The two influenza glycoproteins encode N-terminal targeting sequences that interact with SRP to direct their synthesis at the endoplasmic reticulum. Nordholm et al. show that in addition to coding for endoplasmic reticulum–targeting sequences, these 5′ mRNA regions also function as translational regulatory elements that are controlled by a viral RNA-binding protein.
Abstract
A primary function of 5′ regions in many secretory protein mRNAs is to encode an endoplasmic reticulum (ER) targeting sequence. In this study, we show how the regions coding for the ER-targeting sequences of the influenza glycoproteins NA and HA also function as translational regulatory elements that are controlled by the viral RNA-binding protein (RBP) NS1. The translational increase depends on the nucleotide composition and 5′ positioning of the ER-targeting sequence coding regions and is facilitated by the RNA-binding domain of NS1, which can associate with ER membranes. Inserting the ER-targeting sequence coding region of NA into different 5′ UTRs confirmed that NS1 can promote the translation of secretory protein mRNAs based on the nucleotides within this region rather than the resulting amino acids. By analyzing human protein mRNA sequences, we found evidence that this mechanism of using 5′ coding regions and particular RBPs to achieve gene-specific regulation may extend to human-secreted proteins.
Introduction
Posttranscriptional gene regulation mechanisms are highly dependent on mRNA structure (Jackson et al., 2010; Mortimer et al., 2014). With few exceptions, 5′ UTRs in mammalian mRNAs generally contribute to mRNA export and translation initiation (Muckenthaler et al., 1998; Carmody and Wente, 2009; Lee et al., 2015). In secretory protein mRNAs, the region after the 5′ UTR often encodes an N-terminal targeting sequence for the ER (Blobel and Dobberstein, 1975; Walter and Johnson, 1994). A primary function of ER-targeting sequences is to recruit the signal recognition particle (SRP) for trafficking the nascent secretory protein to the ER translocon, where synthesis resumes (Walter and Blobel, 1981; Walter et al., 1981; Gilmore et al., 1982; Kurzchalia et al., 1986). More recent studies associate the variation in ER-targeting sequences with a regulatory function, as sequence changes can affect SRP binding as well as targeting for degradation (Kang et al., 2006; Karamyshev et al., 2014). However, current data do not exclude the possibility that SRP accommodates an array of ER-targeting sequences to provide flexibility to the 5′ coding regions for posttranscriptional gene regulation, potentially explaining the unique structural profile of these mRNA regions (Kertesz et al., 2010).
Enveloped viruses commonly express their secretory glycoproteins late, likely to prevent premature budding and antibody-mediated recognition of infected cells. Although the late gene distinction is less defined for influenza A viruses (IAVs), the secretory glycoprotein HA has been shown to be synthesized after the viral proteins NP and NS1, but little data exist for when the secretory glycoprotein neuraminidase (NA) is synthesized (Skehel, 1973; Lamb and Choppin, 1976; Shapiro et al., 1987). Because IAVs display little temporal variation in viral gene transcription (Vester et al., 2010; Kawakami et al., 2011), this would imply that HA, and possibly NA, are posttranscriptionally regulated. The early expressed RNA-binding protein (RBP) NS1 has been shown to promote splicing and has also been proposed to confer translational specificity to viral mRNAs by associating with the short 5′ UTRs and multiple translation factors, among many other proteins (Enami et al., 1994; de la Luna et al., 1995; Park and Katze, 1995; Aragón et al., 2000; Lin et al., 2012; Heaton et al., 2016). Although these studies have identified different aspects of IAV gene regulation, it is still unclear how IAVs regulate HA and NA synthesis and what contributions their ER-targeting sequences make to this process.
Results and discussion
Influenza glycoprotein expression is enhanced by NS1
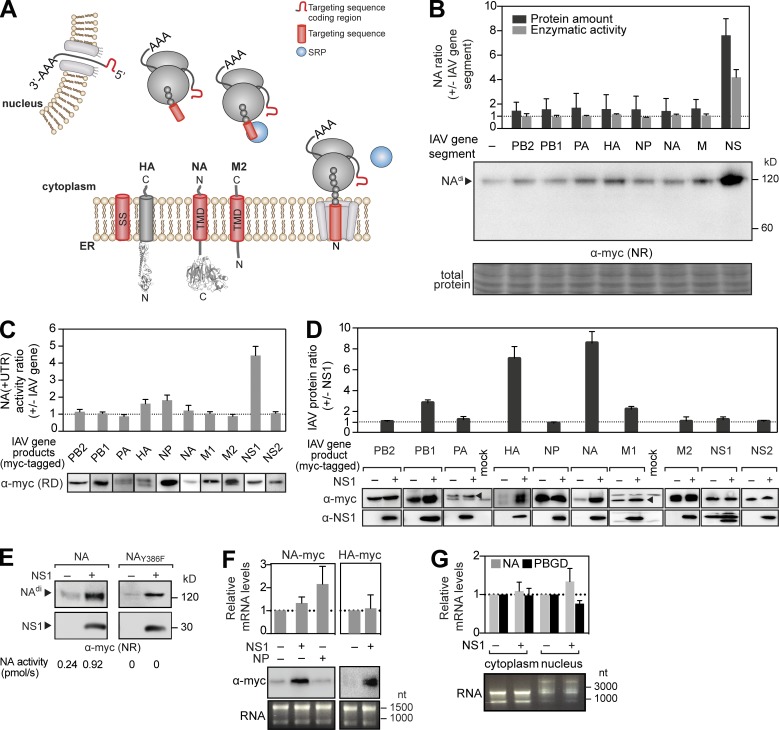
Each IAV membrane protein, HA, NA, and M2, encodes an N-terminal ER-targeting sequence to recruit SRP for trafficking to the ER membrane (Fig. 1 A). Because NS1 is known to contribute to M2 regulation by promoting alternative splicing (Mor et al., 2016), we coexpressed NA with each gene segment from the H1N1 strain A/WSN/33 in HEK 293T cells to determine whether an IAV gene product could also regulate viral secretory glycoprotein expression. Based on protein and sialidase activity levels, only coexpression with the NS segment significantly increased NA protein production (Fig. 1 B). Using a reciprocal approach with C-terminally tagged versions of the 10 major IAV proteins, NS1 was found to be responsible for increasing the NA protein levels (Fig. 1 C), and the increase became more pronounced with the coexpression time (Fig. S1 A).
Figure 1.
Expression of the secretory proteins NA and HA is enhanced by NS1. (A) Topology and ER targeting of the three IAV membrane proteins by their respective N-terminal SS or TMD. (B) An NA expression plasmid lacking the viral UTRs was cotransfected with expression plasmids for the indicated IAV gene segments. NA proteins (from nonreduced [NR] immunoblots) and activity levels at 48 h were normalized to cells expressing only NA. (C) An NA expression plasmid containing viral UTRs was cotransfected with plasmids for the indicated Myc-tagged IAV gene products. NA activity levels were normalized to cells expressing NA alone. IAV gene synthesis was confirmed by reducing (RD) immunoblots. (D) Changes in IAV gene product levels were determined with respect to NS1 coexpression. Arrowheads indicate PA and M1 bands. (E) Immunoblots of NA and inactive NAY386F expressed alone or with NS1. (F) Total NA and HA mRNA levels were determined by quantitative PCR and normalized using the single transfection expression values. Immunoblots and agarose gels showing the RNA amounts used for cDNA synthesis are displayed. (G) mRNA levels of NA and control porphobilinogen-deaminase (PBGD) were measured in the cytoplasmic and nuclear RNA fractions (shown in the agarose gel) and normalized to single transfected samples. Error bars indicate SD. n = 3.
