Abstract
Transcription pause release from gene promoters has been recognized to be a critical point for transcriptional regulation in higher eukaryotes. Recent studies suggest that regulatory RNAs are extensively involved in transcriptional control, which may enlist various RNA binding proteins. We recently showed a key role of SRSF2, a member of the SR family of splicing regulators, in binding to promoter-associated small RNA to mediate transcription pause release, a regulatory strategy akin to the function of the HIV Tat protein via binding to the TAR element in nascent RNA to activate transcription. In this report, we further dissect the structural requirement for SRSF2 to function as a transcription activator and extend the analysis to multiple SR and hnRNP proteins by using the MS2 tethering strategy. Our results reveal that SRSF2 is a unique SR protein that activates transcription in a position-dependent manner while three other SR proteins enhance translation in a position-independent fashion. In contrast, multiple hnRNP proteins appear to negatively influence mRNA levels, especially when tethered in the gene body. These findings suggest broad participation of RNA binding proteins in diverse aspects of regulated gene expression at both the transcriptional and posttranscriptional levels in mammalian cells.
Keywords: SR proteins, hnRNP proteins, transcriptional pause release, transcriptional and posttranscriptional control of gene expression
Introduction
Transcription is known to be a highly regulated process from initial assembly of the pre-initiation complex at gene promoters to transcription pause release from the pausing sites near the promoter proximal region, to elongation within the gene body, and to transcription termination at the end of gene.1,2 Each of these distinct steps in transcription is also coupled with various RNA processing reactions, raising the possibility that transcription and co-transcriptional RNA processing may mutually influence one another, which together may constitute a highly intertwined regulatory circuitry during gene expression in mammalian cells.3-5 Furthermore, increasing evidence suggests the involvement of various non-coding RNAs in transcriptional control, which likely enlists specific RNA binding proteins in the regulation.6
Indeed, various RNA binding factors or protein-RNA complexes (known as RNPs for ribonuclear protein particles) have been implicated in transcription, thus expanding their functions to both transcriptional and posttranscriptional levels. For example, hnRNP L has been shown to interact with a specific component in the large Mediator complex, which appears to assist efficient assembly of the RNAP II complex at gene promoters;7 hnRNP U seems to interact with some key transcription co- activators, such as p300, to induce transcription;8 and hnRNP K has been demonstrated to play a vital role in p53-regulated gene expression.9 These RNA binding proteins likely function as part of RNPs via specific RNA molecules, even though we currently know little about specific mechanisms for these RNA binding proteins to operate in various regulatory gene expression events.
The SR (for serine/arginine-rich) family of proteins is a class of RNA binding proteins with well-established roles in constitutive and alternative pre-mRNA splicing in higher eukaryotes.10,11 Interestingly, SR proteins have also been implicated in diverse pathways in gene expression from pre-mRNA splicing in the nucleus to mRNA nuclear export to translational control in the cytoplasm.12 Pursuing SRSF2, a specific SR protein family member previously implicated to play a direct role in transcription elongation,13 we recently demonstrated that this SR protein is intimately associated with a large number of gene promoters via the 7SK complex where its switch from the 7SK non-coding RNA to nascent RNA induces the relocation of a key RNAP II C-terminal domain (CTD) kinase P-TEFb from the 7SK complex to the RNAP II complex assembled at gene promoters.14 This proves to be a critical event for triggering RNAP II transcriptional pause release from the initial pausing site to enter the gene body. Interestingly, the elucidated mechanism draws multiple parallels to the HIV tat protein, which activates transcription through binding to the TAR element present at the 5′ end of transcribed HIV RNA.15 These findings suggest that SRSF2 may function as a long-sought Tat equivalent for activation of cellular genes through its sequential interaction with different promoter- associated RNPs.
Many questions remain to be addressed with respect to the function of SRSF2 in transcriptional control. In particular, many SR proteins show related functions in the splicing reaction and our previous study only compared between two prototypical SR proteins SRSF1 and SRSF2; it thus remains to be determined whether a direct role in transcription is uniquely associated with SRSF2 or also attributable to other SR protein family members. As SRSF2 appears to carry a sequence to specifically interfere with its ability to shuttle out of the nucleus, it is curious whether this non-shuttling property is related to its function in transcription activation. Given the emerging roles of hnRNP proteins in transcription, we also wonder how SR proteins might compare with hnRNP proteins in transcriptional control. With these questions in mind, we used the MS2 tethering strategy to first determine the structural requirement for SRSF2 to function in transcription activation. As the MS2 strategy permits binding of individual MS2 fusion proteins to an engineered high affinity MS2 binding site in transcribed RNA so that different RNA binding proteins can be compared under the same experimental conditions,16 we also utilized this approach to analyze a large panel of SR and hnRNP proteins relative to SRSF2 in the regulation of gene expression at both the transcriptional and posttranscriptional levels.
Results
The MS2 system for functional studies of RNA binding proteins
We previously used a PUF domain-based strategy to demonstrate the role of SRSF2 in transcription activation.14 In the present study, we wished to take a related, but independent reporter system to further confirm the activity of SRSF2 in transcriptional activation. We decided to take the MS2 tethering strategy for this purpose, because the approach has been widely employed to target individual proteins to specific locations in RNA, thus bypassing the requirement for cis-acting element-dependent recruitment of the protein under investigation for functional analysis.17 Similar to our previous study, we fused a luciferase reporter driven by the HSV promoter (Fig. 1A). The reporter also contains a splicing unit in the front of the luciferase coding sequence, thus permitting us to insert three copies of the MS2 binding sites either in the first or second exon before the translational start.
