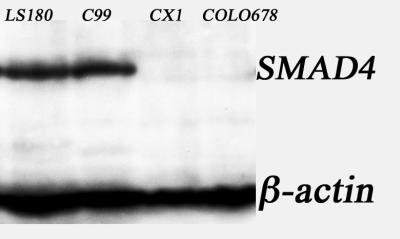Abstract
Loss of chromosome 18q21 is well documented in colorectal cancer, and it has been suggested that this loss targets the DCC, DPC4/SMAD4, and SMAD2 genes. Recently, the importance of SMAD4, a downstream regulator in the TGF-β signaling pathway, in colorectal cancer has been highlighted, although the frequency of SMAD4 mutations appears much lower than that of 18q21 loss. We set out to investigate allele loss, mutations, protein expression, and cytogenetics of chromosome 18 copy number in a collection of 44 colorectal cancer cell lines of known status with respect to microsatellite instability (MSI). Fourteen of thirty-two MSI− lines showed loss of SMAD4 protein expression; usually, one allele was lost and the other was mutated in one of a number of ways, including deletions of various sizes, splice site changes, and missense and nonsense point mutations (although no frameshifts). Of the 18 MSI− cancers with retained SMAD4 expression, four harbored missense mutations in the 3′ part of the gene and showed allele loss. The remaining 14 MSI− lines had no detectable SMAD4 mutation, but all showed allele loss at SMAD4 and/or DCC. SMAD4 mutations can therefore account for about 50–60% of the 18q21 allele loss in colorectal cancer. No MSI+ cancer showed loss of SMAD4 protein or SMAD4 mutation, and very few had allelic loss at SMAD4 or DCC, although many of these MSI+ lines did carry TGFBIIR changes. Although SMAD4 mutations have been associated with late-stage or metastatic disease, our combined molecular and cytogenetic data best fit a model in which SMAD4 mutations occur before colorectal cancers become aneuploid/polyploid, but after the MSI+ and MSI− pathways diverge. Thus, MSI+ cancers may diverge first, followed by CIN+ (chromosomal instability) cancers, leaving other cancers to follow a CIN−MSI− pathway.
Keywords: colorectal cancer cell lines, 18q
Allele loss at 18q21.1 has been demonstrated in up to 60% of colorectal cancers (CRCs; ref. 1). Mapping to this chromosomal band are DCC (Deleted in Colon Carcinoma), DPC4/SMAD4 (Deleted in Pancreatic Cancer 4) and SMAD2. For many years the allele loss observed around 18q21.1 in colorectal cancer was believed to be targeting DCC (2, 3). It has more recently been shown, however, that mutations of DCC and SMAD2 are very rare in colorectal cancer (4). By contrast, changes in SMAD4 are much more common in the pathogenesis and evolution of colorectal cancer (5, 6), as has also been shown in pancreatic cancer (7). In particular, inactivation of SMAD4 has been associated with late stage or metastatic colorectal cancer (6, 8, 9). Additionally, it has recently been shown that constitutional mutations in SMAD4 can cause Juvenile Polyposis Syndrome (JPS; refs. 10 and 11), a disorder characterized by the presence of hamartomatous polyps in the gastrointestinal tract and a markedly increased risk of developing a gastrointestinal malignancy.
The SMAD4 gene codes for a protein involved as a downstream regulator in the transforming growth factor β (TGF-β) signal transduction pathway. The SMAD4 protein acts as a trimer and forms complexes with the receptor-phosphorylated SMAD2 and SMAD3; these heteromeric complexes then translocate from the cytoplasm to the nucleus where association with DNA binding factors facilitates the transcription of target genes. These target genes include cyclin-dependent kinase inhibitors such as p15(ink4B) (12) and the inhibitory SMAD7 (13, 14). Abrogation of SMAD4 function may cause a breakdown in this signaling pathway and loss of transcription of genes critical to cell-cycle control (15). Cells may therefore become TGF-β resistant and escape from TGF-β-mediated growth control and apoptosis (16).
Loss of 18q21 is seen in 80% of pancreatic cancers, with inactivation of SMAD4 shown to result from homozygous deletions in 30% of cases and point mutations/small alterations in 20% of cases (7). Of the 60% of colorectal cancers showing 18q loss, mutations of SMAD4 have been demonstrated in about one third, with the loss thought to be targeting DCC or another nearby gene in the remainder of cases (1).
We have determined how many of 44 colorectal cancer cell lines have loss of the SMAD4 protein and investigated the cause of this loss by mutation analysis and assessment of allele loss. In a subset of lines, we have also determined the copy number of SMAD4, using molecular cytogenetic techniques, and related this to the karyotype of the cell line. The results have implications not just for SMAD4 inactivation in bowel tumors, but also more generally for genetic pathways of colorectal tumorigenesis.