NS1 has previously been implicated in translational regulation of viral mRNAs as it binds to translation factors and viral 5′ UTRs (Enami et al., 1994; de la Luna et al., 1995; Park et al., 1999). However, in our experimental approach, NS1 increased NA levels in the absence (Fig. 1 B) and presence (Fig. 1 C) of the short NA viral UTRs. Therefore, the viral UTRs were excluded when NS1 was coexpressed with other IAV genes. With the exception of NA and HA, most IAV protein levels were largely unaffected by NS1 (Fig. 1 D). The increase was not related to NA sialidase function (Fig. 1 E), changes in HA and NA mRNA levels (Fig. 1 F), or the nuclear and cytoplasmic mRNA distributions (Fig. 1 G), indicating NS1 may regulate synthesis of the secretory glycoproteins NA and HA posttranscriptionally.
NA and HA synthesis is regulated by the NS1 RNA-binding domain (RBD)
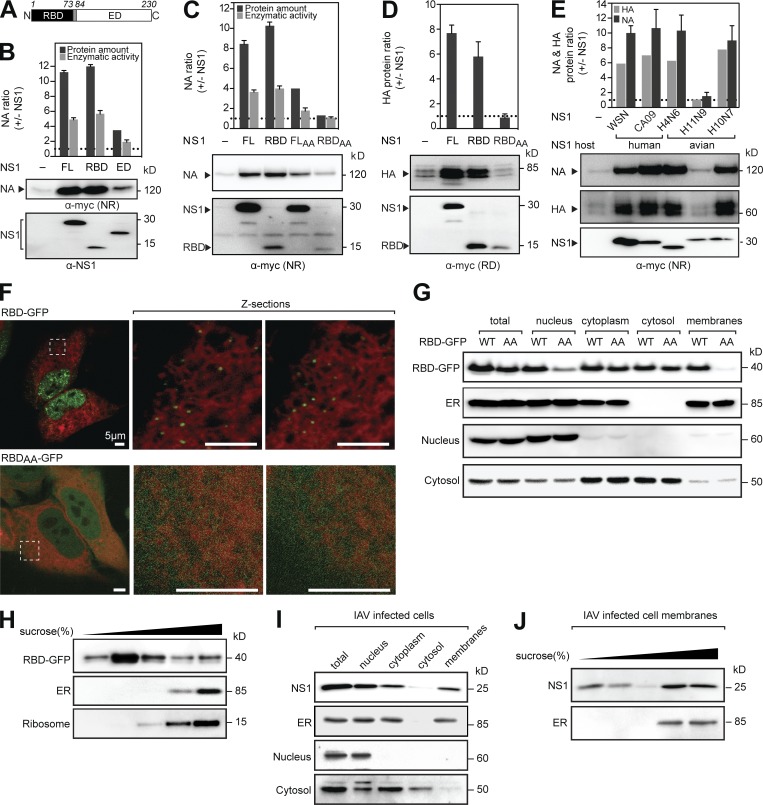
NS1 possesses a conserved two-domain structure (Fig. 2 A). To determine whether one domain is responsible for the synthesis increase, we coexpressed NA with the RBD or the effector domain (ED). Independent of the linker region, the RBD increased NA levels similar to full-length NS1, whereas the ED caused a modest increase (Figs. 2 B and S1 B). Consistent with this result, the R38A/K41A mutations, which abolish NS1 RNA-binding activity (Qian et al., 1995; Wang et al., 1999), also diminished the synthesis increase of NA (Fig. 2 C) and HA (Fig. 2 D), indicating that NS1 positively regulates NA and HA synthesis through an RNA-dependent mechanism.
Figure 2.
NA and HA regulation is controlled by the NS1 RBD, which cofractionates with ER membranes. (A) Diagram of the RBD and ED in NS1WSN. (B and C) Changes in NA protein and activity levels upon coexpression with full-length (FL) NS1, the RBD, and the ED (B) or with the RNA-binding mutants NS1AA or RBDAA (C). (D) Changes in HA protein levels upon coexpression with NS1, the RBD, or RBDAA. RD, reduced. (E) Changes in NAWSN and HAWSN protein levels upon cotransfection with the indicated NS1 genes. NR, nonreduced. (F) Live images of HeLa cells expressing the indicated RBD-GFP construct and the ER marker mCherry-Sec61β (red) are shown with different z sections of the indicated boxed regions. (G and I) Subcellular fractions from cells expressing RBD-GFP (WT) or RBDAA-GFP (AA; G), or infected with WSN for 6 h (I). Immunoblots using antisera against GFP (RBD-GFP), NS1, or the indicated cell compartment markers are shown. (H and J) The total membrane fractions isolated from transfected 293T cells (H) or from infected MDCK cells (J) were separated using a discontinuous sucrose gradient. Equal fraction amounts were immunoblotted using antisera to GFP, NS1, or the indicated markers. Error bars indicate SD. n = 3.
The RBD regulatory function is conserved across NS1 alleles
In IAVs, NS1 genes are phylogenetically diverse (Fig. S1 C; Treanor et al., 1989), which questions whether the ability of NS1 to increase NA and HA synthesis is conserved. Therefore, we coexpressed NA and HA with four NS1 type A alleles and one avian exclusive type B (H10N7) allele (Fig. 2 E). All but one NS1 gene enhanced NA and HA synthesis, and the increase depended more on the presence of a specific NS1 than quantity. Surprisingly, the exception (NS1H11N9) shared the highest amino acid identity to NS1WSN (Fig. S1 D). However, the RBD from NS1H11N9 still increased NA levels (Fig. S1 E), and NS1 chimeras confirmed the EDH11N9 caused the NA regulation loss (Fig. S1 F). Out of several cell lines, NS1WSN only failed to increase NA synthesis in interferon-deficient Vero cells, but the regulatory function was also restored by removing the ED (Fig. S1, G and H). Collectively, these results demonstrate that the RBD-mediated increase of NA and HA synthesis is a conserved function in NS1, which the ED can negatively regulate.
The RBD RNA-binding activity facilitates an association with ER membranes
Secretory proteins are synthesized at the ER. However, NS1 is mainly reported to be a nuclear protein (Greenspan et al., 1988; Hale et al., 2008), which makes it difficult to reconcile how NS1 could regulate secretory protein translation. Because nuclear import is dynamic, we used live-cell imaging to look for potential NS1 association with an ER membrane marker. For visualization, RBD-GFP or RBDAA-GFP, which retained their expected NA-regulatory properties (Fig. S1 I), were coexpressed with the ER marker. In addition to nuclear localization, RBD-GFP and NS1-GFP showed a unique foci pattern near the ER membrane that was absent with RBDAA-GFP (Figs. 2 F and S1 J). Subcellular fractionations confirmed that RBD-GFP, but not RBDAA-GFP, cosedimented with cellular membranes (Fig. 2 G) and, furthermore, comigrated at a density corresponding with ER and ribosomal markers (Fig. 2 H), a partitioning equally evident for NS1 from infected cells (Fig. 2, I and J). The requirement of the RNA-binding activity for ER association and for increasing NA and HA synthesis suggests that NS1 could promote the ER targeting of NA and HA mRNAs or facilitate their translation initiation by ER-associated ribosomes.
