Zaganjor et al. preview work from Webb et al. that shows that the liver-specific isoform of the glycolysis enzyme PFK1 can assemble into filaments and localize to distinct puncta near the plasma membrane.
Abstract
Numerous metabolic enzymes assemble into filamentous structures, which are thought to serve additional regulatory functions. In this issue, Webb et al. (2017. J. Cell Biol. https://doi.org/10.1083/jcb.201701084) show that the liver-specific isoform of phosphofructokinase-1 forms filaments in vitro and localizes as puncta in cells along the plasma membrane. This suggests spatial organization of glycolysis in higher organisms.
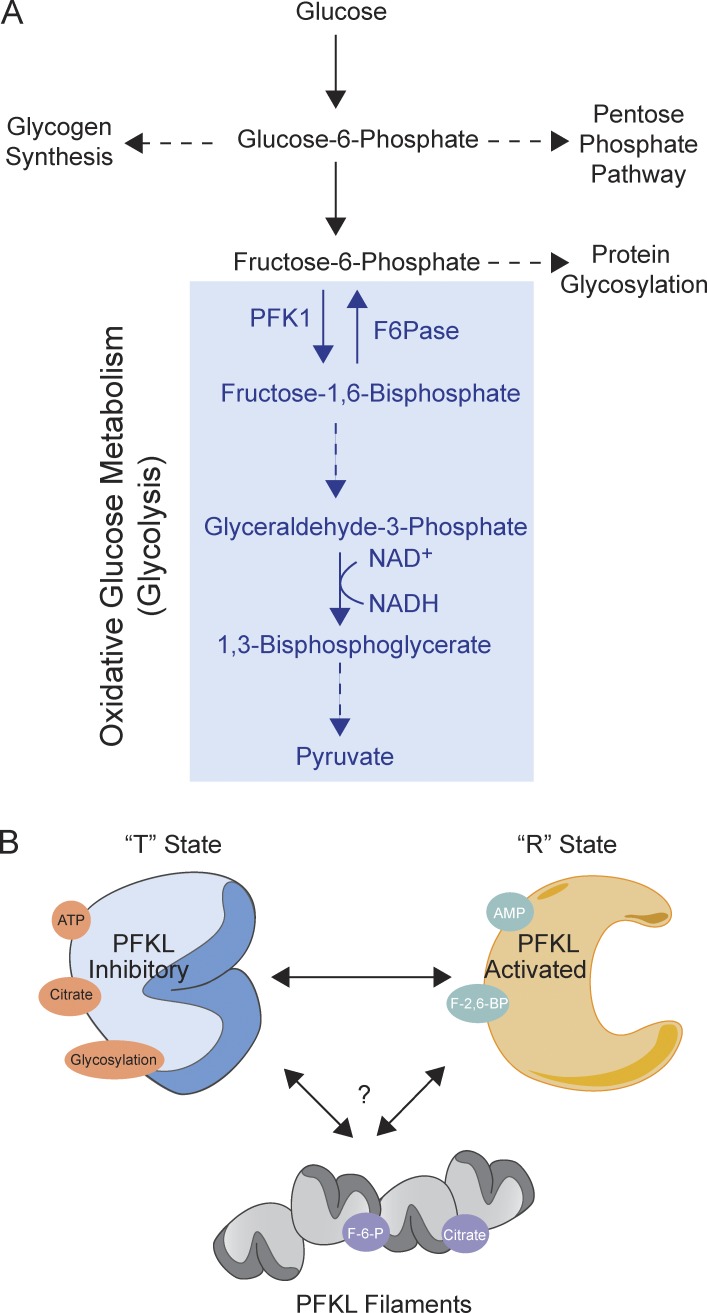
Glycolysis is the core of central carbon metabolism; its intermediates provide precursors important for generating ATP through glucose oxidation, serine for one carbon metabolism, sugars for protein glycosylation, and building blocks for nucleotide synthesis through the pentose phosphate shunt (Fig. 1 A). Although glucose metabolism is highly studied, spatiotemporal aspects of glycolysis remain largely unexplored. Polymerization of metabolic enzymes is one means of spatially regulating cellular processes, and it has been observed for numerous enzymes including acetyl-CoA carboxylase (ACC), cytidine triphosphate (CTP) synthase, glutamate dehydrogenase, and β-glucosidase (O’Connell et al., 2012). A recent study has shown that PFK1, the fate-determining step of glycolysis that converts fructose 6-phosphate (F6P) to fructose 1,6-bisphosphate (F1,6BP) polymerizes in yeast (Shen et al., 2016). However, it is unclear whether PFK1 can polymerize in higher organisms, how PFK1 polymerization is regulated, and the structural and functional features of these fibers. Webb et al. set out to address these uncertainties.
Figure 1.
The liver isoform of PFKL, a gatekeeper of glycolysis, forms filaments. (A) Schematic of glucose usage. Glucose is broken down and used for numerous metabolic processes, including for ATP-generating glycolysis. PFK1 is an irreversible regulator and thus commits glucose to oxidative degradation. (B) PFK1 has two states of quaternary structure: the inhibitory T state and the activating R state. The T state is regulated by ATP, citrate, and glycosylation; the R state is stabilized by AMP and F2,6BP, causing increased glycolytic flux. The liver isoform of PFK1 is further regulated through filament formation.
