFKP
- platelet PFK1
- TEM
- transmission EM
- TIRF
- total internal reflection fluorescence
References
- Albe K.R., Butler M.H., and Wright B.E.. 1990. Cellular concentrations of enzymes and their substrates. J. Theor. Biol. 143:163–195. 10.1016/S0022-5193(05)80266-8 [DOI] [PubMed] [Google Scholar]
- Attanasio F., Caldieri G., Giacchetti G., van Horssen R., Wieringa B., and Buccione R.. 2011. Novel invadopodia components revealed by differential proteomic analysis. Eur. J. Cell Biol. 90:115–127. 10.1016/j.ejcb.2010.05.004 [DOI] [PubMed] [Google Scholar]
- Barry R.M., Bitbol A.-F., Lorestani A., Charles E.J., Habrian C.H., Hansen J.M., Li H.-J., Baldwin E.P., Wingreen N.S., Kollman J.M., and Gitai Z.. 2014. Large-scale filament formation inhibits the activity of CTP synthetase. eLife. 3:e03638 10.7554/eLife.03638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaty N.B., and Lane M.D.. 1983. The polymerization of acetyl-CoA carboxylase. J. Biol. Chem. 258:13051–13055. [PubMed] [Google Scholar]
- Beckner M.E., Stracke M.L., Liotta L.A., and Schiffmann E.. 1990. Glycolysis as primary energy source in tumor cell chemotaxis. J. Natl. Cancer Inst. 82:1836–1840. 10.1093/jnci/82.23.1836 [DOI] [PubMed] [Google Scholar]
- Brüser A., Kirchberger J., Kloos M., Sträter N., and Schöneberg T.. 2012. Functional linkage of adenine nucleotide binding sites in mammalian muscle 6-phosphofructokinase. J. Biol. Chem. 287:17546–17553. 10.1074/jbc.M112.347153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke F.M., and Masters C.J.. 1975. On the association of glycolytic enzymes with structural proteins of skeletal muscle. Biochim. Biophys. Acta. 381:37–46. 10.1016/0304-4165(75)90187-7 [DOI] [PubMed] [Google Scholar]
- Cohen R.J., Jedziniak J.A., and Benedek G.B.. 1976. The functional relationship between polymerization and catalytic activity of beef liver glutamate dehydrogenase: II. Experiment. J. Mol. Biol. 108:179–199. 10.1016/S0022-2836(76)80102-7 [DOI] [PubMed] [Google Scholar]
- De Bock K., Georgiadou M., Schoors S., Kuchnio A., Wong B.W., Cantelmo A.R., Quaegebeur A., Ghesquière B., Cauwenberghs S., Eelen G., et al. . 2013. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 154:651–663. 10.1016/j.cell.2013.06.037 [DOI] [PubMed] [Google Scholar]
- Egelman E.H. 2010. Reconstruction of helical filaments and tubes. Methods Enzymol. 482:167–183. 10.1016/S0076-6879(10)82006-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelman E.H., Xu C., DiMaio F., Magnotti E., Modlin C., Yu X., Wright E., Baker D., and Conticello V.P.. 2015. Structural plasticity of helical nanotubes based on coiled-coil assemblies. Structure. 23:280–289. 10.1016/j.str.2014.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg H., and Tomkins G.M.. 1968. Molecular weight of the subunits, oligomeric and associated forms of bovine liver glutamate dehydrogenase. J. Mol. Biol. 31:37–49. 10.1016/0022-2836(68)90052-1 [DOI] [PubMed] [Google Scholar]
- Foe L.G., and Trujillo J.L.. 1980. Quaternary structure of pig liver phosphofructokinase. J. Biol. Chem. 255:10537–10541. [PubMed] [Google Scholar]