Properties of the NA ER-targeting sequence determine the regulation by NS1
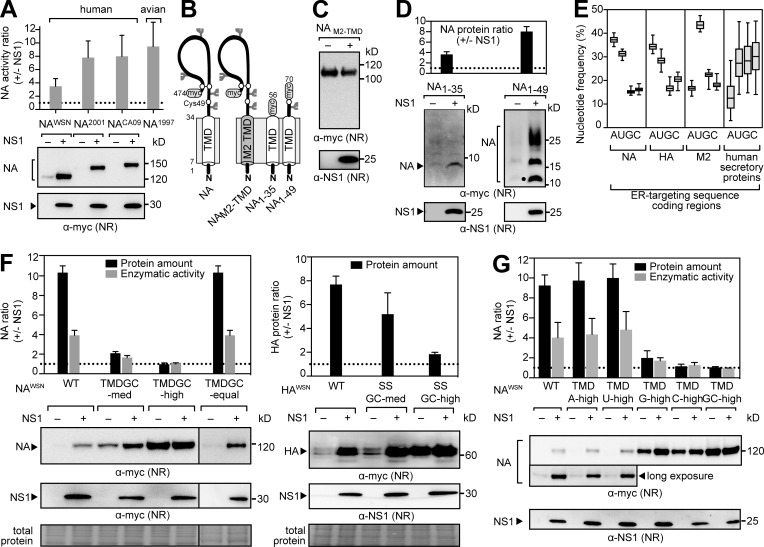
Next, NS1 was coexpressed with NAs derived from human and avian IAV strains to test whether the synthesis increase was NAWSN specific. All the tested NAs expressed better with NS1, indicating that a conserved property in NA, and likely HA, facilitates regulation by NS1 (Fig. 3 A). During synthesis, NA and HA are directed to the ER by N-terminal targeting sequences, which are a signal sequence (SS) for HA and a transmembrane domain (TMD) for NA (Fig. 1 A; Bos et al., 1984; Daniels et al., 2003). Because NS1 does not affect M2, which also uses a TMD for ER targeting (Hull et al., 1988), we exchanged the NA TMD with the M2 TMD (NAM2-TMD; Fig. 3 B). Similar to M2, NAM2-TMD was unaffected by NS1 (Fig. 3 C), suggesting that the regulation depends on a property of the NA TMD. We then examined two short NA constructs with 21-residue C-terminal tags (Fig. 3 B), which were previously shown to be incapable (NA1–35) and capable (NA1–49) of cotranslational ER targeting (Dou et al., 2014). Although NS1 increased the levels of both NAs, the increase was greater for NA1–49 (Fig. 3 D), indicating that NS1 may contribute to NA synthesis initiation and ER targeting through a conserved TMD characteristic.
Figure 3.
NS1 regulation is determined by AU content in the ER-targeting sequence coding region. (A) Changes in protein and activity levels of NAs from human and avian IAVs upon NS1 coexpression. (B) Topology of the NA Myc-tagged constructs with amino acid numbers and N-linked glycan sites (forks). (C and D) Immunoblots of NAM2-TMD (C) or NA1–35 and NA1–49 expressed alone or with NS1 (D). NA1–49 resolved as an unglycosylated (circle) and two higher glycosylated species. (E) Nucleotide frequencies of ER-targeting sequence coding regions for NA, HA, and M2 from H1N1 IAVs and for human secretory proteins (2.5–97.5 percentile). (F) NA (left) and HA (right) containing synonymous substitutions increasing the GC content of their ER-targeting sequence coding regions were expressed alone or with NS1. Representative immunoblots are shown with the stained membrane (total protein). (G) NAs with synonymous substitutions enriching the indicated nucleotide in the TMD coding region were coexpressed with NS1. Changes in protein levels are shown with representative immunoblots. Error bars indicate SD. n = 3. NR, nonreduced.
The 5′ ER-targeting sequence coding region determines the regulation by NS1
Because NA and HA synthesis is regulated by the RBD of NS1 and depends on the ER-targeting sequence, we hypothesized that the 5′ coding regions determine the regulation rather than the resulting amino acids. In support of this hypothesis, the ER-targeting sequence coding regions for NA and HA in the IAV database revealed a high A and U as well as a low G and C nucleotide pattern that was distinct from corresponding regions in M2 and human secretory protein mRNAs (Fig. 3 E). To investigate potential contributions by the nucleotides, we used synonymous substitution schemes to either increase the GC content in the ER-targeting sequence coding regions or enrich for a particular nucleotide (Fig. S2 A). Increasing the GC content in these regions of NA and HA alleviated the translational inhibitory property and NS1 regulation (Fig. 3 F). This suggested that the AU-rich composition determines the NS1 dependence, explaining why NA regulation persisted upon rearranging the A and U nucleotides (NA-TMDGC-equal). The individual nucleotide enrichment further supported the notion that high A or U content in the 5′ ER-targeting sequence coding region inhibits translation and enables regulation by NS1 (Fig. 3 G).
Regulation by the ER-targeting sequence coding region is dependent on the 5′ positioning
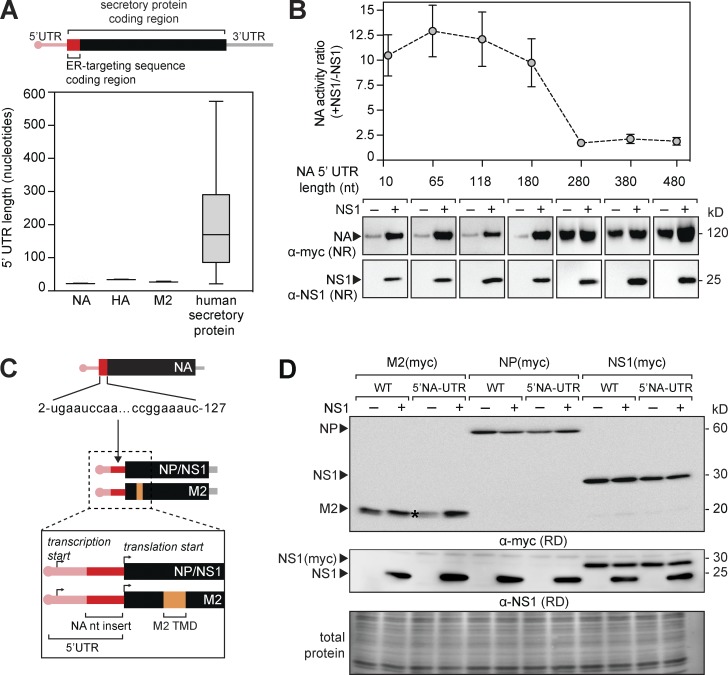
ER-targeting sequence coding regions generally follow 5′ UTRs, an ideal position to function in 5′ posttranscriptional regulation. However, 5′ UTRs are shorter in IAV mRNAs compared with human secretory protein mRNAs (Fig. 4 A). Therefore, we asked how positioning of the ER-targeting sequence coding region from NA affects its regulatory function. Similar to the synonymous substitutions, elongating the 5′ UTR resulted in a length-dependent increase in NA synthesis that corresponded with decreasing NS1 regulation (Fig. 4 B). Surprisingly, optimal NA regulation occurred with 5′ UTRs of <200 nucleotides, the length of most 5′ UTRs in human secretory protein mRNAs. The regulation loss that occurred by increasing the distance between the ER-targeting sequence coding region and the transcription start site further indicates that these regions in NA and likely HA contribute to 5′ mRNA regulation.
