How are membrane proteins in distal dendrites degraded by the lysosome? Goo et al. provide the first evidence that lysosomes are positioned locally at dendritic spines in an activity-dependent manner to facilitate the remodeling of synapses through local degradation.
Abstract
In neurons, lysosomes, which degrade membrane and cytoplasmic components, are thought to primarily reside in somatic and axonal compartments, but there is little understanding of their distribution and function in dendrites. Here, we used conventional and two-photon imaging and electron microscopy to show that lysosomes traffic bidirectionally in dendrites and are present in dendritic spines. We find that lysosome inhibition alters their mobility and also decreases dendritic spine number. Furthermore, perturbing microtubule and actin cytoskeletal dynamics has an inverse relationship on the distribution and motility of lysosomes in dendrites. We also find trafficking of lysosomes is correlated with synaptic α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid–type glutamate receptors. Strikingly, lysosomes traffic to dendritic spines in an activity-dependent manner and can be recruited to individual spines in response to local activation. These data indicate the position of lysosomes is regulated by synaptic activity and thus plays an instructive role in the turnover of synaptic membrane proteins.
Introduction
Neurons are highly polarized cells that have processes spanning tens to hundreds of microns away from the cell body. Dendrites, which receive thousands of synaptic inputs, are capable of synthesizing proteins locally at synapses to maintain normal function (Steward and Schuman, 2003; Alvarez-Castelao and Schuman, 2015). Other cellular components such as mitochondria (Chang et al., 2006), Golgi apparatus (Horton and Ehlers, 2003), and recycling endosomes (Park et al., 2006) have been found in distal dendrites, further suggesting that dendrites can control protein homeostasis independently of the cell body.
Degradation, an important process in maintaining cellular homeostasis, mainly occurs through the proteasome or the lysosome. Although there is a growing body of evidence that indicates the proteasome is an important mediator of activity-dependent remodeling (Bingol and Schuman, 2006; Djakovic et al., 2009, 2012; Bingol et al., 2010; Hamilton et al., 2012), little is known about the regulation and function of lysosomes in dendrites. In fact, until recently, it was thought that dendrites were devoid of lysosomes and that lysosomes were predominantly located in the cell body and axons of neurons (Parton et al., 1992). How then are membrane proteins degraded in distal dendrites? Given the importance of this organelle in membrane protein turnover, it is plausible that lysosomes exist in dendrites and that their trafficking in dendrites is tightly regulated.
The lysosome, a membrane-bound degradative organelle, functions downstream of the endosomal sorting complexes required for transport (ESCRT) pathway to degrade internalized membrane proteins (Henne et al., 2011). The autolysosome, created via the fusion of an autophagosome and a lysosome, is a key player in autophagy (Klionsky and Emr, 2000). The relevance of autophagy in health and disease has dominated the field (Shintani and Klionsky, 2004; Wong and Cuervo, 2010). However, the neuron's elaborate morphology creates a layer of complexity related to the trafficking and function of lysosomes. Interestingly, lysosomal trafficking has primarily been studied in axons and somatic compartments. Until fairly recently, the lysosome was previously thought to be a passive molecular “incinerator” blinded to its surroundings. However, recent studies show that the lysosome can actually play an instructive role in maintaining cellular homeostasis. For example, the lysosome has been shown to serve as a signaling organelle that can sense nutrient availability and regulate energy metabolism (Settembre et al., 2013). Furthermore, we and others previously showed that lysosomes degrade α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPARs) in an activity-dependent manner (Ehlers, 2000; Schwarz et al., 2010), which is important for homeostatic downscaling and amyloid-β–induced loss of AMPARs (Scudder et al., 2014; Rodrigues et al., 2016). However, it remains unclear if lysosomes degrade synaptic membrane cargo exclusively in the cell body or locally at or near synapses in dendrites.
Here, we sought to determine if the trafficking of lysosomes is regulated by neuronal activity to facilitate local protein degradation of membrane proteins at synapses. We found that the distribution and trafficking of lysosomes is highly correlated with synaptic proteins, including AMPARs, and that the trafficking of lysosomes into dendritic spines is regulated by synaptic activity. Moreover, synaptic activation of a single spine could recruit a lysosome to the base of that spine. These results provide the first evidence that lysosomes are positioned locally at dendritic spines in an activity-dependent manner to facilitate the remodeling of synapses through local degradation.
Results
Lysosomes are found in dendrites and dendritic spines
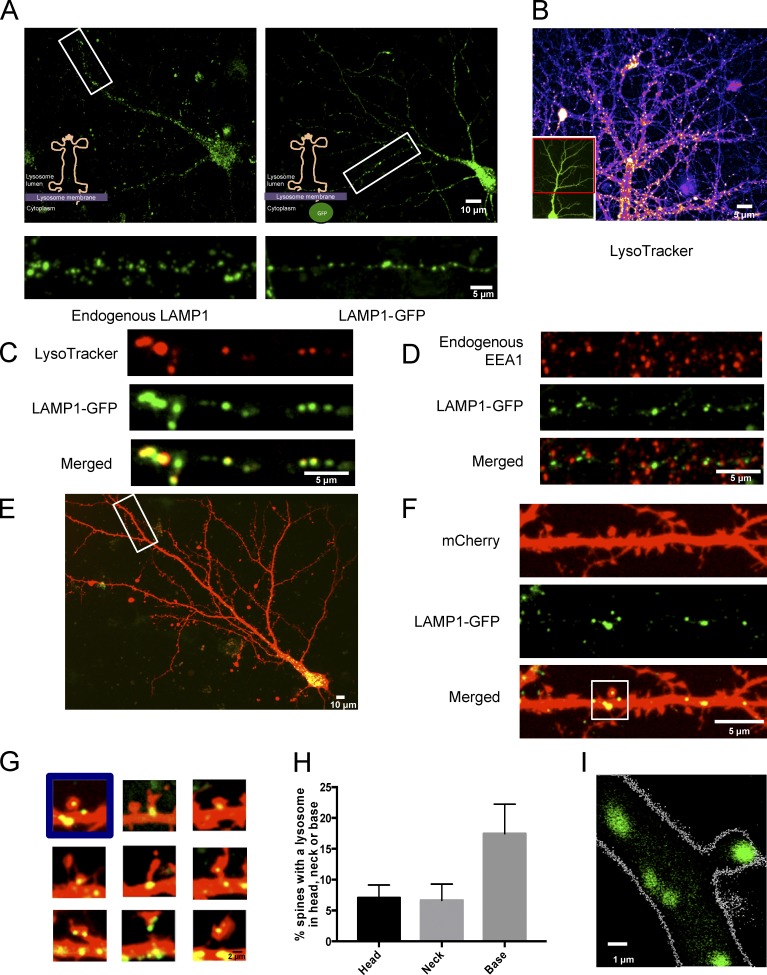
To determine the spatial distribution of lysosomes in dendrites, we performed immunocytochemistry to detect endogenous LAMP1, a membrane protein found on late endosomes and lysosomes, in dissociated hippocampal cultures and imaged using confocal microscopy. LAMP1-positive structures were detected in the soma and throughout primary and secondary dendrites (Fig. 1 A). However, because of the nature of dissociated hippocampal mixed cultures, surrounding glia cells also contain lysosomes and were frequently overlapping neurons, making it difficult distinguish between endogenous LAMP1 in neurons versus glia. To limit our analysis to neuronal lysosomes, we transfected hippocampal cultures with LAMP1-GFP for less than 24 h. The low transfection efficiency allowed us to distinguish LAMP1-GFP expression in neurons versus glia. We found the distribution of LAMP1-GFP was similar to the endogenous staining of LAMP1 (Fig. 1 A).
Figure 1.
Lysosomes are reliably labeled by LAMP1-GFP and are found in distal dendrites and dendritic spines. (A) Endogenous LAMP1 distribution is similar to LAMP1-GFP, as both labeled structures were found in somatic and dendritic compartments. Representative images of dissociated hippocampal neurons (DIV16) either stained for endogenous LAMP1 (left) or transfected with LAMP1-GFP (right) and allowed to express for ≤24 h. (B) Representative image of LysoTracker red–labeled acidic vesicles found in dendritic compartments (DIV16). Inset shows entire cell image of neuron expressing GFP (Sindbis virus). (C) LAMP1-GFP marks acidic vesicles. Representative image of a dendrite from a dissociated hippocampal neuron transfected with LAMP1-GFP and costained with LysoTracker red, which marks acidic compartments. Neurons were imaged live at DIV16. (D) LAMP1-GFP does not colocalize with early endosomes. Representative image of a dendrite from dissociated hippocampal neuron (DIV16) transfected with mCherry and LAMP1-GFP and stained for the endogenous early endosomal protein marker EEA1. (E and F) LAMP1-GFP–positive vesicles are found in distal dendrites. A straightened secondary distal dendrite from a dissociated hippocampal neuron (DIV16) with LAMP1-GFP present throughout the dendrite. Depicted are a representative whole-cell image (E) and straightened distal secondary dendrite (F). (G) LAMP1-GFP is present at the base, neck, and head of dendritic spines. Representative images of dendritic spines in cultured hippocampal neurons (DIV16) transfected with mCherry and LAMP1-GFP. (H) Quantification the percentage of spines with a lysosome either in the head, neck, or base of the spine. Data represent means ± SEM. (I) Representative two-photon image of a CA1 pyramidal neuron from a hippocampal organotypic (DIV8) slice biolistically transfected with LAMP1-GFP and dsRed. LAMP1-GFP–labeled lysosomes are found in dendritic spines.
To evaluate if these LAMP1-positive structures were acidic, we labeled neurons with LysoTracker red, a dye that labels acidic organelles. Consistent with endogenous LAMP1 immunostaining and transfection with LAMP1-GFP, we found LysoTracker-positive structures throughout the dendrites (Fig. 1 B and Video 1). Importantly, LAMP1-GFP–positive structures were also acidic as seen by colocalization with LysoTracker (Fig. 1 C). Furthermore, LAMP1-GFP did not colocalize with EEA1, a marker for early endosomes (Fig. 1 D).