Materials and Methods
DNA was extracted from 44 established CRC cell lines (C10, C32, C70, C80, C84, C99, C106, C125, CAC02, COLO201, COLO205, COLO320, COLO678, COLO741, CX1/HT29, GP2D, HCA46, HCA7, HCT8, DLD1/HCT15, HCT116, HRA19, HT55, LIM1863, LOVO, LS174T, LS180, LS411, LS1034, PC/JW, SKC01, SW48, SW403, SW480, SW620, SW837, SW948, SW1116, SW1222, SW1417, VACO4A, VACO4S, VACO5, and VACO10), using standard methods. The APC mutation status, β-catenin mutation status, and microsatellite instability (MSI) status have been reported (summarized in ref. 17).
For F-SSCP (fluorescent-single stranded conformational polymorphism analysis), exon-by-exon amplification of the 11 exons of SMAD4 (covering all coding sequence and intron/exon boundaries) was performed in each CRC line by using previously reported primers (11) with added fluorescent 5′ and 3′ labels (FAM, HEX, or TET). PCRs were then diluted 1:50 with distilled water and combined with an internal size standard (Tamra 500, Perkin–Elmer Applied Biosystems) and formamide. F-SSCP analysis at 20°C was performed by using an ABI310 sequencer (Perkin–Elmer Applied Biosystems). Fragments showing both aberrant and normal migration were reamplified by using non-fluorescently labeled primers, purified by using Qiaquick columns (Qiagen, Hilden, Germany) and then sequenced in both forward and reverse orientations by using the ABI Big Dye Terminator kit (Perkin–Elmer Applied Biosystems). Repeated failure to amplify any segment of the SMAD4 gene in the PCR, despite successful amplification in control PCRs (17), was taken to denote homozygous deletion of that segment.
For Western blotting, pellets from all 44 CRC cell lines were lysed in 0.1 M DTT/bromophenol blue and separated on a 15% resolving gel. After transfer to the poly(vinylidene difluoride) (PVDF) membrane (Millipore) and blocking in 5% Marvel (Premier Brands, Stafford, United Kingdom), the primary antibodies were exposed to the membrane for 2 hours at room temperature. The anti-SMAD4 mouse monoclonal antibody B8 (Santa Cruz Biotechnology) recognizes an epitope in exon 5 (codons 68–108; M. Howell and C. Hill, personal communication); this antibody was diluted 1/100, and the control anti-β-actin mouse monoclonal antibody (Sigma) was diluted 1/1,000, and exposed to the membrane both simultaneously and separately. After detection of the proteins with enhanced chemiluminescence reagents (ECL, Amersham Pharmacia) according to the manufacturer's instructions, the membrane was exposed to film for 1 and 5 min. After scanning the films by using a Bio-Rad GS-700 densitometer, band intensity ratios were compared for SMAD4 and β-actin. SMAD4 protein was classed simply as “present” or “absent,” based on immunohistochemical data showing absent protein expression in many pancreatic cancers (18), colorectal cancers, and JPS polyps (K.L.W.-R. and I.P.M.T., unpublished data).
For the protein truncation test (PTT), RNA was extracted from a subset of CRC cell lines (SW1222, HRA19, SKC01, SW948, JW, HCT8, COLO205, LS174T, SW48, LOVO, SW620, COLO320, GP2D, HT29, CACO2, and HCA46) by using the Fast-track RNA extraction kit (Invitrogen). cDNA was synthesized by using the First Strand Synthesis kit (Promega). Primer pairs were designed by using Primer 3 (http://www.genome.wi.mit.edu/cgi-bin/primer/primer3 www.cgi) to cover the entire SMAD4 coding sequence (GenBank accession no. U44378). PCRs were performed by using standard conditions with either primer pair A (forward 5′-atggacaatatgtctattacga-3′, reverse 5′-ggaatgcaagctcattgtga-3′) covering codons 1–311 or primer pair B (forward 5′-cagcatccaccaagtaatcg-3′, reverse 5′-aaggttgtgggtctgcaatc-3′) covering codons 182 to 553, with the forward primer tagged with a T7 RNA-polymerase binding site, a ribosome binding site, and an in-frame start codon. In vitro coupled transcription–translation was performed on the tagged PCR products by using the TNT Rabbit Reticulocyte Lysate Kit (Promega), incorporating [35S]methionine, and the resulting “proteins” were separated according to size on a 12.5% polyacrylamide resolving gel. Once fixed and dried, gels were exposed to film overnight and developed.
