DNA topoisomerase I (Top1) is the sole target of camptothecin (CPT) and other Top1 poisons [1] that are widely used drugs for treatment of colon cancer and other malignancies. Our previous studies have shown that Top1 is also an important target for the fluoropyrimidine polymer F10 reviewed in [2], and that Top1 poisoning plays an important, but relatively undefined role in thymineless death (TLD), the process by which inhibiting de novo thymidylate synthesis causes DNA damage and cell death. While Top1 poisoning is an important aspect of TLD, thymidine deprivation may also cause DNA damage by Top1-independent pathways, such as replication fork collapse, and these multiple sources of DNA damage may provide the basis for the very strong potency of F10 relative to alternative DNA damaging agents observed for many types of malignant cells. Elucidating the DNA repair pathways activated in cells treated with F10 relative to CPT can provide insight into how Top1 poisoning induced by fluoropyrimidine drugs differs from traditional Top1 poisons, and also clarify which aspects of the DNA damage response are activated by Top1-independent processes.
Top1 relieves DNA torsional stress generated during replication and transcription by nicking supercoiled DNA allowing relaxation to proceed via controlled rotation [3], and then re-ligating the nick [4] to restore the DNA duplex. CPT forms a ternary complex with Top1 and the nicked DNA resulting in formation of a Top1 cleavage complex (Top1cc) that, if not repaired, can lead to DNA double-strand breaks (DSBs) through collision with advancing replication forks [5]. Top1cc also form upon F10 treatment as Top1 acts upon DNA in which FdU has become incorporated proximal to Top1 cleavage sites [6]. Top1 cleaves FdU-substituted DNA, but FdU inhibits the re-ligation step of Top1 catalysis generating Top1cc. Top1cc are repaired by two major pathways [7]: i) TDP1/PARP-1; and ii) Mus81. TDP1 [8] removes the residual peptide remaining after Top1 undergoes in situ proteasomal degradation. TDP1 requires activation by PARP-1 [9], and the clinical efficacy of PARP inhibitors in combination with Top1 poisons [10,11] indicates that the TDP1/PARP-1 pathway contributes to survival in CPT-treated cells. TDP1 inhibitors are being developed to provide an alternative point of modulation for this pathway [12]. The second major pathway of Top1cc repair involves Mus81-mediated endonucleolytic cleavage of DNA. This pathway is activated in response to prolonged replication fork collapse that occurs upon treatment with CPT [13], and also with drugs that cause dNTP depletion, such as hydroxyurea (HU) [14].
The relative contributions of specific genes and associated pathways for repair of Top1cc and other types of DNA damage may be studied using knockout cells. DT40 cells are widely used to investigate DNA repair in vertebrate cells because specific gene knockouts are readily achieved using these cells. The DT40 B-cell line has also been used to study the effects of reduced Top1 expression on activation-induced deaminase abundance [15], and these cellular models may also be useful to study the effects of reduced TOP1 expression on drug sensitivity. A previous study revealed CPT sensitivity was dependent on TDP1 and PARP1 expression, as expected [16]. The effects of knockout of Top1cc repair enzymes for drugs that cause TLD, such as F10, have not been evaluated previously. However, since Top1 poisoning is an important aspect of TLD, many of the same pathways identified as contributing to sensitivity, or resistance, for Top1 poisons likely also affect response to F10. We evaluated the relative sensitivity of F10, CPT, and 5-FU in DT40 cell lines deficient in TDP1-mediated Top1cc repair (Figures 1 & 2). These studies revealed a marked dependence on Top1cc repair for both F10 and CPT, consistent with both drugs being cytotoxic in a Top1-dependent process. However, the observed effects differed considerably for these two drugs, and for higher concentrations of F10 the effects were opposite to those observed for CPT indicating the role that Top1cc formation and repair is distinct for F10-induced TLD relative to CPT. Our studies also reveal that not only is 5-FU much less potent than F10, but that it is not dependent on Top1cc formation or repair consistent with its role as an RNA, rather than DNA-directed drug [17].