Figure 4.
The 5′ region of NA is a mobile RNA regulatory element for secretory proteins. (A) 5′ UTR lengths in NA, HA, M2, and human-secreted soluble protein mRNAs (5–95 percentile). (B) NA with 5′ UTRs of increasing length were expressed alone or with NS1, and the activity increase is shown with representative immunoblots. Error bars indicate SD. n = 3. (C and D) Diagram (C) of the NA 5′ coding region (nucleotides 2–127) that was inserted into the 5′ UTR to generate 5′NA-UTRM2, 5′NA-UTRNP, and 5′NA-UTRNS1, which were expressed alone or with NS1 (D). Representative immunoblots are shown with the stained membrane (total protein). The asterisk indicates 5′NA-UTRM2, which increases with NS1 coexpression. RD, reduced.
Regulation by the NA RNA element is specific for secretory protein mRNAs
Given the synonymous substitution and 5′ UTR results, the possibility that NS1 assists translation of mRNAs with suboptimal 5′ regions remained. However, NS1 regulation persisted when a more optimal Kozak translation initiation site was inserted before the NA coding region (Fig. S2 B). As a further test, we inserted a short 5′ NA coding region devoid of AUG initiation codons into the 5′ UTR of the membrane protein M2 and the nonsecretory viral proteins NP and NS1 (Fig. 4 C). The insertion in M2 (5′NA-UTRM2) resulted in a decreased protein production that was increased by NS1, whereas the nonsecretory proteins were unaffected by the 5′ UTR insertion or NS1 (Fig. 4 D). In summary, these findings demonstrate that viral 5′ coding regions for ER-targeting sequences can function as translational regulatory elements and that the mechanism used by NA and the RBP NS1 is more specific for secretory protein mRNAs.
Model for NA and HA regulation by NS1
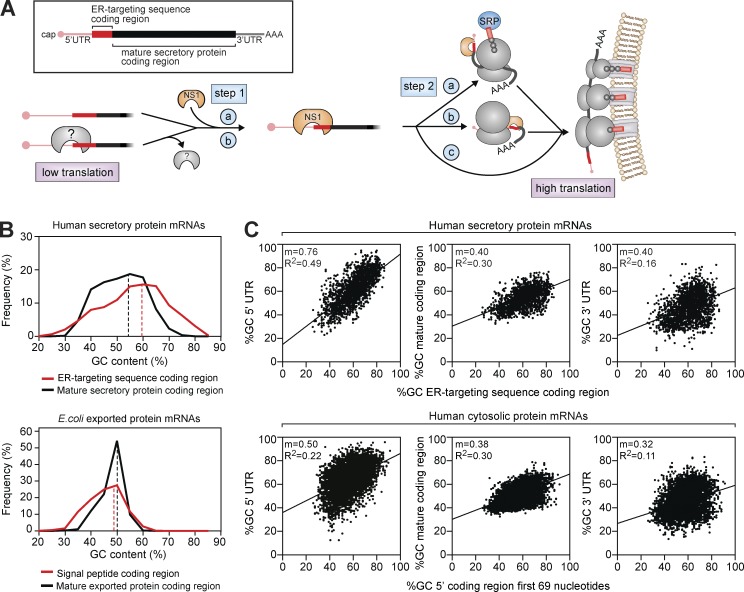
Our results show that the 5′ coding regions of NA and HA act as regulators that limit expression and that this property is not related to proteasomal degradation (Fig. S2 C) or the unfolded protein response (Fig. S2 D). Instead, it is determined by the nucleotide content, 5′ positioning, cell growth (Fig. S2 E), and temperature (Fig. S2, F and G), suggesting that the 5′ regions form translation inhibitory structures or associate with cellular RBPs that suppress translation, such as AU-rich 3′ UTR elements (Barreau et al., 2005). Based on additional data showing that NS1 protects ∼100 nucleotide RNA fragments, binds short 5′ NA mRNAs, and enhances NA synthesis in vitro (Fig. S3, A–F), we propose that NS1 associates with these regions or outcompetes cellular RBPs (Fig. 5 A, step 1). NS1 binding could then promote translation initiation or ER targeting by recruiting translation initiation factors or SRP (Fig. 5 A, step 2) previously found to associate with NS1 (Aragón et al., 2000; Lin et al., 2012; Heaton et al., 2016). Although additional studies are needed to precisely define how NS1 enhances NA and HA synthesis, these results provide a working model for future studies.
Figure 5.
Model for NA and HA regulation by the RBP NS1. (A) The ER-targeting sequence coding region in NA and HA suppresses synthesis by forming a structure that impairs the translation initiation factor recruitment alone or by associating with an inhibitory cellular RBP (gray). When present, NS1 binds the structure (step 1, a), or replaces the cellular RBP (step 1, b), and either helps to recruit the translation and ER-trafficking machinery (step 2, a), transport the nascent chain to the ER (step 2, b), or traffic the mRNA to ER ribosomes (step 2, c). (B) Distribution of human secretory protein mRNAs (top) and E. coli–exported protein mRNAs (bottom) by GC content in coding regions for the mature protein or their respective targeting sequences. Dashed lines show the mean GC content. (C) Correlation plots comparing the GC content in the indicated regions of human mRNAs encoding secreted (top) and cytosolic proteins (bottom). For each mRNA, the GC content in the 5′ coding region (ER-targeting sequences of secreted proteins or the first 69 nucleotides of cytosolic proteins) was plotted with respect to the 5′ UTR (left), protein-coding region (middle), and 3′ UTR (right). mRNAs with 5′ UTRs of <300 nucleotides are shown with Pearson’s correlation coefficient (R2) and the linear regression curve slope (m).
It is possible that the use of 5′ ER-targeting sequence coding regions and an RBP for gene regulation is IAV specific. However, when we examined human secretory protein mRNAs, the nucleotide composition of the ER-targeting sequence coding regions was found to differ from regions encoding for the mature secretory proteins (Fig. 5 B), and this was neither linked to length or hydrophobicity (Fig. S3 G) nor was it observed for Escherichia coli–exported protein genes (Fig. 5 B). Our data also suggest that ER-targeting sequence coding regions act as 5′ UTR extensions, and if the 5′ UTR is too long, its influence on translation initiation at the 5′ region diminishes. Accordingly, we also found a near 1:1 linear correlation between the GC content of ER-targeting sequence coding regions in human-secreted protein mRNAs with their respective 5′ UTRs (Fig. 5 C). The correlation decreased with longer 5′ UTRs (Fig. S3 H) as well as the distance from the 5′ region (e.g. protein coding regions and 3′ UTRs), and it was less in human cytosolic protein mRNAs (Fig. 5 C). Because cells possess many RBPs, which generally bind short stretches of specific nucleic acid bases with low affinity (Ray et al., 2013; Gerstberger et al., 2014; Helder et al., 2016), these correlations suggest that coding regions for human-secreted protein ER-targeting sequences may have evolved with their 5′ UTR regions to enhance recruitment of specific RBPs for posttranscriptional regulation at the ER.
The 5′ regions of IAV mRNAs are generally thought to function as packaging signals, potentially explaining why any involvement in gene-specific regulation has been overlooked. From a mechanistic perspective, regulating HA and NA synthesis by an early expressed protein could help prevent premature budding of empty viral particles, a process that can be driven by NA and HA (Chlanda et al., 2015), limit the length of time cells display these antigens for immune system recognition, and help coordinate the replication process. The potential involvement of NS1 in regulating expression at a key node like the ER could explain why it is associated with numerous functions that are linked to substrates that traverse the secretory pathway, such as antagonizing the interferon response (Hale et al., 2008). However, more direct investigations of NS1 from the different subcellular regions are needed to better define how the protein regulates its RNA substrates and to determine whether NS1 performs two spatially distinct functions.