Figure 1.
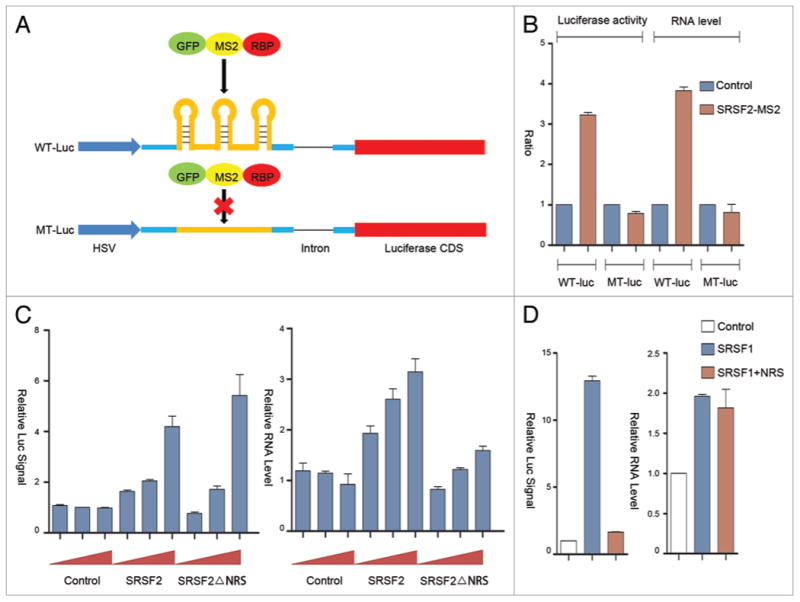
The MS2 tethering strategy to decipher the potential contribution of the nuclear retention signal (NRS) in SRSF2 to transcription activation. (A) Schematic illustration of the MS2 tethering strategy. Three copies of the MS2 binding sites were cloned in the first exon of the reporter. The mutant version deleted the stem-loop from the MS2 binding site. (B) Dual-luciferase assay and RT-qPCR analysis upon co-transfection of the GFP-MS2-SRSF2 fusion protein with the luciferase reporters. (C) Dosage-dependent activation of the wild type luciferase reporter at the enzymatic and RNA levels by the GFP-MS2-SRSF2 fusion with or without the NRS (simply labeled as SRSF2 or SRSF2ΔNRS). (D) Dual-luciferase assay (left panel) and RT-qPCR (right panel) upon expressing the wild type GFP-MS2-SRSF1 protein or the protein further fused with the NRS from SRSF2.
Previous studies showed that a point mutation in the stem region of the MS2 binding motif is sufficient to eliminate sequence specific binding by MS2 fusion proteins.18 However, our initial analysis indicated that the mutant still showed some degree of response to the MS2-SRSF2 fusion protein, likely due to some promiscuous binding activities of the SR protein to the mutant motif (data not show). Therefore, to minimize such potential non-specific effects of this and other RNA binding proteins, we decided to construct a non-responsive reporter to serve as a negative control by using neutral linker sequences to replace the MS2 stem-loop, which allowed us to effectively record the MS2 motif-dependent activities in response to transfected RNA binding proteins.
To verify the responsiveness of this reporter system to SRSF2 fused to a MS2 binding motif, we linked the SRSF2 coding sequence to a GFP-MS2 fusion protein at the C-terminus (Fig. 1A). As expected, we detected the ability of this GFP-MS2-SRSF2 fusion protein to activate the luciferase reporter that carries the wild type, but not mutant MS2 binding site in co-transfected HEK293T cells (Fig. 1B). Such activation of the luciferase activity is correlated with SRSF2-dependent production of the reporter mRNA, consistent with transcriptional activation and thus confirming our previous results from the PUF-based reporter system.14
Contribution of the nuclear retention signal in SRSF2 to transcription activation
Compared with most SR proteins, SRSF2 does not efficiently shuttle between the nucleus and the cytoplasm.19 A previous domain swap study revealed that the C-terminal portion of SRSF2 functions as a nuclear retention signal (NRS), which is transferable to a shuttling SR protein to convert it to a non- shuttling one.19,20 This functional property of the NRS implies a strong nuclear interaction(s) responsible for retarding SRSF2 within the nucleus. We hypothesized that this NRS might be responsible for SRSF2-dependent transcription activation in the nucleus.
To test this hypothesis, we examined the NRS deleted construct and observed that the GFP-MS2-SRSF2 depleted of NRS (simply referred to as SRSF2ΔNRS) was still able to activate the reporter activity (Fig. 1C, left panel). However, the mRNA level was significantly reduced (Fig. 1C, right panel). We detected no effect of the NRS depleted SRSF2 on the mutant reporter at both the luciferase activity or mRNA levels (data not shown). In light of the previous finding that RS domains from several SR proteins are able to enhance translation when tethered to a luciferase reporter,21 we interpret these results to indicate that, while depletion of the NRS reduced the activity of SRSF2 in transcription activation, the removal of NRS permits the mutant SRSF2 to shuttle out of the nucleus to increase translation of the reporter, thereby producing a neutral net effect on the luciferase activity. Importantly, this analysis suggests a significant contribution of the NRS to the activity of SRSF2 in transcription activation.