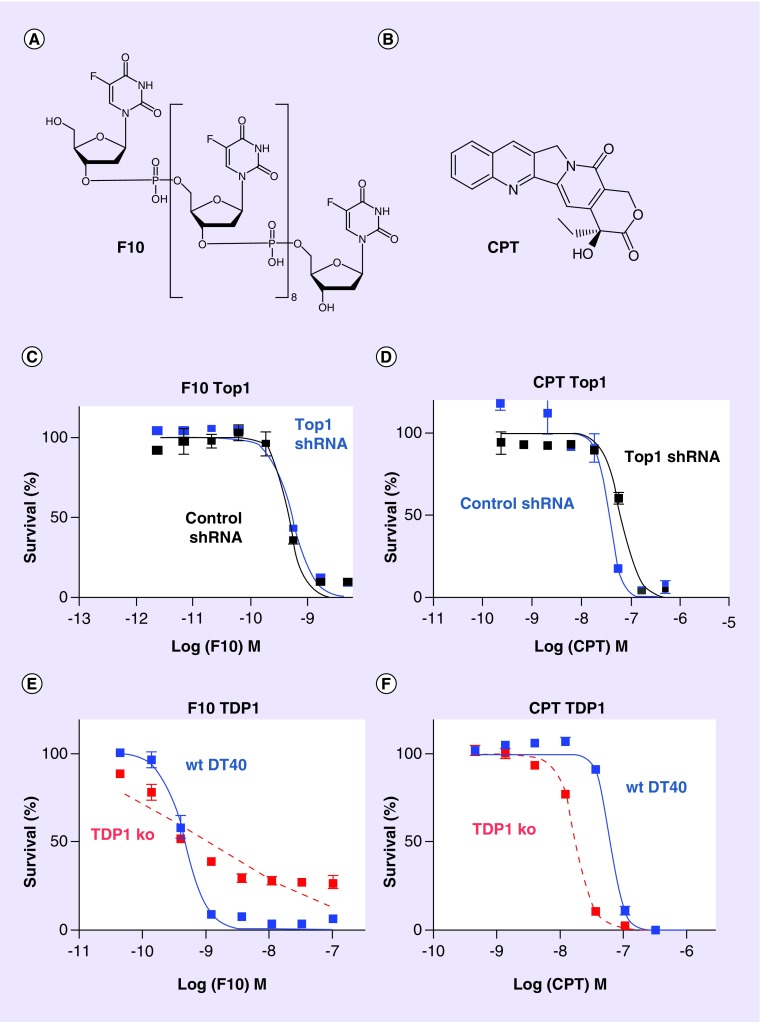
Figure 1. . The Top1 poisons F10 and camptothecin display differential dependence on TOP1 and TDP1 expression.
Structures of (A) F10 and (B) CPT. (C–E) show dose–response curves for (C & E) F10 and (D & F) CPT in DT40 cells. (C & D) show the effects of (C) F10 and (D) CPT in DT40 cells with shRNA knockdown of TOP1 (cells gift of Dr Patricia Gearhart, NIH) relative to DT40 cells treated with a control shRNA (C & D). TOP1 levels are knocked down about 60% in the TOP1 shRNA cells. Response curves for (E) F10 and (F) CPT in TDP1-ko DT40 cells relative to wt are also shown.
CPT: Camptothecin; ko: Knockout; wt: Wild-type.
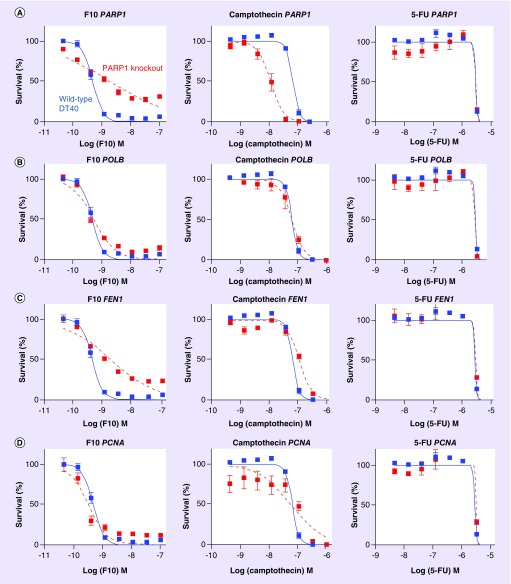
Figure 2. . F10 and camptothecin show differential sensitivity to knockout of genes involved in single-strand break repair while 5-FU sensitivity is not affected by knockout of DNA repair genes.
(A) Data for PARP1 knockout cells are shown in red and wild-type DT40 cells are shown in blue with data for F10 (left), camptothecin (middle), and 5-FU (right). (B–D) Show similar data as for (A) except data are shown for (B) POLB, (C) FEN1 and (D) PCNA.