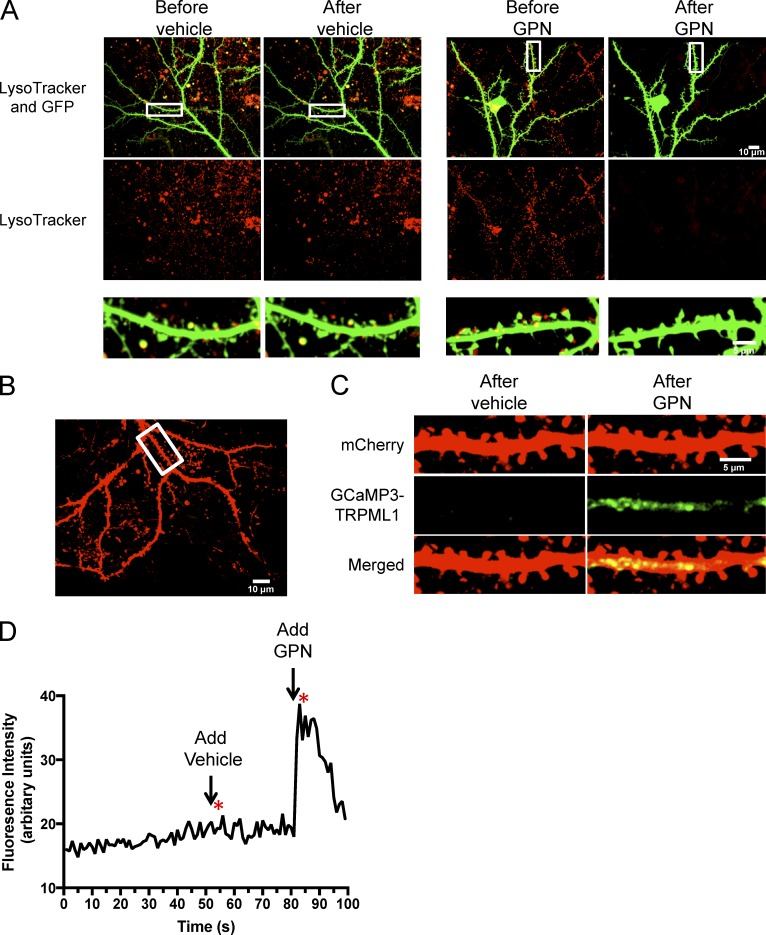
To further evaluate lysosomal distribution in dendrites, cells expressing GFP were incubated with LysoTracker and then treated with 40 µM glycyl-l-phenylalanine 2-naphthylamide (GPN; 5 min). GPN is a substrate of Cathepsin C, which is only found in lysosomes. When GPN is cleaved by cathepsin C, it induces osmotic lysis of lysosomes (Shen et al., 2012). We found that treatment with GPN diminishes LysoTracker fluorescence in neuronal dendrites compared to cells treated with vehicle only (Fig. 2 A). To further determine if these were active lysosomes, we investigated whether lysosomes in dendrites release calcium (Ca2+) upon GPN treatment. It is known that Ca2+ efflux from lysosomes is primarily through mucolipin transient receptor potential channel 1 (TRPML1; Wang et al., 2014). We therefore transfected cells with mCherry and GCaMP3-TRPML1 in primary neurons and measured the GCaMP3 signal to monitor Ca2+ release after GPN treatment. The application of vehicle (DMSO) did not increase TRPML1-GCamP3 fluorescence in dendrites, however the addition of 40 µM GPN dramatically increased TRPML1-GCaMP3 fluorescence suggesting that Ca2+ was released from active lysosomes (Fig. 2, B–D; and Video 2). Therefore, we conclude that functional lysosomes are found in distal dendrites of neurons.
Figure 2.
Active lysosomes are found in distal dendrites. (A) Lysosome membrane disruption by GPN abolishes LysoTracker staining. Representative dissociated hippocampal neurons expressing GFP (Sindbis virus) and costained with LysoTracker. Images were taken before adding either vehicle (DMSO) or 40 µM GPN. Images were taken 5 min after either vehicle or GPN. Regions of interest (boxes) are magnified below. (B) Representative neuron transfected with mCherry and GCaMP3-TRPML1. Neuron was imaged live for 100 s in the 488 and 568 channel. (C) Straightened segment from B showing GCaMP3-TRPML1 signal 2 s after adding either vehicle or 40 µM GPN. (D) Detection of calcium release from lysosome stores by GCaMP3-TRPML1 in representative neuron transfected with mCherry and GCaMP3-TRPML1 from B and C. Vehicle was added 50 s after the start of the recording, and 40 µM GPN was added 80 s after the start of the recording. Asterisks show time where images from D were taken.
We next wanted to determine the spatial distribution of lysosomes in relation to dendritic spines. Dissociated neurons were cotransfected with LAMP1-GFP to mark lysosomes and mCherry to visualize cellular morphology. We found lysosomes not only in the cell body and dendrites (Fig. 1 E) but also in dendritic spines (Fig. 1, F and G). Upon quantification, ∼7% of spines have a lysosome in the head of the spine (Fig. 1 H). We also found that LysoTracker-positive LAMP1-GFP organelles were located in dendritic spines (Fig. 2 A). To examine lysosome distribution in the context of intact circuits, organotypic hippocampal slices were biolistically transfected with LAMP1-GFP and dsRed and imaged live with two-photon microscopy. Again, we found a subset of dendritic spines with a lysosome in the head of the spine (Fig. 1 I).
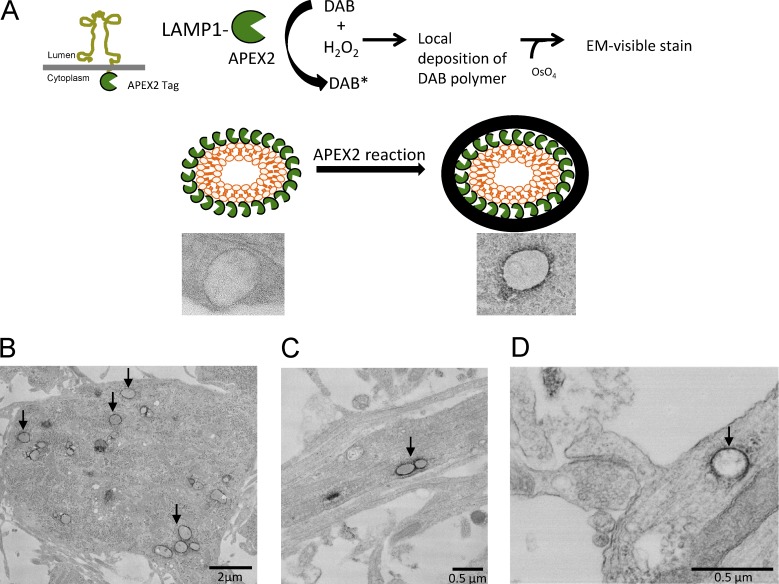
To obtain higher resolution images of lysosomes at or near a dendritic spine, we used EM in combination with APEX2 technology. APEX2 is an engineered peroxidase that can be used for imaging and localizing specific proteins by EM (Lam et al., 2015). APEX2 was cloned onto the C terminus of LAMP1, which places APEX2 on the cytoplasmic side of the lysosomal membrane. Dissociated hippocampal cultures were then transfected with LAMP1-APEX2. The cells were then fixed with glutaraldehyde ≤24 h later and treated with DAB and hydrogen peroxide to produce osmium-sensitive precipitates. Because APEX2 is tagged at the C-terminal domain of LAMP1, we expected a dark stain around the membrane of the lysosome on the cytoplasmic side (Fig. 3 A). To verify our expectations, we performed EM imaging, which showed DAB deposits exclusively on the perimeter of electron-dense vesicular structures. In neurons transfected with LAMP1-APEX2, we observed DAB-labeled structures in both neuronal cell bodies (Fig. 3 B) and dendrites (Fig. 3 C). Importantly, DAB-stained lysosomes were also found near the base of dendritic spines (Fig. 3 D). Together, these data indicate that LAMP1 is a specific marker for lysosomes and that lysosomes are found in dendrites and near dendritic spines.
Figure 3.
APEX2 technology for EM shows ectopic expression of LAMP1 is specific to lysosomes. (A) Schematic for how APEX2 is used to label lysosomes by EM. APEX2 was cloned on the cytoplasmic side of LAMP1 (LAMP1-APEX2) which allowed for the labeling (with minimal spread of the DAB reaction) of the outside perimeter of intact lysosomes. (B and C) Representative transmission EM images of cultured hippocampal neurons (DIV16) transfected with LAMP1-APEX2. APEX2-stained lysosomes are present in the cell body and in dendrites of neurons. (D) LAMP1-APEX2–labeled lysosome found near dendritic spines. EM image of lysosomes near a base of a dendritic spine. Arrows point to LAMP1-APEX2–positive structures.
Lysosomal inhibition alters lysosome trafficking in dendrites and decreases synapse number
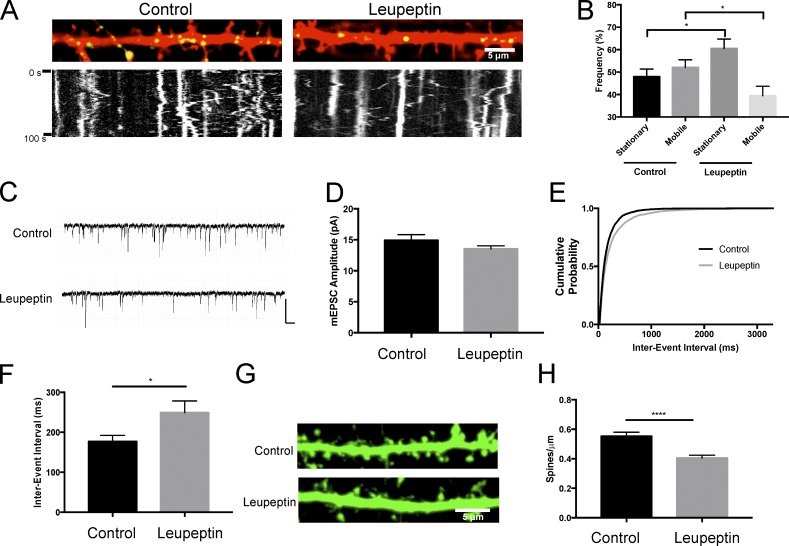
We then wanted to understand the relationship between lysosome function and its motility. To do this, we treated neurons transfected with LAMP1-GFP with 200 µM leupeptin (3 h) to inhibit proteases contained within the lysosome and then quantified lysosomal motility in dendrites. Under control conditions, approximately half of the lysosomes were stationary (47 ± 3.4%), whereas the other half moved bidirectionally (Fig. 4, A and B; and Video 3). However, upon lysosomal inhibition, lysosomes became increasingly immobile (60.5 ± 4.2%; Fig. 4, A and B; and Video 4). We also found that loss of motility was not caused by a compromise in cell health or cytoskeletal integrity in the cells treated with 200 µM leupeptin for 3, 6, and 12 h (Fig. S1). This suggests that the activity of lysosomes is related to their motility.
Figure 4.