To determine whether the NRS activity in transcription is transferrable, we next fused the NRS to SRSF1 to convert this SR protein to a non-shuttling SR protein, which has been previously documented.21 As expected, this largely attenuated SRSF1- dependent translational enhancement of the luciferase reporter (Fig. 1D). By measuring the reporter mRNA, we found that the wild type GFP-MS2-SRSF1 modestly increased the mRNA level, likely due to some indirect effects in transfected HEK293T cells, as we previously observed,14 and importantly, fusion of the SRSF2 NRS to SRSF1 was unable to further increase the mRNA production (Fig. 1D). These data indicate that the NRS alone was insufficient to function as a trans-activation domain in the context of another SR protein.
Domain requirement for SRSF2 to act as a transcription activator
To systematically dissect the domain requirement for the transcription activation activity of SRSF2, we divided the protein into RNA recognition motif (RRM), spacer, the arginine/serine (RS) domain, and NRS, each of which was fused with the GFP-MS2 protein and examined their activities on our reporter system (Fig. 2A). The full-length SRSF2 severed as a positive control. We also constructed and tested a RNA binding deficient SRSF2 expression unit in which the two conserved phenylalanine residues in its RRM were mutated to aspartic acids (FF2DD). It appeared that only the RS domain had a recordable activity in the luciferase reporter assay and extension of the RS by including the N-terminal spacer or C-terminal NRS was able to further increase the reporter activity (Fig. 2B). However, based on the amount of mRNA produced by the reporter, it is evident that only full-length SRSF2 was able to maximally function as a transcription activator. As expected, the RRM mutant SRSF2 functioned like wild type SRSF2 when tethered to the reporter. Collectively, these data strongly suggest that individual parts of the SRSF2 protein function together in transcription activation.
Figure 2.
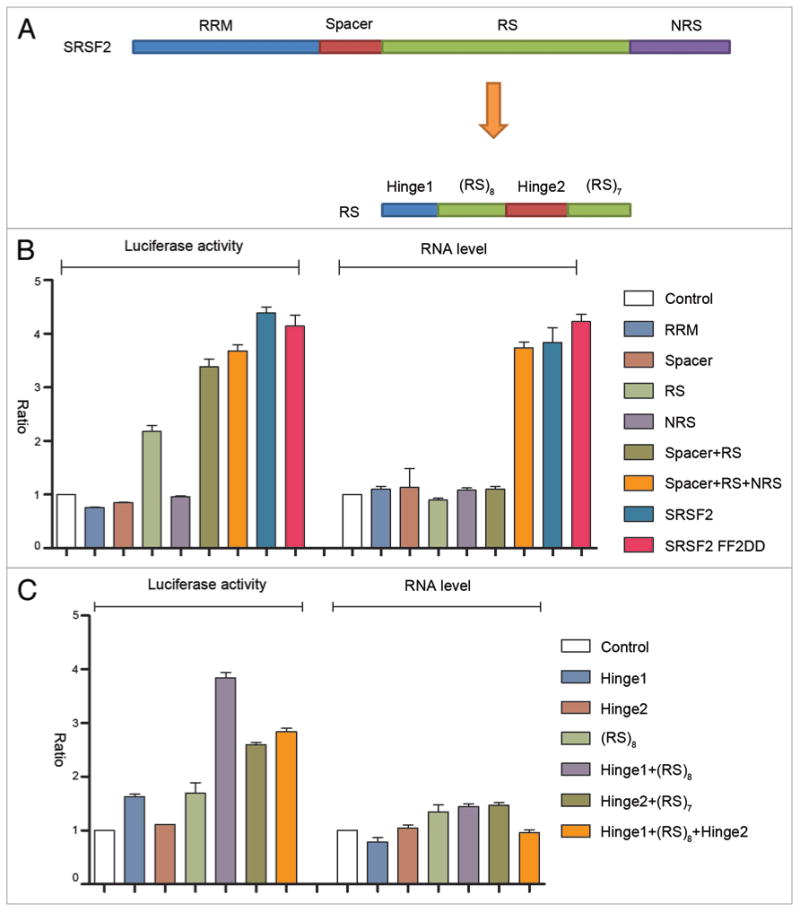
Dissecting the sequence requirement for SRSF2 to act as a transcription activator. (A) Illustration of SRSF2 domains and sub-domains. (B and C) Dual-luciferase and RT-qPCR assays upon expression of different SRSF2 fragments fused to GFP-MS2.
We further dissected the RS domain of SRSF2 into smaller fragments, which could be divided into two hinge regions at the ends and two sub-RS domains consisting of 8 or 7 arginine/serine repeats. The data indicated that a hinge region plus an sub-RS domain had the most activity in reporter activation, and again, none of the subdomains in the RS domain of SRSF2 was able to significantly increase the mRNA level of the reporter (Fig. 2C), which is largely consistent with previous finding that the RS domain is responsible for translational enhancement in the cytoplasm.21 Taken together, these structure-function analyses suggest, although individual domains in SRSF2 may independently affect other aspects of RNA metabolism, the overall structure is essential for SRSF2 to act as a transcription activator.
Systematic comparison among SR and hnRNP proteins
We next wished to extend the analysis to other typical SR protein family members and to multiple hnRNP proteins to determine potential function of these two important classes of RNA binding proteins in transcription activation (or repression) when tethered on the luciferase reporter. We cloned these proteins as GFP-MS2 fusion proteins, all of which were expressed at equivalent levels in transfected HEK293T cells (Fig. 3A and D). Because of potential non-specific effects via other sequences in the reporter, we displayed the effects of individual fusion proteins on both the wild type and mutant reporters, rather than calculating the activity of each fusion protein as a ratio of the wild type over the mutant reporter. This allowed us to evaluate the magnitude of both indirect and MS2 tethering site-dependent effects.