Top1 deficiency & TDP1-mediated SSBR differentially affect F10 & CPT
We utilized TOP1 knockdown [15], and TDP1 [18] and PARP1 knockout DT40 cells [10] to investigate how Top1 levels and TDP1-mediated Top1cc repair affect the sensitivity of cells to F10 and CPT. Top1 is the sole target for CPT [1], and cells with reduced Top1 levels are less sensitive to CPT than wild-type cells [19]. F10 also causes Top1 poisoning [6,20], but through a different mechanism than for CPT, and how changes in Top1 levels affect sensitivity to F10 has not previously been reported. As in our previous studies with numerous cancer cell lines, we found F10 to be highly potent with the IC50 dose for F10 (6.69 × 10-10 M) being 140-fold lower than for CPT (3.56 × 10-8 M) and 4394-fold lower than for 5-FU (2.94 × 10-6 M) in wild-type DT40 cells. In addition to being more potent than CPT, F10 displayed a different dose–response profile in DT40 cells with reduced Top1 levels (Figure 1). DT40 cells with reduced TOP1 expression (˜60% knockdown) were less sensitive to CPT than wild-type, as expected. By contrast, DT40 cells with TOP1 knockdown displayed only a trend to decreased sensitivity to higher doses of F10 but TOP1 knockdown did not have a significant overall effect on F10 sensitivity, consistent with non-Top1-mediated processes also contributing to F10 cytotoxicity. To gain further insight into the role of TOP1 expression and Top1cc formation on the relative cytotoxicity of F10 and CPT to DT40 cells, we investigated the effects of TDP1 knockout on drug sensitivity. TDP1 mediates Top1cc repair in a process that involves proteasomal degradation of Top1 [7], and TDP1 is also involved in removing non-native nucleotides from DNA [8]. Previous studies, also using DT40 cells, demonstrated TDP1 knockout cells were more sensitive to CPT than wild-type [16], a result that is consistent with persistent Top1cc being responsible for the cytotoxic effects of CPT. Increased sensitivity to CPT has also been observed for cancer cell lines upon either shRNA knockdown or pharmacological inhibition of TDP1 [12]. In contrast to the results obtained with CPT, TDP1-knockout cells were hypersensitive only to low doses of F10, but were relatively resistant to higher doses of F10 (Figure 1). The unusual dose–response for F10 in TDP1 knockout cells suggests F10 may act by two different mechanisms that have different relative contributions at high and low F10 doses. Dose–response profiles for F10 in several other DNA repair genes including PARP1 (Figure 2) also showed similar properties. The dose–response of TDP1 knockout cells to F10 plateaued near 20% survival for the highest F10 doses tested demonstrating a particularly strong dependence for F10 cytotoxicity on TDP1 expression at higher drug doses.
PARP-1 activates TDP1 in response to DNA damage and this process is essential for TDP1-mediated Top1cc repair. Previous studies evaluated the effects of TDP1 and PARP1 single- and double-knockouts on CPT sensitivity and concluded that these genes are epistatic in the repair of CPT-induced Top1cc [9]. Our studies also show increased sensitivity to CPT for both PARP1- and TDP1-knockout cells, similar to previous studies and consistent with TDP1/PARP1 epistasis in repair of CPT-induced Top1cc. The response of PARP1 knockout cells to F10 differed considerably from the response to CPT (Figure 2), and was similar to what was observed for F10 in TDP1 knockout cells. Specifically, PARP1 knockout decreased overall sensitivity to F10, with the effects being greater for higher drug doses and with PARP1 knockout actually increasing sensitivity to F10 at lower drug doses. The similarity in dose–response to F10 in both TDP1 and PARP1 knockout cells is also consistent with TDP1/PARP1 epistasis in the repair of F10-induced Top1cc. As PARP-1/TDP1-mediated repair of Top1cc results in nicked DNA that structurally resembles intermediates generated during BER, we also investigated whether a similar dose–response pattern as that observed for TDP1 and PARP1 knockout also occurred for DNA polymerase β (POLB) and FEN1, knockout which would be consistent with single strand break repair (SSBR) contributing to F10 cytotoxicity at higher drug doses while being protective at lower drug doses (Figure 2). A similar dose–response pattern for F10 was observed in the POLB and FEN1 knockout cells as for the TDP1 and PARP1 knockout cells, consistent with SSBR of F10-induced Top1cc contributing to drug cytotoxicity rather than to repair, particularly for higher drug concentrations.