Lysosomal inhibition alters lysosome trafficking and decreases dendritic spine density. (A) Lysosomal inhibition decreases lysosome trafficking in hippocampal dendrites. Representative live images of secondary dendrites from cultured hippocampal neurons (DIV16) transfected with mCherry and LAMP1-GFP with the corresponding kymograph below. Cultures were treated with 200 µM leupeptin for 3 h. Images represent the first image in the time-lapse sequence. Live images were taken every second for 100 s. Vertical lines in kymographs represent stationary structures. (B) Quantification of LAMP1-GFP movement in dendrites from kymographs represented in A. Movement was manually counted in a blinded fashion. 306 (control) and 245 (leupeptin) vesicles from n = 23 and 19 dendrites for control and leupeptin, respectively. *, P < 0.05 between stationary groups; *, P < 0.05 between mobile, unpaired Student’s t test. Data represent mean ± SEM. Experimenter was blinded to condition upon analysis. (C–F) Lysosomal inhibition decreases mEPSC frequency and dendritic spine density. (C) Representative traces of mEPSCs recorded from control and 200 µM leupeptin (2–4 h)–treated cultured hippocampal neurons (DIV 18–24); n = 25 and 31 cells for control and leupeptin, respectively; mean mEPSC amplitude (D); cumulative probability distribution of interevent interval (IEI) of all mEPSCs record from control and leupeptin-treated neurons n = 3679 and 4552 events, respectively (E); inter-event interval (*, P < 0.05 unpaired Student’s t test). (F). Data represent mean ± SEM. Bars: 200 ms; (traces) 20 pA. Experimenter was blinded to condition upon analysis. (G) Representative straightened dendrites after control and 200 µM leupeptin (3 h)–treated cultured hippocampal neurons (DIV15 to DIV16) expressing GFP via Sindbis viral transduction (16 h). (H) Quantification of spine density from conditions displayed in G. ****, P < 0.0001 unpaired Student’s t test. Data represent mean ± SEM with ≥49 dendrites quantified per treatment. Experimenter was blinded to condition upon analysis.
We next examined the effects of altered lysosomal function on basal synaptic transmission. To assess this, we recorded miniature excitatory postsynaptic currents (mEPSCs) from control or 200 µM leupeptin (3 h)–treated dissociated hippocampal neurons. Relative to control neurons, we detected no significant change in mEPSC amplitude, suggesting synaptic strength is unchanged when lysosomes are inhibited (control, amplitude: 14.9 ± 0.9 pA; leupeptin, amplitude: 13.5 ± 0.5 pA; P = 0.19; Fig. 4, C and D). However, leupeptin caused a significant increase in mEPSC interevent interval (IEI; control: 176.9 ± 15.5 ms; leupeptin: 249.2 ± 29.2 ms; P < 0.05; Fig. 4, C, E, and F), which is a 29.0% increase in IEI. Decreased mEPSC frequency in many instances indicates an alteration in presynaptic function but is also a result of decreased synapse number (Turrigiano and Nelson, 2004). Because the majority of excitatory synapses are formed on dendritic spines (Harris, 1999), we quantified dendritic spine number as a proxy for excitatory synapse density to determine if inhibiting lysosomal function altered excitatory synapse number. To visualize neuronal morphology, we expressed GFP in hippocampal neurons by Sindbis virus. We observed a significant decrease in the number of spines after treatment with leupeptin (control: 0.55 ± 0.03 spines/µm; leupeptin: 0.41 ± 0.01 spines/µm; P < 0.0001; Fig. 4, G and H). Interesting, we found that the magnitude of the change in IEI (29.0% increase) and spine density (25.4% decrease) in the presence of leupeptin to be strikingly similar. Collectively, these data indicate that lysosomal inhibition increases mEPSC IEI through loss of excitatory synapses.
Altered microtubule and actin cytoskeletal dynamics inversely affect lysosomal trafficking in dendrites
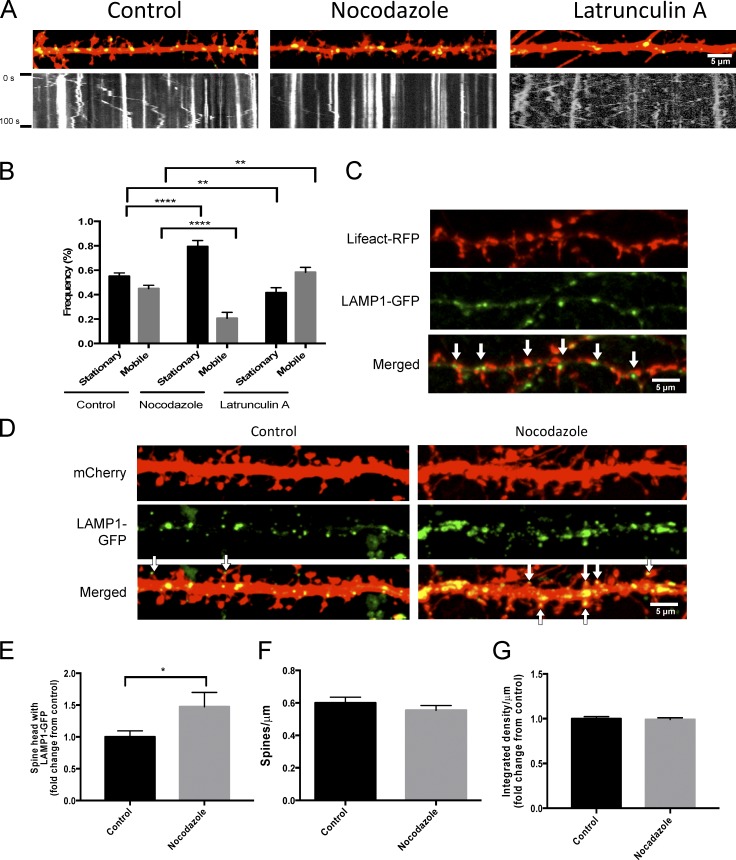
Because we observed that lysosomes move bidirectionally in dendrites and that they can enter dendritic spines, we next investigated how lysosomal motility is controlled in dendrites. Dynein and kinesin-1 and kinesin-2, minus-end– and plus-end–directed microtubule motors, respectively, have been known to associate with late endosomes and lysosomes (Hendricks et al., 2010; Maday et al., 2014). Interactions between lysosomes at both plus- and minus-end motors can partly account for the lysosomes bidirectional motility through a stochastic tug of war (Bananis et al., 2004; Brown et al., 2005; Loubéry et al., 2008). Furthermore, it is thought that kinesin-2 is the primary anterograde motor for Rab7-positive late endosome/lysosome motility in axons. (Hendricks et al., 2010). We decided to examine how lysosomes travel through the complex cytoskeletal environment in dendrites and at or near dendritic spines where actin and microtubules networks interact. To do this, we treated neurons transfected with LAMP1-GFP and mCherry with either 10 µg/ml nocodazole for 1 h, which destabilizes microtubules, or 20 µM latrunculin A for 10 min, which inhibits actin polymerization and disrupts F-actin in spines. As expected, we found that destabilization of microtubules with nocodazole significantly increases the amount of stationary lysosomes compared with control conditions (control: 55 ± 2.7%; nocodazole: 79 ± 5.0%; P < 0.0001; Fig. 5, A and B; and Video 5). However, to our surprise, inhibition of actin polymerization led to a significant increase in lysosomal trafficking, as demonstrated by an increased percentage of mobile lysosomes (control: 44 ± 2.8%; latrunculin: 58 ± 4.1%; P < 0.01; Fig. 5, A and B; and Video 6). Interestingly we found that LAMP1-GFP-labeled lysosomes were codistributed with Lifeact–RFP, which marks F-actin (Fig. 5 C), suggesting that F-actin may be involved in positioning lysosomes near dendritic spines and synapses.
Figure 5.
Perturbations in microtubule and actin dynamics alter trafficking of lysosome. (A and B) Disruption of microtubule and actin dynamics inversely affects lysosome trafficking. (A) Representative images of cultured neurons under control conditions, after treatment with 10 µg/ml nocodazole (1 h) or 20 µM latrunculin A (10 min) with respective kymographs. Straightened dendrites represent the first image in the time-lapse sequence. Live images were taken every second for 100 s. (B) Quantification of LAMP1-GFP movement after nocodazole or latrunculin A treatment from Fig. 4 A. Approximately 435 vesicles from 36, 15, and 24 dendrites in control, nocodazole-, and latrunculin A–treated cells, respectively, were analyzed from four independent experiments. ****, P < 0.0001; **, P < 0.01, unpaired Student’s t test. Data represent mean ± SEM. Experimenter was blinded to condition upon analysis. (C) Representative images of dissociated hippocampal cell expressing LAMP1-GFP and Lifeact-RFP. Arrows point to LAMP1-GFP juxtaposed to Lifeact-RFP. (D–G) Disruption of microtubule dynamics with nocodazole increases lysosomes in dendritic spines. (D) Representative image of a secondary dendrite from a cultured hippocampal neuron (DIV16) transfected with mCherry and LAMP1-GFP under control or 10 µg/ml nocodazole treatment (1 h). Arrows point to LAMP1-GFP in a dendritic spine. (E–G) Quantification of the percentage of spines that have LAMP1-GFP in the head of a spine (E). Number of spines per micrometer (F) and signal intensity of LAMP1-GFP (G) showed no significant difference between treatments. 690 and 512 spines for nocodazole from >25 dendrites for control and nocodazole treated cells, respectively, were analyzed from two independent experiments. *, P < 0.05, unpaired Student’s t test. Data represent mean ± SEM. Experimenter was blinded to condition upon analysis.
Next we examined the effect of microtubule destabilization with nocodazole on lysosomal trafficking into dendritic spines. Could the destabilization of microtubules increase lysosomal interaction with actin at spines? We treated hippocampal cultures that were transfected with LAMP1-GFP and mCherry with 10 µg/ml nocodazole for 1 h then fixed and imaged them on a confocal microscope. We then quantified how many spines had a lysosome in the head of the spine. Interestingly, we observed a significant increase in the percentage of spines that had a lysosome in the head after nocodazole treatment (control: 1 ± 0.09; nocodazole: 1.5 ± 0.21; P < 0.05; Fig. 5, D and E). This suggested that destabilization of microtubules may facilitate increased interaction of lysosomes with F-actin or interactors of F-actin in dendritic spines. The significant change in lysosomes entering spines was not caused by increases in the number of spines or lysosomes, as we did not see any changes in the number of spines per micrometer (Fig. 5 F) or the LAMP1-GFP signal intensity per micrometer between treatments, respectively (Fig. 5 G). Together, these data indicate that microtubule and actin cytoskeletal dynamics play an active role in the trafficking of lysosomes in dendrites and into dendritic spines.