For the assessment of 18q allelic loss, seven microsatellite markers were selected from the Location Database (http://cedar.genetics.soton.ac.uk/pub/chrom18/map.html): for SMAD4 the markers were in the order, D18S479–1.45 Mb–D18S474–0.01 Mb–D18S46–0.35 Mb–(SMAD4); and for DCC (about 1.5 Mb telomeric of SMAD4) the order was D18S484–0.16 Mb–D18S487–0.44 Mb–DCC–0.19 Mb–D18S35. The heterozygosity for the seven markers was reported to be 60%, 82%, 80%, 72%, 81%, 87%, and 70%, respectively. In addition, markers mapping to 18p (D18S481) and the telomeric region of 18q (D18S878) were included. The forward primer was fluorescently labeled with HEX, FAM, or TET. Standard PCR conditions were used, and then the PCR products were combined with the internal size standard Tamra350 (Perkin–Elmer Applied Biosystems) and run on an ABI377 semiautomated sequencer. Results were analyzed by using GENOTYPER software to assign peak sizes. Because no normal tissue was available, allelic loss was assumed to have occurred at DCC or SMAD4 if all microsatellite markers close to that gene were hemizygous/homozygous (corresponding to P < 0.01).
CRC cell line karyotypes were determined by using standard methods. Copy number of 18q21 was assessed by a combination of comparative genomic hybridization (CGH; refs. 19 and 20), spectral karyotyping (SKY; ref. 21), and locus-specific fluorescence in situ hybridization (FISH), using PAC 224 j 22, to which SMAD4 is known to map, as described (22). The samples analyzed by using cytogenetic techniques are shown in Table 1.
Table 1.
Summary of molecular and cytogenetic data for 18q21.1 status in colorectal cancer cell lines
| Cell line | MSI* | B8 Western† | SMAD4 mutation‡ | Predicted effect of mutation§ | Loss at SMAD4¶ | Loss at DCC∥ | Karyotype** | 18q21 status‡‡ | TGFBIIR†† |
|---|---|---|---|---|---|---|---|---|---|
| COLO678 | MSI− | Absent | EX1 − 11del | No protein | Yes | Yes | ? | — | |
| COLO201 | MSI− | Absent | EX1 − 4del | No protein | Yes | Yes | 78 | 2 copies | None |
| COLO205 | MSI− | Absent | EX1 − 4del | No protein | Yes | Yes | 68 | 2 copies | None |
| VACO10 | MSI− | Absent | c.715C>T (Q239X), EX5 | Nonsense | Yes | Yes | 115 | — | None |
| C10 | MSI− | Absent | IVS6 − 1G>T | Splice disruption | Yes | Yes | 49 | — | — |
| HT29/CX1 | MSI− | Absent | c.931C>T (Q311X), EX7 | Nonsense | 2/3 markers | Yes | 71 | 2 copies | None |
| SW480 | MSI− | Absent | IVS7 + 5G>C | Splice disruption | Yes | Yes | 57 | 1 copy | None |
| SW620 | MSI− | Absent | IVS7 + 5G>C | Splice disruption | Yes | Yes | 50 | 1 copy | None |
| CACO2 | MSI− | Present | c.1051G>C (D351H), EX8 | Missense | Yes | Yes | 96 | — | None |
| C80 | MSI− | Present | c.1051G>C (D351H), EX8 | Missense | Yes | Yes | 69 | 2 copies | — |
| SW403 | MSI− | Absent | EX10 − 11del | Truncated protein | Yes | Yes | 68 | 2 copies | — |
| SW948 | MSI− | Present | c.1609G>T (D537Y), EX11 | Missense | Yes | No | 67 | — | — |
| SW1222 | MSI− | Present | c.1619T>G (L540R), EX11 | Missense | Yes | Yes | ? | 18q loss | — |
| SW1116 | MSI− | Absent | None found | Yes | Yes | 63 | — | — | |
| HT55 | MSI− | Absent | None found | Yes | Yes | 80 | 2 copies | — | |
| C106 | MSI− | Absent | None found | Yes | Yes | 79 | 2 copies | — | |
| PC/JW | MSI− | Absent | None found | Yes | Yes | 70 | 2 copies | — | |
| SW1417 | MSI− | Absent | None found | Yes | No | 70 | 2 copies | — | |
| SW837 | MSI− | Present | None found | Yes | Yes | 40 | 1 copy | None | |
| C99 | MSI− | Present | None found | Yes | Yes | 52 | 1 copy | — | |
| C84 | MSI− | Present | None found | No | Yes | 56 | 1 copy | — | |