Our studies demonstrate that F10 is a highly potent and DNA-directed fluoropyrimidine and that Top1-poisoning is an important aspect of F10 cytotoxicity [6,21]. F10 displays marked potency advantages relative to 5-FU [21] and, unlike 5-FU, is highly sensitive to knockout of diverse genes involved in DNA repair consistent with cytotoxicity resulting from DNA damage rather than RNA-mediated effects (Figure 2). While sensitivity changes to F10 displayed only a trend and were not significantly affected by TOP1 knockdown, this may be due to Top1 levels being reduced by only ˜60% which is sufficient to observe significant effects with CPT, for which Top1 is the sole target, but not for F10 which has alternative targets (Figure 1). Significant effects were, however, observed for F10 sensitivity in TDP1 and PARP1 knockout cells that are deficient in Top1cc repair, which is consistent with Top1cc being an important aspect of F10 cytotoxicity. In particular, higher concentrations of F10 were more cytotoxic toward wild-type cells relative to TDP1 knockout cells, consistent with TDP1-mediated repair of F10-induced Top1cc contributing to drug cytotoxicity. By contrast, CPT at all concentrations evaluated is more potent to TDP1 knockout cells than to wild-type, consistent with TDP1-mediated repair of CPT-induced Top1cc attenuating CPT cytotoxicity. The results with TOP1 knockdown and TDP1 knockout cells for CPT are consistent with cells that have higher levels of Top1 are more amenable to CPT-induced Top1cc that persist, increasing the likelihood of either transcription- or replication-mediated DNA DSBs. The effects of Top1cc formation and repair for F10 sensitivity are dose-dependent which could reflect the involvement of at least two pathways in mediating F10 cytotoxicity.
We have identified both similarities and differences in Top1cc repair affecting sensitivity to F10 relative to CPT. These studies further demonstrate the importance of Top1cc formation and repair in F10-induced TLD, but also demonstrate fundamental differences in Top1cc repair in F10-treated cells relative to CPT. These differences may be used advantageously in personalized medicine approaches to cancer treatment as differential sensitivity to Top1, TDP1, PARP-1 and other enzymes may reflect when one type of drug relative to another is selected for optimal response. Our data also indicate DNA repair processes independent of Top1cc formation are also important during the course of TLD providing additional targets for modulating the therapeutic response of polymeric fluoropyrimidines. Future studies will clarify the role of Top1 in mediating TLD and will evaluate new approaches to enhance F10-induced TLD by modulating DNA repair pathways contributing to cell survival.
Footnotes
Financial & competing interests disclosure
WH Gmeiner is involved in commercialization of F10 for cancer treatment and is an inventor on issued patents and pending patent applications filed by Wake Innovations. Studies were supported by the Comprehensive Cancer Center at Wake Forest University, Wake Innovations, NIH-NCI P30 012197 (B Pasche), and NIH P42 ES005948 (J Nakamura) and NIH P30 ES010126 (J Nakamura). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Pommier Y. Drugging topoisomerases: lessons and challenges. ACS Chem. Biol. 2013;8(1):82–95. doi: 10.1021/cb300648v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gmeiner WH, Milligan D, Milligan C, Caudell D, Pardee TS. The applications of the novel polymeric fluoropyrimidine F10 in cancer treatment: current evidence. Future Oncol. 2016 doi: 10.2217/fon-2016-0091. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart L, Redinbo MR, Qiu X, Hol WG, Champoux JJ. A model for the mechanism of human topoisomerase I. Science. 1998;279(5356):1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 4.Seol Y, Zhang H, Pommier Y, Neuman KC. A kinetic clutch governs religation by type IB topoisomerases and determines camptothecin sensitivity. Proc. Natl Acad. Sci. USA. 2012;109(40):16125–16130. doi: 10.1073/pnas.1206480109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seiler JA, Conti C, Syed A, Aladjem MI, Pommier Y. The intra-S-phase checkpoint affects both DNA replication initiation and elongation: single-cell and -DNA fiber analyses. Mol. Cell Biol. 2007;27(16):5806–5818. doi: 10.1128/MCB.02278-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liao ZY, Sordet O, Zhang HL, et al. A novel polypyrimidine antitumor agent FdUMP [10] induces thymineless death with topoisomerase I-DNA complexes. Cancer Res. 2005;65(11):4844–4851. doi: 10.1158/0008-5472.CAN-04-1302. [DOI] [PubMed] [Google Scholar]