Materials and methods
Plasmids and constructs
The pHW2000 reverse genetics plasmids for influenza virus A/WSN/33 (Hoffmann et al., 2000) were used to express the eight genome segments and NA with their respective viral 5′ and 3′ UTRs. For individual IAV gene expression without the viral UTR regions, PB1, PB2, PA, NP, NA, HA, M1, M2, NS1, and NS2 coding sequences from WSN were fused to the 3′ coding region for the 21–amino acid Myc-His tag in pcDNA3.1A-Myc-His (Invitrogen) by PCR using primers with overlapping extensions (Mellroth et al., 2012). For HA visualization, the His tag was replaced with a second Myc epitope. Plasmids encoding M2 and NS2 were created by splicing out the M1 and NS1 coding regions from the M and NS gene segments by overlapping PCR. The NS1WSN RBD and ED constructs were generated similarly by removing the regions corresponding with residues 85–230 (RBD) or 1–80 (ED). The R38A and K41A mutations that abolish RNA-binding activity in NS1 (NS1AA) and the RBD (RBDAA) were introduced by site-directed mutagenesis.
The NS1 A and B alleles were cloned into pcDNA3.1A-Myc-His using the following IAV genomic RNA provided by R. Webster (St. Jude Children’s Research Hospital, Memphis, TN) as a template: A/Chicken/Germany/49 (H10N7), A/Duck/HK/P50/97 (H11N9), A/Duck/Czech/56 (H4N6), and A/human/England53/2009 (CA09). The additional NA genes corresponded with the following IAV sequences: A/Waikato/7/2001 H1N1 (NA2001), A/California/4/2009 H1N1 (NACA09), and A/Teal/Hong Kong/W312/97 H6N1 (avian NA1997; provided by R. Webster). The NS1 RBD-ED chimeras were created by exchanging the variable ED amino acid coding regions (residues 102–230) of NS1WSN and NS1H11N9.
NAWSN GC-medium and GC-high TMD coding variants (residues 7–34) and HAWSN SS coding variants (residues 1–17) were designed with the Codon Juggling algorithm (Richardson et al., 2006). NA-TMD GC-equal, A-high, T-high, G-high, and C-high were all generated using synonymous mutations that either maintained the NAWSN nucleotide frequency (GC-equal) or increased the indicated nucleotide (Fig. S2 A). The enzymatically inactive NA mutant (Y386F) was created by site-directed mutagenesis (da Silva et al., 2013). NA in pcDNA3.1A− was C-terminally truncated to generate NA1–35, and NA1–49 (56 and 70 residues including the Myc-His tag, respectively; Nordholm et al., 2013). To produce NAM2-TMD, the NA TMD coding region (residues 7–34) was replaced with the M2 TMD coding region (residues 26–48) while keeping the NA cytoplasmic tail (residues 1–6) to maintain the N-in, C-out orientation. To increase the 5′ UTR length, a nontranslated stretch of nucleotides containing a balanced GC content of ∼60% (a minimum of 53% and a maximum of 65% on a sliding 100-nucleotide window) was inserted between the cytomegalovirus promoter and the NA initiation codon in the expression plasmid. To create 5′NA-UTRM2, 5′NA-UTRNP, and 5′NA-UTRNS1, nucleotides 2–127 from an NAWSN aN3 TMD chimera, which does not encode in-frame or out-of-frame initiation codons, were inserted before the start codon of each gene in the pcDNA3.1A− vector. For live imaging, GFP was fused to the C termini of the NS1WSN constructs with a GSSG linker sequence. All constructs were verified by sequencing before use (Eurofins MWG Operon).
Cell culture, transfections, and cell lysate preparation
All cell lines were obtained from ATCC. Cells were cultured in F12 media (A549) or DMEM (HEK 293T, HeLa, COS-1, Vero, and DF-1) supplemented with 10% FBS and 100 U/ml of penicillin and streptomycin, all acquired from Invitrogen. The cell lines were maintained in a humidified 5% CO2 incubator at 37°C unless specified otherwise. DF-1 cells were maintained at 39°C. For single transfections, 1.5 µg of plasmid DNA was mixed with 1 ml of opti-MEM (Invitrogen) and 5 µg polyethylenimine (PEI) for 20 min. Trypsinized 293T cells were resuspended in opti-MEM 10% FBS to a density of ∼106 cells/ml, and 1 ml of cells was added to each transfection mixture before plating on a 3.5-cm dish. For cotransfections, 1.5 µg of the second plasmid DNA was included before mixing with 5 µg of PEI. HeLa, A549, COS-1, Vero, and DF-1 cell transfections were performed with 5 µl of LT-1 (Mirius) in place of PEI. Unless otherwise stated, cells were collected 48 h after transfection by scraping in 150 µl of lysis buffer (50 mM Tris, pH 8, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 10 mM N-ethylmaleimide, and 1× protease inhibitor [Sigma-Aldrich]), sonicated on ice for 30 s, and stored at −80°C.
Immunoblotting and quantification of protein expression and NA activity
Equal protein amounts of each transfected cell lysate were mixed with nonreducing or reducing (100 mM DTT) Laemmli sample buffer, heated to 37°C for 10 min, resolved by Tris-glycine SDS-PAGE, and transferred to a 0.45-µm pore PVDF membrane at 15 V for 60 min. Lysates containing NS2, RBD, RPS20, NA1–35, or NA1–49 were mixed with nonreducing or reducing (100 mM DTT) Tris-tricine sample buffer (Bis-Tris 360 mM, bicine 160 mM, 1% SDS, and 15% glycerol), resolved by 13% Tris-tricine SDS-PAGE, and transferred to a 0.2-µm pore PVDF membrane at 15 V for 30 min or 10 min for NA1–35. Primary antibodies used in the study are mouse anti-myc (9B11; Cell Signaling Technology), anti-NS1WSN, and anti-GFP (raised rabbit antisera to recombinant purified proteins expressed in E. coli). Cellular fractionations were validated with mouse antisera to cytosol (anti–β-tubulin; Sigma Aldrich) or rabbit antisera to ER (anticalnexin; HPA009433), ribosome (anti-RPS20; HPA003570), and nucleus (antilamin B1; HPA050524) markers. Band intensities of the indicated IAV proteins were quantified from the same blot and exposure conditions using Multi Gauge software (Fujifilm). The protein expression change was determined by normalizing intensities from the coexpressed samples to the independently expressed sample. Equal loading of total protein is shown with amido black staining of the PVDF membranes. For sialidase activity measurements, equal total protein amounts of transfected cell lysate were diluted to 195 µl using 37°C reaction buffer (0.1 M potassium phosphate, pH 6.0, and 1 mM CaCl2), after which 5 µl of 2 mM 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (Sigma-Aldrich) was added. The activity was monitored at 30-s intervals at 37°C in a SpectraMax Gemini EM (365-nm excitation and 450-nm emission; da Silva et al., 2013). Activity rates were determined using a methylumbelliferyl standard. The ratios were calculated by normalizing the rates from the coexpressed samples to samples where NA was independently expressed.