Distribution and trafficking of lysosomes is correlated with internalized membrane proteins near synapses, including synaptic AMPARs
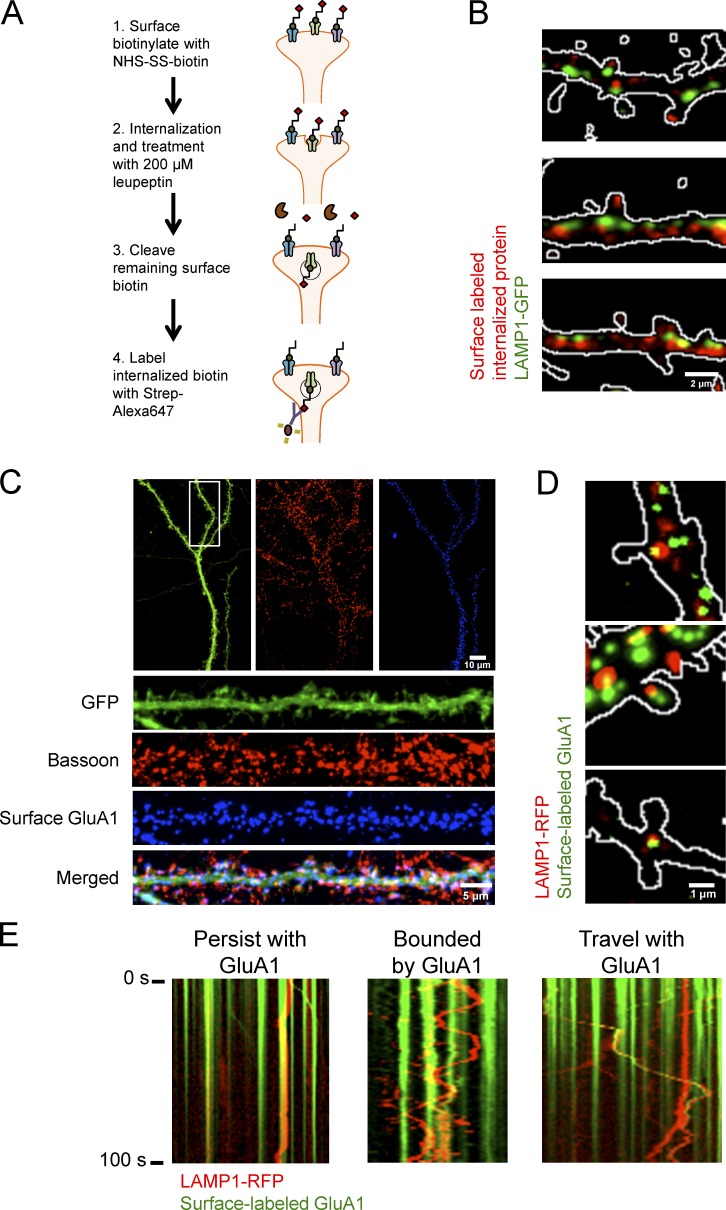
Given the evidence that lysosomes are in dendrites we next wanted to determine if synaptic proteins are degraded by lysosomes near synapses. To evaluate this, we developed a bulk surface membrane internalization assay. In this assay, we transfected neurons with LAMP1-GFP to mark lysosomes and biotinylated cell surface proteins with NHS-SS biotin for 10 min, and we then washed off excess NHS-SS biotin and allowed membrane proteins to be internalized. Cells were also treated with 200 µM leupeptin to prevent lysosomal degradation and to facilitate intracellular accumulation. Remaining biotin was then cleaved with glutathione and cells were fixed, permeabilized, and stained with Streptavidin–Alexa 647 (Fig. 6 A). In HEK293T cells, we saw that the assay effectively labels internalized membrane proteins previously labeled with NHS-SS biotin at the cell surface (Fig. S2). In dissociated hippocampal neurons, we found that LAMP1-GFP colocalizes with and is juxtaposed to internalized membrane proteins (Fig. 6 B), which suggest that degradation of internalized membrane proteins can occur at or near synapses.
Figure 6.
Distribution and trafficking of lysosomes is highly correlated with internalized membrane proteins and synaptic AMPARs. (A) Model of bulk surface membrane internalization assay. (B) Representative images of dissociated hippocampal neurons expressing LAMP1-GFP (green) and internalized membrane proteins (red). Outlines of dendritic spines were generated by outlining the mCherry signal. (C) Representative immunofluorescent images of dissociated hippocampal neurons (16 DIV) expressing GFP and GFP-GluA1. Surface GFP-GluA1 was labeled with Alexa Fluor 647 and subsequently stained for Bassoon, a presynaptic marker. Surface GluA1 is juxtaposed to Bassoon. (D) Representative images of dissociated hippocampal cultures (DIV16) transfected with GFP, GFP-GluA1, and LAMP1-RFP. GFP-GluA1 can colocalize with a lysosome in and at the base of a dendritic spine. Cultures were treated with 100 µg/ml leupeptin and live-labeled with GFP antibody conjugated to Alexa Fluor 647. After washout, cultures were treated with 100 µM AMPA for 10 min to promote endocytosis. Outlines of dendritic spines were generated by outlining the GFP signal. Surface GFP-GluA1 labeled with anti-GFP Alexa Fluor 647 was false-colored green to show colocalization with LAMP1-RFP. (E) Live imaging of labeled surface GluA1 with LAMP1-RFP after 100 µM AMPA treatment. LAMP1-RFP labeled lysosomes persist at a location of surface-labeled AMPARs, rapidly move bidirectionally between two sites of surface-labeled AMPARs, and cotraffic with surface-labeled AMPARs likely destined for degradation.
Our laboratory has previously shown that AMPARs are ubiquitinated and targeted for degradation by the lysosome in response to specific synaptic stimuli (Schwarz et al., 2010; Scudder et al., 2014). Therefore, we then wanted to determine if lysosomal trafficking in dendrites was correlated with surface AMPARs that may be undergoing internalization and degradation. First, we showed that a majority of surface-labeled AMPARs are found at synapses by their juxtaposition with Bassoon, a presynaptic protein marker, by surface labeling GFP-GluA1, a subunit of AMPARs, and staining for endogenous Bassoon (Fig. 6 C). To evaluate the trafficking of lysosomes near synapses and surface AMPARs, we transfected dissociated hippocampal neurons with LAMP1-RFP to mark lysosomes, GFP-GluA1 to mark surface AMPARs, and GFP to fill the cell. Before imaging, neurons were preincubated with 200 µM leupeptin for 1 h to prevent loss of surface-labeled AMPARs signal caused by degradation. The neurons were live-labeled with anti–GFP antibodies conjugated to Alexa Fluor 647 for 20 min to label surface GFP-GluA1 receptors. GFP, used as cell fill, was not detected by live labeling with anti-GFP Alexa Fluor 647 conjugate as indicated by its lack of punctate signal. After washing out the unbound anti–GFP antibody, we bath applied 100 µM AMPA, which induces AMPAR internalization and degradation by the lysosome (Schwarz et al., 2010; Scudder et al., 2014), and then subsequently imaged LAMP1-RFP and GFP-GluA1 (anti-GFP Alexa Fluor 647 conjugate). We found in a subset of spines, surface-labeled GFP-GluA1 colocalized with LAMP1-RFP in and at the base of dendritic spines (Fig. 6 D). Surface-labeled GFP-GluA1 (marked by anti-GFP Alexa Fluor 647 conjugate) was not found in all spines, and this is most likely because AMPA was bath applied to induced internalization and sorting to a degradative fate by the lysosome (Schwarz et al., 2010; Scudder et al., 2014). Using the same experimental setup and treatment paradigm, we live-imaged neurons transfected with LAMP1-RFP, GFP-GluA1 (tagged with Alexa Fluor 647), and GFP and found that a subset of lysosomes have little to no motility and are colocalized with GFP-GluA1 surface AMPARs (Fig. 6 E). The motility of other lysosomes, however, was confined between GFP-GluA1 puncta (Fig. 6 E), and on occasion, we observed correlated motility of LAMP1-RFP and surface-labeled GFP-GluA1, suggesting that they are in the same compartment (Fig. 6 E). These stereotyped motilities indicate that lysosomes are positioned in place to facilitate the degradation of membrane protein cargo, such as AMPARs.
Synaptic activation of AMPA and N-methyl-d-aspartate receptors (NMDARs) regulates the trafficking of lysosomes in dendritic spines
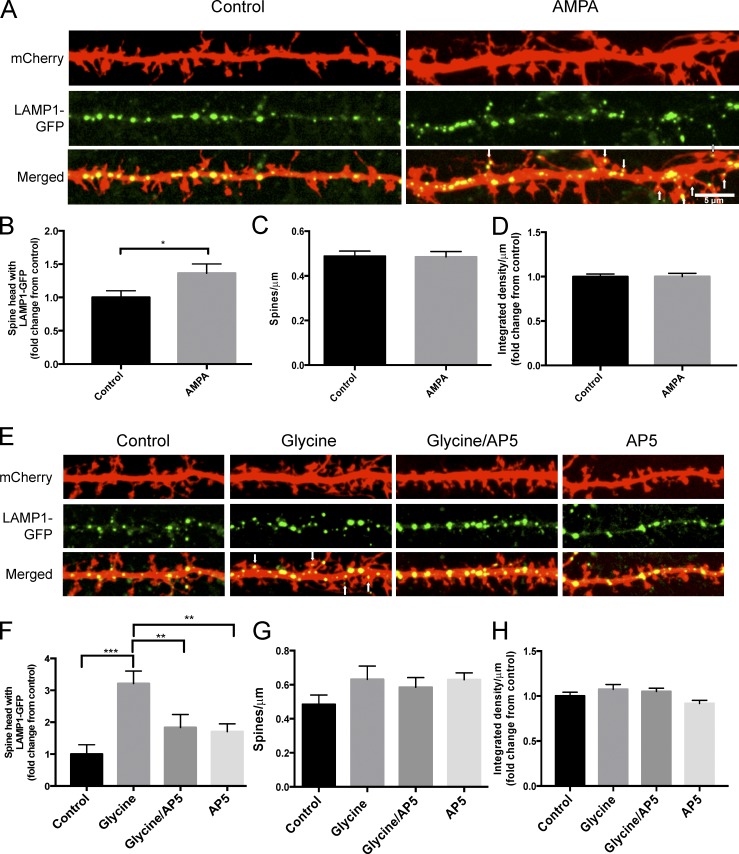
The trafficking of membrane-bound organelles at synapses, such as recycling endosomes and the endoplasmic reticulum, has been shown to be regulated by synaptic activity (Park et al., 2006; Cui-Wang et al., 2012). Because the internalization and degradation of membrane proteins like AMPARs is regulated by synaptic activity, and because we previously observed that a subset of dendritic spines contained lysosomes, we investigated whether the trafficking of lysosomes to dendritic spines could also be regulated by synaptic activity. To evaluate this, we treated hippocampal neurons with 200 µM AMPA for 2 min and found a significant increase in the number of spines that had a lysosome in the spine head (control: 1.0 ± 0.10; AMPA: 1.4 ± 0.14; P < 0.05; Fig. 7, A and B), with no change in spine number (Fig. 7 C) or lysosome number (Fig. 7 D).