Figure 3.
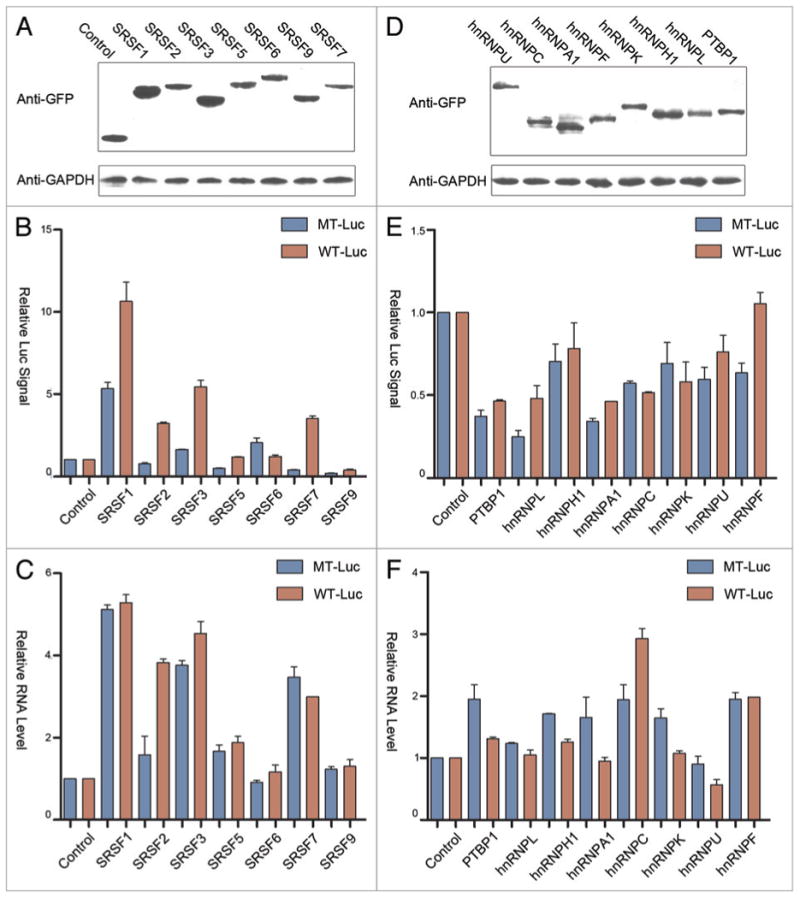
Comparison among SR and hnRNP proteins. (A and D) western blot analysis of MS2-GFP fused to individual SR (panel A) and hnRNP (panel D) proteins detected by anti-GFP antibody. GAPDH protein was probed as the loading control. (B and E) Dual luciferase assays for SR (panel B) and hnRNP (panel E) proteins on wild type (red) and mutant (blue) luciferase reporters. (C and F) RT-qPCR analysis of luciferase mRNA from cells transfected with individual SR (panel C) and hnRNP (panel F) proteins.
Interestingly, besides SRSF2, we found that SRSF1 (SF2/ASF), SRSF3 (SRp20) and SRSF7 (9G8) were able to stimulate the luciferase activity that depended on their tethering to the MS2 binding sites in the reporter (Fig. 3B). By monitoring reporter mRNA production, we found that overexpressed SRSF1, 3, and 7 appear to indirectly increase the mRNA level, as their effects were not dependent on the MS2 binding sites in the reporter (Fig. 3C). These findings imply that the MS2 binding-dependent induction of the luciferase activity may be due to the function of these shuttling SR proteins in enhancing translation in the cytoplasm. Because the reporter contains a small intron, we checked potential additional impact of tethered SR proteins on mRNA splicing. The data indicate that none of the first exon tethered SR proteins showed further enhancement on this constitutive splicing event (Fig. S1A and B). These data indicate that some minor changes in splicing made minimal impact on the observed changes in both luciferase activity and steady-state mRNA.
In contrast to SR proteins, most hnRNP proteins appear to modulate both the reporter activity (Fig. 3E) and mRNA levels (Fig. 3F) to various degrees, but in general, the effects are relatively minor compared with SR proteins. At the luciferase activity level, all hnRNP proteins showed some inhibitory effects, but not dependent on binding to the MS2 site. We also detected some changes in the mRNA level in hnRNP protein co-transfected cells, which may reflect experimental variations and/or complex effects of these RNA binding proteins in modulation of mRNA production and/or stability, as none of the detected effects was MS2-dependent. In contrast, the tethered PTBP1, hnRNP K, U, and F each showed a degree of inhibition of reporter mRNA splicing (Fig. S1A and B), in line with the known influence of exon-bound hnRNP proteins on splicing, despite the fact that not all hnRNP proteins tested showed recordable effects in our current reporter system. Importantly, the comparison between SR and hnRNP proteins indicates that SRSF2 appears to play a unique role in transcription activation, suggesting that it has separately evolved to acquire such additional function in higher eukaryotes.