Using both transmission electron microscopy and 90° light scattering, Webb et al. (2017) demonstrated that a recombinant liver-specific isoform of PFK1 (liver PFK1; PFKL) forms filaments in vitro. However, filament assembly was not observed in platelet PFK1 (PFKP)- or muscle PFK1–specific isoforms. Remarkably, the PFKL filaments were an average of 65.4 nm in length, approximately six tetramers long. Binding of F6P, which is a substrate of PFK1, strongly induced PFKL polymerization, and mutagenesis of the F6P binding pocket revealed that F6P binding was crucial for filament formation.
In addition to identifying PFKL polymerization, Webb et al. (2017) examined the filament structure with negative-stain EM and iterative helical real-space reconstruction. They found that PFKL filaments are distinct from other filamentous enzymes. PFKL filaments formed through interaction of the C-terminal regulatory domains of tetramers. In the filament structure, each tetramer is composed of two structurally distinct dimers, dimers A and B. Each dimer has two interfaces, in which interface 1 of dimer A interacts with interface 1 of dimer A on the adjacent tetramer, and interface 2 of dimer B is solvent exposed. Furthermore, longitudinal contacts alternate, with interface 2 of dimer A interacting with interface 1 of dimer B on the adjacent tetramer. This creates an unusual helical symmetry such that the last subunit can either (a) bind another subunit in a linear manner or (b) bind a subunit and introduce a kink into the filament. The kinks within the PFKL filaments appeared abundant and random.
Because the C-terminal regulatory domain enables tetramers to interact within PFKL filaments, Webb et al. (2017) assessed whether this domain is sufficient for assembly. Using a chimera of the PFKL C-terminal regulatory domain and PFKP catalytic region, they identified that the C-terminal regulatory domain of PFKL is sufficient to form filaments. This result underscores the importance of isoform specificity in filament formation.
The liver is a critical anabolic organ that supports systemic metabolism (Rui, 2014). In particular, the liver senses the levels of glucose in an organism and uses the processes of glycogenolysis and gluconeogenesis to generate glucose, which is subsequently delivered to other tissues in the body. The process of glycolysis (glucose breakdown) and gluconeogenesis (glucose synthesis) are opposing. Therefore, most of the enzymes in glucose metabolism are bidirectional to support both anabolic and catabolic glucose metabolism. The catalytic activity of most enzymes involved in glucose metabolism is allosterically regulated by metabolites. PFK1 is distinct, however, because it is not reversible, and it is the fate-determining step of oxidative glucose metabolism (Fig. 1 A).
PFK1 has two states of quaternary structure: the inhibitory T state and the activated R state (Fig. 1 B; Webb et al., 2015). ATP, citrate, and phosphoenolpyruvate bind PFK1, stabilizing the T state and therefore inhibiting catalytic activity. These metabolites are elevated in a cell when there is sufficient energy production through TCA cycle flux and glycolysis, respectively. Alternatively, metabolites such as AMP and F2,6BP stabilize the PFK1 R state, activating it to increase glycolytic flux in low-energy conditions. In addition to allosteric modifications, PFK1 is regulated by posttranslational modifications such as glycosylation. Glycosylation inhibits PFK1 activity and rewires glucose metabolism to the pentose phosphate pathway (Yi et al., 2012). Whether PFK1 filament formation is a novel mechanism of enzyme regulation for unique glucose metabolism in the liver is still unclear. The isoform specificity of PFK1 polymerization shown in this study strongly implies that this structural feature may dictate differing PFK1 regulation of glycolysis in tissues of higher organisms. Mechanistically, because the liver performs gluconeogenesis, it may be the only organ that can produce a high enough local concentration of F6P to activate PFKL filament formation. Although we can speculate about different organ functions and unique requirements for their enzyme regulation, the question of how and why PFKL assembles into the filament, whereas PFKP and PFKM do not, remains to be determined.
What is the significance of filament formation for metabolic enzymes? Currently, there is no unifying view for why metabolic enzymes organize in filaments. Indeed, filament formation can enhance or inhibit activity of a metabolic enzyme. For example, starvation in yeast results in a drop in pH, which subsequently causes the formation of enzymatically inactive glutamine synthetase filaments (Petrovska et al., 2014). Alternatively, ACC is activated by polymerization into filaments (Beaty and Lane, 1983). Although Webb et al. (2017) do not definitively provide evidence that filament formation of PFKL alters its activity, they raise this hypothesis as a possibility for two reasons. First, PFKL assembly into filaments is substrate dependent in vitro, suggesting regulation of activity. However, the concentration of F6P that induces PFKL filament formation in vitro is not present in the cell, where additional factors may be required to promote filament formation. Second, the PFKL catalytic site remains exposed upon filament formation, suggesting that the enzyme may be in the active state. Future studies are required to determine whether the filamentous structure of PFKL regulates its activity and whether PFKL filaments form during distinct physiological conditions in vivo.