- 7.Tomicic MT, Kaina B. Topoisomerase degradation, DSB repair, p53 and IAPs in cancer cell resistance to camptothecin-like topoisomerase I inhibitors. Biochim. Biophys. Acta. 2013;1835(1):11–27. doi: 10.1016/j.bbcan.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Huang SY, Murai J, Dalla Rosa I, et al. TDP1 repairs nuclear and mitochondrial DNA damage induced by chain-terminating anticancer and antiviral nucleoside analogs. Nucleic Acids Res. 2013;41(16):7793–7803. doi: 10.1093/nar/gkt483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Das BB, Huang SY, Murai J, et al. PARP1-TDP1 coupling for the repair of topoisomerase I-induced DNA damage. Nucleic Acids Res. 2014;42(7):4435–4449. doi: 10.1093/nar/gku088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murai J, Huang SY, Das BB, et al. Trapping of PARP1 and PARP2 by clinical PARP inhibitors. Cancer Res. 2012;72(21):5588–5599. doi: 10.1158/0008-5472.CAN-12-2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murai J, Zhang Y, Morris J, et al. Rationale for PARP inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J. Pharmacol. Exp. Ther. 2014;349(3):408–416. doi: 10.1124/jpet.113.210146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchand C, Lea WA, Jadhav A, et al. Identification of phosphotyrosine mimetic inhibitors of human tyrosyl-DNA phosphodiesterase I by a novel AlphaScreen high-throughput assay. Mol. Cancer Ther. 2009;8(1):240–248. doi: 10.1158/1535-7163.MCT-08-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Regairaz M, Zhang YW, Fu H, et al. Mus81-mediated DNA cleavage resolves replication forks stalled by topoisomerase I-DNA complexes. J. Cell Biol. 2011;195(5):739–749. doi: 10.1083/jcb.201104003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanada K, Budzowska M, Davies SL, et al. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 2007;14(11):1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 15.Maul RW, Saribasak H, Cao Z, Gearhart PJ. Topoisomerase I deficiency causes RNA polymerase II accumulation and increases AID abundance in immunoglobulin variable genes. DNA Repair (Amst.) 2015;30:46–52. doi: 10.1016/j.dnarep.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maede Y, Shimizu H, Fukushima T, et al. Differential and common DNA repair pathways for topoisomerase I- and II-targeted drugs in a genetic DT40 repair cell screen panel. Mol. Cancer Ther. 2014;13(1):214–220. doi: 10.1158/1535-7163.MCT-13-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brody JR, Hucl T, Costantino CL, et al. Limits to thymidylate synthase and TP53 genes as predictivedeterminants for fluoropyrimidine sensitivity and further evidence forRNA-based toxicity as a major influence. Cancer Res. 2009;69(3):984–991. doi: 10.1158/0008-5472.CAN-08-3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murai J, Huang SY, Das BB, Dexheimer TS, Takeda S, Pommier Y. Tyrosyl-DNA phosphodiesterase 1 (TDP1) repairs DNA damage induced by topoisomerases I and II and base alkylation in vertebrate cells. J. Biol. Chem. 2012;287(16):12848–12857. doi: 10.1074/jbc.M111.333963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burgess DJ, Doles J, Zender L, et al. Topoisomerase levels determine chemotherapy response in vitro and in vivo . Proc. Natl Acad. Sci. USA. 2008;105(26):9053–9058. doi: 10.1073/pnas.0803513105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pardee TS, Gomes E, Jennings-Gee J, Caudell D, Gmeiner WH. Unique dual targeting of thymidylate synthase and topoisomerase 1 by FdUMP [10] results in high efficacy against AML and low toxicity. Blood. 2012;119:3561–3570. doi: 10.1182/blood-2011-06-362442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gmeiner WH, Reinhold WC, Pommier Y. Genome-wide mRNA and microRNA profiling of the NCI 60 cell-line screen and comparison of FdUMP [10] with fluorouracil, floxuridine, and topoisomerase 1 poisons. Mol. Cancer Ther. 2010;9(12):3105–3114. doi: 10.1158/1535-7163.MCT-10-0674. [DOI] [PMC free article] [PubMed] [Google Scholar]