| C125 | MSI− | Present | None found | Yes | Yes | 60 | 1 copy | — | |
| VACO4A | MSI− | Present | None found | Yes | Yes | 60 | 2 copies | — | |
| HCA46 | MSI− | Present | None found | Yes | Yes | 71 | 2 copies | None | |
| C32 | MSI− | Present | None found | Yes | Yes | 74 | 2 copies | — | |
| LIM21-1863 | MSI− | Present | None found | Yes | Yes | 80 | 3 copies | — | |
| C70 | MSI− | Present | None found | Yes | Yes | 127 | 4 copies | — | |
| COLO320 | MSI− | Present | None found | Yes | Yes | 53 | — | None | |
| VACO4S | MSI− | Present | None found | Yes | Yes | 64 | — | — | |
| LS1034 | MSI− | Present | None found | Yes | Yes | 77 | — | — | |
| HRA19 | MSI− | Present | None found | Yes | Yes | ? | — | — | |
| SKC01 | MSI− | Present | None found | No | Yes | hypertriploid | — | — | |
| HCT8 | MSI+ | Present | None found | No | No | ? | — | — | |
| LS180 | MSI+ | Present | None found | No | No | 45 | — | — | |
| SW48 | MSI+ | Present | None found | No | No | 47 | — | Mutant × 2 | |
| LS174T | MSI+ | Present | None found | No | No | 45 | 2 copies | Mutant × 2 | |
| DLD1/HCT15 | MSI+ | Present | None found | No | No | 46 | 2 copies | Mutant × 2 | |
| LOVO | MSI+ | Present | None found | No | No | 49 | 2 copies | Mutant × 2 | |
| VACO5 | MSI+ | Present | None found | No | No | 47 | 2 copies | Mutant × 2 | |
| HCA7 | MSI+ | Present | None found | No | No | 43 | 2 copies | Mutant × 2 | |
| HCT116 | MSI+ | Present | None found | No | No | 45 | 2 copies | Mutant × 2 | |
| LS411 | MSI+ | Present | None found | No | No | 75 | 2 copies | Mutant × ? | |
| GP2D | MSI+ | Present | None found | No | No | 46 | 2 copies | — | |
| COLO741 | MSI+ | Present | None found | Yes | Yes | ? | 2 copies | — |
COLO201/COLO205, HT29/CX1, and DLD1/HCT8/HCT15 are essentially identical cell lines.
MSI status.
SMAD4 protein expression as assessed by using B8 and western blotting.
Identified SMAD4 mutations.
Predicted effect of SMAD4 mutations.
∥ Allelic loss as inferred from homozygosity at microsatellite markers near SMAD4 and at DCC, respectively.
Modal chromosome number of cell line (?, not known).
18q21.1 status as determined by CGH, SKY, and FISH (—, not done).
TGFBIIR mutation status (—, not done).
Results
Overall, 32% of the CRC cell lines showed loss of SMAD4 expression (Table 1, Fig. 1), comprising 14 of 32 (44%) MSI− lines and 0 of 14 MSI+ lines (P < 0.004, Fisher's exact test). In no case was a truncated protein band observed (Table 1). Several different types of mutation were found to account for the loss of expression (Table 1). Importantly, sequencing showed that all SMAD4 mutations were present in the homozygous or hemizygous state (Fig. 2), with no underlying wild-type sequence.
Figure 1.
Western blot analysis of SMAD4 and β-actin in colorectal cancer cell lines. Shown are the results of Western blot analysis using the B8 antibody against SMAD4, and a monoclonal anti-β-actin for four cell lines (LS180, C99, CX1, and COLO678). Note the complete absence of SMAD4 expression in CX1 and COLO678.
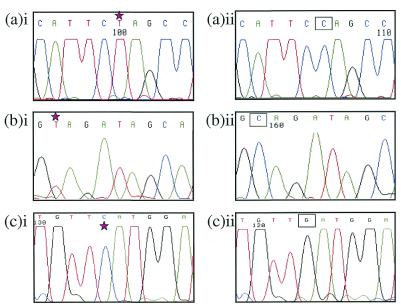
Figure 2.
Sequencing results of SMAD4 for HT29/CX1, VACO10, and CACO2. (a)i shows the mutated cell line HT29/CX1 (c.931C>T transition causing Q311X amino acid truncation) compared with (a)ii wild type. (b)i SMAD4 exon 5 of VACO10 showing the c.715C>T transition causing Q239X truncation, compared with (b)ii wild type. (c)i SMAD4 exon 8 of CACO2 showing the c.1051G>C nucleotide change, which results in a D351H missense change, compared with (c)ii wild type. Note the lack of a wild-type sequence underlying a mutated (starred) sequence. The corresponding wild-type base is shown in a box.