Positional effects of SR and hnRNP proteins in transcriptional and posttranscriptional control
One of the striking properties of SRSF2 in transcription activation is the requirement for the RNA binding protein to interact with nascent RNA near the 5′ end in order to effectively trigger transcription pause release from the initial pausing site.14 Our functional analysis and comparison with other RNA binding proteins had been performed on the reporters with the engineered MS2 binding sites at the optional position in the first exon similar to our previous study.14 While this binding site configuration ensures the optional function of SRSF2 in transcription activation, it might not be generally applicable to other RNA binding proteins. To address this possibility, we moved the MS2 binding sites to the second exon, but still before the open reading frame of the luciferase gene (Fig. 4A). Our previous study showed that SRSF2 activates transcription in a highly position sensitive manner, which we now further confirmed on the current MS2 tethering system, as when tethered to the second exon, we only noted a minor effect in enhancing the luciferase reporter, but the effect was unrelated to increased transcription (Fig. 4B and C).
Figure 4.
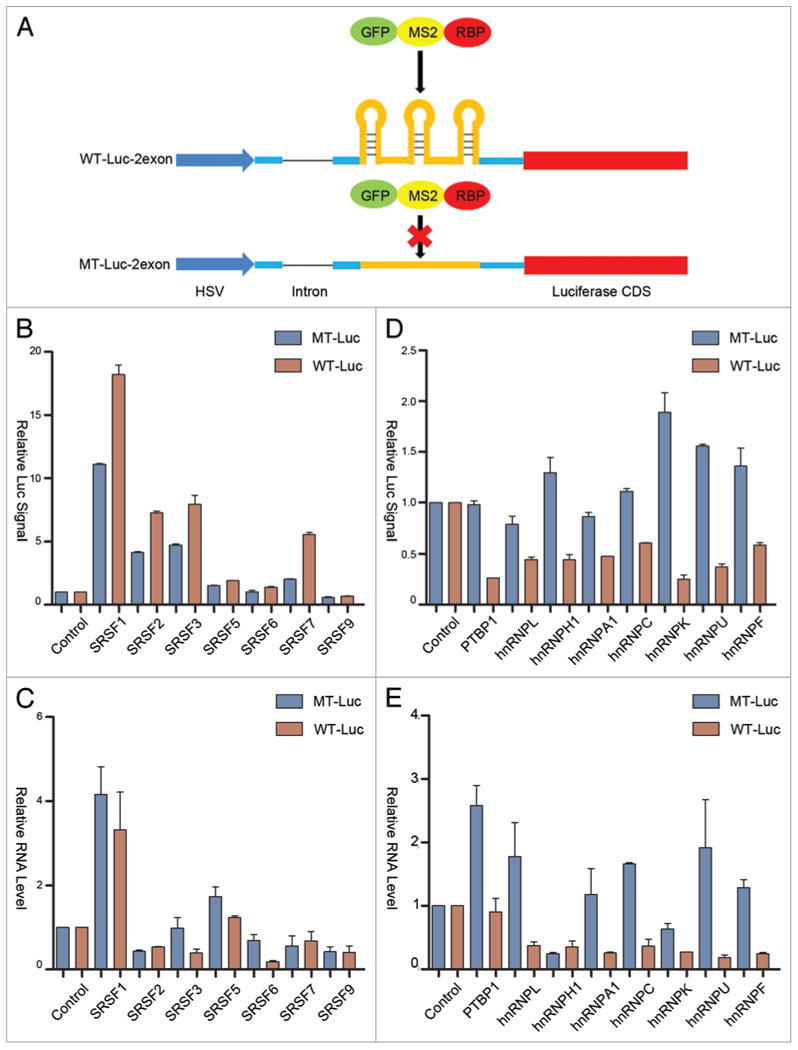
Position-dependent effects of SR and hnRNP proteins in transcriptional and posttranscriptional control. (A) Schematic diagram of the constructs with and without carrying the MS2 binding sites in the second exon of the luciferase reporter. (B and D) Dual luciferase assays for the response to SR (panel B) and hnRNP (panel D) proteins tethered to the second exon of wild type (red) and mutant (blue) luciferase reporters. (C and E) RT-qPCR analysis of reporter RNA in cells transfected with individual SR (panel C) and hnRNP (panel E) proteins fused to GFP-MS2.
Interestingly, we observed that SRSF1, 3, and 7 were still able to activate the luciferase activity when tethered on the second exon, indicating that SR protein-mediated translational enhancement is relatively positional independent (Fig. 4B). As with its tethering to the first exon, SRSF1 also showed some indirect enhancement of the reporter mRNA (Fig. 4C). Interestingly, compared with their effects on the first exon, we observed that individual hnRNPs tethered to the second exon showed more obvious MS2-dependent reduction of the reporter activity (Fig. 4D). These inhibitory effects are also reflected at the level of the reporter mRNA with the exception of hnRNP H1 (Fig. 4E). It is also interesting to note that, relative to their activities on the first exon, the hnRNP proteins appear to be more effective in modulating both reporter mRNA production and translation when were tethered to the second exon. Again, such responses appear to be unrelated to potential impact on the splicing of the intron when these RNA binding proteins were tethered to the second exon, as we only detected some minor inhibition of splicing with PTBP1 and hnRNP K (Fig. S1C and D). We suspect that the location of the MS2 binding sites in the second exon might allow more efficient tethering, perhaps due to less competition with other molecular interactions near the transcription start. These findings suggest an intriguing possibility that hnRNP proteins may be more effective in modulating RNA metabolism when binding on the gene body in mammalian cells.
Discussion
This work extends our recent discovery of SRSF2 as a transcription activator at the promoter proximal region,14 which together highlights the unexpected function of a typical splicing regulator in transcription. Here we discuss both our published work and the new data with emphasis on the emerging role of SRSF2 in transcriptional control as well as other specific RNA binding proteins in diverse aspects of RNA metabolism in mammalian cells.