- Frey T.G., Eisenberg D., and Eiserling F.A.. 1975. Glutamine synthetase forms three- and seven-stranded helical cables. Proc. Natl. Acad. Sci. USA. 72:3402–3406. 10.1073/pnas.72.9.3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning B.E. 1965. The fine structure of chloroplast stroma following aldehyde osmium-tetroxide fixation. J. Cell Biol. 24:79–93. 10.1083/jcb.24.1.79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habrian C., Chandrasekhara A., Shahrvini B., Hua B., Lee J., Jesinghaus R., Barry R., Gitai Z., Kollman J., and Baldwin E.P.. 2016. Inhibition of Escherichia coli CTP synthetase by NADH and other nicotinamides and their mutual interactions with CTP and GTP. Biochemistry. 55:5554–5565. 10.1021/acs.biochem.6b00383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerson-Mahar M., Briegel A., Werner J.N., Jensen G.J., and Gitai Z.. 2010. The metabolic enzyme CTP synthase forms cytoskeletal filaments. Nat. Cell Biol. 12:739–746. 10.1038/ncb2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang S., Nelson J.C., Bend E.G., Rodríguez-Laureano L., Tueros F.G., Cartagenova L., Underwood K., Jorgensen E.M., and Colón-Ramos D.A.. 2016. Glycolytic enzymes localize to synapses under energy stress to support synaptic function. Neuron. 90:278–291. 10.1016/j.neuron.2016.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z., Barbier L., Stuart H., Amraei M., Pelech S., Dennis J.W., Metalnikov P., O’Donnell P., and Nabi I.R.. 2005. Tumor cell pseudopodial protrusions. Localized signaling domains coordinating cytoskeleton remodeling, cell adhesion, glycolysis, RNA translocation, and protein translation. J. Biol. Chem. 280:30564–30573. 10.1074/jbc.M501754200 [DOI] [PubMed] [Google Scholar]
- Kemp R.G. 1971. Rabbit liver phosphofructokinase. Comparison of some properties with those of muscle phosphofructokinase. J. Biol. Chem. 246:245–252. [PubMed] [Google Scholar]
- Kim S.-Y., Kim Y.-W., Hegerl R., Cyrklaff M., and Kim I.-S.. 2005. Novel type of enzyme multimerization enhances substrate affinity of oat β-glucosidase. J. Struct. Biol. 150:1–10. 10.1016/j.jsb.2004.07.007 [DOI] [PubMed] [Google Scholar]
- Lander G.C., Stagg S.M., Voss N.R., Cheng A., Fellmann D., Pulokas J., Yoshioka C., Irving C., Mulder A., Lau P.-W., et al. . 2009. Appion: An integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166:95–102. 10.1016/j.jsb.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Banjade S., Cheng H.-C., Kim S., Chen B., Guo L., Llaguno M., Hollingsworth J.V., King D.S., Banani S.F., et al. . 2012. Phase transitions in the assembly of multivalent signalling proteins. Nature. 483:336–340. 10.1038/nature10879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.-L. 2010. Intracellular compartmentation of CTP synthase in Drosophila. J. Genet. Genomics. 37:281–296. 10.1016/S1673-8527(09)60046-1 [DOI] [PubMed] [Google Scholar]
- Mahboubi H., and Stochaj U.. 2017. Cytoplasmic stress granules: Dynamic modulators of cell signaling and disease. Biochim. Biophys. Acta. 1863:884–895. 10.1016/j.bbadis.2016.12.022 [DOI] [PubMed] [Google Scholar]
- Meredith M.J., and Lane M.D.. 1978. Acetyl-CoA carboxylase. Evidence for polymeric filament to protomer transition in the intact avian liver cell. J. Biol. Chem. 253:3381–3383. [PubMed] [Google Scholar]
- Noree C., Sato B.K., Broyer R.M., and Wilhelm J.E.. 2010. Identification of novel filament-forming proteins in Saccharomyces cerevisiae and Drosophila melanogaster. J. Cell Biol. 190:541–551. 10.1083/jcb.201003001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell J.D., Zhao A., Ellington A.D., and Marcotte E.M.. 2012. Dynamic reorganization of metabolic enzymes into intracellular bodies. Annu. Rev. Cell Dev. Biol. 28:89–111. 10.1146/annurev-cellbio-101011-155841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B.R., Svenneby G., Kvamme E., Tveit B., and Eskeland T.. 1970. Formation and ultrastructure of enzymically active polymers of pig renal glutaminase. J. Mol. Biol. 52:239–245. 10.1016/0022-2836(70)90028-8 [DOI] [PubMed] [Google Scholar]