For one cell line (COLO678), the whole gene was homozygously deleted, whereas other cell lines showed partial homozygous deletions (exons 1–4 in COLO201/COLO205, and exons 10–11 in SW403). All these mutations were accompanied by allelic loss at SMAD4 (Table 1).
Putative splicing mutations were detected in three cell lines, SW480 and SW620 (derived from a primary tumor and metastasis) and C10. All these lines showed absent SMAD4 protein and allelic loss at SMAD4. Of these three lines, SW620 was tested by using the protein truncation test (PTT) and only normal-length mRNA was detected. Thus, although the mutation in SW620 does not lead to detectable abnormal mRNA splicing, we cannot exclude the possibility that it has pathogenic effects through reduction of normal mRNA levels, which could be assessed by using RNase protection assay.
Nonsense SMAD4 mutations, accompanied by allele loss and absent protein, were detected in two cell lines: HT29/CX1 (Q311X, exon 7) and VACO10MS (Q239X, exon 5). Missense mutations, plus allelic loss, were found in four lines: CACO2 (D351H), C80 (D351H), SW948 (D537Y), and SW1222 (L540R). SMAD4 protein was present in all of these four lines.
Four MSI− cell lines had absent SMAD4 protein and allelic loss, but no detectable SMAD4 mutation (Table 1). None of the 14 MSI+ cell lines possessed a pathogenic SMAD4 mutation, a highly significant difference from the MSI− lines (P < 0.005, Fisher's exact test). Known SMAD4 intronic polymorphisms (23) were also detected (data not shown) in MSI− and MSI+ lines.
Overall, there was a striking level of allele loss at SMAD4 and DCC in all of the MSI− lines, with 30/32 (94%) showing loss at SMAD4 and 30/32 (94%) with loss at DCC (Table 1). In accordance with the sequencing data, all lines with allelic loss showed complete absence of one microsatellite allele; thus, all copies of 18q21 in each cell were derived from the same chromosome-18 homologue, even where the cancer was polyploid and had more than one copy of 18q. In contrast, 11/12 (92%) MSI+ lines showed heterozygosity for at least one marker each at SMAD4 and DCC, indicating that loss of 18q material is not critical for the development and progression of these tumors. The different levels of 18q21 loss in MSI− and MSI+ tumors were confirmed in a set of 61 sporadic colorectal tumors, in which mean loss at four markers close to SMAD4 and DCC was 4% in 14 MSI+ cancers and 53% in 47 MSI− cancers (P < 0.003, Fisher's exact test; data not shown).
The molecular cytogenetic and karyotype data are summarized in Table 1. In the lines with 18q21 loss, the amount of chromosome 18q21 material was always less than the overall ploidy. For example MSI− cell lines with a modal chromosome number of less than 60 (C84, SW837, C99, C125, SW480, and SW620) had only one copy of chromosome 18q21, whereas cell lines with near-triploid karyotypes (SW403, HT29/CX1, HT55, COLO201, COLO205, C32, C106, C80, PC/JW, SW1417, VACO4A, and HCA46) had two copies of chromosome 18q21. Two lines with hypotetraploid (LIM1863) and hypohexaploid (C70) karyotypes had three and four copies of chromosome 18q21, respectively. Typing of extra markers mapping to the p-arm and 18q telomeric region confirmed comparative genomic hybridization (CGH)/spectral karyotyping (SKY) observations (data not shown).
In most cases, the cytogenetic analysis showed the whole 18q arm to be deleted, but there were exceptions. SW1417 was homozygous for microsatellites at SMAD4 and showed loss of SMAD4 protein, but was heterozygous for markers near DCC. The microsatellite data for SW1417 were consistent with an interstitial deletion that targeted SMAD4, but left DCC intact. CGH data for SW1417 also showed a deletion around SMAD4, rather than loss of the whole arm. Conversely, C84 and SKC01 were heterozygous for microsatellites at SMAD4, but homozygous for microsatellites around DCC. Again this was substantiated by CGH data, which showed loss of chromosome 18 distal to 18q21. In these lines, SMAD4 expression was retained and it appeared that DCC or another tumor suppressor was being targeted, leaving SMAD4 intact. The line HT29/CX1 had two apparently identical copies of chromosome 18 with SMAD4 mutations, plus one deleted 18q. This line was heterozygous at D18S474, but homozygous at the rest of the microsatellites and for the SMAD4 mutation, showing that the breakpoint for the deletion almost certainly lay just centromeric to SMAD4. Finally, cell line COLO678 was found to be homozygous for markers mapping to both the SMAD4 and DCC regions, but heterozygous for markers on the p-arm and 18q telomeric region. Cytogenetic data were not available for this line, but presumably it contains an interstitial deletion targeting SMAD4 and DCC without loss of the whole q-arm.