Real-time quantitative PCR analysis
293T cells were trypsinized 48 h after transfection, sedimented (2,000 g at 2 min), washed twice with PBS, pH 7.4, and then total RNA was isolated from the cells using the RNeasy mini kit (QIAGEN) with a DNase digestion step according to the manufacturer’s instructions. To separate cytoplasmic and nuclear RNA, cells were incubated on ice for 30 min with PBS containing 0.5% Triton X-100 and then sedimented (2,000 g at 2 min). The RNA was extracted from the supernatant (cytoplasmic fraction) and the pelleted nuclei separately. For all comparisons, cDNAs were generated for each sample with the iScript cDNA Synthesis kit (Bio-Rad Laboratories) by using 1 µg of total RNA and an oligo(dT) primer. To determine relative mRNA values, equal volumes of cDNA were mixed with iTaq universal SYBR green Supermix (Bio-Rad Laboratories) and gene-specific primers (NA forward, 5′-GAGAACACAAGAGTCTGAATGTAC-3′, and reverse, 5′-CTTCTCGATCTTGAAAATTTTGTACG-3′; HA forward, 5′-CCCAAAGCTGACCAATTCCTATGTG3-′, and reverse, 5′-GCTATTTCCGGGGTGAATCTCCTG-3′; and porphobilinogen-deaminase forward, 5′-CTGGTAACGGCAATGCGGCT-3′, and reverse, 5′-GCAGATGGCTCCGATGGTGA-3′) and then were analyzed with a CFX96 real-time system (Bio-Rad Laboratories) using Bio-Rad CFX Manager 3.1 software.
Imaging of live cells
Approximately 105 HeLa cells or 2 × 105 293T cells were cotransfected with plasmids expressing the indicated C-terminal GFP-tagged versions of NS1 and the ER membrane marker mCherry-Sec61β (Zurek et al., 2011) and then plated into one of the four chambers on a glass-bottomed 3.5-cm culture dish (Greiner Bio-One). At 48 h after transfection, regions from 1 µm below the nucleus to 1 µm above the nucleus were imaged in 500-nm steps using a 63× 1.4 numerical aperture oil emersion objective. Images were acquired using Zen software connected to an inverted microscope (LSM700; ZEISS) with 405-, 488-, and 555-nm solid-state lasers.
Subcellular fractionation
RBD-GFP and RBDAA-GFP were transfected into separate 10-cm dishes containing 6 × 106 293T cells. For infections, A/WSN/33 virus (multiplicity of infection, ∼0.5) was bound to 6 × 106 MDCK cells in a 10-cm dish at 4°C for 30 min using infection media (DMEM containing 0.1% FBS, 0.3% BSA, and 0.1% penicillin/streptomycin). After binding, the infection media was replaced, and the cells were shifted back to 37°C. 48 h after transfection or 6 h after infection, cells were washed with PBS, scraped in 500 µl homogenization buffer (50 mM triethanolamine, pH 7.5, 50 mM KOAc, 6 mM Mg(OAc)2, 0.25 M sucrose, and 1× protease inhibitor) and then homogenized by 45 strokes in a 2-ml dounce homogenizer on ice followed by 30 passages through a 27-gauge needle. The homogenate (total fraction) was centrifuged (1,000 g for 10 min) to isolate nuclei and unbroken cells (pellet) from the cytoplasm (supernatant). The cytoplasm was centrifuged (150,000 g for 1 h 30 min) at 4°C in a TLA100 rotor (Beckman Coulter) to separate cell membranes (pellet) from the cytosol (supernatant). Samples were retained from each fraction, and equal portions were analyzed by immunoblotting. Cell membrane fractions were further separated by density equilibrium centrifugation (250,000 g for 16 h) using a discontinuous 20–60% sucrose gradient (in homogenization buffer). The resulting fractions were concentrated by trichloroacetic acid (20%) precipitation, washed once with 95% acetone, and solubilized using sample buffer.
Sequence analysis
Unique human mRNAs (5′ UTRs, coding regions, and 3′ UTRs) encoding soluble secreted proteins (n = 3,589) or cytosolic proteins (n = 8,029) were obtained from the Ensembl database. To avoid extreme length-dependent nucleotide content calculations, mRNAs with 5′ UTR lengths from 1–10 nucleotides were excluded from the analysis (n = 104). Unique coding regions for predicted E. coli–exported proteins (n = 1,198), human-secreted proteins (n = 1,603), and type II human membrane proteins (n = 387) were all obtained from Swiss-Prot. Coding regions corresponding with the cleaved targeting sequences (SSs or signal peptides) were determined using the predicted signal peptidase cleavage sites (Bendtsen et al., 2004; Daniels et al., 2010), whereas the coding regions corresponding with the targeting sequences in type II membrane proteins (TMDs) were identified with the ΔG predictor using a 19–33-amino-acid window (Hessa et al., 2007; Dou et al., 2014). Sequences were grouped in 5% GC intervals for Fig. 5 B. Unique coding sequences for the H1N1 human IAV genes NA (n = 3,363), HA (n = 5,237), and M2 (n = 1,073) were all obtained from the NCBI Influenza Virus Resource. For calculating the IAV mRNA 5′ UTR lengths, sequences lacking the conserved sequence 5′-AGCAAAAGCAGG-3′ in the 5′ UTR were excluded, and we did not account for additional length caused by the 5′ cap addition.
Protein purification and RNA analysis
BL21 Rosetta 2 E.coli (Novagen) cells containing a pET21 expression vector encoding for RBDWSN with a C-terminal 6×His tag were grown to OD600 = 0.6 at 37°C and induced with 0.15 mM IPTG for 16 h at 20°C. Cells were sedimented, resuspended in Hepes–KOAc buffer (0.01 M Hepes, pH 8, 0.15 M KOAc, and 5 mM NaCl) supplemented with 0.1 mM MgCl2, 10 µg DNase, 100 µg of RNase A, and lysozyme. After incubation for 1 h at 25°C, cells were passed through an EmulsiFlex (Avestin) and sedimented, and the RBDWSN was extracted from the soluble fraction using a His-Trap HP column. The protein was dialyzed against Hepes–KOAc buffer containing 1 mM EDTA and 0.5 mM DTT and adjusted to a final concentration of ∼1 mg/ml. Size-exclusion chromatography was performed on an ÄKTA purifier using a Superdex 200 column (GE Healthcare) and Hepes–KOAc buffer. The protein was analyzed by blue-native PAGE using 4–16% bistricine gels (Invitrogen) according to the manufacturer’s instructions. The bound RNA was isolated using an RNeasy mini kit, and the size was determined by resolution on a 10% urea-PAGE gel stained with SYBR safe (Invitrogen).
mRNA synthesis, electrophoretic mobility shift assay, and in vitro translation
PCR products from the NAWSN pcDNA3.1A− vector that included ∼130 nucleotides upstream of the start codon were used as templates to transcribe the full-length (nucleotides 1–1,359) and short (nucleotides 1–405) NA mRNAs with the mMESSAGE mMACHINE T7 kit (Ambion). For electrophoretic mobility shift assays, mRNAs were labeled using the LabelIT fluorescein kit (Mirus) at a 1:5 dilution from the manufacturer’s instructions, and equivalent amounts of labeled RNA were incubated with RBD protein ranging from 0.5–50 pmol in a 10-µl reaction containing 5 U RNAsin (Promega), 1 mM Hepes, pH 8, and 5 mM KCl for 10 min at 37°C. The products were immediately resolved on a 1% TBE (89 mM Tris, pH 7.6, 89 mM boric acid, and 2 mM EDTA) agarose gel and imaged using a LAS1000 charge-coupled device camera (Fujifilm). The in vitro translations using microsomes were performed as previously described using 8-µl reactions (4.95 µl treated rabbit reticulocyte lysate, 0.25 µl 100 mM DTT, 0.2 µl 1 mM amino acid mixtures lacking methionine, 0.2 µl mCi/ml [35S]methionine, 0.4 µl 1 equivalent/µl nuclease-treated canine pancreatic microsomes, and 2 µl 150 ng/µl mRNA; Francis et al., 2002) supplemented with 2 µl of Hepes–KOAc buffer or 2 µl of purified RBD in Hepes–KOAc to obtain the indicated concentration. Translations were performed at 30°C for 30 min, mixed with 40 µl reducing Laemmli sample buffer, and resolved by SDS-PAGE. The gels were dried and developed using a phosphor imager (FLA-9000; Fujifilm), and the [35S]-labeled protein was quantified with Multi Gauge V3.0 software.