Transcriptional pause release has been recognized in recent years as a major step in regulated gene expression,2 but how the transcriptionally engaged RNAP II complex moves beyond the initial pausing site has remained a mechanistically challenging question. Most of our existing knowledge is derived from studying HIV Tat-mediated transcription activation.22 In this seemly unique viral strategy, the highly positively charged Tat protein appears to bind the 7SK non-coding RNA, leading to extraction of a key RNAP II CTD kinase P-TEFb from the 7SK complex. The released P-TEFb has been thought to first join the cytoplasmic pool of the active kinase, which is then recruited to the promoter- proximal region via sequence-specific binding of Tat to the TAR element at the 5′ end of nascent viral RNA.23 This series of events likely involves various protein-protein interactions, including the interactions between Tat and the Cyclin T subunit of P-TEFb and between the chromatin-bound Brd4 and the CDK9 subunit of P-TEFb. These highly orchestrated protein-RNA and protein-protein interactions eventually trigger the transition of transcriptionally engaged RNAP II from the pausing to the elongating state.
Relative to such detailed understanding of Tat-mediated HIV transcription, we know little about how cellular genes are activated at this critical step. We recently discovered that SRSF2, a well-characterized splicing factor/regulator, binds to RNA in exonic regions,24 but to DNA around gene promoters.14 By pursuing these distinct genomic interactions, we found that SRSF2 is initially associated with the 7SK complex, which appears to localize together near active gene promoters, instead of being distributed in separate storage sites in the nucleus. Importantly, when a specific binding site for SRSF2 emerges from nascent RNA near the transcription start, it recruits the RNA binding protein, which then triggers the release of P-TEFb from the 7SK complex to transcriptionally engaged RNAP II complex and induces the transition of the RNAPII complex into the gene body. This process is highly reminiscent of HIV Tat-mediated transcription activation, suggesting that SRSF2 may be a long-sought Tat equivalent in activating cellular genes. In this regard, we note that SRSF2 is highly enriched with arginine residues in its RS domain, a feature essential for Tat to activate transcription.25 Our findings also assign a function to promoter-associated small RNAs, which have been detected in various genomic studies,26 indicating that these promoter-associated RNA provide critical signals for transcriptional pause release, much like the HIV TAR element. Therefore, like nearly all other viral strategies, HIV Tat-mediated transcription also resembles a widely used cellular mechanism.
As SRSF2 is a member of the SR family of RNA proteins with overlapping functions in pre-mRNA splicing, the question is whether the transcription activation function of SRSF2 is unique to this SR protein or more generally shared among SR proteins. In our published work,14 we compared SRSF2 with another prototypical SR protein SRSF1 and found that, while this related SR protein shows similar RNA and DNA interaction patterns in the mouse genome as SRSF2, it does not directly activate transcription when tethered to a nascent RNA near the 5′ end. In the present study, we further confirmed this observation by using the MS2 system, and more importantly, extended the analysis to most core SR family members. These results suggest that SRSF2 appears to be a unique SR protein with a direct role in transcription.
We initially suspect that such unique function of SRSF2 may be due to its nuclear retention signal, which is only present in this SR protein. Through domain dissection, we now show that this NRS indeed contributes to the SRSF2-depedent transcription activation. However, this domain alone is insufficient to activate transcription when fused to SRSF1. It is also interesting to observe the requirement of the RRM in SRSF2 for its activity in transcription, even though the protein has been tethered to the MS2 binding site. This indicates that the RNA binding domain may contribute to additional protein-RNA interactions as well as protein-protein interactions, because the RRM in SR proteins has been shown to be able to engage in simultaneous protein-protein and protein-RNA interactions.27 There is only one RRM in SRSF2, but most SR proteins contain two RRMs. However, SRSF3 is another single RRM SR protein, but it lacks the ability to activate transcription based on our reporter assays. These observations imply that both the RNA binding specificity and the RRM configuration may contribute to the transcription activity of SRSF2. We also observed that the spacer region between the RRM and the RS domain contributes to the ability of SRSF2 in transcription activation. Previous studies showed that a similar spacer region in SRSF1 plays a docking function for SR protein specific kinases to efficiently catalyze SR protein phosphorylation.28,29 This suggests a possibility that the spacer region in SRSF2 may have a similar role in mediating its RS domain phosphorylation, pointing to a potential regulatory function of SRSF2 phosphorylation in modulating its activity in transcription. In this regard, we recently established a key role of SR protein specific kinases in transducing external growth signals to regulate the splicing activities of SR proteins in the nucleus,30 raising an intriguing possibility that this newly elucidated signal transduction pathway may modulate SRSF2-dependent and signal-induced transcription.
Finally, we have also extended our analysis to multiple hnRNP proteins in the current work. Since various hnRNP proteins have been implicated as co-factors in transcriptional control, it has been unclear whether individual hnRNP proteins function in transcription via specific non-coding RNAs and/or through their interactions with nascent RNA. Our MS2 tethering assay results indicate that the only recordable activity of hnRNP proteins is the repression of the reporter mRNA. However, we have not yet further distinguished between transcriptional repression and reduced mRNA stability, which deserves a close look at the levels of endogenous gene transcripts in future studies. In any case, these initial observations at least show that hnRNP proteins are unable to activate transcription via binding to nascent transcripts. If these RNA binding proteins have a capacity to modulate transcription, they are more likely involved in transcriptional repression, rather than activation. Clearly, we have much more to learn with respect to the involvement of specific RNA binding proteins in transcriptional control in mammalian cells.