Figure 7.
Activity-dependent trafficking of lysosomes to dendritic spines. (A) Representative image of secondary dendrites from dissociated hippocampal neurons (DIV 16) transfected with mCherry and LAMP1-GFP under control conditions or after 200 µM AMPA treatment (2 min). Arrows point to LAMP1-GFP in a dendritic spine. (B–D) Quantification of the percentage of spines that have LAMP1-GFP-labeled lysosomes in the head of a spine (B). Number of spines per micrometer (C) and signal intensity of LAMP1-GFP (D) showed no significant difference between treatments. 830 (control) and 833 (AMPA) dendritic spines were analyzed (n = 41 for control and AMPA) over three independent experiments, *, P < 0.05, unpaired Student’s t test. Experimenter was blinded to condition upon analysis. (E) Representative image of secondary dendrites (DIV16) transfected with mCherry and LAMP1-GFP under control conditions or after 200 µM glycine (10 min), 200 µM glycine (10 min) with 50 µM AP5 (60-min treatment pretreatment), or 50 µM AP5 alone (60 min) in HBS containing 0 mM Mg+2. Arrows point to LAMP1-GFP in a dendritic spine. (F–H) Quantification of the percentage of spines that have LAMP1-GFP-labeled lysosomes in the head of a spine (F). Number of spines per micrometer (G) Signal intensity of LAMP1-GFP (H) showed no significant difference between treatments. 211 dendritic spines for control, 246 for glycine, 356 for glycine/AP5, and 573 for AP5 were analyzed (n = 14 for control, 14 for glycine, 18 for glycine/AP5, and 28 for AP5) over three independent experiments. ***, P < 0.001; **, P < 0.01; *, P < 0.05, one-way analysis of variance with Tukey’s post hoc analysis. Experimenter was blinded to condition upon analysis. All data represent mean ± SEM.
Because lysosomes mediate the degradation of a large amount of membrane and endocytosed material, it is possible that other activity-dependent paradigms can recruit lysosomes to spines. To evaluate this, we treated hippocampal neurons expressing LAMP1-GFP with high concentrations of glycine, a treatment paradigm previously shown to potentiate neurons through the activation of synaptic NMDARs (Lu et al., 2001; Park et al., 2004) and asked if the trafficking of lysosomes into dendritic spines was altered. We found that a brief (10 min) application of 200 µM glycine in cultures bathed in Mg+2-free extracellular solution markedly increased the number of spines that had a lysosome in the spine head (control: 1.0 ± 0.29; glycine: 3.2 ± 0.39; P < 0.001; Fig. 7, E and F). However, glycine-induced redistribution of lysosomes to spines was blocked by application of the NMDAR antagonist AP5 (control: 1 ± 0.29; glycine/AP5: 1.8 ± 0.41; AP5: 1.7 ± 0.24; P = 0.37 and P = 0.45 for control to glycine/AP5 and AP5, respectively; Fig. 7, E and F). Collectively, these data indicate that the increase of lysosomes in the head of spines was dependent on NMDAR activation. There was no significant change in the number of spines or lysosomes in any condition (Fig. 7, G and H). Although there are likely other synaptic cues that regulate the trafficking of lysosomes throughout dendrites and into dendritic spines, these data suggest that NMDAR activity plays a significant role.
Activation of single spines recruits lysosomes to the base
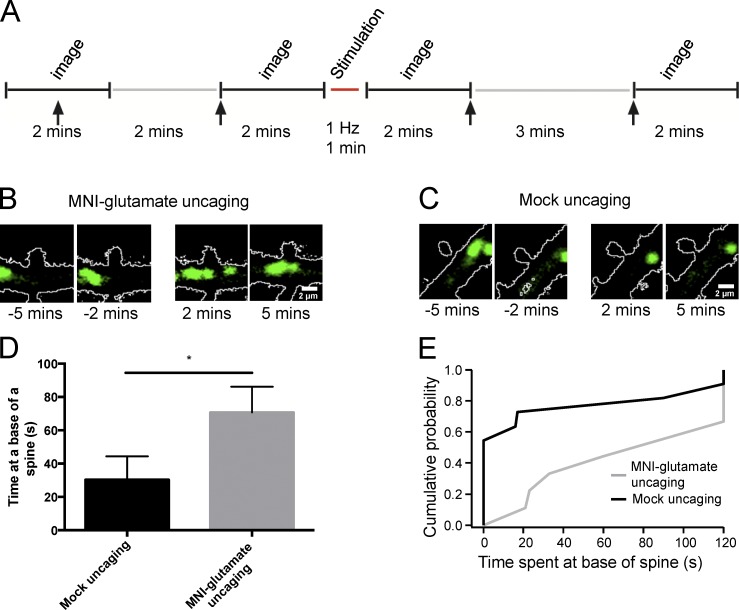
Given that activation of synaptic NMDARs recruits lysosomes to dendritic spines, we next probed whether activation of a single synapse was sufficient for this recruitment. Organotypic hippocampal slice cultures were prepared from Sprague-Dawley pups (postnatal day 7). 2 d later, cultures were biolistically transfected with DsRed to highlight the morphology of the transfected neurons and LAMP1-GFP to mark and visualize lysosomes. 7 to 10 d after transfection, pyramidal neurons were imaged live with two-photon laser-scanning microscopy. Neurons with motile LAMP1-GFP–positive structures were imaged, indicating healthy cultures. We found, in some instances, lysosomes that abruptly stop at the base of dendritic spines (Video 7). In our experimental paradigm, lysosomes were imaged intermittently for 4 min (two 2-min epochs, with images acquired at 1 Hz) followed by two-photon uncaging of MNI-glutamate for 1 min (1 Hz; Matsuzaki et al., 2004) at a visually identified spine (Fig. 8 A). Immediately after stimulation, or mock uncaging where no MNI-glutamate was included in the bath, dendritic segments were imaged for another 4 min (Fig. 8 A). Strikingly, we observed that in 9 out of 10 (90%) spines stimulated with glutamate uncaging, a lysosome paused at the base of the spine (Fig. 8 B and Video 8). Spines that received mock uncaging were notably less likely to have a lysosome pause in the dendrites at the base of the spine (5 out of 12 [42%]; Fig. 8 C and Video 9). Furthermore, lysosomes that paused at the base of MNI-glutamate–stimulated spines had significantly longer dwell times than mock-stimulated spines (mock uncaging: 30.3 ± 14.2 s; glutamate: 70.7 ± 15.4 s; P ≤ 0.05; Fig. 8, D and E). Collectively, these data indicate that the trafficking of lysosomes to dendritic spines can be regulated by neuronal activity. In addition, activation of a single spine recruits a lysosome to its base, supporting the idea that lysosomal trafficking can be precisely regulated by synaptic activity and that tight spatial regulation of degradation organelles is maintained in neuronal dendrites.
Figure 8.
Activation of a single spine recruits a lysosome to the base of the spine. (A) Experimental timeline of two-photon imaging and MNI-glutamate uncaging in hippocampal organotypic cultures. Individual spines on secondary dendrites of CA1 pyramidal neurons were visualized with a laser tuned to 900 nm. A 720-nm laser was used to stimulate individual spines, using 0.5-ms pulses at 1 Hz for 1 min. Stimulation was done with MNI-glutamate (2.5 mM MNI-glutamate) or without (“mock uncaging”) as a control in ACSF containing 0 mM Mg2+. Arrows signal the time points at which the representative images from B and C were taken. (B and C) Representative images of a secondary dendrite in a CA1 pyramidal neuron in a rat organotypic hippocampal slice. Time points are 5 and 2 min before uncaging and 2 min and 5 min after uncaging. (D) Mean dwell time for a lysosome at the base of a spine in either control or MNI-glutamate conditions. *, P ≤ 0.05, Mann–Whitney U test. Data represent mean ± SEM. (E) Cumulative probability distribution for each condition with and without (mock uncaging) MNI-glutamate. n = 12 for control, 10 for uncaging. P ≤ 0.01, Kolmogorov–Smirnov test. Experimenter was blinded to condition upon analysis.
Discussion
Lysosomes are found throughout proximal and distal dendrites and in dendritic spines
Lysosomes have been primarily thought to exist exclusively in somatic and axonal compartments. As such, little attention has been given to the function and trafficking of lysosomes in neuronal dendrites. A priori, there is no clear evidence that supports the notion that lysosomes should be excluded from dendrites. In very recent studies, however, the regulation of lysosome motility in dendrites has begun to be described (Schwenk et al., 2014; Tsuruta and Dolmetsch, 2015). In this study, we provide detailed characterization of lysosome trafficking dynamics in dendrites and the first evidence of their activity-dependent recruitment to dendritic spines. Our study was predicated on one simple question: how do lysosomes degrade membrane proteins in distal regions of dendrites? We considered three models: (1) internalized membrane protein cargo is transported back toward the cell body, where there is a high density of lysosomes; (2) a population of lysosomes traffic away from the cell body to distal regions; or (3) lysosomes travel between stationary outposts throughout dendrites. Our data provide compelling evidence that lysosomes travel along microtubules to distal dendritic compartments including to the base of dendritic spines and, on occasion, into the spine head, whereas others remain stationary. We determined this by expressing the late endosomal/lysosomal protein LAMP1 fused to GFP (LAMP1-GFP) in hippocampal neurons. We verified that LAMP1-GFP labeled lysosomes by demonstrating that LAMP1-GFP reliably labels the acidified membrane-bound organelles as indicated by colocalization with low pH–sensing LysoTracker (Fig. 1). Furthermore, these LAMP1-labeled structures in dendrites lose their acidity and release calcium upon GPN treatment, providing compelling evidences that these structures are active lysosomes (Fig. 2). Moreover, applying the APEX2 technology allowed us to validate that ectopic expression of LAMP1-labeled lysosomes (Fig. 3).