Discussion
This study aimed to clarify the role of SMAD4 in the development of 44 colorectal cancer cell lines. Loss of SMAD4 expression and/or mutation were found in about half of the MSI− lines, but in no MSI+ lines. A broad spectrum of SMAD4 mutations was seen; although, for unknown reasons, there were no frameshift changes. No association was made between the presence of a SMAD4 mutation and tumor grade. Mutations appeared to occur more frequently in the C-terminal MH2 domain of SMAD4, although missense mutations in the N-terminal MH1 domain have been shown to cause in vitro protein instability (8, 24). Again, the reasons for this observation are unclear. In five of the fourteen CRC lines with absence of SMAD4 protein, no mutation was detected even after sequencing all exons in forward and reverse orientations, suggesting that cryptic SMAD4 mutations or some other means of inactivating SMAD4 may have occurred. Inactivation of SMAD4 due to hypermethylation has been shown not to occur (25) and, consistent with this finding, we detected SMAD4 mRNA in PC/JW which had absent protein, but no detectable mutation. It is also possible that changes in a protein upstream of SMAD4 can sometimes lead to loss of SMAD4 expression.
In addition to mutations causing loss of protein, four cell lines with retained protein expression had missense SMAD4 mutations in the MH2 region (Table 1). The missense mutations of C80 and CACO2 (D351H) occurred in the three loop–helix of the MH2 and have known pathogenic effects (26). The nearby R361C mutation, found not only in sporadic cancer, but also in the germline of JPS patients, is associated with undetectable levels of protein in JPS polyps, as assessed by using immunohistochemistry (data not shown). Mutations in the three-helix bundle of the MH2, such as R537Y (SW948) and L540R (SW1222), appear not to lead to unstable protein, but, like D351H, are predicted to abrogate protein function (26–28) by impairing the ability of SMAD4 to act as a homo- or heterooligomer and so interfering with signal transduction.
All lines with SMAD4 abnormalities were uniformly hemi- or homozygous at both SMAD4 and DCC microsatellites and for the SMAD4 mutation, showing that all copies of 18q were derived from the same chromosome 18 homologue, even where more than one copy of 18q21 was present in the cell. However, the dosage of 18q21 was always decreased relative to the overall level of ploidy. We found no cancer with more than one independent intragenic mutation in SMAD4. The simplest model to explain these data (Fig. 3) is one in which SMAD4 mutations occur before colorectal tumors acquire chromosomal instability (CIN) and polyploidy/aneuploidy, and are not late changes, as has been suggested (6, 8, 9). We believe that alternative explanations to this model are less plausible, because they would require several independent mutations to inactivate all copies of SMAD4 and/or frequent, independent occurrence of identical SMAD4 mutations in the same cell. However, Fodde et al. (29) have suggested that tetraploidy is a very early event in APC-mutant colorectal tumors, contrary to our favored model. Were the data of Fodde et al. (29) to be confirmed, we would be forced to conclude that some CRCs had lost three copies of 18q, acquired an intragenic SMAD4 mutation, and reduplicated the remaining SMAD4 mutant chromosome.
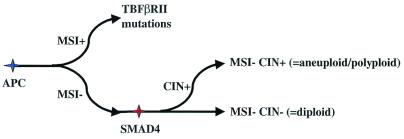
Figure 3.
Pathway showing possible sequence of events in tumorigenesis. Following mutations in the APC gene, a subset of tumors have inactivation of mismatch repair genes and so diverge along a pathway that includes mutations of the TGFβRII gene, and is characterized by microsatellite instability (MSI+). A subset of the remaining tumors acquire mutations in the SMAD4 gene, accompanied by loss of the wild-type chromosome 18, and this either precedes or causes another subset of tumors to diverge along a chromosomal instability (CIN+) pathway with aneuploidy/polyploidy. The remaining tumors are MSI−/CIN−.