Methods
Plasmid construction
All SR and hnRNP proteins were fused in frame with the common GFP-MS2 polypeptide at the C-terminus. To construct this series of plasmids, the pEGFP-C1 vector was digested with BglII and Hind III and then ligated to the DNA fragment encoding for the MS2 coat protein amplified from the MS2-MBP plasmid digested with the same restriction enzymes. The coding sequences for individual SR and hnRNP proteins were amplified from a cDNA library and fused to the same reading frame of the pEGFP-MS2 expression plasmid.
To engineer the MS2 binding sites in the first or second exon in the Renilla luciferase reporter, new restriction sites were first introduced into the pRL-TK vector (Promega) by PCR followed by insertion of the PCR-amplified DNA fragment containing the wild type or mutant MS2 binding sites, as follows: wt sequences: gatatccgta caccatcagg gtacgagcta gcccttggcg tacaccatca gggtacgact agtagatctc gtacaccatc agggtacgga attctctaga gcgtcgacct caagctcgag; mutant sequences: gatatcagct agcccttgga ctagtagatc tgaattctct agagcgtcga cctcaagctc gag.
Cell culture and dual luciferase assay
HEK293T cells were cultured in DMEM, supplemented with 10% (v/v) FCS and penicillin/streptomycin under humid 5% CO2 at 37°C. The cells were grown to 90% confluence in 48-well plates for transfection. A mixture of 0.82 μg of plasmid DNA consisting of 800 ng pEGFP-MS2-RBP, 10 ng pRL-TK- Renilla Luciferase and 10 ng of the pCMV-Firefly expression vector, and Lipofectamine 2000 (Invitrogen) or Turbofect (Fermentas) was used to transfect cells following the manufacturers' instructions. The transfection medium was replaced with fresh medium after 12 h incubation, and cells were then incubated for another 36 h. For dual luciferase assays, HEK293T cells were lysed using passive lysis buffer (Promega) and the levels of Firefly and Renilla luciferase activities were assayed using the Promega's Dual Luciferase Assay Kit. The luminescence was measured with a GLOMAX luminometer (Promega).
RT-qPCR and western blotting analysis
To monitor reporter RNA levels in transfected cells, total RNA was isolated from transfected HEK293T cells using the TRIzol Reagent (Invitrogen). Following digestion with RQ1 DNase (Promega), cDNA was synthesized using the M-MLV Reverse Transcriptase (Promega) with oligo dT. PCR was performed on the CFX384 Touch (Bio-rad) using SYBR Green fluorescence. Each 20 μL PCR reaction was composed of 1 μL cDNA, 10 μL of 2 × SYBR Green mix (Toyobo), 1 μL of primer mix (0.5 nM final concentration), and 9 μL of water. Amplification conditions were: 95°C for 30 s; 60°C for 20 s; 72°C for 20 s; 50 cycles. PCR products were analyzed using the CFX manager (Bio-Rad) software.
Protein samples were resolved by SDS-PAGE and then transferred to PVDF membranes (PALL). Nonspecific binding sites were blocked by incubation of the membrane with 5% non-fat dry milk in TBST (20 mM Tris at pH 7.5, 137mM NaCl, and 0.1% Tween 20). Proteins were detected using the following primary antibodies (described below) diluted in 1% no fat milk in TBST: mouse monoclonal anti-GFP (1:5000 Proteintech), mouse monoclonal anti-GAPDH (1:1000 Abnova). Following washing in TBST, blots were incubated with the appropriate secondary antibodies conjugated to horseradish peroxidase (Pierce) and detected with Super Signal West Pico detection reagent (Pierce).
Supplementary Material
Acknowledgments
This work was supported by the China 973 program (2011CB811300, 2012CB910800), the Chinese 111 program grant (B06018) and NIH grants (GM049369, HG004659, HG007005) to X-DF.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
References
- 1.Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119–43. doi: 10.1146/annurev-biochem-052610-095910. http://dx.doi.org/10.1146/annurev-biochem-052610-095910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nat Rev Genet. 2012;13:720–31. doi: 10.1038/nrg3293. http://dx.doi.org/10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–5. doi: 10.1016/j.ceb.2008.03.001. http://dx.doi.org/10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luco RF, Allo M, Schor IE, Kornblihtt AR, Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. http://dx.doi.org/10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braunschweig U, Gueroussov S, Plocik AM, Graveley BR, Blencowe BJ. Dynamic integration of splicing within gene regulatory pathways. Cell. 2013;152:1252–69. doi: 10.1016/j.cell.2013.02.034. http://dx.doi.org/10.1016/j.cell.2013.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. http://dx.doi.org/10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y, Li W, Yao X, Lin QJ, Yin JW, Liang Y, Heiner M, Tian B, Hui J, Wang G. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell. 2012;45:459–69. doi: 10.1016/j.molcel.2011.12.022. http://dx.doi.org/10.1016/j.molcel.2011.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Obrdlik A, Kukalev A, Louvet E, Farrants AKO, Caputo L, Percipalle P. The histone acetyltransferase PCAF associates with actin and hnRNP U for RNA polymerase II transcription. Mol Cell Biol. 2008;28:6342–57. doi: 10.1128/MCB.00766-08. http://dx.doi.org/10.1128/MCB.00766-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–19. doi: 10.1016/j.cell.2010.06.040. http://dx.doi.org/10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou Z, Fu XD. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma. 2013;122:191–207. doi: 10.1007/s00412-013-0407-z. http://dx.doi.org/10.1007/s00412-013-0407-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long JC, Caceres JF. The SR protein family of splicing factors: master regulators of gene expression. Biochem J. 2009;417:15–27. doi: 10.1042/BJ20081501. http://dx.doi.org/10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 12.Zhong XY, Wang P, Han J, Rosenfeld MG, Fu XD. SR proteins in vertical integration of gene expression from transcription to RNA processing to translation. Mol Cell. 2009;35:1–10. doi: 10.1016/j.molcel.2009.06.016. http://dx.doi.org/10.1016/j.molcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu XD. The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol. 2008;15:819–26. doi: 10.1038/nsmb.1461. http://dx.doi.org/10.1038/nsmb.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, Xiao R, Burge CB, Fu XD. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell. 2013;153:855–68. doi: 10.1016/j.cell.2013.04.028. http://dx.doi.org/10.1016/j.cell.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ott M, Geyer M, Zhou Q. The control of HIV transcription: keeping RNA polymerase II on track. Cell Host Microbe. 2011;10:426–35. doi: 10.1016/j.chom.2011.11.002. http://dx.doi.org/10.1016/j.chom.2011.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Z, Reed R. Purification of functional RNA- protein complexes using MS2-MBP. Current Protocols in Molecular Biology. 2003;Chapter 27(Unit 3) doi: 10.1002/0471142727.mb2703s63. [DOI] [PubMed] [Google Scholar]
- 17.Erkelenz S, Mueller WF, Evans MS, Busch A, Schöneweis K, Hertel KJ, Schaal H. Position- dependent splicing activation and repression by SR and hnRNP proteins rely on common mechanisms. RNA. 2013;19:96–102. doi: 10.1261/rna.037044.112. http://dx.doi.org/10.1261/rna.037044.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witherell GW, Gott JM, Uhlenbeck OC. Specific interaction between RNA phage coat proteins and RNA. Prog Nucleic Acid Res Mol Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. http://dx.doi.org/10.1016/S0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]
- 19.Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Cáceres JF. Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol. 2002;22:6871–82. doi: 10.1128/MCB.22.19.6871-6882.2002. http://dx.doi.org/10.1128/MCB.22.19.6871-6882.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin SR, Xiao R, Sun PQ, Xu XD, Fu XD. Dephosphorylation-dependent sorting of SR splicing factors during mRNP maturation. Mol Cell. 2005;20:413–25. doi: 10.1016/j.molcel.2005.09.015. http://dx.doi.org/10.1016/j.molcel.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 21.Sanford JR, Gray NK, Beckmann K, Cáceres JF. A novel role for shuttling SR proteins in mRNA translation. Genes Dev. 2004;18:755–68. doi: 10.1101/gad.286404. http://dx.doi.org/10.1101/gad.286404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. http://dx.doi.org/10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 23.Krueger BJ, Varzavand K, Cooper JJ, Price DH. The mechanism of release of P-TEFb and HEXIM1 from the 7SK snRNP by viral and cellular activators includes a conformational change in 7SK. PLoS One. 2010;5:e12335. doi: 10.1371/journal.pone.0012335. http://dx.doi.org/10.1371/journal.pone.0012335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pandit S, Zhou Y, Shiue L, Coutinho-Mansfield G, Li H, Qiu J, Huang J, Yeo GW, Ares M, Jr, Fu XD. Genome-wide analysis reveals SR protein cooperation and competition in regulated splicing. Mol Cell. 2013;50:223–35. doi: 10.1016/j.molcel.2013.03.001. http://dx.doi.org/10.1016/j.molcel.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Calnan BJ, Biancalana S, Hudson D, Frankel AD. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991;5:201–10. doi: 10.1101/gad.5.2.201. http://dx.doi.org/10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- 26.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. doi: 10.1038/nrg3074. http://dx.doi.org/10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 27.Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, Ghosh G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1-70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci U S A. 2011;108:8233–8. doi: 10.1073/pnas.1017700108. http://dx.doi.org/10.1073/pnas.1017700108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hagopian JC, Ma CT, Meade BR, Albuquerque CP, Ngo JCK, Ghosh G, Jennings PA, Fu XD, Adams JA. Adaptable molecular interactions guide phosphorylation of the SR protein ASF/SF2 by SRPK1. J Mol Biol. 2008;382:894–909. doi: 10.1016/j.jmb.2008.07.055. http://dx.doi.org/10.1016/j.jmb.2008.07.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngo JCK, Chakrabarti S, Ding JH, Velazquez-Dones A, Nolen B, Aubol BE, Adams JA, Fu XD, Ghosh G. Interplay between SRPK and Clk/Sty kinases in phosphorylation of the splicing factor ASF/SF2 is regulated by a docking motif in ASF/SF2. Mol Cell. 2005;20:77–89. doi: 10.1016/j.molcel.2005.08.025. http://dx.doi.org/10.1016/j.molcel.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Z, Qiu J, Liu W, Zhou Y, Plocinik RM, Li H, Hu Q, Ghosh G, Adams JA, Rosenfeld MG, et al. The Akt-SRPK-SR axis constitutes a major pathway in transducing EGF signaling to regulate alternative splicing in the nucleus. Mol Cell. 2012;47:422–33. doi: 10.1016/j.molcel.2012.05.014. http://dx.doi.org/10.1016/j.molcel.2012.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