Aside from modulating the activity of an enzyme, filament formation may allow for localized production of metabolic intermediates. Webb et al. (2017) found PFKL particles localized near the plasma membrane. This localized F1,6BP production could potentially affect cell protrusions, endocytosis, or cytoskeletal remodeling that supports migration. Indeed, the cytoskeletal dynamics are intimately linked with glycolysis. Aldolase, which is a glycolytic enzyme downstream of PFK1 and converts F1,6BP into dihydroxyacetone-phosphate and glyceraldehyde 3-phosphate, is sequestered and inhibited by actin filaments (F-actin; Arnold and Pette, 1970). Phosphoinositide 3-kinase–mediated cytoskeletal turnover allows for release of aldolase and increased glycolysis (Hu et al., 2016). In this case, it is possible that localized ATP production from glycolysis could increase F-actin formation, whereas a drop in energy and ATP hydrolysis would release and activate glycolytic enzymes to drive ATP production. Although PFK1 has been shown to associate with F-actin (Clarke and Masters, 1975), it is unclear whether it is filamentous and how this may affect its activity.
Metabolic enzymes have evolved to rapidly respond to ever-changing cellular conditions. Therefore, a high order of regulation is required to fine-tune their activity. Advances in metabolomics have allowed us to assess global changes in cellular metabolic profiles, and yet the spatiotemporal aspects of metabolism are still largely unexplored and represent an exciting area for future studies. The existence of metabolic enzyme filaments implies the importance of localized metabolism and suggests an additional layer of complexity and regulation of glycolysis.
Acknowledgments
E. Zaganjor was supported by the American Heart Association. J.B. Spinelli was supported by the National Science Foundation Graduate Research Fellowship Program (grant DGE1144152). M.C. Haigis was funded by the Ludwig Center at Harvard Medical School.
The authors declare no competing financial interests.
References
- Arnold H., and Pette D.. 1970. Binding of aldolase and triosephosphate dehydrogenase to F-actin and modification of catalytic properties of aldolase. Eur. J. Biochem. 15:360–366. 10.1111/j.1432-1033.1970.tb01016.x [DOI] [PubMed] [Google Scholar]
- Beaty N.B., and Lane M.D.. 1983. Kinetics of activation of acetyl-CoA carboxylase by citrate. Relationship to the rate of polymerization of the enzyme. J. Biol. Chem. 258:13043–13050. [PubMed] [Google Scholar]
- Clarke F.M., and Masters C.J.. 1975. On the association of glycolytic enzymes with structural proteins of skeletal muscle. Biochim. Biophys. Acta. 381:37–46. 10.1016/0304-4165(75)90187-7 [DOI] [PubMed] [Google Scholar]
- Hu H., Juvekar A., Lyssiotis C.A., Lien E.C., Albeck J.G., Oh D., Varma G., Hung Y.P., Ullas S., Lauring J., et al. . 2016. Phosphoinositide 3-kinase regulates glycolysis through mobilization of aldolase from the actin cytoskeleton. Cell. 164:433–446. 10.1016/j.cell.2015.12.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell J.D., Zhao A., Ellington A.D., and Marcotte E.M.. 2012. Dynamic reorganization of metabolic enzymes into intracellular bodies. Annu. Rev. Cell Dev. Biol. 28:89–111. 10.1146/annurev-cellbio-101011-155841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovska I., Nüske E., Munder M.C., Kulasegaran G., Malinovska L., Kroschwald S., Richter D., Fahmy K., Gibson K., Verbavatz J.M., and Alberti S.. 2014. Filament formation by metabolic enzymes is a specific adaptation to an advanced state of cellular starvation. eLife. 3e02409 10.7554/eLife.02409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L. 2014. Energy metabolism in the liver. Compr. Physiol. 4:177–197. 10.1002/cphy.c130024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q.J., Kassim H., Huang Y., Li H., Zhang J., Li G., Wang P.Y., Yan J., Ye F., and Liu J.L.. 2016. Filamentation of metabolic enzymes in Saccharomyces cerevisiae. J. Genet. Genomics. 43:393–404. 10.1016/j.jgg.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B.A., Forouhar F., Szu F.E., Seetharaman J., Tong L., and Barber D.L.. 2015. Structures of human phosphofructokinase-1 and atomic basis of cancer-associated mutations. Nature. 523:111–114. 10.1038/nature14405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B.A., Dosey A.M., Wittmann T., Kollman J.M., and Barber D.L.. 2017. The glycolytic enzyme phosphofructokinase-1 assembles into filaments. J. Cell Biol. 10.1083/jcb.201706005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi W., Clark P.M., Mason D.E., Keenan M.C., Hill C., Goddard W.A. III, Peters E.C., Driggers E.M., and Hsieh-Wilson L.C.. 2012. Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science. 337:975–980. 10.1126/science.1222278 [DOI] [PMC free article] [PubMed] [Google Scholar]