Lysosomal inhibition decreases lysosome motility and synapse number
Our findings show that inhibiting lysosome protease activity with leupeptin for 3 h decreases the motility of lysosomes in dendrites (Fig. 4). We also found that this same treatment caused an increase in mEPSC IEI, whereas mEPSC amplitude was unchanged. Interestingly, changes in frequency have also been reported in miniature inhibitory postsynaptic currents (Arancibia-Cárcamo et al., 2009). We found that the change in mEPSC frequency is likely caused by a loss in the number of excitatory synapses. These data are consistent with a recently published paper claiming that activity-dependent exocytosis of lysosomes regulates the structural plasticity of dendritic spines (Padamsey et al., 2017). Padamsey et al. found that inhibition of either lysosomal calcium signaling or cathepsin B release prevented the maintenance of dendritic spine growth in response to long-term potentiation–inducing stimuli. Together, our findings highlight the importance of intact lysosome function on synaptic structure and function.
An interplay between microtubule and actin cytoskeletal dynamics controls lysosomal trafficking in dendrites
Lysosomes travel bidirectionally on microtubules, presumably by interacting with motor proteins (Maday et al., 2014). The direction of movement is also likely facilitated by a mixture of microtubules in dendrites with plus or minus ends orientated toward or away from the cell body. When we perturbed microtubule dynamics with nocodazole, we found that trafficking of lysosomes was halted (Fig. 5). When actin dynamics were disrupted by latrunculin A, there was an increase in lysosome motility (Fig. 5). To our surprise, however, we see that lysosomes are juxtaposed to F-actin found in dendrites and that the number of dendritic spines containing lysosomes significantly increased when neurons were treated with nocodazole (Fig. 5). It is possible that disrupting microtubule dynamics may facilitate lysosomal interaction with proteins at dendritic spines, such as F-actin and F-actin–binding proteins, sequestering lysosomes to spines. Conversely, when F-actin is disrupted by latrunculin A, lysosomal motility in the dendritic shaft increases. This suggests that lysosome motility occurs through its dynamic interactions between microtubules or the actin cytoskeleton motility.
The distribution and trafficking of lysosomes is correlated with internalized membrane proteins near synapses, including synaptic AMPARs
We previously demonstrated that AMPARs are ubiquitinated by the ubiquitin ligase Nedd4-1 in response to certain synaptic cues and are targeted for degradation by the lysosome (Schwarz et al., 2010; Scudder et al., 2014). Importantly, we showed that this mechanism is critical for homeostatic downscaling and Αβ-induced decrease in synaptic strength and the number of dendritic spines (Scudder et al., 2014; Rodrigues et al., 2016). Here, we provided evidence that lysosomal movement in dendritic shafts is correlated with the distribution of internalized membrane proteins and synaptic AMPARs (Fig. 6). By imaging previously surface-labeled AMPARs together with LAMP1-RFP, we found that lysosomes move between sites of surface-labeled AMPARs and persist at sites correlated with surface-labeled AMPARs. In some cases, we observed surface-labeled AMPARs moving together with LAMP1-GFP–labeled lysosomes, suggesting that structures containing internalized AMPARs have been delivered to the lysosome for degradation (Fig. 6). It should be noted that we cannot unequivocally determine if these once-labeled surface AMPARs have been internalized and are in route to being degraded. Nonetheless, the degree of correlated movement between and with once surface-labeled AMPARs provides compelling evidence that mechanisms exist to target lysosomes to membrane cargo at synapses.
Lysosomes traffic to dendritic spines in an activity-dependent manner
It has long been hypothesized that protein synthesis and protein degradation machinery receive instructional cues from synaptic signals to facilitate dynamic and proper protein half-life control. Indeed, we and others have shown that the ubiquitin proteasome system can traffic to synapses in response to synaptic activity (Bingol and Schuman, 2006; Djakovic et al., 2009, 2012). Interestingly, we see that a lysosome can stop abruptly at a base of a dendritic spine, suggesting that there is a mechanism for capturing lysosomes at a spine (Video 7). To determine if lysosomes traffic to dendritic spines in response to synaptic activity, we treated dissociated hippocampal neurons expressing mCherry and LAMP1-GFP with either AMPA or high glycine. AMPA, which is known to promote the ubiquitination and degradation of AMPARs by the lysosome, recruits lysosomes to dendritic spine heads. This positions them nicely to degrade AMPAR locally at synapses. Conversely, treatment with high glycine, which is known to potentiate synapses (Lu et al., 2001; Park et al., 2004), also increases the number of lysosomes in dendritic spines in response to synaptic activity, which, in part, involves the activation of NMDARs, because APV blocks this effect (Fig. 7). Strikingly, we demonstrated that two-photon glutamate uncaging at a single spine could recruit a lysosome to the base of that spine (Fig. 8). Thus, lysosomes can respond to very specific and spatially restricted synaptic input. The exact synaptic signals and molecular mechanisms will be of great interest to uncover.
Conclusions
How is the selective degradation of membrane proteins at individual synapses in dendrites accomplished? We suggest that, in part, the regulation of lysosome trafficking in dendrites is a key determinant. Our results describe the activity-dependent control of lysosome trafficking in dendrites and the recruitment to dendritic spines, indicating that lysosomal trafficking is under tight spatial control. The presence of lysosomes in the spine suggests that localized degradation can play an active role in the remodeling of synapses to facilitate plasticity events and to help locally maintain cellular homeostasis. Identifying the exact transport machinery and defining the signaling mechanisms that couple synaptic cues to lysosomal trafficking and function specifically in dendrites and at spines will be an important next step to understand. Activity-dependent trafficking of lysosomes to spines provides an attractive cellular mechanism for how synaptic protein turnover can occur selectively at one group of synapses, but not at another group of synapses on the same neuron. This work helps to lay the foundation for future studies involving local degradation of synaptic membrane proteins by lysosomes and the mechanisms that are in place to synergize the trafficking and function of lysosomes with membrane cargo previously internalized and destined for destruction. Moreover, because lysosomal dysfunction is thought to play an integral role in neurodegenerative disease, it will be of interest to determine if altered lysosome trafficking and function, specifically in dendrites, is a contributing factor.
Materials and methods
DNA constructs and antibodies and reagents
GFP and dsRed was purchased from the Takara Bio Inc. or Addgene DNA repository, respectively. LAMP1-GFP, LAMP1-RFP and mCherry was a gift from S. Roy (University of California, San Diego, La Jolla, CA). GFP-GluA1 was a gift from R. Malinow (University of California, San Diego, La Jolla, CA). GCaMP3-TRPML1 was a gift from H. Xu (University of Michigan, Ann Arbor, MI). LAMP1-APEX2 was made by removing GFP tag from LAMP1-GFP vector, and the APEX2 tag was subcloned at the C-terminal end of LAMP1. APEX2 was PCR amplified from the APEX2 construct. Using the forward primer 5′-ATCTCAGGATCCATGGGGAAATCATACCCAACAG-3′ and reverse primer 5′-CCGACGCCTAAGCGGCCGCATAATG-3′, the APEX2 tag was inserted in frame into LAMP1 at the BamHI and NotI cut sites.
The antibodies used were LAMP1 (1:400; ab24170; Abcam) and EEA1 (1:10,000; BD610456; BD) GFP tag antibody Alexa Fluor 647 conjugate (1:300; A-31852; Thermo Fisher Scientific), GFP (1:2,000; Thermo Fisher Scientific), MAP2 (1:10,000; ab5392; Abcam), DAPI (1:10,000; D1306; Thermo Fisher Scientific) and Bassoon (1:400; SAP7F407; Enzo Life Sciences). Reagents were as follows: D-AP5 (0106; Tocris Bioscience), Glycine (BP381-5; Thermo Fisher Scientific), latrunculin A (L5163; Sigma-Aldrich), nocodazole (2190S; Cell Signaling Technology), AMPA (1074; Tocris Bioscience), leupeptin (EI8; EMD Millipore), GPN (Cayman Chemical), LysoTracker red (Invitrogen), EZ-Link sulfo-NHS-SS biotin (Thermo Fisher Scientific), and MNI-caged l-glutamate (1490; Tocris Bioscience).
Neuronal cultures, transfections, and infections
Dissociated hippocampal neurons from postnatal day 1 Sprague-Dawley of either sex as described previously (Djakovic et al., 2009, 2012; Schwarz et al., 2010). Hippocampal neurons were transfected using a calcium phosphate transfection kit (Takara Bio Inc.). Transfection solution was applied for ≤4 h to avoid cell death, and expression was allowed for ≤24 h to avoid overexpression. For infections, hippocampal cultures were infected with Sindbis virus expressing GFP at day in vitro (DIV) 15 and allowed to express for 16 to 22 h.
Immunostaining
After transfection and drug treatments, neurons were washed with PBS with 1 mM MgCl2 and 0.1 mM CaCl2 (PBS-MC) and fixed with a solution containing 4% paraformaldehyde and 4% sucrose for 10 min. Cells were permeabilized in PBS containing 2% normal goat serum, 1% BSA, and 0.1% saponin for 1 h at room temperature. Primary antibody was diluted in 2% BSA in PBS-MC and applied to neurons overnight at 4°C, and then secondary antibody was diluted in 2% BSA and applied to neurons for 1 h at room temperature. Coverslips were mounted onto glass slides for confocal imaging.
LysoTracker staining
Dissociated hippocampal neurons transfected with LAMP1-GFP for <24 h (for colocalization experiments) then incubated with 1 µM LysoTracker red in Neurobasal medium for 30 to 60 min at 37°C followed by washing with fresh Neurobasal medium (two times). Subsequently, Neurobasal medium was replaced with prewarmed Hepes-buffered saline (HBS) containing 119 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 30 mM glucose, and 10 mM Hepes, pH 7.2, to reduce fluorescent background. All images were taken at DIV16.
Organotypic slice preparation and transfection
Slice preparation and transfection was prepared as previously described previously (Bloodgood et al., 2013). In brief, P8 Sprague-Dawley rat hippocampi were rapidly dissected in ice-cold dissection media consisting of 1 mM CaCl2, 5 mM MgCl2, 10 mM glucose, 4 mM KCl, 26 mM NaHCO3, 218 mM sucrose, 1.3 mM NaH2PO4 H2O, and 30 mM Hepes. On the second day, in vitro cultures were biolistically (Helios Gene Gun; Bio-Rad Laboratories) cotransfected with 1 µm gold particles coated with dsRed and LAMP1-GFP (particles prepared with 5 µg dsRed and 50 µg LAMP1-GFP).