Given that none of 12 MSI+ cell lines had SMAD4 mutations or loss of SMAD4 protein, it is most likely that the MSI+ pathway had diverged before SMAD4 inactivation occurred (Fig. 3). All eight MSI+ lines previously studied have been reported to have TGFBIIR mutations (30, 31), whereas none of the ten MSI− lines studied had TGFBIIR mutations (Fisher's exact test, P < 0.000003). Although inactivation of SMAD4 and TGFBIIR may well not be functionally equivalent, these data certainly suggest that mutations of TGFBIIR occur after the divergence of the MSI−/+ lineages. Thus, our model shows initial divergence of the MSI+ pathway, followed by 18q loss and SMAD4 mutation in the MSI− cancers. Given that limited data suggest a high frequency of 18q loss in MSI−CIN− CRCs (32), divergence of the CIN− and CIN+ pathways probably occurs after 18q loss and SMAD4 mutation.
Almost all MSI− cell lines showed allelic loss, and even in MSI− lines without defects in SMAD4, the dosage of 18q21 was always decreased relative to the overall level of ploidy: the question therefore remains as to the cause of 18q loss in lines with no evidence of SMAD4 inactivation. Although SMAD2 remains an unlikely target, haplo-insufficiency of SMAD4 is possible and DCC changes, although unlikely, cannot be excluded with certainty. Any other target genes on 18q must, like SMAD4, be altered after the MSI+ pathway had diverged [although mutations in these gene(s) might, of course, be common to the MSI+ and MSI− pathways]. Given data which suggest that 18q21 loss occurs in colorectal adenomas or close to the adenoma–carcinoma transition (33), but that SMAD4 expression is lost only in CRCs (9), we speculate that 18q21 loss sometimes precedes mutation or silencing of the other copy of one or more target genes in the region.
Acknowledgments
We thank the Equipment Park, Imperial Cancer Research Fund, and those who have very kindly provided cell lines for this work.
Abbreviations
- MSI
microsatellite instability
- CRC
colorectal cancer
- CIN
chromosomal instability
References
- 1.Thiagalingam S, Lengauer C, Leach F S, Schutte M, Hahn S A, Overhauser J, Willson J K, Markowitz S, Hamilton S R, Kern S E, et al. Nat Genet. 1996;13:343–346. doi: 10.1038/ng0796-343. [DOI] [PubMed] [Google Scholar]
- 2.Fearon E R, Vogelstein B. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 3.Hamilton S R. Cancer. 1992;70, Suppl 5:1216–1221. doi: 10.1002/1097-0142(19920901)70:3+<1216::aid-cncr2820701505>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 4.Riggins G J, Thiagalingam S, Rozenblum E, Weinstein C L, Kern S E, Hamilton S R, Willson J K, Markowitz S D, Kinzler K W, Vogelstein B. Nat Genet. 1996;13:347–349. doi: 10.1038/ng0796-347. [DOI] [PubMed] [Google Scholar]
- 5.Tagaki Y, Kohmura H, Futamura M, Kida H, Tanemura H, Shimokawa K, Saji S. Gastroenterology. 1996;111:1369–1372. doi: 10.1053/gast.1996.v111.pm8898652. [DOI] [PubMed] [Google Scholar]
- 6.Miyaki M, Iijima T, Konishi M, Sakai K, Ishii A, Yasuno M, Hishima T, Koike M, Shitara N, Iwama T, et al. Oncogene. 1999;18:3098–3103. doi: 10.1038/sj.onc.1202642. [DOI] [PubMed] [Google Scholar]
- 7.Hahn S A, Hoque A T, Moskaluk C A, da Costa L T, Schutte M, Rozenblum E, Seymour A B, Weinstein C L, Yeo C J, Hruban R H, Kern S E. Cancer Res. 1996;56:490–494. [PubMed] [Google Scholar]
- 8.Koyama M, Ito M, Nagai H, Emi M, Moriyama Y. Mutat Res. 1999;406:71–77. doi: 10.1016/s1383-5726(99)00003-5. [DOI] [PubMed] [Google Scholar]
- 9.Maitra A, Molberg K, Albores-Saavedra J, Lindberg G. Am J Pathol. 2000;157:1105–1111. doi: 10.1016/S0002-9440(10)64625-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howe J R, Roth S, Ringold J C, Summers R W, Jarvinen H, Sistonen P, Tomlinson I P M, Houlston R S, Bevan S, Mitros F A, Stone E M, Aaltonen L A. Science. 1998;280:1086–1088. doi: 10.1126/science.280.5366.1086. [DOI] [PubMed] [Google Scholar]