Online supplemental material
Fig. S1 shows additional NS1 properties that affect NA regulation (time of cotransfection, cell line dependence, and the ED) and shows that the regulation by NS1 is retained in the presence of a C-terminal GFP tag. Fig. S2 shows the nucleotide sequences of the synonymous NA TMD and HA SS substitutions and also shows that NA regulation is dependent on cell growth and temperature and independent of the unfolded protein response and proteasomal degradation. Fig. S3 shows the properties of the recombinant NS1 RBD (isolated from E. coli), which was found to bind NA mRNA and stimulate NA synthesis in vitro; it also has data for human mRNAs with 5′ UTRs that are >300 nucleotides, which complements the nucleotide analysis in Fig. 5 C.
Supplementary Material
Acknowledgments
We thank Robert Webster for IAV RNA used to isolate the NS1 genes and the NA1997 expression plasmid, and Tara Hessa, Jan-Willem de Gier, IngMarie Nilsson, and Gunnar von Heijne for providing reagents and helpful discussions.
This work was supported by grants from the Swedish Research Council, Carl Trygger Foundation, and Harald Jeanssons Stiftelse to R. Daniels as well as the Royal Swedish Academy of Sciences, Lindhés Advokatbyrå AB, and the Sven and Lilly Lawskis Fund to J. Nordholm.
The authors declare no competing financial interests.
Author contributions: R. Daniels and J. Nordholm conceived and wrote the study with input from all other authors. J. Nordholm, J. Petitou, H. Östbye, D.V. da Silva, D. Dou, H. Wang, and R. Daniels performed experiments and interpreted data.
Footnotes
Abbreviations used:
- ED
- effector domain
- IAV
- influenza A virus
- NA
- neuraminidase
- RBD
- RNA-binding domain
- SRP
- signal recognition particle
- SS
- signal sequence
- TMD
- transmembrane domain
References
- Aragón T., de la Luna S., Novoa I., Carrasco L., Ortín J., and Nieto A.. 2000. Eukaryotic translation initiation factor 4GI is a cellular target for NS1 protein, a translational activator of influenza virus. Mol. Cell. Biol. 20:6259–6268. 10.1128/MCB.20.17.6259-6268.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreau C., Paillard L., and Osborne H.B.. 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 33:7138–7150. 10.1093/nar/gki1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen J.D., Nielsen H., von Heijne G., and Brunak S.. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795. 10.1016/j.jmb.2004.05.028 [DOI] [PubMed] [Google Scholar]
- Blobel G., and Dobberstein B.. 1975. Transfer of proteins across membranes. I. Presence of proteolytically processed and unprocessed nascent immunoglobulin light chains on membrane-bound ribosomes of murine myeloma. J. Cell Biol. 67:835–851. 10.1083/jcb.67.3.835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos T.J., Davis A.R., and Nayak D.P.. 1984. NH2-terminal hydrophobic region of influenza virus neuraminidase provides the signal function in translocation. Proc. Natl. Acad. Sci. USA. 81:2327–2331. 10.1073/pnas.81.8.2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmody S.R., and Wente S.R.. 2009. mRNA nuclear export at a glance. J. Cell Sci. 122:1933–1937. 10.1242/jcs.041236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chlanda P., Schraidt O., Kummer S., Riches J., Oberwinkler H., Prinz S., Kräusslich H.G., and Briggs J.A.. 2015. Structural analysis of the roles of influenza A virus membrane-associated proteins in assembly and morphology. J. Virol. 89:8957–8966. 10.1128/JVI.00592-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniels R., Kurowski B., Johnson A.E., and Hebert D.N.. 2003. N-linked glycans direct the cotranslational folding pathway of Influenza hemagglutinin. Mol. Cell. 11:79–90. 10.1016/S1097-2765(02)00821-3 [DOI] [PubMed] [Google Scholar]
- Daniels R., Mellroth P., Bernsel A., Neiers F., Normark S., von Heijne G., and Henriques-Normark B.. 2010. Disulfide bond formation and cysteine exclusion in gram-positive bacteria. J. Biol. Chem. 285:3300–3309. 10.1074/jbc.M109.081398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Silva D.V., Nordholm J., Madjo U., Pfeiffer A., and Daniels R.. 2013. Assembly of subtype 1 influenza neuraminidase is driven by both the transmembrane and head domains. J. Biol. Chem. 288:644–653. 10.1074/jbc.M112.424150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Luna S., Fortes P., Beloso A., and Ortín J.. 1995. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J. Virol. 69:2427–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou D., da Silva D.V., Nordholm J., Wang H., and Daniels R.. 2014. Type II transmembrane domain hydrophobicity dictates the cotranslational dependence for inversion. Mol. Biol. Cell. 25:3363–3374. 10.1091/mbc.E14-04-0874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enami K., Sato T.A., Nakada S., and Enami M.. 1994. Influenza virus NS1 protein stimulates translation of the M1 protein. J. Virol. 68:1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis E., Daniels R., and Hebert D.N.. 2002. Analysis of protein folding and oxidation in the endoplasmic reticulum. Curr. Protoc. Cell Biol. 15:16 10.1002/047114303.cb1506s14 [DOI] [PubMed] [Google Scholar]
- Gerstberger S., Hafner M., and Tuschl T.. 2014. A census of human RNA-binding proteins. Nat. Rev. Genet. 15:829–845. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R., Walter P., and Blobel G.. 1982. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J. Cell Biol. 95:470–477. 10.1083/jcb.95.2.470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan D., Palese P., and Krystal M.. 1988. Two nuclear location signals in the influenza virus NS1 nonstructural protein. J. Virol. 62:3020–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale B.G., Randall R.E., Ortín J., and Jackson D.. 2008. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 89:2359–2376. 10.1099/vir.0.2008/004606-0 [DOI] [PubMed] [Google Scholar]
- Heaton N.S., Moshkina N., Fenouil R., Gardner T.J., Aguirre S., Shah P.S., Zhao N., Manganaro L., Hultquist J.F., Noel J., et al. . 2016. Targeting viral proteostasis limits influenza virus, HIV, and dengue virus infection. Immunity. 44:46–58. (published erratum appears in Immunity 44:438) 10.1016/j.immuni.2015.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helder S., Blythe A.J., Bond C.S., and Mackay J.P.. 2016. Determinants of affinity and specificity in RNA-binding proteins. Curr. Opin. Struct. Biol. 38:83–91. 10.1016/j.sbi.2016.05.005 [DOI] [PubMed] [Google Scholar]
- Hessa T., Meindl-Beinker N.M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S.H., and von Heijne G.. 2007. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 450:1026–1030. 10.1038/nature06387 [DOI] [PubMed] [Google Scholar]