- Ranjit S., Dvornikov A., Holland D.A., Reinhart G.D., Jameson D.M., and Gratton E.. 2014. Application of three-photon excitation FCS to the study of protein oligomerization. J. Phys. Chem. B. 118:14627–14631. 10.1021/jp511126x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart G.D., and Lardy H.A.. 1980. Rat liver phosphofructokinase: use of fluorescence polarization to study aggregation at low protein concentration. Biochemistry. 19:1484–1490. 10.1021/bi00548a035 [DOI] [PubMed] [Google Scholar]
- Rohou A., and Grigorieff N.. 2015. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192:216–221. 10.1016/j.jsb.2015.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H.W. 2012. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180:519–530. 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres S.H.W., and Chen S.. 2012. Prevention of overfitting in cryo-EM structure determination. Nat. Methods. 9:853–854. 10.1038/nmeth.2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. . 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segall J.E., Tyerech S., Boselli L., Masseling S., Helft J., Chan A., Jones J., and Condeelis J.. 1996. EGF stimulates lamellipod extension in metastatic mammary adenocarcinoma cells by an actin-dependent mechanism. Clin. Exp. Metastasis. 14:61–72. 10.1007/BF00157687 [DOI] [PubMed] [Google Scholar]
- Shaikh T.R., Gao H., Baxter W.T., Asturias F.J., Boisset N., Leith A., and Frank J.. 2008. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat. Protoc. 3:1941–1974. 10.1038/nprot.2008.156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.-J., Kassim H., Huang Y., Li H., Zhang J., Li G., Wang P.-Y., Yan J., Ye F., and Liu J.-L.. 2016. Filamentation of metabolic enzymes in Saccharomyces cerevisiae. J. Genet. Genomics. 43:393–404. 10.1016/j.jgg.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J., et al. . 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S., Pemble H., Murrow L., and Wittmann T.. 2012. Imaging intracellular protein dynamics by spinning disk confocal microscopy. Methods Enzymol. 504:293–313. 10.1016/B978-0-12-391857-4.00015-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stehbens S.J., Paszek M., Pemble H., Ettinger A., Gierke S., and Wittmann T.. 2014. CLASPs link focal-adhesion-associated microtubule capture to localized exocytosis and adhesion site turnover. Nat. Cell Biol. 16:558–570. 10.1038/ncb2975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suloway C., Pulokas J., Fellmann D., Cheng A., Guerra F., Quispe J., Stagg S., Potter C.S., and Carragher B.. 2005. Automated molecular microscopy: The new Leginon system. J. Struct. Biol. 151:41–60. 10.1016/j.jsb.2005.03.010 [DOI] [PubMed] [Google Scholar]
- Trujillo J.L., and Deal W.C. Jr. 1977. Metabolic control and structure of glycolytic enzymes. 17. Pig liver phosphofructokinase: asymmetry properties, proof of rapid association-dissociation equilibriums, and effect of temperature and protein concentration on the equilibriums. Biochemistry. 16:3098–3104. 10.1021/bi00633a009 [DOI] [PubMed] [Google Scholar]
- Wang J., Morris A.J., Tolan D.R., and Pagliaro L.. 1996. The molecular nature of the F-actin binding activity of aldolase revealed with site-directed mutants. J. Biol. Chem. 271:6861–6865. 10.1074/jbc.271.12.6861 [DOI] [PubMed] [Google Scholar]
- Webb B.A., Forouhar F., Szu F.-E., Seetharaman J., Tong L., and Barber D.L.. 2015. Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations. Nature. 523:111–114. 10.1038/nature14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.