Imaging for confocal images (static and time-lapse imaging)
Images were acquired with a DMI6000 inverted microscope (Leica) equipped with a Yokogawa Electric Corporation Nipkon spinning disk confocal head, an Orca ER high-resolution black and white cooled CCD camera (6.45 µm/pixel at 1×), Plan Apochromat 63×/1.0 or 40×/1.0 numerical aperture objective, and an argon/krypton 100 mW air-cooled laser for 488/568/647 nm excitations. Cells were transfected with the desired constructs and distal regions of the primary dendrites or first-order branches of secondary dendrites were selected for imaging. For experiments with Glycine and/or AP5 treatment, cells were placed in HBS containing no magnesium (Mg+2). For live imaging, cultures were placed in a humidified chamber maintained at 37°C. For live imaging, cells were transferred to either HBS or Hibernate-E low-fluorescence medium (BrainBits) at 37°C. Single z-planes were captured using a 40× or 63× objective with consistent imaging parameters (500 ms exposure at either 1 × 1 binning or 2 × 2 binning). Kymographs were generated in ImageJ using the multiple kymograph plugin. For fixed imaging, maximum projected confocal z-stacks (0.4 µm z-step) were analyzed with ImageJ (National Institutes of Health). Analysis of kymographs and fixed images was done manually in a blinded fashion. All images were taken at DIV16.
For spine density analysis, dissociated hippocampal cultures were infected with Sindbis virus expressing GFP for 16 h before treatments. After fixation, dendrites were straightened using ImageJ, and spine density was determined by manually counting spines. Experimenters were blinded to condition during data collection and analysis, and statistical significance was determined through unpaired t tests using Prism software (GraphPad Software).
Live imaging before and after GPN treatment
DIV14 to DIV16 low-density dissociated hippocampal neurons were treated with Sindbis virus expressing GFP for <16 h. On the day of the experiment, cells were treated with 1 µM LysoTracker red for 15 to 20 min. Cells were washed with Hibernate-E low-fluorescence imaging media and images taken on a confocal spinning disk. z-stack images were taken at time 0 and either diluted DMSO or 40 µM GPN was added to cells. After 5 min, another z-stack image was taken, and LysoTracker fluorescence was compared.
For GCaMP3-TRPML1 studies, DIV14 to DIV15 dissociated hippocampal neurons were transfected with GCaMP3-TRPML1 and mCherry using calcium phosphate transfection protocol. Less than 24 h later, cells were live imaged on a confocal spinning disk. Single z-planes were taken for 100 s. Vehicle (diluted DMSO) was added at 50 s, and 40 µM GPN was added at 80 s.
DAB staining and preparation of cultured cells for EM
Cultured hippocampal neurons transfected with LAMP1-APEX2 were prepared as previously described (Martell et al., 2012). Images were taken on a JEOL 1200EX transmission EM and developed on film.
Electrophysiology
For whole-cell voltage-clamp recordings of mEPSCs, dissociated hippocampal neurons were incubated in room temperature HBS recording solution containing 119 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 30 mM glucose, and 10 mM Hepes, pH 7.2, along with 1 µM TTX and 10 µM bicuculline. The electrode recording solution contained 10 mM CsCl, 105 mM CsMeSO3, 0.5 mM ATP, 0.3 mM GTP, 10 mM Hepes, 5 mM glucose, 2 mM MgCl2, and 1 mM EGTA, pH 7.2. Electrode resistances ranged from 3.5 to 4.5 MΩ, and access resistances ranged from 10 to 25 MΩ. Signals were amplified, filtered to 2 or 5 kHz, and digitized at 10 kHz sampling frequency. Holding potential for all traces was −70 mV. mEPSCs were analyzed using ClampFit 10.3 (Molecular Devices). Pyramidal-like neurons were chosen. Experimenters were blinded to condition during analysis.
Two-photon imaging and uncaging
Combined two-photon uncaging of MNI-l-glutamate and two-photon imaging of organotypic slice cultures was done using a custom-built two-photon laser-scanning microscope. Organotypic inserts containing three or four slices were placed in a recording chamber constantly perfused with artificial cerebrospinal fluid (ACSF) containing 125 mM NaCl, 2.5 mM KCl, 21.4 mM NaHCO3, 1.25 mM NaH2PO4, 2.0 mM CaCl2, and 11.1 mM glucose and equilibrated with 95% O2/5% CO2. ACSF did not include Mg+2 to maximize NMDAR-mediated conductance. DsRed and LAMP1-GFP were excited at a wavelength of 900 nm, and MNI-caged-l-glutamate was uncaged with 500-µs pulses of 720-nm light. MNI-caged-L-glutamate was used at a concentration of 2.5 mM at 31°C.
Spine analysis was restricted to spines located along an apical dendrite after the first branching point. Two baseline images were acquired at 1 Hz for 2 min, separated by a 2 min break to avoid photo-bleaching and photo-damage, at a 256 × 256 resolution, with a 2-ms line scan. After baseline image acquisition, uncaging laser power was calibrated for each spine using 30% to 40% photobleaching of the red channel (DsRed) as previously described (Bloodgood and Sabatini, 2007). Photobleaching was a direct readout of the laser power at the uncaging site that was independent of depth of the spine and tissue aberrations affecting refraction. After laser power calibration, 500-µs pulses delivered at 1 Hz for 1 min just off the spine head to avoid photodamage was performed (Fig. S3; Matsuzaki et al., 2004; Tanaka et al., 2008).
After the high-frequency uncaging, images were then immediately collected at 1 Hz for 2 min (same imaging parameters as for baseline imaging). Images were acquired for two 2-min intervals, with a 3-min interval of no acquisition to minimize any photobleaching or photodamage. Images were processed on ImageJ and visually assessed, in a blinded fashion, to determine if a lysosome was at the base of a spine. Healthy neurons were determined by morphology and the presence of moving lysosomes. Time at the base of the spine was quantified by counting the number of frames a LAMP1-GFP vesicle was present at a base of a spine.
Bulk surface membrane internalization assay
HEK293T or dissociated hippocampal neurons (DIV14–16) were transfected with GFP-LAMP1 and mCherry. Cells were washed twice with PBS-MC and surface labeled with 0.5 mg/ml sulfo-NHS-SS biotin (Thermo Fisher Scientific) in PBS at 37°C for 10 min. Biotin was washed out with 0.1% BSA in PBS-MC twice and media was added back. Cells were then treated with 200 µM leupeptin for 1 h. Biotin still on cell surface was cleaved with cleaving buffer (20 mM glutathione in 75 mM NaCl and 10 mM EDTA with 1% BSA and 0.075 N NaOH) twice at 2 to 5 min each. Cells were then fixed 4% paraformaldehyde and 4% sucrose for 10 min. Cells were permeabilized with 0.2% Triton X-100 and 2% BSA in PBS-MC for 20 min, followed by a 3 h block in 5% BSA in PBS-MC. GFP antibody was diluted in 2% BSA in PBS-MC and applied to neurons for 1 h at room temperature then secondary antibody along with Streptavidin conjugated to either Alexa Flor 568 or 647 was diluted in 2% BSA and applied to neurons for 1 h at room temperature. Coverslips were mounted onto glass slides for confocal imaging.
Live labeling of surface GluA1 with RFP-LAMP1 expression
Dissociated neurons transfected with GFP, GFP-GluA1, and LAMP1-RFP for ≤24 h were pretreated with 100 µg/ml leupeptin for 1 h to block lysosomal degradation. Cultures were simultaneously treated with GFP tag antibody, Alexa Fluor 647 conjugate treated for 20 min to label surface GluA1. Antibody was washed out with two rinses of PBS and replaced with conditioned B27-supplemented Neurobasal media. Cultures were then treated with 100 µM AMPA for 10 min for fixed images. For live imaging, cultures were treated briefly (5 min) with 100 µM AMPA and then immediately imaged at 37°C. All Alexa Fluor 647 images are displayed in green. Kymographs were generated using a modified custom macro, which was a gift from G. Pekkurnaz (University of California, San Diego, La Jolla, CA).
Online supplemental material
Fig. S1 shows that cell health is not compromised upon leupeptin treatment. Fig. S2 shows biotin-streptavidin staining of internalized membrane proteins in HEK293T cells. Fig. S3 shows calibration of 2P uncaging power and a 2D image of a CA1 pyramidal neuron dendrite. Video 1 shows a time lapse of LysoTracker-labeled structures trafficking in a hippocampal neuron. Video 2 shows a time lapse of hippocampal neuron expressing mCherry and GCaMP3-TRPML1. Video 3 shows a time lapse of LAMP1-GFP–labeled lysosomes in a dendrite under basal conditions. Video 4 shows a time lapse of LAMP1-GFP–labeled lysosomes in a dendrite after leupeptin treatment (200 µM for 3 h). Video 5 shows a time lapse of LAMP1-GFP–labeled lysosomes in a dendrite after nocodazole treatment. Video 6 shows a time lapse of LAMP1-GFP–labeled lysosomes in a dendrite after latrunculin A treatment. Video 7 shows a time lapse of LAMP1-GFP–labeled lysosomes in an organotypic hippocampal slice. Video 8 shows a time lapse of LAMP1-GFP-labeled lysosomes in an organotypic hippocampal slice before and after glutamate uncaging. Video 9 shows a time lapse of LAMP1-GFP–labeled lysosomes in an organotypic hippocampal slice before and after mock uncaging.
Supplementary Material
Acknowledgments
We are grateful to Subhojit Roy, Utpal Das, and members of the Patrick laboratory for helpful discussions. We thank Lara Dozier, Kelly Martyniuk, Pei-Ann Lin Acosta, and Andrea Thor for technical assistance.
This work was supported by a National Science Foundation graduate research fellowship (M.S. Goo), a Ford Foundation predoctoral fellowship (M.S. Goo), National Institutes of Health grant NS060847 (G.N. Patrick), University of California, San Diego funds (G.N. Patrick), Searle Scholars program grant 14-SSP-184 (B.L. Bloodgood), and Pew Charitable Trusts grant 00028631 (B.L. Bloodgood). The National Center for Microscopy and Imaging Research at the University of California, San Diego is supported by the National Institutes of Health grants P41GM103412 (M.H. Ellisman) and GM086197 (Stephen Adams/M.H. Ellisman/D. Boassa).
The authors declare no competing financial interests.
Author contributions: M.S. Goo, L. Sancho, N. Slepak, and T.J. Deerinck, collected and analyzed data. M.S. Goo, L. Sancho, D. Boassa, M.H. Ellisman, B.L. Bloodgood, and G.N. Patrick contributed to experimental design. M.S. Goo, L. Sancho, B.L. Bloodgood, and G.N. Patrick prepared the manuscript.