- Hoffmann E., Neumann G., Kawaoka Y., Hobom G., and Webster R.G.. 2000. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc. Natl. Acad. Sci. USA. 97:6108–6113. 10.1073/pnas.100133697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull J.D., Gilmore R., and Lamb R.A.. 1988. Integration of a small integral membrane protein, M2, of influenza virus into the endoplasmic reticulum: analysis of the internal signal-anchor domain of a protein with an ectoplasmic NH2 terminus. J. Cell Biol. 106:1489–1498. 10.1083/jcb.106.5.1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R.J., Hellen C.U., and Pestova T.V.. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11:113–127. 10.1038/nrm2838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.W., Rane N.S., Kim S.J., Garrison J.L., Taunton J., and Hegde R.S.. 2006. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 127:999–1013. 10.1016/j.cell.2006.10.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamyshev A.L., Patrick A.E., Karamysheva Z.N., Griesemer D.S., Hudson H., Tjon-Kon-Sang S., Nilsson I., Otto H., Liu Q., Rospert S., et al. . 2014. Inefficient SRP interaction with a nascent chain triggers a mRNA quality control pathway. Cell. 156:146–157. 10.1016/j.cell.2013.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami E., Watanabe T., Fujii K., Goto H., Watanabe S., Noda T., and Kawaoka Y.. 2011. Strand-specific real-time RT-PCR for distinguishing influenza vRNA, cRNA, and mRNA. J. Virol. Methods. 173:1–6. 10.1016/j.jviromet.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M., Wan Y., Mazor E., Rinn J.L., Nutter R.C., Chang H.Y., and Segal E.. 2010. Genome-wide measurement of RNA secondary structure in yeast. Nature. 467:103–107. 10.1038/nature09322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzchalia T.V., Wiedmann M., Girshovich A.S., Bochkareva E.S., Bielka H., and Rapoport T.A.. 1986. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 320:634–636. 10.1038/320634a0 [DOI] [PubMed] [Google Scholar]
- Lamb R.A., and Choppin P.W.. 1976. Synthesis of influenza virus proteins in infected cells: Translation of viral polypeptides, including three P polypeptides, from RNA produced by primary transcription. Virology. 74:504–519. 10.1016/0042-6822(76)90356-1 [DOI] [PubMed] [Google Scholar]
- Lee A.S., Kranzusch P.J., and Cate J.H.. 2015. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature. 522:111–114. 10.1038/nature14267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L., Li Y., Pyo H.M., Lu X., Raman S.N., Liu Q., Brown E.G., and Zhou Y.. 2012. Identification of RNA helicase A as a cellular factor that interacts with influenza A virus NS1 protein and its role in the virus life cycle. J. Virol. 86:1942–1954. 10.1128/JVI.06362-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellroth P., Daniels R., Eberhardt A., Rönnlund D., Blom H., Widengren J., Normark S., and Henriques-Normark B.. 2012. LytA, major autolysin of Streptococcus pneumoniae, requires access to nascent peptidoglycan. J. Biol. Chem. 287:11018–11029. 10.1074/jbc.M111.318584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor A., White A., Zhang K., Thompson M., Esparza M., Muñoz-Moreno R., Koide K., Lynch K.W., García-Sastre A., and Fontoura B.M.. 2016. Influenza virus mRNA trafficking through host nuclear speckles. Nat. Microbiol. 1:16069 10.1038/nmicrobiol.2016.69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer S.A., Kidwell M.A., and Doudna J.A.. 2014. Insights into RNA structure and function from genome-wide studies. Nat. Rev. Genet. 15:469–479. 10.1038/nrg3681 [DOI] [PubMed] [Google Scholar]
- Muckenthaler M., Gray N.K., and Hentze M.W.. 1998. IRP-1 binding to ferritin mRNA prevents the recruitment of the small ribosomal subunit by the cap-binding complex eIF4F. Mol. Cell. 2:383–388. 10.1016/S1097-2765(00)80282-8 [DOI] [PubMed] [Google Scholar]
- Nordholm J., da Silva D.V., Damjanovic J., Dou D., and Daniels R.. 2013. Polar residues and their positional context dictate the transmembrane domain interactions of influenza A neuraminidases. J. Biol. Chem. 288:10652–10660. 10.1074/jbc.M112.440230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.W., and Katze M.G.. 1995. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J. Biol. Chem. 270:28433–28439. [DOI] [PubMed] [Google Scholar]
- Park Y.W., Wilusz J., and Katze M.G.. 1999. Regulation of eukaryotic protein synthesis: Selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1. Proc. Natl. Acad. Sci. USA. 96:6694–6699. 10.1073/pnas.96.12.6694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X.Y., Chien C.Y., Lu Y., Montelione G.T., and Krug R.M.. 1995. An amino-terminal polypeptide fragment of the influenza virus NS1 protein possesses specific RNA-binding activity and largely helical backbone structure. RNA. 1:948–956. [PMC free article] [PubMed] [Google Scholar]
- Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X., Gueroussov S., Albu M., Zheng H., Yang A., et al. . 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 499:172–177. 10.1038/nature12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S.M., Wheelan S.J., Yarrington R.M., and Boeke J.D.. 2006. GeneDesign: Rapid, automated design of multikilobase synthetic genes. Genome Res. 16:550–556. 10.1101/gr.4431306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro G.I., Gurney T. Jr., and Krug R.M.. 1987. Influenza virus gene expression: control mechanisms at early and late times of infection and nuclear-cytoplasmic transport of virus-specific RNAs. J. Virol. 61:764–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel J.J. 1973. Early polypeptide synthesis in influenza virus-infected cells. Virology. 56:394–399. 10.1016/0042-6822(73)90320-6 [DOI] [PubMed] [Google Scholar]
- Treanor J.J., Snyder M.H., London W.T., and Murphy B.R.. 1989. The B allele of the NS gene of avian influenza viruses, but not the A allele, attenuates a human influenza A virus for squirrel monkeys. Virology. 171:1–9. 10.1016/0042-6822(89)90504-7 [DOI] [PubMed] [Google Scholar]
- Vester D., Lagoda A., Hoffmann D., Seitz C., Heldt S., Bettenbrock K., Genzel Y., and Reichl U.. 2010. Real-time RT-qPCR assay for the analysis of human influenza A virus transcription and replication dynamics. J. Virol. Methods. 168:63–71. 10.1016/j.jviromet.2010.04.017 [DOI] [PubMed] [Google Scholar]
- Walter P., and Blobel G.. 1981. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 91:557–561. 10.1083/jcb.91.2.557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., and Johnson A.E.. 1994. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu. Rev. Cell Biol. 10:87–119. 10.1146/annurev.cb.10.110194.000511 [DOI] [PubMed] [Google Scholar]
- Walter P., Ibrahimi I., and Blobel G.. 1981. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 91:545–550. 10.1083/jcb.91.2.545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Riedel K., Lynch P., Chien C.Y., Montelione G.T., and Krug R.M.. 1999. RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA. 5:195–205. 10.1017/S1355838299981621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek N., Sparks L., and Voeltz G.. 2011. Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic. 12:28–41. 10.1111/j.1600-0854.2010.01134.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.