- 11.Houlston R, Bevan S, Williams A, Young J, Dunlop M, Rozen P, Eng C, Markie D, Woodford-Richens K, Rodriguez-Bigas M, et al. Hum Mol Genet. 1998;7:1907–1912. doi: 10.1093/hmg/7.12.1907. [DOI] [PubMed] [Google Scholar]
- 12.Feng X H, Lin X, Derynck R. EMBO J. 2000;19:5178–5193. doi: 10.1093/emboj/19.19.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brodin G, Ahgren A, ten Dijke P, Heldin C H, Heuchel R. J Biol Chem. 2000;275:29023–29030. doi: 10.1074/jbc.M002815200. [DOI] [PubMed] [Google Scholar]
- 14.von Gersdorff G, Susztak K, Rezvani F, Bitzer M, Liang D, Bottinger E P. J Biol Chem. 2000;275:11320–11326. doi: 10.1074/jbc.275.15.11320. [DOI] [PubMed] [Google Scholar]
- 15.de Caestecker M P, Piek E, Roberts A B. J Natl Cancer Inst. 2000;92:1388–1402. doi: 10.1093/jnci/92.17.1388. [DOI] [PubMed] [Google Scholar]
- 16.Moren A, Itoh S, Moustakas A, Dijke P, Heldin C H. Oncogene. 2000;19:4396–4404. doi: 10.1038/sj.onc.1203798. [DOI] [PubMed] [Google Scholar]
- 17.Rowan A J, Lamlum H, Ilyas M, Wheeler J, Straub J, Papadopoulou A, Bicknell D, Bodmer W F, Tomlinson I. Proc Natl Acad Sci USA. 2000;97:3352–3357. doi: 10.1073/pnas.97.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilentz R E, Su G H, Dai J L, Sparks A B, Argani P, Sohn T A, Yeo C J, Kern S E, Hruban R H. Am J Pathol. 2000;156:37–43. doi: 10.1016/S0002-9440(10)64703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kallioniemi A, Kallioniemi O P, Sudar D, Rutovitz D, Gray J W, Waldman F, Pinkel D. Science. 1992;258:818–821. doi: 10.1126/science.1359641. [DOI] [PubMed] [Google Scholar]
- 20.Weiss M M, Hermsen M A, Meijer G A, van Grieken N C, Baak J P, Kuipers E J, van Diest P J. Mol Pathol. 1999;52:243–251. doi: 10.1136/mp.52.5.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abdel-Rahman W M, Katsura K, Rens W, Gorman P A, Sheer D, Bicknell D, Bodmer W F, Arends M J, Wyllie A H, Edwards P A. Proc Natl Acad Sci USA. 2001;98:2538–2543. doi: 10.1073/pnas.041603298. . (First Published February 20, 2001; 10.1073/pnas.041603298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woodford-Richens K, Williamson J, Bevan S, Young J, Leggett B, Frayling I, Thway Y, Hodgson S, Kim J C, Iwama T, et al. Cancer Res. 2000;60:2477–2482. [PubMed] [Google Scholar]
- 23.MacGrogan D, Pegram M, Slamon D, Bookstein R. Oncogene. 1997;15:1111–1114. doi: 10.1038/sj.onc.1201232. [DOI] [PubMed] [Google Scholar]
- 24.Xu J, Attisano L. Proc Natl Acad Sci USA. 2000;97:4820–4825. doi: 10.1073/pnas.97.9.4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roth S, Laiho P, Salovaara R, Launonen V, Aaltonen L A. Br J Cancer. 2000;83:1015–1019. doi: 10.1054/bjoc.2000.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi Y, Hata A, Lo R S, Massague J, Pavletich N P. Nature (London) 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- 27.Eppert K, Scherer S W, Ozcelik H, Pirone R, Woodless P, Kim H, Tsui L C, Bapat B, Gallinger S, Andrulis I L, et al. Cell. 1996;86:543–552. doi: 10.1016/s0092-8674(00)80128-2. [DOI] [PubMed] [Google Scholar]
- 28.Schutte M. Ann Oncol. 1999;10, Suppl. 4:56–59. [PubMed] [Google Scholar]
- 29.Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gaspar C, van Es J H, Breukel C, Wiegant J, Giles R H, Clevers H. Nat Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- 30.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan R S, Zborowska E, Kinzler K W, Vogelstein B, Brattain M, Willson J K V. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 31.Ilyas M, Efstathiou J A, Straub J, Kim H C, Bodmer W F. Proc Natl Acad Sci USA. 1999;96:3087–3091. doi: 10.1073/pnas.96.6.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Georgiades I B, Curtis L J, Morris R M, Bird C C, Wyllie A H. Oncogene. 1999;18:7933–7940. doi: 10.1038/sj.onc.1203368. [DOI] [PubMed] [Google Scholar]
- 33.Yashiro M, Carethers J M, Laghi L, Saito K, Slezak P, Jaramillo E, Rubio C, Koizumi K, Hirakawa K, Boland C R. Cancer Res. 2001;61:2676–2683. [PubMed] [Google Scholar]