Footnotes
Abbreviations used:
- ACSF
- artificial cerebrospinal fluid
- AMPA
- α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid
- AMPAR
- AMPA receptor
- DIV
- day in vitro
- GPN
- glycyl-l-phenylalanine 2-naphthylamide
- HBS
- Hepes-buffered saline
- IEI
- interevent interval
- mEPSC
- miniature excitatory postsynaptic current
- NMDAR
- N-methyl-d-aspartate receptor
References
- Alvarez-Castelao B., and Schuman E.M.. 2015. The regulation of synaptic protein turnover. J. Biol. Chem. 290:28623–28630. 10.1074/jbc.R115.657130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arancibia-Cárcamo I.L., Yuen E.Y., Muir J., Lumb M.J., Michels G., Saliba R.S., Smart T.G., Yan Z., Kittler J.T., and Moss S.J.. 2009. Ubiquitin-dependent lysosomal targeting of GABA(A) receptors regulates neuronal inhibition. Proc. Natl. Acad. Sci. USA. 106:17552–17557. 10.1073/pnas.0905502106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bananis E., Nath S., Gordon K., Satir P., Stockert R.J., Murray J.W., and Wolkoff A.W.. 2004. Microtubule-dependent movement of late endocytic vesicles in vitro: Requirements for dynein and kinesin. Mol. Biol. Cell. 15:3688–3697. 10.1091/mbc.E04-04-0278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B., and Schuman E.M.. 2006. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 441:1144–1148. 10.1038/nature04769 [DOI] [PubMed] [Google Scholar]
- Bingol B., Wang C.F., Arnott D., Cheng D., Peng J., and Sheng M.. 2010. Autophosphorylated CaMKIIalpha acts as a scaffold to recruit proteasomes to dendritic spines. Cell. 140:567–578. 10.1016/j.cell.2010.01.024 [DOI] [PubMed] [Google Scholar]
- Bloodgood B.L., and Sabatini B.L.. 2007. Nonlinear regulation of unitary synaptic signals by CaV(2.3) voltage-sensitive calcium channels located in dendritic spines. Neuron. 53:249–260. 10.1016/j.neuron.2006.12.017 [DOI] [PubMed] [Google Scholar]
- Bloodgood B.L., Sharma N., Browne H.A., Trepman A.Z., and Greenberg M.E.. 2013. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 503:121–125. 10.1038/nature12743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.L., Maier K.C., Stauber T., Ginkel L.M., Wordeman L., Vernos I., and Schroer T.A.. 2005. Kinesin-2 is a motor for late endosomes and lysosomes. Traffic. 6:1114–1124. 10.1111/j.1600-0854.2005.00347.x [DOI] [PubMed] [Google Scholar]
- Chang D.T., Honick A.S., and Reynolds I.J.. 2006. Mitochondrial trafficking to synapses in cultured primary cortical neurons. J. Neurosci. 26:7035–7045. 10.1523/JNEUROSCI.1012-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui-Wang T., Hanus C., Cui T., Helton T., Bourne J., Watson D., Harris K.M., and Ehlers M.D.. 2012. Local zones of endoplasmic reticulum complexity confine cargo in neuronal dendrites. Cell. 148:309–321. 10.1016/j.cell.2011.11.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic S.N., Schwarz L.A., Barylko B., DeMartino G.N., and Patrick G.N.. 2009. Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 284:26655–26665. 10.1074/jbc.M109.021956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djakovic S.N., Marquez-Lona E.M., Jakawich S.K., Wright R., Chu C., Sutton M.A., and Patrick G.N.. 2012. Phosphorylation of Rpt6 regulates synaptic strength in hippocampal neurons. J. Neurosci. 32:5126–5131. 10.1523/JNEUROSCI.4427-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers M.D. 2000. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 28:511–525. 10.1016/S0896-6273(00)00129-X [DOI] [PubMed] [Google Scholar]
- Hamilton A.M., Oh W.C., Vega-Ramirez H., Stein I.S., Hell J.W., Patrick G.N., and Zito K.. 2012. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 74:1023–1030. 10.1016/j.neuron.2012.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K.M. 1999. Structure, development, and plasticity of dendritic spines. Curr. Opin. Neurobiol. 9:343–348. 10.1016/S0959-4388(99)80050-6 [DOI] [PubMed] [Google Scholar]
- Hendricks A.G., Perlson E., Ross J.L., Schroeder H.W. III, Tokito M., and Holzbaur E.L.. 2010. Motor coordination via a tug-of-war mechanism drives bidirectional vesicle transport. Curr. Biol. 20:697–702. 10.1016/j.cub.2010.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne W.M., Buchkovich N.J., and Emr S.D.. 2011. The ESCRT pathway. Dev. Cell. 21:77–91. 10.1016/j.devcel.2011.05.015 [DOI] [PubMed] [Google Scholar]
- Horton A.C., and Ehlers M.D.. 2003. Dual modes of endoplasmic reticulum-to-Golgi transport in dendrites revealed by live-cell imaging. J. Neurosci. 23:6188–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., and Emr S.D.. 2000. Autophagy as a regulated pathway of cellular degradation. Science. 290:1717–1721. 10.1126/science.290.5497.1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S.S., Martell J.D., Kamer K.J., Deerinck T.J., Ellisman M.H., Mootha V.K., and Ting A.Y.. 2015. Directed evolution of APEX2 for electron microscopy and proximity labeling. Nat. Methods. 12:51–54. 10.1038/nmeth.3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loubéry S., Wilhelm C., Hurbain I., Neveu S., Louvard D., and Coudrier E.. 2008. Different microtubule motors move early and late endocytic compartments. Traffic. 9:492–509. 10.1111/j.1600-0854.2008.00704.x [DOI] [PubMed] [Google Scholar]
- Lu W., Man H., Ju W., Trimble W.S., MacDonald J.F., and Wang Y.T.. 2001. Activation of synaptic NMDA receptors induces membrane insertion of new AMPA receptors and LTP in cultured hippocampal neurons. Neuron. 29:243–254. 10.1016/S0896-6273(01)00194-5 [DOI] [PubMed] [Google Scholar]
- Maday S., Twelvetrees A.E., Moughamian A.J., and Holzbaur E.L.. 2014. Axonal transport: Cargo-specific mechanisms of motility and regulation. Neuron. 84:292–309. 10.1016/j.neuron.2014.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martell J.D., Deerinck T.J., Sancak Y., Poulos T.L., Mootha V.K., Sosinsky G.E., Ellisman M.H., and Ting A.Y.. 2012. Engineered ascorbate peroxidase as a genetically encoded reporter for electron microscopy. Nat. Biotechnol. 30:1143–1148. 10.1038/nbt.2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M., Honkura N., Ellis-Davies G.C., and Kasai H.. 2004. Structural basis of long-term potentiation in single dendritic spines. Nature. 429:761–766. 10.1038/nature02617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padamsey Z., McGuinness L., Bardo S.J., Reinhart M., Tong R., Hedegaard A., Hart M.L., and Emptage N.J.. 2017. Activity-dependent exocytosis of lysosomes regulates the structural plasticity of dendritic spines. Neuron. 93:132–146. 10.1016/j.neuron.2016.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Penick E.C., Edwards J.G., Kauer J.A., and Ehlers M.D.. 2004. Recycling endosomes supply AMPA receptors for LTP. Science. 305:1972–1975. 10.1126/science.1102026 [DOI] [PubMed] [Google Scholar]
- Park M., Salgado J.M., Ostroff L., Helton T.D., Robinson C.G., Harris K.M., and Ehlers M.D.. 2006. Plasticity-induced growth of dendritic spines by exocytic trafficking from recycling endosomes. Neuron. 52:817–830. 10.1016/j.neuron.2006.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parton R.G., Simons K., and Dotti C.G.. 1992. Axonal and dendritic endocytic pathways in cultured neurons. J. Cell Biol. 119:123–137. 10.1083/jcb.119.1.123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues E.M., Scudder S.L., Goo M.S., and Patrick G.N.. 2016. Aβ-induced synaptic alterations require the E3 ubiquitin ligase Nedd4-1. J. Neurosci. 36:1590–1595. 10.1523/JNEUROSCI.2964-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz L.A., Hall B.J., and Patrick G.N.. 2010. Activity-dependent ubiquitination of GluA1 mediates a distinct AMPA receptor endocytosis and sorting pathway. J. Neurosci. 30:16718–16729. 10.1523/JNEUROSCI.3686-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk B.M., Lang C.M., Hogl S., Tahirovic S., Orozco D., Rentzsch K., Lichtenthaler S.F., Hoogenraad C.C., Capell A., Haass C., and Edbauer D.. 2014. The FTLD risk factor TMEM106B and MAP6 control dendritic trafficking of lysosomes. EMBO J. 33:450–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scudder S.L., Goo M.S., Cartier A.E., Molteni A., Schwarz L.A., Wright R., and Patrick G.N.. 2014. Synaptic strength is bidirectionally controlled by opposing activity-dependent regulation of Nedd4-1 and USP8. J. Neurosci. 34:16637–16649. 10.1523/JNEUROSCI.2452-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C., Fraldi A., Medina D.L., and Ballabio A.. 2013. Signals from the lysosome: A control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14:283–296. 10.1038/nrm3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen D., Wang X., Li X., Zhang X., Yao Z., Dibble S., Dong X.P., Yu T., Lieberman A.P., Showalter H.D., and Xu H.. 2012. Lipid storage disorders block lysosomal trafficking by inhibiting a TRP channel and lysosomal calcium release. Nat. Commun. 3:731 10.1038/ncomms1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., and Klionsky D.J.. 2004. Autophagy in health and disease: A double-edged sword. Science. 306:990–995. 10.1126/science.1099993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward O., and Schuman E.M.. 2003. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 40:347–359. 10.1016/S0896-6273(03)00635-4 [DOI] [PubMed] [Google Scholar]
- Tanaka J., Horiike Y., Matsuzaki M., Miyazaki T., Ellis-Davies G.C., and Kasai H.. 2008. Protein synthesis and neurotrophin-dependent structural plasticity of single dendritic spines. Science. 319:1683–1687. 10.1126/science.1152864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuruta F., and Dolmetsch R.E.. 2015. PIKfyve mediates the motility of late endosomes and lysosomes in neuronal dendrites. Neurosci. Lett. 605:18–23. 10.1016/j.neulet.2015.07.021 [DOI] [PubMed] [Google Scholar]
- Turrigiano G.G., and Nelson S.B.. 2004. Homeostatic plasticity in the developing nervous system. Nat. Rev. Neurosci. 5:97–107. 10.1038/nrn1327 [DOI] [PubMed] [Google Scholar]
- Wang W., Zhang X., Gao Q., and Xu H.. 2014. TRPML1: An ion channel in the lysosome. Handb. Exp. Pharmacol. 222:631–645. 10.1007/978-3-642-54215-2_24 [DOI] [PubMed] [Google Scholar]
- Wong E., and Cuervo A.M.. 2010. Autophagy gone awry in neurodegenerative diseases. Nat. Neurosci. 13:805–811. 10.1038/nn.2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.